Abstract
Quantitative assessment of growth of filamentous microorganisms, such as streptomycetes, is generally restricted to determination of dry weight. Here, we describe a straightforward methylene blue-based sorption assay to monitor microbial growth quantitatively, simply, and rapidly. The assay is equally applicable to unicellular and filamentous bacterial and eukaryotic microorganisms.
TEXT
The quantification of small cell amounts of filamentous and aggregative microorganisms, like the saprophytic bacterium Streptomyces coelicolor, is methodologically limited (1). Turbidity (optical density) is often used to determine bacterial density, but it is usually restricted to nonaggregative unicellular microorganisms (2). Generally, quantitative assessment of growth of filamentous microorganisms typically involves dry-weight determination (3, 4). Nevertheless, it remains a challenge to monitor growth of pure cultures of Streptomyces below 1 mg (dry weight) of material under laboratory conditions. A potential strategy to measure microbial growth indirectly involves the use of a dye-based method. Methylene blue is a tricyclic phenothiazine cationic dye (5, 6) that reversibly adsorbs to anionic biological material (7, 8). A recent study showed the utility of methylene blue to visualize small differences in the amount of S. coelicolor mycelium growing in minimal medium (9). Here we have developed this method further to determine cell amounts/biomass of filamentous microorganisms quantitatively by determining the amount of methylene blue that adsorbs to, or desorbs from, cells. We demonstrate the applicability of this methylene blue-based assay for Streptomyces coelicolor, Penicillium chrysogenum, and both unicellular and filamentous cyanobacterial species.
Two parameters that give a quantitative assessment of methylene blue bound to anionic cellular structures can be monitored. The first parameter is the amount of dye remaining in solution after adsorption of dye by cells; the second parameter is the amount of dye desorbed from those cells by altering the buffer or pH (7).
Streptomyces coelicolor cells adsorb quantifiable amounts of methylene blue.
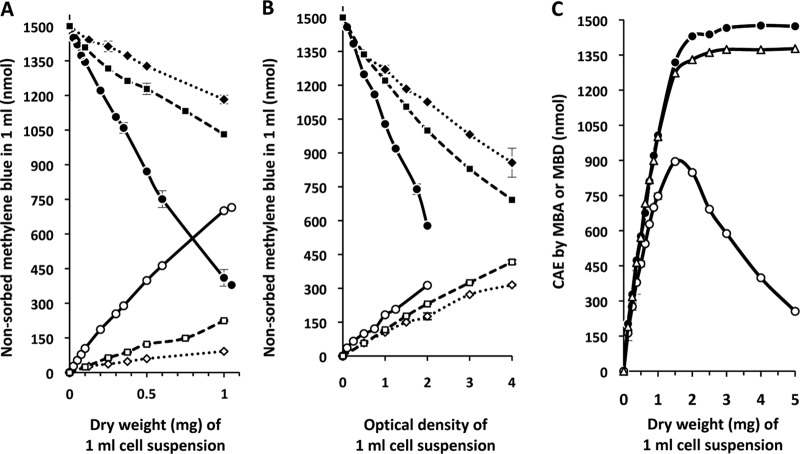
To develop a general method to determine the amount of methylene blue bound by S. coelicolor, unwashed cells from 1 ml of an exponentially growing culture in tryptic soy broth (TSB) (10) were mixed with 1 ml of a filtered, 1.5 mM aqueous methylene blue solution and heat inactivated, and the amount of dye adsorbed by the cells was calculated after subtracting the dye remaining in the supernatant after centrifugation (see the supplemental material for details of the method). Optimal staining conditions and analysis of factors such as medium components, ionic strength, and pH, which influence adsorption, were determined empirically (see Table S1 in the supplemental material). The analysis revealed a direct correlation between the amount of dye adsorbed and the dry weight of S. coelicolor cell material (Fig. 1A). The method proved to be applicable for other filamentous microbes, including cyanobacteria and Penicillium chrysogenum, as well as for unicellular microbes, such as Escherichia coli and Saccharomyces cerevisiae (Fig. 1A and B).
Fig 1.
Characterization of methylene blue binding by different microorganisms. (A and B) Linear correlations between adsorbed (filled symbols) or desorbed (open symbols) methylene blue (measured at 660 nm) and dry weight (mg) (A) or optical density value (measured at 600 nm or 750 nm) (B) for a variety of microorganisms are shown. (A) The organisms analyzed were Streptomyces coelicolor M145 (full line; circles), Penicillium chrysogenum Ib (dashed line; squares), and Anabaena sp. PCC7120 (dotted line; diamonds). (B) The organisms analyzed were Escherichia coli K-12 (full line; circles), Synechococcus elongatus PCC7942 (dashed line; squares), and Saccharomyces cerevisiae (dotted line; diamonds). The optical density of S. elongatus PCC7942 was measured at 750 nm, and that of the other species was measured at 600 nm. (C) Methylene blue adsorption and desorption curves for S. coelicolor at 25°C. Methylene blue adsorption (filled circles) and desorption (open symbols) curves (measured at 660 nm) are shown in relation to the cell amount equivalents (CAE) determined by dry weight. Desorption curves with either 250 mM HCl (open triangles) or 500 mM potassium phosphate buffer, pH 7 (open circles), as the solvent are shown. All assays were performed with a staining solution of 1,500 nmol ml-1 methylene blue solution, and experiments were repeated at least three times.
Quantitation of methylene blue desorption.
Desorption of methylene blue from cell material can be achieved by treatment of stained cell pellets with acid (7, 8). Quantitative desorption of methylene blue bound to S. coelicolor cells was able to be demonstrated with 250 mM HCl (see Fig. S1 in the supplemental material). The assay revealed a linear correlation between the dry weight of cell material and the amount of dye released. Other potential desorption solutions were empirically tested (see the supplemental material). Five hundred millimolar potassium phosphate buffer and 500 mM MOPS (morpholinepropanesulfonic acid)-NaOH proved equally effective at desorbing methylene blue bound to cells, while ethanol was ineffective (see Fig. S1 in the supplemental material). These data indicate that desorption with either HCl or 500 mM potassium phosphate buffer can provide a quantitative assessment of the amount of cell material present in small volumes of mycelial cultures.
Adsorption/desorption capacities of different model microorganisms.
The methylene blue quotient (MBQ) is the ratio of methylene blue desorption (MBD) with mild solvent (KP buffer) to methylene blue adsorption (MBA), and for exponentially growing cells of S. coelicolor in TSB, it was determined to be 0.69 (Table 1). Remarkably, regardless of which medium (TSB, YEME, or MAP) (10) was used for growth of S. coelicolor, the MBQ remained unchanged; only negligible changes in the absolute values for adsorption and desorption occurred. Only after growth of S. coelicolor in minimal medium was a small reduction in the desorption capacity observed (Table 1). This finding suggested that each microorganism should have a defined MBQ which is governed by its complement of anionic biomolecules. The MBQ values for E. coli and S. cerevisiae in relation to the cell mass contained in 1 ml of an exponentially growing cell suspension with an optical density at 600 nm (OD600) of 1, as well as for P. chrysogenum with 1 mg (dry weight) of cell material, were determined. For all three organisms, it was possible to quantify reproducibly the adsorption and desorption capacities by using the same assay conditions (Table 1). Notably, each organism exhibited a different percentage of dye that was able to be adsorbed to, or desorbed with KP buffer from, its cell material. This probably reflects differences in the cell composition between the species. For example, S. coelicolor appears to have a large number of adsorption sites with a low energy of adsorption, so that nearly 70% of the adsorbed dye was able to be detached by 500 mM potassium phosphate. For E. coli, the quotient of adsorption and desorption was 50%, whereas for the eukaryotes S. cerevisiae and P. chrysogenum, only 40% of the adsorbed methylene blue was able to be desorbed under the conditions used (Table 1).
Table 1.
Methylene blue adsorption/desorption capacity of different microorganisms determined with a standardized protocola
| Organism | Mediumb | MBAc |
MBDd |
MBQ (MBD/MBA)d | ||
|---|---|---|---|---|---|---|
| nmol/OD | nmol/mgdw | nmol/OD | nmol/mgdw | |||
| Escherichia coli | DNB | 354 ± 7 | 827 ± 63 | 177 ± 16 | 413 ± 37 | 0.5 |
| Saccharomyces cerevisiae | YPD | 237 ± 31 | 667 ± 87 | 100 ± 14 | 281 ± 39 | 0.42 |
| Streptomyces coelicolor M145 | TSB | NDe | 1,029 ± 82 | ND | 715 ± 47 | 0.69 |
| YEME | ND | 891 ± 58 | ND | 609 ± 43 | 0.68 | |
| MAP | ND | 985 ± 77 | ND | 677 ± 51 | 0.69 | |
| Minimal | ND | 1,030 ± 17 | ND | 623 ± 6 | 0.6 | |
| Penicillium chrysogenum | MAP | ND | 389 ± 45 | ND | 148 ± 39 | 0.38 |
Desorption was carried out with 500 mM potassium phosphate buffer, pH 7. MBA, methylene blue adsorption; MBD, methylene blue desorption; MBQ, quotient of methylene blue desorption/adsorption. Experiments were repeated at least four times, and the standard deviations are indicated.
DNB, Difco nutrient broth; YPD, yeast extract peptone dextrose medium; TSB, tryptone soy broth; YEME, yeast extract malt extract medium; MAP, malt extract peptone medium. See the supplemental material for medium composition.
The optical density was measured at 600 nm, the absorbance of methylene blue was measured at 660 nm, and the dry weight of cell material is presented in mg (mgdw).
The solvent was 500 mM KH2PO4/K2HPO4 buffer, pH 7.
ND, not determined.
Correlation between methylene blue assay and microbial biomass.
To demonstrate whether the described methylene blue adsorption/desorption assay is applicable to monitor growth of a variety of microorganisms, we compared the linear correlations between methylene blue adsorption (MBA) and desorption (MBD) and established cell amount equivalent (CAE) values (equivalent to bacterial density) obtained by determining dry weight or performing optical density measurements (Fig. 1A and B). Exponential cultures of S. coelicolor, P. chrysogenum, and Anabaena (Fig. 1A) and E. coli, Synechococcus, and S. cerevisiae (Fig. 1B) were analyzed using the methylene blue assay. Regardless of whether the microorganism was unicellular or filamentous, bacterial or eukaryotic, the CAE correlation proved to be linear.
Using either adsorption or desorption with 250 mM HCl, this method was able to accurately assess S. coelicolor cell amounts in a dry weight range between 0 and 1 mg (Fig. 1C). Accurate biomass determination above this level was limited only by the low concentration (1,500 nmol) of methylene blue used in the assay. Notably, desorption of incompletely stained cells with the mild solvent 0.5 M potassium phosphate revealed that less dye was able to be desorbed, probably reflecting interaction of the dye with strong adsorption sites (Fig. 1C).
Because filamentous microorganisms like S. coelicolor also grow as aggregates (11), we wished to demonstrate the robustness of the methylene blue assay for the analysis of different-sized S. coelicolor aggregates. Remarkably, the methylene blue assay also allowed the determination of the cell amounts of individual filamentous aggregates of both S. coelicolor, with pellet sizes ranging between 1.5 and 5 mm (dry weight between 0.1 and 1.4 mg) (Fig. 2A), and P. chrysogenum, with pellet sizes ranging between 2 and 8 mm (dry weight between 0.7 and 6 mg) (Fig. 2B; see also the supplemental material for details of the methods used).
Fig 2.
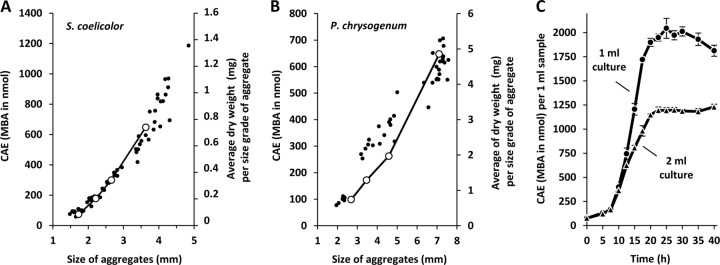
Application of the methylene blue adsorption assay for quantitative growth determination. (A and B) Absolute CAE of individual filamentous aggregates/pellets of S. coelicolor (A) and Penicillium chrysogenum (B) with various sizes (open circles) were determined by methylene blue adsorption and compared with the dry weights (full circles) determined for the individual aggregates. The latter was calculated as the average of several aggregates dried together to a minimal measureable level of 5 mg. (C) Aerobic growth of S. coelicolor at 30°C and in 1 ml (circles) or 2 ml (triangles) of TSB (30 g/liter, containing 100 mM MOPS-NaOH, pH 7.0) is shown. Cell amount equivalents (CAE) were measured by methylene blue adsorption (MBA). Results with standard deviations of four independent experiments are shown.
Finally, to demonstrate that different growth phases do not significantly affect the methylene blue assay, we showed for E. coli (see Fig. S2 in the supplemental material) and for S. cerevisiae (data not shown) that the relationship between the indirect CAE of optical density and MBA and MBD remained the same throughout the growth curve.
Proof of principle: small-scale growth curves of Streptomyces coelicolor.
Due to the high resolution and sensitivity of the methylene blue assay for the determination of cell amounts compared with measurement of dry weight, we tested the efficacy of the assay by reducing the culture volume from the flask scale (minimal volume of 20 ml) (12) to the 1- to 2-ml scale in microtiter plates (3-ml wells). In this case, each well represented an individual sample point (see the supplemental material for details). In order to ensure dispersed growth of filamentous S. coelicolor, all wells in the microtiter plates were filled with 5 glass beads to facilitate stirring, and the plates contained either 1 ml or 2 ml of TSB medium for comparison. The whole growth curve was performed independently with three different spore suspensions in triplicate (Fig. 2C). Growth occurred mainly between 10 and 20 h, and subsequent to 20 h, the amount of methylene blue adsorbed to cells had reached its maximum value. This correlated with the appearance of red coloring of the cells in the well-aerated 1-ml wells, suggesting the initiation of undecylprodigiosin production (13, 14). Production of antibiotics did not interfere with the methylene blue assay. The amounts of oxygen available to the cells in the 1- and 2-ml wells were comparable immediately after spore inoculation (same density) and in early growth. After further growth, however, and because the air-to-water interface (in stasis, 186 mm2) was the same in both wells, the oxygen in the 2-ml wells was more rapidly reduced than that in the 1-ml wells. This implies that as growth proceeded, the oxygen available per CAE (cell amount equivalent) became limiting more rapidly in the 2-ml well. Consequently, for an aerobic organism, the growth of individual cells slowed down, and in this case, the maximum value of the CAE (concentration of ca. 1,200 nmol/ml) was roughly half of that adsorbed in the 1-ml wells (Fig. 2C).
The data presented in this study highlight a facile methylene blue assay applicable to both unicellular and filamentous microbes, regardless of their phylogeny. The assay allows accurate determination of growth parameters at high resolution for S. coelicolor using only 1-ml cultures. The assay is quantitative, incurs a minimal cost, and requires only standard laboratory equipment. Each microorganism has its own reproducible methylene blue quotient (ratio of adsorbed to desorbed dye), and therefore, the assay may be used to analyze cell surface mutants or for the study of growth of biofilms.
Supplementary Material
ACKNOWLEDGMENTS
We thank Karl Forchhammer (University of Tübingen, Germany) for providing cell material of the cyanobacterial species.
This work was supported by the Deutsche Forschungsgemeinschaft (grant Sa 494/4-1) and the region of Saxony-Anhalt.
Footnotes
Published ahead of print 10 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00778-13.
REFERENCES
- 1. Hopwood DA. 2006. Soil to genomics: the Streptomyces chromosome. Annu. Rev. Genet. 40: 1–23 [DOI] [PubMed] [Google Scholar]
- 2. Dalgaard P, Ross T, Kamperman L, Neumeyer K, McMeekin TA. 1994. Estimation of bacterial growth rates from turbidimetric and viable count data. Int. J. Food Microbiol. 23: 391–404 [DOI] [PubMed] [Google Scholar]
- 3. Monod J. 1949. The growth of bacterial cultures. Annu. Rev. Microbiol. 3: 371–394 [Google Scholar]
- 4. Mallette MF. 1969. Evaluation of growth by physical and chemical means. Methods Microbiol. 1: 521–566 [Google Scholar]
- 5. Bergmann K, O'Konski CT. 1963. A spectroscopic study of methylene blue monomer, dimer and complexes with montmorillonite. J. Phys. Chem. 67: 2169–2177 [Google Scholar]
- 6. Heger D, Jirkovsky J, Klan P. 2005. Aggregation of methylene blue in frozen aqueous solutions studied by absorption spectroscopy. J. Phys. Chem. A 109: 6702–6709 [DOI] [PubMed] [Google Scholar]
- 7. Acemiolu B, Kertmen M, Dirak M, Alma MH. 2010. Use of Aspergillus wentii for biosorption of methylene blue from aqueous solution. Afr. J. Biotechnol. 9: 874–881 [Google Scholar]
- 8. Vijayaraghavan K, Mao J, Yun Y-S. 2008. Biosorption of methylene blue from aqueous solution using free and polysulfone-immobilized Corynebacterium glutamicum: batch and column studies. Bioresour. Technol. 99: 2864–2871 [DOI] [PubMed] [Google Scholar]
- 9. Fischer M, Schmidt C, Falke D, Sawers RG. 2012. Terminal reduction reactions of nitrate and sulfate assimilation in Streptomyces coelicolor A3(2): identification of genes encoding nitrite and sulfite reductases. Res. Microbiol. 163: 340–348 [DOI] [PubMed] [Google Scholar]
- 10. Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom [Google Scholar]
- 11. Kim Y-M, Kim J. 2004. Formation and dispersion of mycelial pellets of Streptomyces coelicolor A3(2). J. Microbiol. 42: 64–67 [PubMed] [Google Scholar]
- 12. Fischer M, Alderson J, Van Keulen G, White J, Sawers RG. 2010. The obligate aerobe Streptomyces coelicolor A3(2) synthesizes three active respiratory nitrate reductases. Microbiology 156: 3166–3179 [DOI] [PubMed] [Google Scholar]
- 13. Nieselt K, Battke F, Herbig A, Bruheim P, Wentzel A, Jakobsen Ø M, Sletta H, Alam MT, Merlo ME, Moore J, Omara WA, Morrissey ER, Juarez-Hermosillo MA, Rodríguez-García A, Nentwich M, Thomas L, Iqbal M, Legaie R, Gaze WH, Challis GL, Jansen RC, Dijkhuizen L, Rand DA, Wild DL, Bonin M, Reuther J, Wohlleben W, Smith MC, Burroughs NJ, Martín JF, Hodgson DA, Takano E, Breitling R, Ellingsen TE, Wellington EM. 2010. The dynamic architecture of the metabolic switch in Streptomyces coelicolor. BMC Genomics 11: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yegneswaran PK, Gray MR, Thompson BG. 1991. Effect of dissolved oxygen control on growth and antibiotic production in Streptomyces clavuligerus fermentations. Biotechnol. Prog. 7: 246–250 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.