Abstract
Lipids can be anaerobically digested to methane, but methanogens are often considered to be highly sensitive to the long-chain fatty acids (LCFA) deriving from lipids hydrolysis. In this study, the effect of unsaturated (oleate [C18:1]) and saturated (stearate [C18:0] and palmitate [C16:0]) LCFA toward methanogenic archaea was studied in batch enrichments and in pure cultures. Overall, oleate had a more stringent effect on methanogens than saturated LCFA, and the degree of tolerance to LCFA was different among distinct species of methanogens. Methanobacterium formicicum was able to grow in both oleate- and palmitate-degrading enrichments (OM and PM cultures, respectively), whereas Methanospirillum hungatei only survived in a PM culture. The two acetoclastic methanogens tested, Methanosarcina mazei and Methanosaeta concilii, could be detected in both enrichment cultures, with better survival in PM cultures than in OM cultures. Viability tests using live/dead staining further confirmed that exponential growth-phase cultures of M. hungatei are more sensitive to oleate than are M. formicicum cultures; exposure to 0.5 mM oleate damaged 99% ± 1% of the cell membranes of M. hungatei and 53% ± 10% of the cell membranes of M. formicicum. In terms of methanogenic activity, M. hungatei was inhibited for 50% by 0.3, 0.4, and 1 mM oleate, stearate, and palmitate, respectively. M. formicicum was more resilient, since 1 mM oleate and >4 mM stearate or palmitate was needed to cause 50% inhibition on methanogenic activity.
INTRODUCTION
Anaerobic degradation of long-chain fatty acids (LCFA) is essential for efficient biogas production from complex lipid-containing wastewaters (1–3). However, there is still concern regarding the potential toxic effect of LCFA toward methanogenic communities. Toxicity of LCFA is the main reason for insufficient treatment results of LCFA-containing wastewaters (4–8). Studies of methanogenic inhibition in the rumen confirm the toxicity of these compounds. Soliva et al. (9) showed the antimethanogenic potential of saturated fatty acids of medium chain length, specifically myristate (C14:0) and laurate (C12:0). The adverse effect of fatty acids toward methanogens appears to be more pronounced at longer chain lengths and more unsaturated bonds (10, 11). Moreover, methanogens inhibition by fatty acids is a rapid phenomenon with long recuperation time, as shown by Koster (12): a 50% loss of methanogenic activity was observed after exposing methanogenic sludge to 7.5 mM laurate for only 7.5 min.
Sensitivity of microorganisms to LCFA seems to be related to their cell wall structure, with Gram-positive bacteria and methanogens being more easily inhibited than Gram-negative bacteria (13). Inhibition of pure cultures of methanogens by fatty acids is reported in literature (11, 14). Nevertheless, methanogens are ubiquitous in anaerobic bioreactors treating LCFA-rich wastewaters, even over prolonged continuous LCFA loading (15, 16). Sousa et al. (16) identified three predominant genera of methanogens in LCFA-degrading sludges: Methanobacterium, Methanosaeta, and Methanosarcina. Methanobacterium-related species were the predominant hydrogenotrophs in continuous bioreactors, together with acetoclastic methanogens closely related to Methanosaeta concilli. Batch incubation of sludges containing high concentrations of LCFA stimulated the development of Methanosarcina, whereas Methanosaeta did not develop. The hydrogenotrophic methanogen Methanospirillum hungatei was identified in LCFA enrichment cultures (13). Recently, Salvador et al. (15) reported the endurance of methanogenic archaea in continuously fed reactors treating oleate-based effluent, with Methanobacterium and Methanosaeta being the predominant genera of hydrogenotrophic and acetoclastic methanogens, respectively.
Understanding the effects of LCFA on growth of methanogens may provide insight into overcoming potential problems occurring during the anaerobic digestion of lipid-rich wastewater. Both hydrogenotrophic and acetoclastic methanogens are essential for methane formation from complex lipid-containing wastewaters. If methanogenesis is inhibited, LCFA conversion by anaerobic bacteria is no longer possible due to the syntrophic nature of LCFA conversion (17, 18). In the present study, the effects of unsaturated and saturated LCFA on methanogenic archaea were studied in two different experiments. First, the prevalence of pure cultures of acetoclastic and hydrogenotrophic methanogens in enrichment cultures degrading oleate (C18:1, unsaturated LCFA) or palmitate (C16:0, saturated LCFA) was investigated. In a second experiment, the effect of LCFA on the methanogenic activity and membrane integrity of pure cultures of two hydrogenotrophic methanogens, Methanobacterium formicicum and Methanospirillum hungatei, was studied.
MATERIALS AND METHODS
Sources of enrichment cultures and microorganisms.
Methanospirillum hungatei (DSM 864T), Methanobacterium formicicum (DSM 1535T), and Methanosarcina mazei (DSM 2053T) were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany. Methanosaeta concilii strain GP-6 (DSM 3671T) was kindly provided by Caroline Plugge, Laboratory of Microbiology, Wageningen University, Wageningen, Netherlands.
The oleate- and palmitate-degrading enrichment cultures (OM and PM cultures, respectively) used in the present study have been previously described by Sousa et al. (19). OM and PM cultures were anaerobically grown in mineral medium containing 1 mM oleate (C18:1, unsaturated LCFA) or 1 mM palmitate (C16:0, saturated LCFA) as the sole carbon and energy sources. Both enrichment cultures were started from the same inoculum sludge originating from an anaerobic lab-scale reactor fed with LCFA-based effluent (19).
Medium composition and cultivation.
Methanogenic pure cultures and enrichment cultures were cultivated under strictly anaerobic conditions in a bicarbonate-buffered mineral salt medium (20). The medium was dispensed into serum bottles that were subsequently sealed with butyl rubber septa and aluminum crimp caps. The headspace of serum bottles was flushed with a mixture of H2/CO2 or N2/CO2 (80:20; 1.7 × 105 Pa). Bottles used for growing hydrogenotrophic methanogens (M. hungatei and M. formicicum) were flushed with H2/CO2, and bottles for growing acetoclastic methanogens (M. concilii and M. mazei) and the enrichment cultures were flushed with N2/CO2. The bottles were autoclaved for 20 min at 121°C. Before inoculation, mineral medium was reduced with 0.8 mM sodium sulfide (Na2S·7-9H2O). For growing the acetoclastic methanogens, sodium acetate was added to the medium to a final concentration of 50 mM. Oleate and palmitate (sodium salts) were added to the enrichment cultures to a final concentration of 1 mM. All of the inoculations and transfers were performed aseptically using sterile syringes and needles. Cultures were incubated statically at 37°C and in the dark.
Selection of methanogenic partners in LCFA enrichments.
To evaluate the selection of hydrogenotrophic partners in the enrichments, M. hungatei and M. formicicum (5% [vol/vol], indicated by “Mh” and “Mf”, respectively, in the following culture designations) were separately added to OM and PM enrichments as hydrogen scavengers. Cultures OM-Mh/PM-Mh and OM-Mf/PM-Mf were subcultured five times with 1 mM oleate-palmitate (underlining indicates the organism that was added to the enrichment cultures). Subsequently, to study the selection of acetoclastic methanogens in the enrichment cultures, OM-Mf and PM-Mh cultures were amended with M. concilii and M. mazei (indicated by “Mc” and “Mm”, respectively, in the following culture designations) in four independent enrichment series: OM-Mf-Mc, PM-Mh-Mc, OM-Mf-Mm, and PM-Mh-Mm. These cultures were incubated with 1 mM oleate-palmitate and subcultured two successive times. Culture samples (10 ml) were withdrawn from all of the enrichments before each transfer to evaluate the prevalence of methanogens by denaturing gradient gel electrophoresis (DGGE).
Methane production by pure cultures of hydrogenotrophic methanogens in the presence of LCFA.
Batch toxicity assays were performed to study the effect of unsaturated and saturated LCFA on pure cultures of M. hungatei and M. formicicum. Pure cultures of M. hungatei and M. formicicum were grown in 50 ml of basal medium using 120 ml serum bottles. The headspace was flushed and pressurized to 1.7 × 105 Pa with a mixture of H2 and CO2 (80:20). After autoclaving, the medium was inoculated with 10 ml of active methanogenic strains. Oleate, stearate (C18:0, saturated LCFA), and palmitate were added to exponentially growing cultures of M. hungatei and M. formicicum at different final concentrations (0.5, 1.0, 2.0, and 4.0 mM). Before LCFA addition, methane accumulated in the headspace was aseptically replaced by H2/CO2 (80:20; 1.7 × 105 Pa) in order to guarantee no substrate limitations during exposure to LCFA. A control assay with no LCFA addition was also performed. Experiments were conducted in triplicates. The vials were incubated at 37°C without agitation. Methane production was measured over time. Inhibition caused by the LCFA was measured by comparing the differences in methane production rate (Rm) between cultures with LCFA (Rm–LCFA) to that of control cultures (Rm–control). Inhibition of different concentrations of the LCFA was defined as the percentage of the methane production rate of cultures with LCFA compared to control cultures without LCFA addition.
| (1) |
Relative methane production rates were plotted against the concentration of the LCFA for calculation of the concentration of LCFA leading to 50% inhibition on methane production rate (50% inhibitory concentration [IC50]).
Effect of unsaturated LCFA on the membrane integrity of hydrogenotrophic methanogens.
The membrane integrity of M. hungatei and M. formicicum grown with H2/CO2 in the presence of 0.5 and 1 mM oleate was analyzed using a Live/Dead BacLight bacterial viability kit (Invitrogen Molecular Probes). Oleate was added to cultures growing on H2/CO2, at the beginning of the exponential growth phase; the bottle headspace was flushed with H2/CO2 (80:20, 1.7 × 105 Pa) before oleate addition. A control assay in the absence of oleate was performed. Cells were stained by dual fluorescent dyes, a green fluorescent nucleic acid stain, SYTO9, and a red fluorescent nucleic acid stain, propidium iodide (PI). SYTO9 stains all cells, but PI only stains cells with damaged membranes. For this reason, microorganisms with intact cell membranes stain green fluorescent, whereas microorganisms with damaged membranes stain red fluorescent. A 1-ml portion of culture sample was mixed in an Eppendorf tube with 500 μl of 0.85% NaCl and 0.5 μl of each stain, SYTO9 and PI, followed by incubation for 15 min at room temperature and in the dark. The mixture was then filtered through 0.2-μm-pore-size polycarbonate black filters (Whatman, Kent, United Kingdom). Filters were mounted with low-fluorescence immersion oil on glass microscope slides and observed on an epifluorescence microscope (Olympus BX51, ×60 magnification oil immersion objective lens) using fluorescein isothiocyanate (FITC) and Cy3 long-pass filters. The FITC filter reveals SYTO9-stained cells, corresponding to all cells, and the Cy3 filter reveals cells that are stained by both dyes and thus damaged. From each membrane, 10 to 20 randomly selected microscopic fields, each 0.0158 mm2, were photographed with a charge-coupled device camera (Olympus DP71) using an image acquisition software (Olympus Cell B). Images obtained with both filters and corresponding to the same microscopic fields were superimposed, and cells with damage and intact membranes were counted. All experiments were performed in duplicate. The percentage of membrane-damaged cells was calculated in relation to the total number of cells.
Analytical methods.
Methane was measured using a Pye Unicam GC-TCD gas chromatograph (Cambridge, England), with a Porapack Q (100 to 180 mesh) column. Helium was the carrier gas (30 ml min−1), and the temperatures of the injection port, column, and detector were 110, 35, and 110°C, respectively. Volatile fatty acids (VFA) were analyzed by high-pressure liquid chromatography from centrifuged (10,000 × g, 10 min) samples of the culture media. VFA were measured with a Polyspher OA HY column (300 by 6.5 mm; Merck, Darmstadt, Germany) and an RI SE-61 refractive index detector (Shodex, Tokyo, Japan). The mobile phase was 0.01 N H2SO4 at a flow rate of 0.6 ml min−1. The column temperature was 60°C.
DNA extraction and amplification.
Inoculum sludge used to start up the OM and PM enrichment series (5 ml), and pure cultures of methanogens (10 ml) and enrichment cultures OM and PM amended with methanogens (10 ml) were concentrated by centrifugation (10,509 × g, 10 min) at the time of sampling, frozen, and stored at −20°C. Total genomic DNA was extracted using a FastDNA Spin kit for soil (MP Biomedicals, USA) in accordance with the manufacturer's instructions. Archaeal 16S rRNA genes were amplified by PCR using a Taq DNA polymerase kit (Invitrogen, Carlsbad, CA); reaction mixtures and PCR programs used were as described elsewhere (16). The primer set A109(T)-f/515-r (21) was used for archaea16S rRNA gene amplification for DGGE. A 40-bp GC-clamp was added at the 5′-end sequence of the primer 515-r (22). Archaeal 16S rRNA genes were selectively amplified for cloning using the primer set Arch109-f/Uni1492-r (21, 23). Size and yield of PCR products were estimated by using a 100-bp DNA ladder (MBI Fermentas, Vilnius, Lithuania) via 1% (wt/vol) agarose gel electrophoresis and ethidium bromide staining.
DGGE analysis.
DGGE analysis of the PCR products was performed with the DCode system (Bio-Rad, Hercules, CA). Gels containing 8% (wt/vol) polyacrylamide (37.5:1 acrylamide-bisacrylamide) were used with a linear denaturing gradient of 30 to 50%, with 100% of denaturant corresponding to 7 M urea and 40% (vol/vol) formamide. Electrophoresis was performed for 16 h at 85 V and 60°C in a 0.5× Tris-acetate-EDTA buffer. DGGE gels were stained with silver nitrate (24) and scanned at 400 dpi.
Cloning and sequencing.
A 16S rRNA gene PCR-based clone library was constructed using the DNA extracted from the enrichments inoculum sludge. The PCR product was purified with Nucleo Spin Extract II kit (Clontech Laboratories) and cloned into Escherichia coli JM109 (NZYTech, Lisbon, Portugal) by using the Promega pGEM-T Easy vector system (Promega, Madison, WI) as previously described (16). Clones with the correct size insert were further amplified for DGGE comparison to the inoculum sample profile and enrichment cultures amended with methanogens. Plasmids of transformants (corresponding to predominant bands in the DGGE community fingerprint) were purified using the Nucleo Spin Extract II kit (Clontech Laboratories) and subjected to DNA sequence analysis. Sequencing reactions were performed at Macrogene (Amsterdam, Netherlands). The consensus sequences obtained were checked for potential chimera artifacts using Pintail v1.1 software (25).
Phylogenetic analysis.
Similarity searches for the 16S rRNA gene sequences were performed using the NCBI BLAST search program within the GenBank database (http://www.ncbi.nlm.nih.gov/blast/) (26).
Statistical analysis.
A modified Gompertz equation was used to describe the progress of cumulative methane production by pure culture methanogens (27). Equation 2 was used to calculate the maximum methane production rate in the toxicity assays:
| (2) |
where M(t) is the cumulative methane production (in mM) at time t, P is the maximum methane production (in mM), Rm is the maximum methane production rate (in mM day−1), e is 2.7182818, and λ is the lag-phase time (in days). For each assay, all of the individual measurements performed in the three replicates were utilized independently. R2 values and the standard errors for each variable were calculated. Significant differences between biological samples, during exposure to different concentrations of oleate, were determined using SPSS 19.0 statistic software. The Mann-Whitney U or Kruskal-Wallis tests were applied to nonviable cell counting data if two or more samples were compared at a time, respectively. The significance threshold was set at P < 0.05.
Nucleotide accession numbers.
The nucleotide sequences obtained in the present study have been deposited in the European Nucleotide Archive under accession numbers HF955499 to HF955506.
RESULTS
Selection of methanogenic partners in LCFA enrichments.
Predominant methanogens in the inoculum sludge used in OM and PM were identified by 16S rRNA gene cloning and sequencing (Table 1). Hydrogenotrophic community was dominated by microorganisms clustering within the Methanobacteriales and Methanomicrobiales orders. Methanosaeta- and Methanosarcina-related species were the acetoclastic predominant methanogens.
Table 1.
Affiliation of archaeal clones retrieved from OM and PM enrichments inoculum sludge samples
| Band | Clone | Closest relativea | % identity |
|---|---|---|---|
| 1 | C8-lcfa | Methanobacterium sp. strain OM15 | 99 |
| Methanobacterium formicicum MG-134 | 99 | ||
| 2 | G1-lcfa | Methanosaeta concilii GP-6 | 99 |
| 3 | A9-lcfa | Uncultured archaeon clone MCSArc_B6 | 98 |
| Methanoculleus bourgensis MS2 | 98 | ||
| 4 | A5-lcfa | Methanosaeta concilii GP-6 | 99 |
| 5 | A1-lcfa | Uncultured Methanospirillum sp. clone sagar115 | 99 |
| Methanospirillum hungatei JF-1 | 99 | ||
| 6 | A6-lcfa | Methanosarcina mazei Tuc01 | 96 |
| 7 | A2-lcfa | Methanosarcina mazei Tuc01 | 100 |
| 8 | G2-lcfa | Methanosarcina mazei Tuc01 | 99 |
Closest relative species of the selected clones determined by an NCBI Blast search. In cases where the first closest database hit was an uncultured microorganism, the first hit of a cultured microorganism is also given.
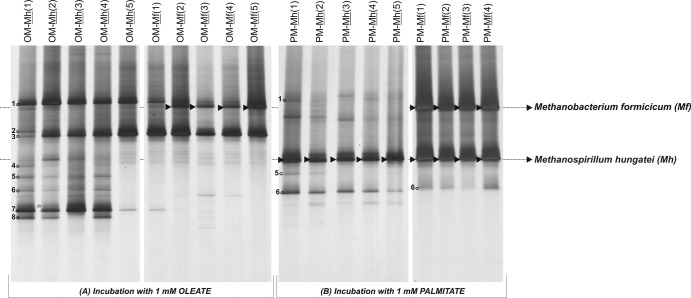
LCFA conversion relies on syntrophic interactions between acetogenic LCFA degraders and hydrogenotrophic methanogens (28). To guarantee high numbers of hydrogen-consuming methanogens in the enrichment procedure, it is current practice to supplement the enrichment cultures with pure cultures of hydrogenotrophs upon transfers. Methanospirillum hungatei and Methanobacterium formicicum are normally chosen as hydrogenotrophic partners for the enrichment and isolation of fatty acid-degrading bacteria (29). In the present study the prevalence of M. hungatei DSM 864T and M. formicicum DSM 1535T in OM and PM cultures was studied during five subsequent transfers of the LCFA-degrading enrichments. The presence or absence of these methanogens in enrichment cultures, after the degradation of 1 mM oleate or palmitate, was assessed by 16S rRNA gene PCR-DGGE (Fig. 1). M. hungatei succeeded to coexist with M. formicicum in the PM enrichment culture, even when M. formicicum was the amended methanogenic partner [Fig. 1B, lanes PM-Mf(1) to PM-Mf(4)]. However, in OM enrichments, M. hungatei did not prevail following its repeated addition to the enrichment cultures [Fig. 1A, lanes OM-Mh(1) to OM-Mh(5)]. M. formicicum sustained in the presence of oleate and grew in the OM enrichment cultures, as revealed by the presence of a predominant DGGE-band corresponding to this methanogen after the second transfer in oleate [Fig. 1A, lanes OM-Mf(2) to OM-Mf(5)].
Fig 1.
DGGE pattern of archaeal 16S rRNA gene fragments present in OM and PM enrichment cultures during successive transfers with addition of Methanospirillum hungatei (Mh) or Methanobacterium formicicum (Mf) as hydrogen consumers. Arrows indicate the presence of added methanogens in the enrichment cultures. Numbers 1 to 8 indicate the DGGE bands identified by cloning and sequencing (see the phylogenetic affiliations in Table 1); clones were retrieved from the inoculum sludge used to start up the enrichment series OM and PM.
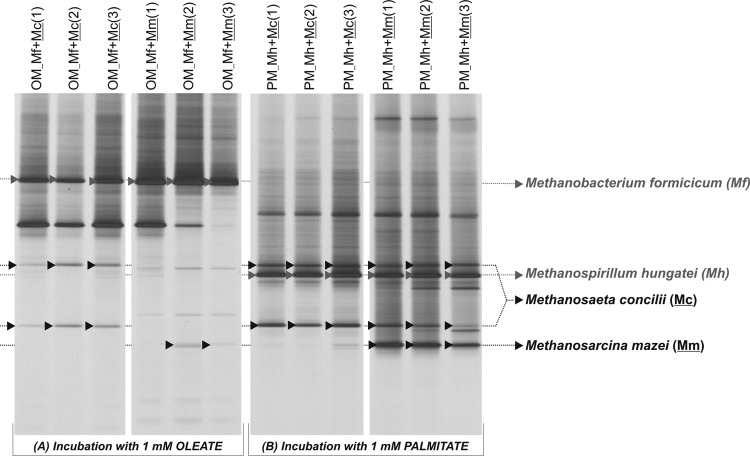
A similar approach was used to investigate the prevalence of the acetoclastic methanogens M. concilii and M. mazei in the OM and PM cultures. The hydrogenotrophic methanogens M. formicicum and M. hungatei were also added to the OM and PM enrichments, respectively (and in view of the previous results), to guarantee efficient hydrogen consumption. M. concilii and M. mazei were detected in OM and PM cultures as shown in Fig. 2. Nevertheless, DGGE bands corresponding to M. concilii and M. mazei showed higher relative intensity (compared to other bands in the same samples) in the PM than in the OM culture DGGE profile, which suggests that these methanogens were more abundant in the PM culture than in the OM culture (Fig. 2). No acetate was detected in PM cultures amended with M. concilii, which shows that this acetoclast could efficiently consume acetate derived from palmitate oxidation. M. mazei was not as efficient in maintaining acetate at low concentrations, likely due to its higher half-saturation coefficient (Ks) for acetate (30), which accumulated in the medium of cultures PM-Mh-Mm at a ratio of 6.6 mol of acetate/mol of palmitate added. In OM cultures, acetate accumulated in the presence of both M. concilii and M. mazei in amounts of 6.5 to 8.0 mol of acetate/mol of oleate added, which are close to stoichiometric values considering complete oleate oxidation. This indicates that acetoclastic activity was affected by oleate, which is coherent with the DGGE results. Methane production in OM cultures, 1.7 ± 0.2 mol of CH4/mol of oleate added, could be justified just by hydrogenotrophic activity.
Fig 2.
DGGE pattern of archaeal 16S rRNA gene fragments present in OM and PM enrichment cultures during successive transfers with addition of Methanosaeta concilii (Mc) or Methanosarcina mazei (Mm) as acetate consumers. Methanospirillum hungatei (Mh) and Methanobacterium formicicum (Mf) were added to PM and OM cultures, respectively, to ensure low hydrogen concentration during LCFA degradation. Arrows indicate the presence of added methanogens in the enrichment cultures.
Methane production by pure cultures of hydrogenotrophic methanogens in the presence of LCFA.
The hydrogenotrophic methanogens, M. hungatei and M. formicicum, were selected to further evaluate the differential toxic effect of unsaturated and saturated LCFA. For comparison, two C18 LCFA with different degree of saturation were used: oleate (unsaturated LCFA, C18:1) and stearate (saturated LCFA, C18:0). Palmitate (saturated LCFA, C16:0) was also selected to evaluate the potential effect of the hydrocarbon chain length. Methane production rate from H2/CO2 by pure cultures of M. hungatei and M. formicicum, in the presence of unsaturated and saturated LCFA, and respective half-inhibitory concentrations (IC50), are shown in Table 2. Overall, LCFA affected both M. hungatei and M. formicicum, as translated by a decrease in methane production rate in the presence of LCFA, even for LCFA concentrations as low as 0.5 mM (see Fig. S1 in the supplemental material).
Table 2.
Methane production rate by hydrogenotrophic methanogens in the presence of different concentrations of LCFA and the concentration required for 50% inhibition (IC50) of the hydrogenotrophic methanogens exposed to LCFA
| LCFA | Concn (mM) | MPR (mM CH4 day−1)a and IC50 (mM) |
|||
|---|---|---|---|---|---|
|
Methanospirillum hungatei |
Methanobacterium formicicum |
||||
| MPR | IC50 | MPR | IC50 | ||
| No LCFA | 4.24 ± 0.27 | 1.81 ± 0.13 | |||
| Oleate | 0.5 | 1.58 ± 0.12 | 0.3 | 1.82 ± 0.10 | 1.0 |
| 1.0 | 1.49 ± 0.18 | 0.3 | 0.86 ± 0.04 | 1.0 | |
| 2.0 | 1.31 ± 0.13 | 0.3 | 0.29 ± 0.06 | 1.0 | |
| 4.0 | 1.01 ± 0.15 | 0.3 | 0.12 ± 0.01 | 1.0 | |
| Stearate | 0.5 | 1.81 ± 0.35 | 0.4 | 1.78 ± 0.12 | >4.0 |
| 1.0 | 2.38 ± 0.33 | 0.4 | 1.79 ± 0.15 | >4.0 | |
| 2.0 | 2.04 ± 0.20 | 0.4 | 1.44 ± 0.16 | >4.0 | |
| 4.0 | 1.11 ± 0.14 | 0.4 | 1.05 ± 0.09 | >4.0 | |
| Palmitate | 0.5 | 2.69 ± 0.32 | 1.0 | 1.62 ± 0.13 | >4.0 |
| 1.0 | 2.39 ± 0.27 | 1.0 | 1.08 ± 0.17 | >4.0 | |
| 2.0 | 0.67 ± 0.07 | 1.0 | 0.95 ± 0.04 | >4.0 | |
| 4.0 | 0.50 ± 0.07 | 1.0 | 1.05 ± 0.09 | >4.0 | |
The methane production rate (MPR) values were calculated by fitting experimental data (triplicates) to the Gompertz model (27): 0.901 < R2 < 0.987.
The effect of oleate on M. hungatei was severe, with only 0.5 mM oleate causing a decrease of ca. 60% in methane production rate from H2/CO2. The same oleate concentration had no major effect on the methane production rate of M. formicicum, but concentrations of >1 mM had a strong effect on this methanogen as well. A reduction in methane production rate of 84 to 93% was observed in the presence of an oleate concentration of ≥2 mM (i.e., from 1.81 ± 0.13 mM CH4 day−1 [in the control assay] to 0.29 ± 0.06 and 0.12 ± 0.01 mM CH4 day−1 in the presence of 2 and 4 mM oleate, respectively [Table 2]). Stearate and palmitate were less inhibitory to M. formicicum than to M. hungatei cultures. A stearate or palmitate concentration of 4 mM caused <50% inhibition of M. formicicum pure cultures, whereas the M. hungatei IC50s were 0.4 and 1 mM for stearate and palmitate, respectively.
Effect of unsaturated LCFA on membrane integrity of hydrogenotrophic methanogens.
The effect of oleate on membrane integrity of M. hungatei and M. formicicum was studied in order to further evaluate the resistance of these two microorganisms toward the presence of unsaturated LCFA (Fig. 3).
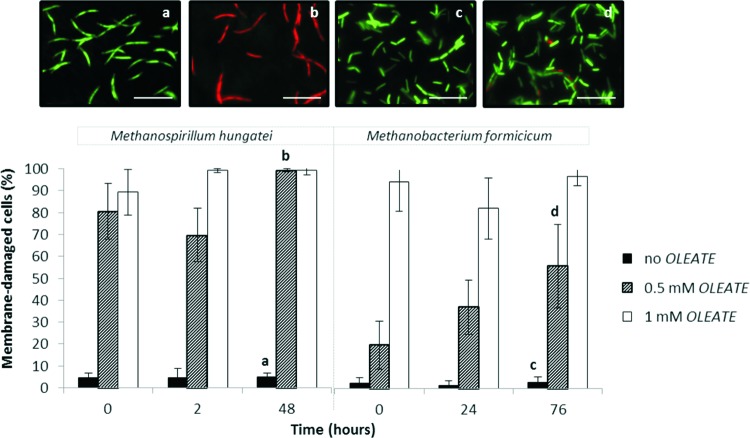
Fig 3.
Percentage of membrane damage cells of M. hungatei and M. formicicum in pure cultures when grown on H2/CO2, without LCFA addition and in the presence of 0.5 and 1 mM oleate. Live/dead staining images of M. hungatei and M. formicicum are shown in subpanels a to d: the letters correlate the images with the assay and the sampling time indicated by the corresponding letter in the graph. Scale bar, 10 μm.
Membrane integrity of M. hungatei and M. formicicum cells was affected by the presence of oleate. The adverse effect of oleate on membrane integrity of M. hungatei is obvious, even for the lowest oleate concentration tested and immediately after the LCFA addition. The percentage of M. hungatei damaged cells just after the supplementation of 0.5 mM oleate to the medium was as high as 81% ± 13% compared to 5% ± 2% of cells damaged in the control assay (P = 0.003). After 48 h of exposure of this microorganism to 0.5 or 1 mM oleate, ca. 99% of the cells were damaged, whereas in the control assay damaged cell membranes represented <7% of all of the cells (P < 0.001). M. formicicum could endure better the lower concentration of oleate (0.5 mM), with just 56% ± 19% cells damaged after 76 h of incubation (this value is significantly different from the percentage of M. hungatei damaged cells measured after 48 h contact with 0.5 mM oleate [P < 0.001]). Nevertheless, oleate at a final concentration of 1 mM had a similar fast and severe injuring effect on the cell membranes of both methanogens with ca. 90% of the cells damaged just after the addition of the inhibitor.
DISCUSSION
This research shows that the response of methanogens to saturated and unsaturated LCFA is diverse. The prevalence of M. hungatei and M. formicicum in LCFA-degrading enrichments was dependent on the saturation degree of the fatty acids (Fig. 1). M. formicicum could prevail in OM and PM enrichments, but M. hungatei was only detected in PM enrichments (Fig. 1). Considering the absolute methane production rate, H2/CO2 conversion to methane by M. hungatei is faster than by M. formicicum, even in the presence of LCFA (Table 2). However, the decrease in the methane production rate in cultures incubated with LCFA compared to the control assays (without LCFA) is noticeably sharper in M. hungatei cultures. As a consequence, a lower IC50 for oleate was determined for M. hungatei (0.3 mM) than for M. formicicum (1 mM). OM enrichments were amended with 1 mM oleate, which can partially explain the endurance of M. formicicum in the OM enrichments. Other factors might affect the structure and methanogenic composition of LCFA-degrading enrichment mixed cultures; the existence of syntrophic interactions probably has an important role in the selection of certain methanogens. M. hungatei has been isolated from sewage sludge (31) and, although frequently detected in anaerobic reactors, Methanobacterium species seem to have the highest predominance in digesters sludge (32). Moreover, Methanobacterium-related microorganisms have been detected as the predominant hydrogen scavengers in oleate-contacting sludges (15, 16, 33). Microorganisms closely related to both of these species were also detected in the inoculum sludge used in OM and PM enrichments (Table 1), which has previously been in contact with LCFA in a continuous-batch reactor, indicating their importance during the degradation of these substrates. M. hungatei and M. formicicum are metabolically similar, but their cell envelope properties are different (34). M. formicicum cells have a rigid pseudomurein wall, whereas M. hungatei cells have a proteinaceous cell surface structure and are normally covered by a sheath that encloses several cells and makes the cells in the middle of the filament less permeable (35, 36). The membrane lipid composition in M. hungatei and M. formicicum membranes is also different (36–38). M. formicicum has four different phospholipids (inositol, ethanolamine, serine, and aminopentanetetrol) compared to only two in M. hungatei (glycerol and aminopentanetetrol). Differences in the membrane composition between the two tested hydrogenotrophs may be related to the differential vulnerability to LCFA. In fact, membrane damage in M. hungatei was more severe than in M. formicicum, as evidenced in the cell viability assays that we performed (Fig. 3). Jarrell et al. (39) verified that, when exposed to ammonia, butyrate, and propionate, M. hungatei was more sensitive than M. formicicum, a finding that is in line with the higher susceptibility of M. hungatei to LCFA, as observed in our work. It should be noted that toxicity and live/dead assays were performed with pure cultures of methanogens; in a bioreactor, and as part of complex syntrophic communities, methanogens might have extra protection to endure higher LCFA concentrations.
The acetoclastic methanogens, M. mazei and M. concilii, were both successfully integrated in PM cultures (Fig. 2), but in OM cultures acetate accumulated to almost stoichiometric concentrations to the corresponding complete oleate oxidation. Also, for the tested acetoclasts, oleate seems to have a more stringent effect than palmitate. This could be related to a higher susceptibility of acetoclastic methanogens to oleate, as happens with hydrogenotrophic methanogens. The study by Sousa et al. (16) describes OM and PM enrichments. Throughout a 2-year period, OM and PM cultures were transferred to enrich for oleate and palmitate degraders; at the moment of the transfers, no acetoclastic methanogens were supplemented. Stable OM enrichments could convert oleate, but acetate accumulated in the medium and was not further converted to methane. Conversely, microbial communities in PM enrichments could convert acetate completely to methane. This indicates that acetoclasts were eliminated from OM cultures over the enrichment process, which can be due to low growth rates of these microorganisms in the presence of oleate. Live/dead assays could give further insights on membrane damage of acetoclasts by LCFA. However, both Methanosaeta and Methanosarcina species form dense aggregates, and we were not able to perform single cell counting. Nevertheless, it was clear from the OM and PM incubations that oleate is more toxic than palmitate to both M. concilii and M. mazei. It has been reported that the toxicity of fatty acids increases with the degree of unsaturation of the molecule (11). Prins et al. (11) determined the IC50 values for pure cultures of rumen methanogens in the presence of linolenate (C18:3) and linoleate (C18:2), observing IC50 values ∼1.8-fold higher for linolenate. However, the mechanisms underlying this differential effect are not completely understood. The bent conformation of unsaturated and polyunsaturated LCFA might influence the way that these acids interact with cell membranes, potentiating their toxicity. The results presented here show that LCFA have an effect on membrane integrity of methanogens, and this effect is more pronounced when considering unsaturated LCFA. These findings should stimulate further investigation on the mechanisms of methanogenic inhibition by LCFA.
Supplementary Material
ACKNOWLEDGMENTS
This study was financially supported by FEDER funds through the Operational Competitiveness Programme (COMPETE) and by national funds through the Portuguese Foundation for Science and Technology (FCT) in the frame of the projects FCOMP-01-0124-FEDER-007087 and FCOMP-01-0124-FEDER-014784. Financial support from the FCT and the European Social Fund (POPH-QREN) through Ph.D. grant SFRH/BD/48960/2008 to A.F.S. is also acknowledged. A.J.M.S. was supported by grants from ALW-TOP (project 700.55.343) and ERC (project 323009).
Footnotes
Published ahead of print 3 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00035-13.
REFERENCES
- 1. Alves MM, Pereira MA, Sousa DZ, Cavaleiro AJ, Picavet M, Smidt H, Stams AJM. 2009. Waste lipids to energy: how to optimize methane production from long-chain fatty acids. Microb. Biotechnol. 2:538–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cavaleiro AJ, Salvador AF, Alves JI, Alves M. 2009. Continuous high rate anaerobic treatment of oleic acid based wastewater is possible after a step feeding start-up. Environ. Sci. Technol. 43:2931–2936 [DOI] [PubMed] [Google Scholar]
- 3. Pereira MA, Sousa DZ, Mota M, Alves MM. 2004. Mineralization of LCFA associated with anaerobic sludge: kinetics, enhancement of methanogenic activity, and effect of VFA. Biotechnol. Bioeng. 88:502–511 [DOI] [PubMed] [Google Scholar]
- 4. Hanaki K, Nagase M, Matsuo T. 1981. Mechanism of inhibition caused by long-chain fatty acids in anaerobic digestion process. Biotechnol. Bioeng. 23:1591–1610 [Google Scholar]
- 5. Koster IW, Cramer A. 1987. Inhibition of methanogenesis from acetate in granular sludge by long-chain fatty acids. Appl. Environ. Microbiol. 53:403–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hwu C-S, Lettinga G. 1997. Acute toxicity of oleate to acetate-utilizing methanogens in mesophilic and thermophilic anaerobic sludges. Enzyme. Microb. Technol. 21:297–301 [Google Scholar]
- 7. Angelidaki I, Ahring BK. 1992. Effects of free long-chain fatty acids on thermophilic anaerobic digestion. Appl. Microbiol. Biotechnol. 37:808–812 [DOI] [PubMed] [Google Scholar]
- 8. Rinzema A, Boone M, van Knippenberg K, Lettinga G. 1994. Bactericidal effect of long chain fatty acids in anaerobic digestion. Water Environ. Res. 66:40–49 [Google Scholar]
- 9. Soliva CR, Meile L, Hindrichsen IK, Kreuzer M, Machmuller A. 2004. Myristic acid supports the immediate inhibitory effect of lauric acid on ruminal methanogens and methane release. Anaerobe 10:269–276 [DOI] [PubMed] [Google Scholar]
- 10. Demeyer DI, Henderic HK. 1967. Effect of C18 unsaturated fatty acids on methane production in vitro by mixed rumen bacteria. Biochim. Biophys. Acta 137:484–497 [DOI] [PubMed] [Google Scholar]
- 11. Prins RA, van Nevel CJ, Demeyer DI. 1972. Pure culture studies of inhibitors for methanogenic bacteria. Antonie van Leeuwenhoek 38:281–287 [DOI] [PubMed] [Google Scholar]
- 12. Koster IW. 1987. Abatement of long-chain fatty acid inhibition of methanogenesis by calcium addition. Biol. Waste 22:295–301 [Google Scholar]
- 13. Roy F, Albagnac G, Samain E. 1985. Influence of calcium addition on growth of highly purified syntrophic cultures degrading long-chain fatty acids. Appl. Environ. Microbiol. 49:702–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Soliva CR, Bucher S, Meile L, Kreuzer M. 2009. Methanogenesis archaea: conditions for inhibitory action of four saturated fatty acids when added to pure cultures, abstr. 242039. Abstr. IOP Conf. Ser. Earth Environ. Sci. [Google Scholar]
- 15. Salvador AF, Cavaleiro AJ, Sousa DZ, Alves MM, Pereira MA. 2013. Endurance of methanogenic archaea in anaerobic bioreactors treating oleate-based wastewater. Appl. Microbiol. Biotechnol. 97:2211–2218 [DOI] [PubMed] [Google Scholar]
- 16. Sousa DZ, Pereira MA, Smidt H, Stams AJM, Alves MM. 2007. Molecular assessment of complex microbial communities degrading long chain fatty acids in methanogenic bioreactors. FEMS Microbiol. Ecol. 60:252–265 [DOI] [PubMed] [Google Scholar]
- 17. McInerney MJ, Struchtemeyer CG, Sieber J, Mouttaki H, Stams AJM, Schink B, Rohlin L, Gunsalus RP. 2008. Physiology, ecology, phylogeny, and genomics of microorganisms capable of syntrophic metabolism. Ann. N. Y. Acad. Sci. 1125:58–72 [DOI] [PubMed] [Google Scholar]
- 18. Schink B, Stams AJM. 2006. Syntrophism among prokaryotes, p 309–335 In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E. (ed), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed, vol 2 Springer-Verlag, New York, NY [Google Scholar]
- 19. Sousa DZ, Pereira MA, Stams AJM, Alves MM, Smidt H. 2007. Microbial communities involved in anaerobic degradation of unsaturated or saturated long chain fatty acids (LCFA). Appl. Environ. Microbiol. 73:1054–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stams AJM, van Dijk JB, Dijkema C, Plugge CM. 1993. Growth of syntrophic propionate-oxidizing bacteria with fumarate in the absence of methanogenic bacteria. Appl. Environ. Microbiol. 59:1114–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grosskopf R, Janssen PH, Liesack W. 1998. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl. Environ. Microbiol. 64:960–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Muyzer G, de Waal EC, Uitterlinden AG. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175 In Stackebrandt E, Goodfellow M. (ed), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom [Google Scholar]
- 24. Sanguinetti CJ, Dias Neto E, Simpson AJ. 1994. Rapid silver staining and recovery of PCR products separated on polyacrylamide gels. Biotechniques 17:914–921 [PubMed] [Google Scholar]
- 25. Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71:7724–7736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Altschul SF, Gish W, Miller W, Meyers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 27. Lay JJ, Li YY, Noike T. 1998. Mathematical model for methane production from landfill bioreactor. J. Environ. Eng. ASCE 124:730–736 [Google Scholar]
- 28. Sousa DZ, Smidt H, Alves MM, Stams AJM. 2009. Ecophysiology of syntrophic communities that degrade saturated and unsaturated long-chain fatty acids. FEMS Microbiol. Ecol. 68:257–272 [DOI] [PubMed] [Google Scholar]
- 29. Stams AJM, Sousa DZ, Kleerebezem R, Plugge CM. 2012. Role of syntrophic microbial communities in high-rate methanogenic bioreactors. Water. Sci. Technol. 66:352–362 [DOI] [PubMed] [Google Scholar]
- 30. Jetten MSM, Stams AJM, Zehnder AJB. 1992. Methanogenesis from acetate: a comparison of the acetate metabolism in Methanothrix soehngenii and Methanosarcina spp. FEMS Microbiol. Rev. 88:181–197 [Google Scholar]
- 31. Ferry JG, Smith PH, Wolfe RS. 1974. Methanospirillum, a new genus of methanogenic bacteria, and characterization of Methanospirillum hungatii sp. nov. Int. J. Syst. Bacteriol. 24:465–469 [Google Scholar]
- 32. Leclerc M, Delgenes JP, Godon JJ. 2004. Diversity of the archaeal community in 44 anaerobic digesters as determined by single strand conformation polymorphism analysis and 16S rDNA sequencing. Environ. Microbiol. 6:809–819 [DOI] [PubMed] [Google Scholar]
- 33. Pereira MA, Roest K, Stams AJM, Mota M, Alves M, Akkermans ADL. 2002. Molecular monitoring of microbial diversity in expanded granular sludge bed (EGSB) reactors treating oleic acid. FEMS Microbiol. Ecol. 41:95–103 [DOI] [PubMed] [Google Scholar]
- 34. Whitman WB, Bowen TL, Boone DR. 2006. The methanogenic bacteria, p 165–207 In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E. (ed), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed, vol 3 Springer-Verlag, New York, NY [Google Scholar]
- 35. Beveridge TJ, Sprott GD, Whippey P. 1991. Ultrastructure, inferred porosity, and Gram staining character of Methanospirillum hungatei filament termini describe a unique cell permeability for this archaeobacterium. J. Bacteriol. 173:130–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Koga Y, Nishihara M, Morii H, Akagawamatsushita M. 1993. Ether polar lipids of methanogenic bacteria: structures, comparative aspects, and biosyntheses. Microbiol. Rev. 57:164–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Koga Y, Morii H. 2005. Recent advances in structural research on ether lipids from archaea including comparative and physiological aspects. Biosci. Biotechnol. Biochem. 69:2019–2034 [DOI] [PubMed] [Google Scholar]
- 38. Koga Y, Morii H, Akagawa-Matsushita M, Ohga I. 1998. Correlation of polar lipid composition with 16S rRNA phylogeny in methanogens: further analysis of lipid component parts. Biosci. Biotechnol. Biochem. 62:230–236 [DOI] [PubMed] [Google Scholar]
- 39. Jarrell KF, Saulnier M, Ley A. 1987. Inhibition of methanogenesis in pure cultures by ammonia, fatty acids, and heavy metals, and protection against heavy metal toxicity by sewage sludge. Can. J. Microbiol. 33:551–554 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.