Abstract
Two homologous transcription factors, CueR and GolS, that belong to the MerR metalloregulatory family are responsible for Salmonella Cu and Au sensing and resistance, respectively. They share similarities not only in their sequences, but also in their target transcription binding sites. While CueR responds similarly to Au, Ag, or Cu to induce the expression of its target genes, GolS shows higher activation by Au than by Ag or Cu. We showed that the ability of GolS to distinguish Au from Cu resides in the metal-binding loop motif. Here, we identify the amino acids within the motif that determine in vivo metal selectivity. We show that residues at positions 113 and 118 within the metal-binding loop are the main contributors to metal selectivity. The presence of a Pro residue at position 113 favors the detection of Cu, while the presence of Pro at position 118 disfavors it. Our results highlight the molecular bases that allow these regulators to coordinate the correct metal ion directing the response to a particular metal injury.
INTRODUCTION
Transition metal homeostasis influences many fundamental aspects of bacterial cell physiology and pathogenesis (1–3). The intracellular concentration of essential metals or the presence of harmful elements is monitored by a set of transcriptional regulators that modulate the expression of factors that rapidly restore metal homeostasis (4, 5). A large class of these metalloregulators belongs to the MerR family, a group of proteins that share similarity at the N-terminal DNA-binding domain (6–9). According to the current model, dimeric metal-sensing MerR regulators control gene transcription via a DNA distortion mechanism. Both the apo- and the metal-bound regulator recognize and interact with their target operators (a dyad-symmetric DNA sequence at the promoter region of their target genes). Binding of the metal ion at the C-terminal inductor-binding site would provoke an allosteric change at the N-terminal DNA binding region of the protein, which in turn transduces changes in the promoter structure resulting in transcription activation of the expression of genes coding mostly for efflux or detoxification systems (10–12).
Most of the metal ion sensors of the MerR family are poorly selective because they cannot distinguish between cognate metals with similar physicochemical properties, including charge and coordination chemistry. For example, the Cu sensor CueR can discriminate between metal ions with +1 and +2 charges, but it cannot distinguish between monovalent metal ions of group 1B—i.e., Cu(I), Ag(I), and Au(I) (13). Structural studies indicate that CueR has only two coordinating ligands, the S-atoms of the conserved C112 and C120 residues which are appropriate for the interaction with the +1 metal ion in a linear array but not to coordinate metal ions with a +2 charge, which requires higher number of ligands (14). The recent identification of two Au-selective MerR sensors, first in the bacterial pathogen Salmonella enterica serovar Typhimurium (S. Typhimurium) and then in Cupriavidus metallidurans, a betaproteobacterium associated with Au grains (15–17), is useful to understand the molecular bases that allow metalloregulators of the MerR family to distinguish between cognate metal ions. Although there is no structural information available for the Au sensors, a number of studies suggest that these sensors coordinate the metal by using the conserved C112 and C120 residues of the metal-binding loop (MBL) apparently in a bis-thiolate geometry, similar to CueR, but unlike the Cu sensor, they can distinguish Au(I) from Cu(I) or Ag(I) (15, 16). Both Salmonella GolS and Cupriavidus CupR activate the expression of their target genes mainly in the presence of Au(I) ions. Recently reported in vitro experiments show that GolS and CueR have similar affinities for Cu(I) (18); nevertheless, in vivo evidence shows that GolS distinguishes Au(I) from Cu(I) or Ag(I) in the induction of its target genes (15, 19–21). Mutant strains with deletions in genes controlled by GolS, including the transcriptional regulator itself, which is autoregulated in Salmonella, have increased susceptibility to gold ions while retaining wild-type copper resistance in the presence of the intact copper resistance cue regulon (15, 21, 22). In this case, it became clear that metal selectivity of GolS is achieved by the combination of subtle modifications in the sensing domain of the gold sensor and the presence of an efficient copper resistance system operating in the cell. Indeed, GolS is activated by copper in a mutant strain deleted in both the copper sensor CueR and the copper transporter CopA, inducing the expression of part of its regulon (20–22). Expression of the P-type ATPase GolT and probably the metal-binding protein GolB serves to alleviate the toxic effect of Cu excess in the absence of CopA and/or CueR (18, 20, 22, 23). Interestingly, one of the components of the gol regulon, the gesABC operon, is solely induced by gold and not by copper, even in a strain deleted of the entire copper resistance cue regulon (21; our unpublished results). These observations strengthened the physiological role of the gol regulon in gold sensing and resistance and prompted us to investigate the determinants of metal selectivity in GolS.
Previously, we showed that the expression of the GolS-regulated golB gene is similarly induced by Au, Cu, or Ag in a mutant that codes for a GolS chimeric protein with the metal-binding loop of CueR (from I109 to C120 and encompassing the two conserved cysteine residues directly involved in metal coordination), resembling the metal response of the wild-type Cu sensor (15). In this study, we dissect the metal-binding loop of GolS and CueR to identify the amino acid residues that direct metal discrimination. We show that residues at positions 113 and 118 within the metal-binding loop are the main contributors to metal selectivity, while other nonconserved amino acids within the loop also cooperate to fine-tune metal selectivity.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains (all derivatives of Salmonella enterica serovar Typhimurium 14028s, except when indicated), plasmids, and oligonucleotides used in this study are listed in Tables S1 and S2 in the supplemental material. Bacterial strains were grown at 37°C in Luria-Bertani (LB) broth, except when indicated. Ampicillin, kanamycin, chloramphenicol, and tetracycline were used at 100, 25, 10, and 15 μg ml−1, respectively. The cell culture medium reagents, chemicals, and oligonucleotides were from Sigma, except for the LB culture medium, which was from Difco.
Genetic and molecular biology techniques.
Gene disruptions, point mutations, or lacZ reporter fusions to promoters were carried out using previously described protocols (24, 25). The constructions were initially done on Salmonella strain LB5010 (26) or the indicated mutant derivative of this strain and transferred to the wild-type ATCC 14028s strain by P22 transduction (27). When necessary, the antibiotic resistance cassette was removed using the temperature-sensitive plasmid pCP20 carrying the FLP recombinase (24).
The desired mutations were introduced on golS or cueR by PCR overlap extension using appropriate primers containing either mismatched bases or regions. To construct golS mutant alleles, a megaprimer was generated using primer golS-F and the corresponding reverse primer for each mutant version. The megaprimer was extended using the oligonucleotide RvP1-golB-R, and the final product of 556 bp was fused to a chloramphenicol resistance cassette (amplified from the plasmid pKD3 using golB-P1-F and golB-P2-R oligonucleotides) by splicing by overlap extension (SOE)-PCR (28). The final product of the SOE-PCR was introduced by linear transformation in the chromosome of Salmonella LB5010 Δ(golS-golB)::kan by the one-step method described by Datsenko et al. (24). To generate the golB::lacZ transcriptional fusion, the chloramphenicol resistance cassette was removed using FLP-mediated recombination followed by site-specific integration of a lac fusion plasmid, pKG136 (see Table S1 in the supplemental material) into the remaining FLP recombination target (FRT) site.
In order to construct cueR mutant alleles, a megaprimer was amplified using primer cueR-F and the corresponding reverse primer for each mutant version. The extension of the megaprimer was done using the oligonucleotide RvP1-cueR-R, and the final product of 577 bp was fused to a chloramphenicol resistance cassette (amplified from plasmid pKD3 using cueR-P1-F and cueR-P2-R). The cueR mutant fused to the antibiotic resistance gene was introduced in the chromosome of Salmonella LB5010 ΔcueR::kan using the one-step method.
The mutant golSP118A allele (golS with a P-to-A change at position 118) was PCR amplified from the Salmonella chromosome using the golS-ORF-F and golS-ORF-R primers and cloned into pUH21-2laqIq to generate the pPB1353 expression plasmid. The plasmids and the linear constructions were introduced into Salmonella strains by electroporation using a Bio-Rad device following the manufacturer‘s recommendations. All constructs were verified by DNA sequencing.
Au and Cu induction assays.
β-Galactosidase assays were carried out essentially as described previously (29). Metal induction assays of GolS- and CueR-controlled genes were done on mid-log-phase (not shown) or overnight cell cultures in LB medium with 40 μM AuHCl4 or 1 mM CuSO4 or without metal (15). When indicated, induction assays were done in SM9 medium or M9 minimal medium without NaCl (15) or with increasing amounts of AuHCl4 or CuSO4. Statistical analysis was performed by one-way analysis of variance and the Holm-Sidak test.
Protein stability assays.
S. Typhimurium 14028s strains PB7116 and PB7171, which carried either the wild-type golS or the golSP118A allele, respectively, were grown at 37°C with shaking in LB medium with 40 μM AuHCl4 overnight. Cells were washed twice with LB medium, resuspended in fresh LB medium, and incubated at 37°C with shaking. Samples were collected after 0, 4, and 12 h, resuspended in a mixture of 50 mM Tris-HCl (pH 7), 2 mM EDTA, and 5 mM dithiothreitol (DTT), and disrupted by sonication. In each case, 20 μg of total soluble protein was subjected to SDS-PAGE. Detection of GolS or GolSP118A in whole-cell extracts was performed by Western blotting using rabbit polyclonal anti-GolS antibodies as described previously (15). The protein concentration was determined by Bradford assay, using bovine serum albumin as the standard.
Protein-DNA interaction analysis.
Electrophoretic gel mobility shift assays were performed essentially as previously described (15). Approximately 6 fmol of labeled DNA fragment containing the golB promoter region was incubated at room temperature for 20 min with purified GolS or GolSP118A. Both proteins were overexpressed and purified from the Salmonella PB5257 strain carrying the corresponding expression plasmids as described previously (15).
Bioinformatics.
The GolS or CueR homologs shown in Table 1 were extracted from the phylogenetic analysis previously performed (20). Complete protein sequences of the GolS or CueR homologues, as well as accession numbers and species names, are shown in a previous publication (20). The alignment of the C-terminal inducer-binding domains was performed using MEGA version 3.0 software (30).
Table 1.
The amino acid residues at positions 113 and 118 are highly conserved within the GolS- and CueR-like groups
| Locus tag or product | Bacterial species | Metal-binding loop sequencea |
|---|---|---|
| GolS-like | ||
| GolS (STM0354) | Salmonella enterica serovar Typhimurium LT2 | CAGDALPDC |
| XCV2319 | Xanthomonas campestris pv. vesicatoria strain 85-10 | CAGDDRPDC |
| Ajs_1373 | Acidovorax sp. strain JS42 | CAGDDRPDC |
| Sputw3181_1111 | Shewanella sp. strain W3-18-1 | CAGDERSEC |
| Smal_1771 | Stenotrophomonas maltophilia R551-3 | CAGDERPEC |
| Sputw3181_1120 | Shewanella sp. strain W3-18-1 | CAGNEKPDC |
| Meso_3892 | Mesorhizobium sp. strain BNC1 | CSGDNRPDC |
| Neut_0068 | Nitrosomonas eutropha C91 | CSGDDRPDC |
| Oant_4049 | Ochrobactrum anthropi ATCC 49188 | CHGDHRPHC |
| CupR (Rmet_3523) | Cupriavidus metallidurans CH34 | CTGDDRPDC |
| CueR-like | ||
| CueR (b0487) | Escherichia coli K-12 MG1655 | CPGDDSADC |
| CueR (STM0499) | Salmonella enterica serovar Typhimurium LT2 | CPGDDSADC |
| CKO_02653 | Citrobacter koseri ATCC BAA-895 | CPGDDSADC |
| ECA1194 | Erwinia carotovora subsp. atroseptica SCRI1043 | CPGDGGSEC |
| plu3823 | Photorhabdus luminescens subsp. laumondii TTO1 | CPGDDGAAC |
| CueR (y1094) | Yersinia pestis KIM | CPGDEGADC |
| CueR (peg.5396) | Serratia marcescens subsp. marcescens Db11 | CPGDEGAEC |
| MS0886 | Mannheimia succiniciproducens MBEL55E | CPGDGSEHC |
| PBPRA2823 | Photobacterium profundum SS9 | CPGNEGAAC |
| CueR (VC0974) | Vibrio cholerae O1 biovar El Tor strain N16961 | CPGDQGSDC |
| VSAK1_08833 | Vibrio shilonii AK1 | CPGDTNSAC |
The amino acid residues at positions 113 and 118 are underlined.
RESULTS
The GolS metal-binding loop is essential for Au selectivity.
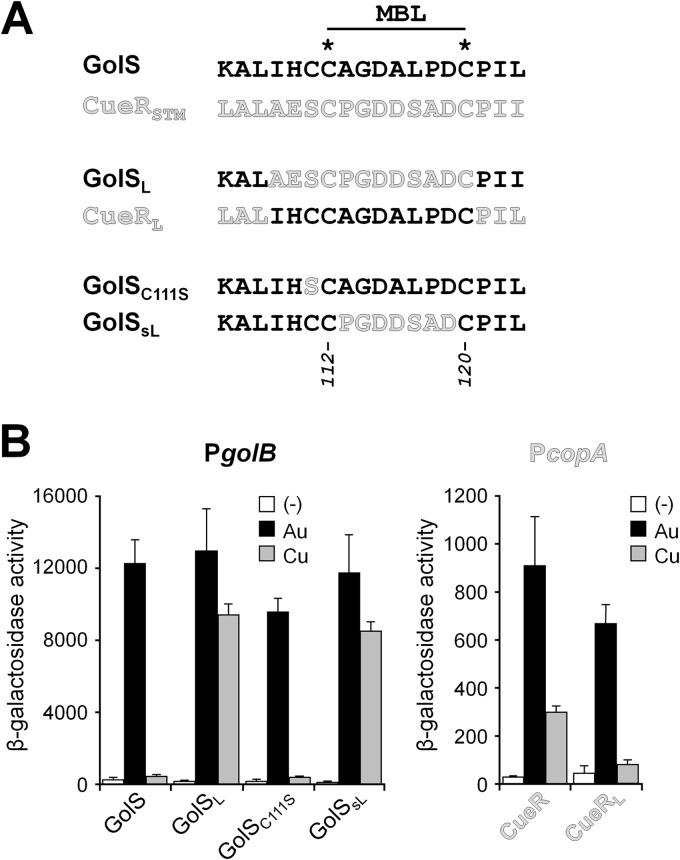
We have previously demonstrated that the Au sensor GolS shows higher activation by Au than by Ag or Cu and that a chimeric protein with a replacement of the region between I109 and C120 of GolS for the same region of CueR (GolSL) induces the transcription of the GolS-controlled golB gene in the presence of either Au or Cu ions, achieving similar transcription levels with both metal ions, resembling the activation pattern of CueR-dependent genes (15) (Fig. 1). To further examine the role of this region in metal selectivity, we modified cueR to code for a chimeric sensor, CueRL, in which the region between A109 and C120 was replaced by the equivalent region from GolS (Fig. 1A). cueRL was introduced into the chromosome of the S. Typhimurium 14028 strain replacing wild-type cueR, and the expression of the CueR-dependent copA::lacZ reporter in cells grown in Luria-Bertani (LB) medium was determined in the presence or absence of either 40 μM AuHCl4 or 1 mM CuSO4 (Fig. 1B). We noticed that the response to Cu decreased in cells with the cueRL allele, in comparison with the induction observed in the wild-type strain, while conserving similar levels of activation by Au ions. These results confirmed that specific amino acid residues within the region from positions 109 to 120 of CueR and GolS determine the ability of these sensor proteins to respond to Cu and in consequence to discern between Au and Cu.
Fig 1.
The metal-binding loop is responsible for metal selectivity in Salmonella GolS and CueR. (A) Schematic representation of Salmonella Typhimurium GolS, CueR (CueRSTM), and the mutant proteins GolSL, CueRL, GolSC111S, and GolSsL. The MBL sequence is shown in each case, and the conserved cysteine residues are depicted. (B) β-Galactosidase activity from GolS- or CueR-regulated transcriptional fusions golB::lacZ (PgolB) or copA-lacZ (PcopA), respectively, on cells carrying either wild-type golS and cueR, or golSL, golSsL, or golSC111S, replacing the wild-type copy of golS, or cueRL, replacing cueR. The lacZ reporter gene was inserted as an operon fusion 3′ of copA, so the expression of the CopA transporter was not affected (22). Cells were grown overnight in LB broth without (−) or with the addition of 40 μM AuHCl4 (Au) or 1 mM CuSO4 (Cu). The data correspond to mean values of four independent experiments performed in duplicate. Error bars correspond to the standard deviations. The β-galactosidase activity values of the GolSL, GolSsL, or CueRL strains grown in the presence of Cu differ significantly (P < 0.001) from those carrying the wild-type alleles. No significant differences were observed in the presence of Au.
GolS and CueR metal-binding loops differ in four out of the seven residues between C112 and C120, the two cysteine residues responsible for the metal binding. Besides, GolS has an additional cysteine residue at position 111, while S. Typhimurium CueR has a Ser and Escherichia coli CueR has an Ala at this position. To investigate the role in the metal response of the residue at position 111, two additional mutant versions of GolS were constructed and the genes coding for them introduced into the S. Typhimurium chromosome to replace the wild-type gene. These are GolSC111S, which only differs from GolS in the residue at position 111, and GolSsL, which keeps the C111, but has the 7 residues between C112 and C120 present in CueR (Fig. 1A). Like GolSL, GolSsL induced transcription of golB::lacZ in the presence of either Au or Cu ions (Fig. 1B). In contrast, GolSC111S exhibited the wild-type activation pattern, confirming that residue 111 is involved in neither metal recognition nor metal selectivity.
These results allowed us to focus on the residues within the metal-binding loop that are not conserved between GolS and CueR (i.e., the residues at positions 113, 116, 117, and 118) to search for determinants of metal selectivity.
The amino acid residue at position 113 plays a key role in copper sensitivity.
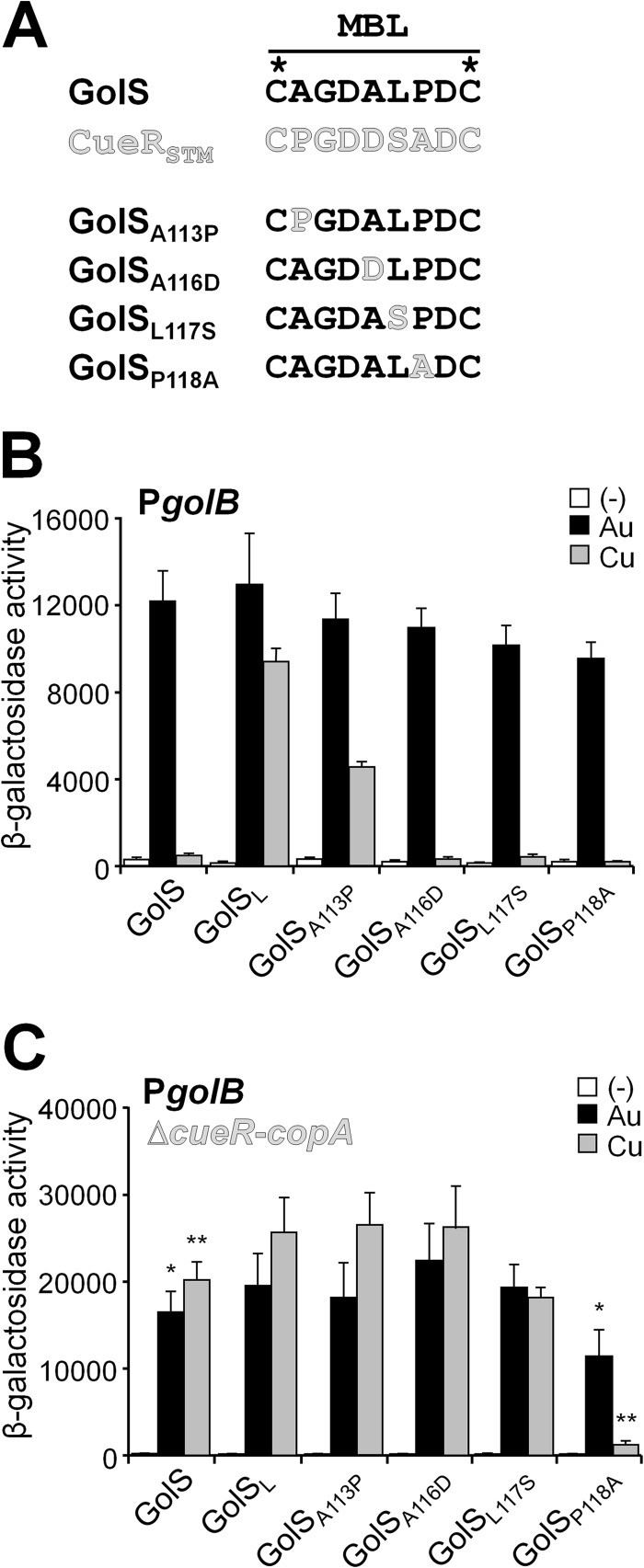
To analyze the molecular bases that favor the activation of GolS by Au over Cu, the amino acid residues at position 113, 116, 117, or 118 in the Au sensor were individually replaced for the corresponding residues present in CueR (Fig. 2A). The mutant alleles were separately introduced into the S. Typhimurium chromosome, replacing the wild-type golS, and the response of each mutant sensor to Au or Cu ions was evaluated, measuring the metal-dependent expression of golB::lacZ. Almost all mutant sensors have wild-type response to Au, both in rich medium (Fig. 2B) and in minimal medium (Fig. 3A), except for GolSP118A, which had a slightly reduced response to the metal ion (Fig. 3A).
Fig 2.
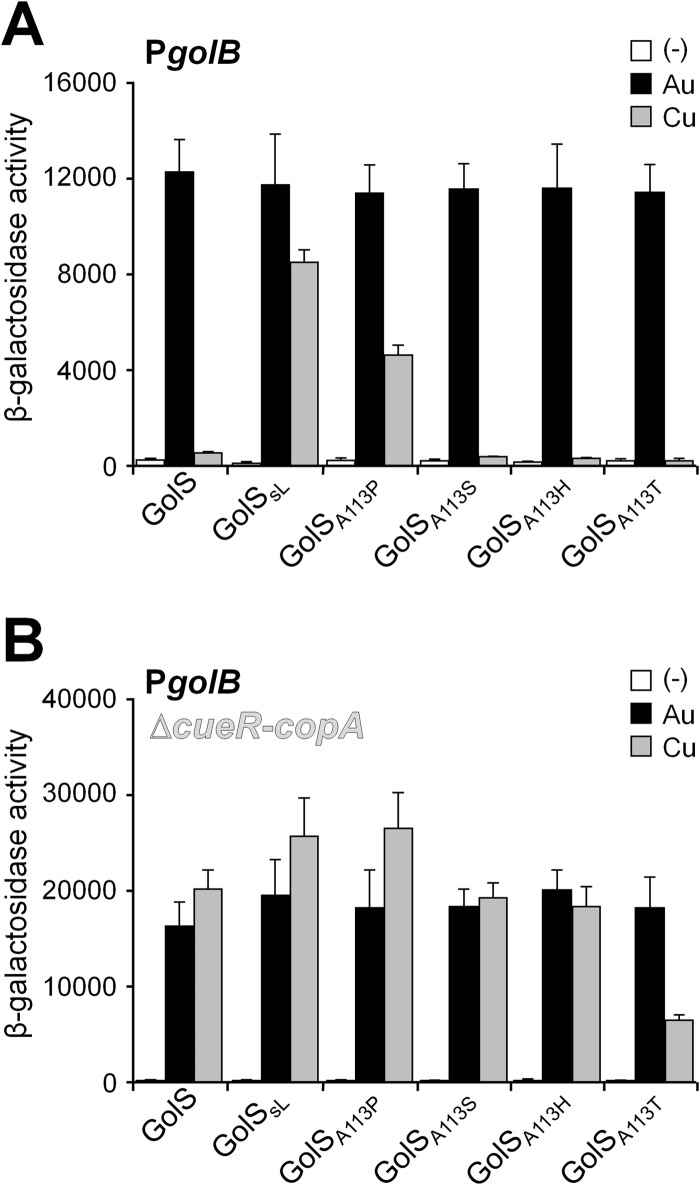
Contribution to metal selectivity of different residues within the GolS metal binding loop. (A) Sequence of the MBL of GolS, CueR, and the mutants GolSA113P, GolSA116D, GolSL117S, and GolSP118A. (B and C) β-Galactosidase activity from a golB::lacZ transcriptional fusion expressed by cells carrying either golS (GolS) or the mutant alleles coding for GolSL, GolSA113P, GolSA116D, GolSL117S, and GolSP118A in otherwise wild-type (B) or Δ(cueR-copA) (C) genetic backgrounds. The cells were grown overnight in LB broth without (−) or with the addition of 40 μM AuHCl4 (Au) or 1 mM CuSO4 (Cu). The data correspond to mean values of four independent experiments performed in duplicate. Error bars correspond to standard deviations. β-Galactosidase activity values of the GolSP118A strain grown in the presence of Au or Cu differ significantly (*, P = 0.02; **, P < 0.001) from the wild-type strain.
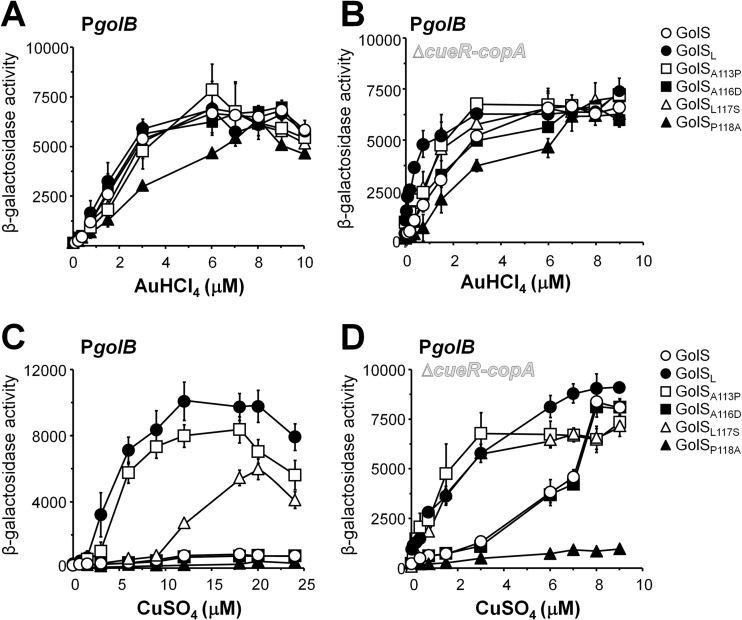
Fig 3.
The residue at position 113 affects the copper response in GolS. β-Galactosidase activity from a golB::lacZ transcriptional fusion expressed by cells carrying either golS (GolS) or the mutant alleles coding for GolSL, GolSA113P, GolSA116D, GolSL117S, or GolSP118A, in an otherwise wild-type genetic background (A and C) or in a Δ(cueR-copA) genetic background (B and D). Cell cultures were grown overnight in SM9 containing the indicated concentrations of AuHCl4 (A and B) or CuSO4 (C and D). The data correspond to mean values of four independent experiments performed in duplicate. Error bars correspond to the standard deviations.
Conversely, GolSA113P showed an increased response to Cu ions compared to wild-type GolS, resembling the response attained by GolSL, although the level of expression of golB::lacZ was always lower in the golSA113P strain than in the golSL strain (Fig. 2B and 3C). The GolSL117S sensor also showed an increased response to Cu compared to the wild-type GolS, but it was only appreciable when concentrations higher than 9 μM CuSO4 were used in the culture medium (Fig. 3C), suggesting that this substitution also favors the detection of the metal ion, although to a lesser extent than the A113P substitution. Under these conditions, neither GolSA116D nor GolSP118A was able to induce the expression of golB::lacZ even in high Cu concentrations, similar to wild-type GolS (Fig. 3C).
We have previously shown that in a strain deleted of both cueR and copA, GolS induces the expression of part of the gol regulon in the presence of Cu, reaching expression levels comparable to those attained by Au (21, 22). Although not determined, this suggests that in the absence of CueR and CopA the intracellular level of Cu increases, reaching the detection threshold of GolS for this metal ion. Therefore, this strain can be used to expand the sensitivity of the mutant sensors to Cu. It is also worth noting here that deletion of cueR and copA only slightly affected the response of all the mutant sensors to Au ions (Fig. 2C and B). This background allowed us to observe that GolSA116D has a wild-type response to Cu (Fig. 2C and 3D). Even under this condition, GolSP118A was unable to induce golB::lacZ expression in the presence of Cu (Fig. 2C and D). This observation and the reduced response to Au ions observed for GolSP118A both in wild-type cue and in the Δ(cueR-copA) backgrounds (Fig. 2 and 3) suggest that the presence of an Ala at position 118 in the metal-binding loop is detrimental either for metal binding or for the intramolecular transduction of the activation state from the metal-binding site to the DNA-binding domain. The possibility that this substitution could affect the stability of the sensor was discarded by comparing the kinetics of decay of the wild-type GolS and GolSP118A expressed from the PgolTS chromosomal promoter after removal of the inducer, Au, from the medium (Fig. 4A). We also compared the abilities of GolS and GolSP118A to interact with the PgolB promoter (Fig. 4B). Neither differences in relative protein stability nor differences in DNA affinity were observed.
Fig 4.
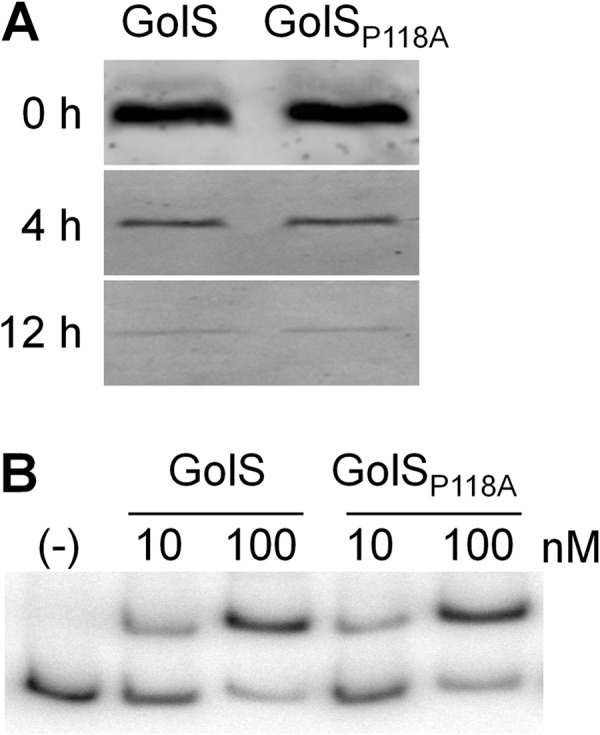
The P118A replacement in GolS does not affect its stability or its DNA affinity. (A) GolS or GolSP118A was expressed from the GolS-dependent PgolTS promoter in the chromosome by the addition of 40 μM AuHCl4 to the growth medium (LB). Overnight cultures were washed and suspended in with fresh LB without AuHCl4 and incubated for 0, 4, or 12 h. Detection of intracellular GolS or GolSP118A was carried out by Western blotting in whole-cell extracts. (B) Electrophoretic gel mobility shift assay analysis using 6 fmol of 32P 3′-end-labeled PCR fragment from the golB promoter region and the purified GolS variant at the indicated concentrations. (−), no protein addition.
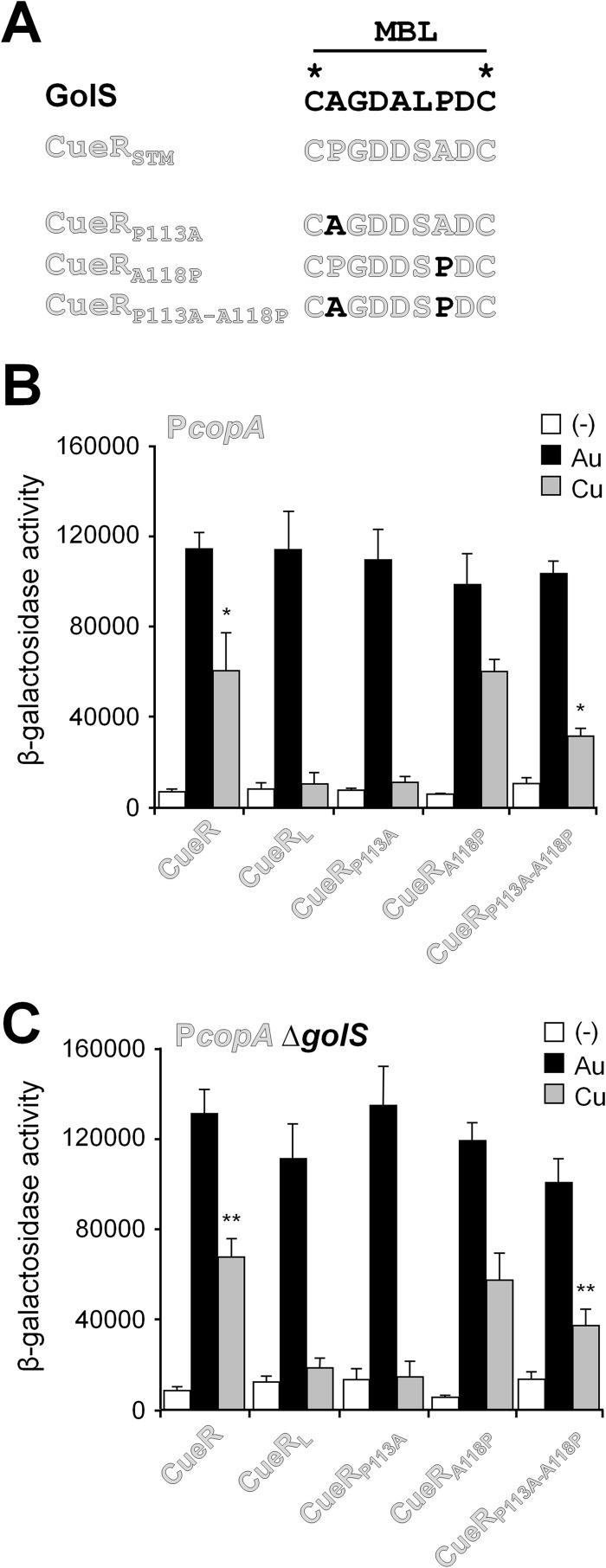
To verify the relevance of residue 113 in directing signal discrimination, the S. Typhimurium CueRP113A and CueRA118P mutants were constructed and tested (Fig. 5). The CueRP113A mutant sensor is expected to be less responsive to Cu than the native CueR sensor, while CueRA118P would have little or no effect on the response of the sensor protein to the metals. The mutant genes were inserted into the chromosome of the S. Typhimurium 14028 strain, replacing the wild-type cueR. The sensor proteins' response was followed by measuring the expression of the CueR-dependent copA::lacZ reporter in LB medium in the absence or presence of either Au or Cu. As expected, the CueRP113A sensor exhibited a metal activation pattern similar to the chimeric CueRL sensor; that is, there was less response to Cu than that observed with wild-type CueR, but there were similar levels of Au-mediated induction (Fig. 5B). Introduction of the A118P mutation in CueR showed a wild-type response to the metal ions. Similar results were obtained in a strain deleted in the Au sensor GolS (Fig. 5C).
Fig 5.
Analysis of the roles of residues at positions 113 and 118 in Salmonella CueR. (A) Sequence of the MBL of GolS, CueR, and the mutant alleles CueRP113A, CueRA118P, and CueRP113A A118P. (B and C) β-Galactosidase activity from a copA::lacZ transcriptional fusion expressed from a reported plasmid by cells carrying cueR (CueR) or the mutant alleles coding for CueRL, CueRP113A, CueRA118P, or CueRP113A A118P in an otherwise wild-type (B) or ΔgolS (C) genetic background. Cells were grown overnight in LB broth without (−) or with the addition of 40 μM AuHCl4 (Au) or 1 mM CuSO4 (Cu). The data correspond to mean values of four independent experiments performed in duplicate. Error bars correspond to the standard deviations. Differences in β-galactosidase activity values from the wild-type CueR and CueRP113A A118P in the presence of Cu were statistically significant (*, P = 0.029; **, P = 0.001). No significant differences were observed from wild-type CueR and CueRA118P in the presence of Cu.
Most sensors that are phylogenetically linked to GolS (15, 20) have Ala, Ser, or His at position 113, and the recently characterized gold sensor CupR from C. metallidurans has a Thr at this position (Table 1). We constructed GolS variants with Ser, His, or Thr at this position and determined their ability to induce the expression of golB::lacZ in the presence of Au or Cu, either in the wild-type cue background or in the Δ(cueR-copA) mutant strain (Fig. 6). The metal-dependent golB induction pattern attained with GolSA113S and GolSA113H variants was similar to that of the wild-type GolS in both genetic backgrounds (Fig. 6A and B). Interestingly, GolSA113T, the C. metallidurans CupR-like variant, responded to Au like the wild-type GolS but exhibited even a lower response to Cu in a Δ(cueR-copA) strain (Fig. 6B).
Fig 6.
Proline at position 113 in GolS enhances the response to copper. β-Galactosidase activity from a golB::lacZ transcriptional fusion expressed by cells carrying the gene coding either for GolS, GolSsL, GolSA113P, GolSA113S, GolSA113H, or GolSA113T in a wild-type (A) or Δ(cueR-copA) (B) background. Cells were grown overnight in LB broth without (−) or with the addition of 40 μM AuHCl4 (Au) or 1 mM CuSO4 (Cu). The data correspond to mean values of four independent experiments performed in duplicate. Error bars correspond to the standard deviations. β-Galactosidase activity values from golSA113T in a Δ(cueR-copA) strain grown in the presence of Cu differ significantly (P < 0.001) from that of the wild-type golS in the same genetic background.
Residues at positions 116, 117, and 118 contribute to fine-tuning the metal response.
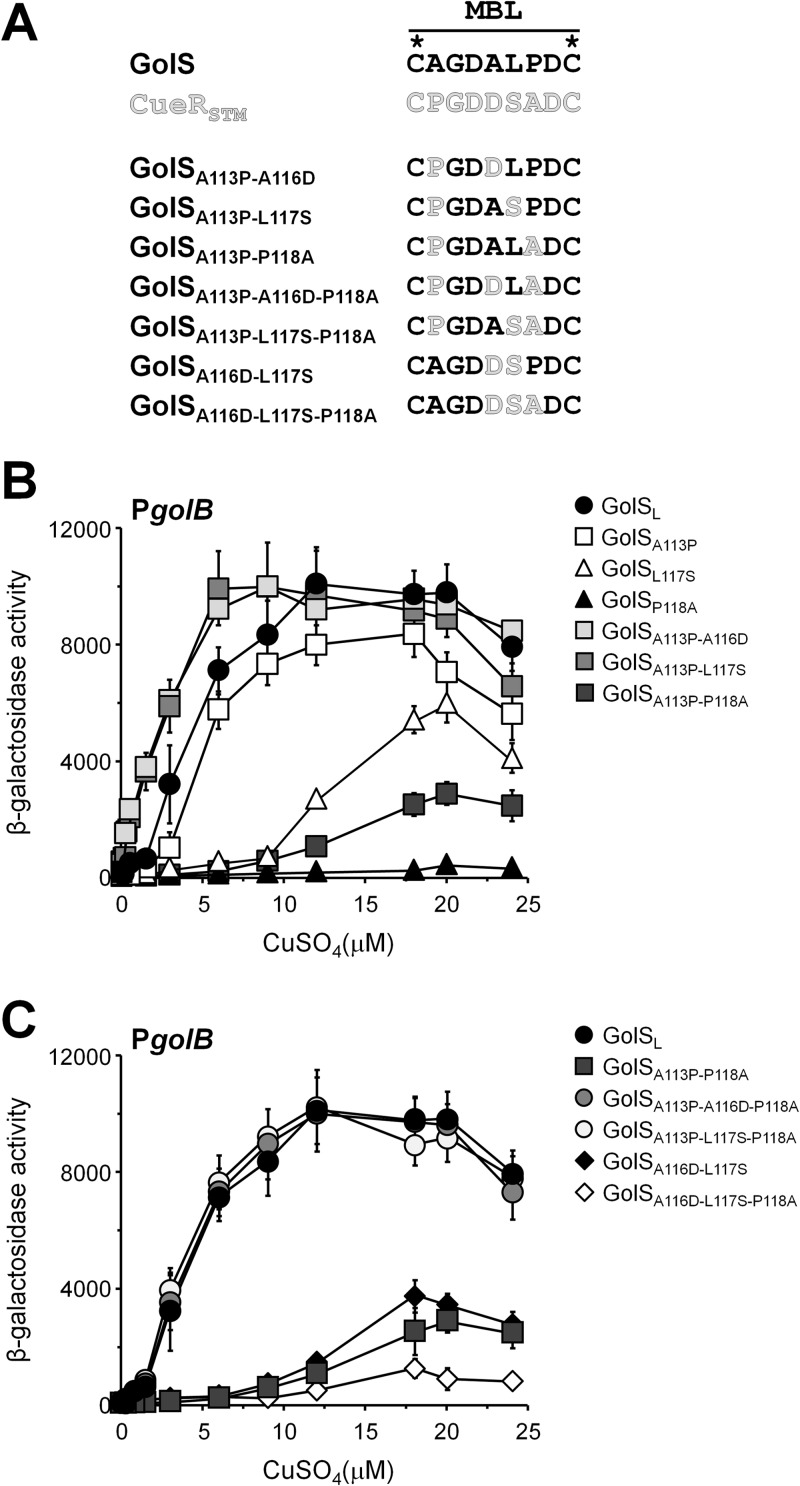
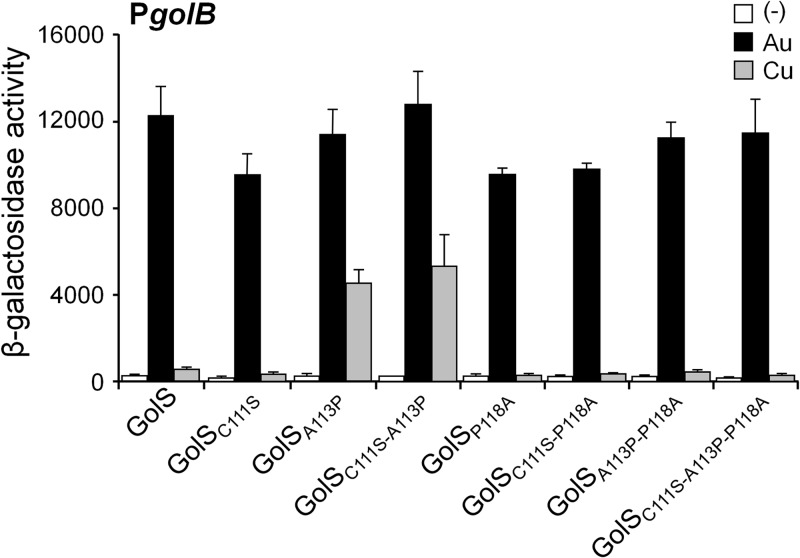
The above observations suggest that the GolS residue 113 plays a key role in the response to copper, and residues at positions 117 and 118 may also modulate the metal selectivity. We combined the above mutations to generate double and triple mutant forms of GolS and tested their response to CuSO4 (Fig. 7). As predicted by the single mutations, the simultaneous modification of the residues at positions 113 and 117 increased the response to copper of GolSA113P L117S (Fig. 7B). Interestingly, the introduction of the apparently neutral replacement at position 116 (A116D) in GolSA113P also enhanced the response to Cu to levels similar to those attained by GolSA113P L117S (Fig. 7B). Both the GolSA113P L117S and GolSA113P A116D reached the maximal golB::lacZ expression at lower Cu concentrations compared to GolSL, suggesting that other amino acids in the GolSL loop negatively affect the Cu response. Indeed, introduction of Ala at position 118 in GolSA113P L117S or in GolSA113P A116D sensors lowered their response to copper to the level observed with GolSL (compare in Fig. 7B and C, GolSA113P L117S with GolSA113P L117S P118A and GolSA113P A116D with GolSA113P A116D P118A) and in all of the loop mutant sensors tested (compare also GolSA113P with GolSA113P P118A and GolSA116D L117S with GolSA116D L117S P118A in Fig. 7B and C). In contrast, the introduction of the C111S mutation in GolS with the simultaneous modification of the residues at position 113, 118, or both did not affect the metal response of the mutant sensors (Fig. 8), confirming that the C111 of GolS is dispensable for both its activity and for metal recognition.
Fig 7.
Role of residues at positions 116, 117, and 118 in tuning metal-induced activation of the gol regulon. (A) Sequence of the MBL of the different GolS alleles analyzed in this figure. (B and C) β-Galactosidase activity from a golB::lacZ transcriptional fusion expressed by cells carrying the different GolS alleles depicted in panel A. Cell cultures were grown overnight in SM9 containing the indicated concentrations of CuSO4. The data correspond to mean values of four independent experiments performed in duplicate. Error bars correspond to the standard deviations.
Fig 8.
The residue Cys111 of GolS is dispensable for metal recognition. β-Galactosidase activity from a golB::lacZ transcriptional fusion expressed by cells carrying the wild-type copy of golS (GolS) or the allele coding for the indicated variant of GolS. Cells were grown overnight in LB broth without (−) or with the addition of 40 μM AuHCl4 (Au) or 1 mM CuSO4 (Cu). The data correspond to mean values of four independent experiments performed in duplicate. Error bars correspond to standard deviations.
Similar results were observed for CueR. The A118P replacement in the CueRP113A mutant partially counteracted the effect of the P113A mutation (Fig. 5B), resulting in a sensor protein with a pattern of induction in the presence of Cu intermediate between wild-type CueR or CueRA118P and CueRL or CueRP113A.
Overall, these results indicate that the Pro residues present at position 113 or 118 are the main contributors of the copper detection ability of the Salmonella MerR monovalent metal ion regulators, while residues at position 117 and, to a lesser extent, at position 116 are required to fine tune the metal-induced activation of these sensors.
DISCUSSION
Metal-binding specificity in MerR sensors is mainly influenced by the intrinsic properties of a metal ion—i.e., charge, size, and coordination chemistry, its intracellular availability, as well as by the protein environment (that is, the array of ligands forming the first and second coordination spheres within the folded protein). MerR-like metal sensors, like CueR, cannot differentiate between metal ions with similar charges and coordination chemistries, like Cu(I), Ag(I), or Au(I) ions (14). In contrast, its homologs, GolS from Salmonella Typhimurium and CupR from Cupriavidus metallidurans, were shown to be distinctively activated by Au(I) (15, 16). Here, we demonstrated the role of individual residues within the C112-to-C120 metal-binding loop in the metal-induced activation of GolS and CueR, the two MerR homologs that detect monovalent metal ions in Salmonella.
As modules of evolutionary exchange, loops play important roles in molecular function and biological recognition. It has been postulated that the primary sequence of a loop affects not only the dynamics of folding of the nascent polypeptide chain but also the mobility and flexibility of the region in the folded protein. In fact, the modification of the length or sequence of a protein loop in immunoglobulins and enzymes has been shown to alter their substrates' specificity (31–34). In all monovalent metal ion sensors of the MerR family, the two conserved cysteine residues that coordinate the metal ion are separated by a 7-residue-long loop (15) (Table 1). Our results allow us to postulate that the loop in CueR must be sufficiently flexible to accommodate cognate monovalent metal ions that greatly differ in ionic ratio (from the smallest Cu to the larger Au). In contrast, in GolS, and probably in CupR, the loop might not have the same plasticity, and in this way it interacts better with Au(I), which exhibits a softer nature and a larger ionic size than Cu(I) (35). In this context, we observed a change in metal preference of the Salmonella MerR monovalent metal ion sensors just by swapping their metal-binding loop (between C112 and C120); i.e., GolSsL, which harbors the CueR loop region, responds to either Au(I) or Cu(I), while CueRL, the CueR sensor with the metal-binding loop of GolS, is less responsive to Cu ions but conserves its Au induction capability (Fig. 1).
A recent report on the redox-sensing members of the MerR family (36) supports our observation about the role of the loop in signal selectivity. It is well known that the SoxR sensors from enteric bacteria are able to trigger a global stress response by sensing a broad spectrum of redox-cycling compounds. In contrast, orthologues from soil-dwelling organisms, such as Pseudomonas aeruginosa and Streptomyces coelicolor, only regulate the expression of a small set of genes in response to a reduced number of redox-active small molecules. In order to understand the molecular bases of these differences, the latter sensors increased their sensitivity to other redox compounds by replacing three amino acid residues present within the 2Fe-2S cluster binding region (36), which is equivalent to the metal-binding loop of the monovalent metal ion sensors.
Using a competitive Cu(I) binding approach, Osman and colleagues recently reported that GolS and CueR share similar in vitro affinities for Cu(I) (18). In view of our results, we proposed that the differences in Cu sensitivity between these sensors observed in living cells would not rely in the metal affinity but rather in the ability of the metal ion to properly activate the sensor protein (4, 37, 38), differences subtle enough that can only be perceived in vivo. Our analysis of the contribution to Cu sensitivity of each of the four nonconserved amino acids within the metal-binding loop between GolS and CueR clearly revealed the importance of the residue at position 113. Replacement of the Ala113 residue by Pro, present in CueR as well as in its xenologues (Table 1), rendered a GolS mutant sensor with an increased sensitivity to Cu, without affecting the response to Au ions (Fig. 2 and 3). Conversely, replacement of Pro113 by Ala in CueR lowered its response to Cu without affecting its activation by Au (Fig. 5). Replacement of the GolS Ala113 residue by Ser, His, or Thr, the residues present in other GolS xenologues (20) (Table 1) had either no effect on metal response compared with wild-type GolS (Fig. 6) or, as observed for GolSA113T, with the Thr residue present in C. metallidurans CupR, becomes less sensitive to Cu ions (Fig. 6B). The latter observation could be interpreted as an adaptation in the case of CupR to better discriminate between monovalent metal ions, although as we have demonstrated here, there are other residues in the loop (residues 116 and 117, for example) not conserved between GolS and CupR (Table 1) that can also influence the metal selectivity.
The presence of an Ala at position 118 in GolS is of note because of its negative effect on the response to Cu. The GolSP118A sensor showed a slight decrease in its response to Au, but it became almost insensitive to Cu, even in the Δ(cueR-copA) strain (Fig. 2 and 3). Although we showed that the P118A substitution in GolS does not have any detectable effect on the protein stability or its ability to bind a GolS operator (Fig. 4), this replacement was always detrimental for Cu sensing in the different GolS variants tested (Fig. 7). Furthermore, the A118P substitution increased the sensitivity of CueRP113A to Cu (Fig. 5B and C), strengthening the role of position 118 residue in metal selectivity. Remarkably, almost all of the putative monovalent metal ion sensors detected in bacterial genomes sequenced to date harbor a Pro residue at either position 113 or 118, but none harbors both simultaneously (20) (Table 1). CueR-like proteins have Pro at position 113, while GolS xenologues harbor a conserved P118 (Table 1). Proline is frequently found in turns and loop structures of proteins and, together with glycine residues, is predicted to affect chain compaction early in folding (39). Also, the side chain of proline can wrap around to form a covalent interaction with the backbone, restricting its flexibility and limiting the conformation of neighboring residues (40). Therefore, proline has a unique role in determining local conformation and movement freedom in the folded protein, functioning as a molecular switch in the regulation of a number of cellular processes (39, 41, 42). In the metal sensors, the restricted set of conformations adopted by Pro at position 113 or 118 may differentially affect chain dynamics and the range of conformations the metal-binding loop can assume. This would alter the flexibility of the region, and in consequence, the protein environment around the metal ion that determines metal selectivity. Nevertheless, structural studies are needed to confirm this hypothesis.
Concerning the other amino acids of the metal-binding region that could modulate the inducer's selectivity or accessibility, we show that individual replacements of the amino acids at position 116 or 117 had minor effects on the metal-induced expression of the reporter genes (Fig. 2 and 3). Only the replacement of Leu117 for Ser increased the response to Cu compared with wild-type GolS, and this was only detectable at high Cu concentrations. Interestingly, most GolS-like sensors have Arg at this position, while all CueR-like sensors have either Ser or Gly, but not Arg at this position (Table 1). High variability is also extensive to the amino acid residues present at position 116. Most sensors have Asp at this position, including homologues to both GolS and CueR (Table 1). Nevertheless, the replacement of the Ala for Asp at this position in GolS (GolSA116D) did not affect its response to either Cu or Au (Fig. 2 and 3), although it increased the response to Cu when combined with the A113P replacement (Fig. 7B). In contrast, mutations at residue 111 of GolS did not alter metal selectivity either alone or in combination with other mutations within the loop (Fig. 1 and 8).
In sum, our results demonstrate that the metal ion response in monovalent metal sensors of the MerR family is affected by the combination and arrangement of the different residues that constitute the metal-binding loop. Furthermore, these studies will also provide information to help understand how an existing allosteric metal-binding site can be reengineered to allow the design of customized metal sensors with different ranges of selectivity and sensitivity against cognate metal ions.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. M. Slauch for the kind gift of plasmid pKG136.
This work was supported by grants from Agencia Nacional de Promoción Científica y Tecnológica and from the National Scientific and Technical Research Council (CONICET) to S.K.C. and F.C.S. M.M.I. and S.C. are fellows of the CONICET. S.K.C. and F.C.S. are career investigators of CONICET. F.C.S. is also a career investigator of the Rosario National University Research Council (CIUNR).
Footnotes
Published ahead of print 3 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00153-13.
REFERENCES
- 1. Festa RA, Jones MB, Butler-Wu S, Sinsimer D, Gerads R, Bishai WR, Peterson SN, Darwin KH. 2011. A novel copper-responsive regulon in Mycobacterium tuberculosis. Mol. Microbiol. 79:133–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ward SK, Abomoelak B, Hoye EA, Steinberg H, Talaat AM. 2010. CtpV: a putative copper exporter required for full virulence of Mycobacterium tuberculosis. Mol. Microbiol. 77:1096–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wolschendorf F, Ackart D, Shrestha TB, Hascall-Dove L, Nolan S, Lamichhane G, Wang Y, Bossmann SH, Basaraba RJ, Niederweis M. 2011. Copper resistance is essential for virulence of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 108:1621–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Finney LA, O'Halloran TV. 2003. Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science 300:931–936 [DOI] [PubMed] [Google Scholar]
- 5. Wosten MM, Kox LF, Chamnongpol S, Soncini FC, Groisman EA. 2000. A signal transduction system that responds to extracellular iron. Cell 103:113–125 [DOI] [PubMed] [Google Scholar]
- 6. Brown NL, Stoyanov JV, Kidd SP, Hobman JL. 2003. The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 27:145–163 [DOI] [PubMed] [Google Scholar]
- 7. Helmann JD, Soonsanga S, Gabriel S. 2007. Metalloregulators: arbiters of metal sufficiency, p 37–71 In Nies DH, Silver S. (ed), Molecular microbiology of heavy metals. Springer-Verlag, Berlin, Germany [Google Scholar]
- 8. Hobman JL, Wilkie J, Brown NL. 2005. A design for life: prokaryotic metal-binding MerR family regulators. Biometals 18:429–436 [DOI] [PubMed] [Google Scholar]
- 9. Summers AO. 2009. Damage control: regulating defenses against toxic metals and metalloids. Curr. Opin. Microbiol. 12:138–144 [DOI] [PubMed] [Google Scholar]
- 10. Lee PE, Demple B, Barton JK. 2009. DNA-mediated redox signaling for transcriptional activation of SoxR. Proc. Natl. Acad. Sci. U. S. A. 106:13164–13168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. O'Halloran TV, Frantz B, Shin MK, Ralston DM, Wright JG. 1989. The MerR heavy metal receptor mediates positive activation in a topologically novel transcription complex. Cell 56:119–129 [DOI] [PubMed] [Google Scholar]
- 12. Outten CE, Outten FW, O'Halloran TV. 1999. DNA distortion mechanism for transcriptional activation by ZntR, a Zn(II)-responsive MerR homologue in Escherichia coli. J. Biol. Chem. 274:37517–37524 [DOI] [PubMed] [Google Scholar]
- 13. Stoyanov JV, Brown NL. 2003. The Escherichia coli copper-responsive copA promoter is activated by gold. J. Biol. Chem. 278:1407–1410 [DOI] [PubMed] [Google Scholar]
- 14. Changela A, Chen K, Xue Y, Holschen J, Outten CE, O'Halloran TV, Mondragon A. 2003. Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science 301:1383–1387 [DOI] [PubMed] [Google Scholar]
- 15. Checa SK, Espariz M, Pérez Audero ME, Botta PE, Spinelli SV, Soncini FC. 2007. Bacterial sensing of and resistance to gold salts. Mol. Microbiol. 63:1307–1318 [DOI] [PubMed] [Google Scholar]
- 16. Jian X, Wasinger EC, Lockard JV, Chen LX, He C. 2009. Highly sensitive and selective gold(I) recognition by a metalloregulator in Ralstonia metallidurans. J. Am. Chem. Soc. 131:10869–10871 [DOI] [PubMed] [Google Scholar]
- 17. Reith F, Etschmann B, Grosse C, Moors H, Benotmane MA, Monsieurs P, Grass G, Doonan C, Vogt S, Lai B, Martinez-Criado G, George GN, Nies DH, Mergeay M, Pring A, Southam G, Brugger J. 2009. Mechanisms of gold biomineralization in the bacterium Cupriavidus metallidurans. Proc. Natl. Acad. Sci. U. S. A. 106:17757–17762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Osman D, Patterson CJ, Bailey K, Fisher K, Robinson NJ, Rigby SE, Cavet JS. 2013. The copper supply pathway to a Salmonella Cu,Zn-superoxide dismutase (SodCII) involves P(1B)-type ATPase copper efflux and periplasmic CueP. Mol. Microbiol. 87:466–477 [DOI] [PubMed] [Google Scholar]
- 19. Cerminati S, Soncini FC, Checa SK. 2011. Selective detection of gold using genetically engineered bacterial reporters. Biotechnol. Bioeng. 108:2553–2560 [DOI] [PubMed] [Google Scholar]
- 20. Pérez Audero ME, Podoroska BM, Ibáñez MM, Cauerhff A, Checa SK, Soncini FC. 2010. Target transcription binding sites differentiate two groups of MerR-monovalent metal ion sensors. Mol. Microbiol. 78:853–865 [DOI] [PubMed] [Google Scholar]
- 21. Pontel LB, Pérez Audero ME, Espariz M, Checa SK, Soncini FC. 2007. GolS controls the response to gold by the hierarchical induction of Salmonella-specific genes that include a CBA efflux-coding operon. Mol. Microbiol. 66:814–825 [DOI] [PubMed] [Google Scholar]
- 22. Espariz M, Checa SK, Pérez Audero ME, Pontel LB, Soncini FC. 2007. Dissecting the Salmonella response to copper. Microbiology 153:2989–2997 [DOI] [PubMed] [Google Scholar]
- 23. Osman D, Waldron KJ, Denton H, Taylor CM, Grant AJ, Mastroeni P, Robinson NJ, Cavet JS. 2010. Copper homeostasis in Salmonella is atypical and copper-CueP is a major periplasmic metal complex. J. Biol. Chem. 285:25259–25268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ellermeier CD, Janakiraman A, Slauch JM. 2002. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290:153–161 [DOI] [PubMed] [Google Scholar]
- 26. Bullas LR, Ryu JI. 1983. Salmonella typhimurium LT2 strains which are r− m+ for all three chromosomally located systems of DNA restriction and modification. J. Bacteriol. 156:471–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Davis RW, Bolstein D, Roth JR. 1980. Advanced bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 28. Aiyar A, Xiang Y, Leis J. 1996. Site-directed mutagenesis using overlap extension PCR. Methods Mol. Biol. 57:177–191 [DOI] [PubMed] [Google Scholar]
- 29. Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 30. Kumar S, Tamura K, Nei M. 2004. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 5:150–163 [DOI] [PubMed] [Google Scholar]
- 31. Fetrow JS. 1995. Omega loops: nonregular secondary structures significant in protein function and stability. FASEB J. 9:708–717 [PubMed] [Google Scholar]
- 32. Hu X, Wang H, Ke H, Kuhlman B. 2007. High-resolution design of a protein loop. Proc. Natl. Acad. Sci. U. S. A. 104:17668–17673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McMahon C, Studer SM, Clendinen C, Dann GP, Jeffrey PD, Hughson FM. 2012. The structure of Sec12 implicates potassium ion coordination in Sar1 activation. J. Biol. Chem. 287:43599–43606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tadwal VS, Sundararaman L, Manimekalai MS, Hunke C, Gruber G. 2012. Relevance of the conserved histidine and asparagine residues in the phosphate-binding loop of the nucleotide binding subunit B of A(1)A(O) ATP synthases. J. Struct. Biol. 180:509–518 [DOI] [PubMed] [Google Scholar]
- 35. Parr RG, Yang W. 1989. Density-functional theory of atoms and molecules. Oxford University Press, Oxford, United Kingdom [Google Scholar]
- 36. Sheplock R, Recinos DA, Mackow N, Dietrich LE, Chander M. 2013. Species-specific residues calibrate SoxR sensitivity to redox-active molecules. Mol. Microbiol. 87:368–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Waldron KJ, Robinson NJ. 2009. How do bacterial cells ensure that metalloproteins get the correct metal? Nat. Rev. Microbiol. 7:25–35 [DOI] [PubMed] [Google Scholar]
- 38. Waldron KJ, Rutherford JC, Ford D, Robinson NJ. 2009. Metalloproteins and metal sensing. Nature 460:823–830 [DOI] [PubMed] [Google Scholar]
- 39. Krieger F, Moglich A, Kiefhaber T. 2005. Effect of proline and glycine residues on dynamics and barriers of loop formation in polypeptide chains. J. Am. Chem. Soc. 127:3346–3352 [DOI] [PubMed] [Google Scholar]
- 40. Chakrabarti P, Pal D. 2001. The interrelationships of side-chain and main-chain conformations in proteins. Prog. Biophys. Mol. Biol. 76:1–102 [DOI] [PubMed] [Google Scholar]
- 41. Bauer F, Sticht H. 2007. A proline to glycine mutation in the Lck SH3-domain affects conformational sampling and increases ligand binding affinity. FEBS Lett. 581:1555–1560 [DOI] [PubMed] [Google Scholar]
- 42. Pavlicek J, Coon SL, Ganguly S, Weller JL, Hassan SA, Sackett DL, Klein DC. 2008. Evidence that proline focuses movement of the floppy loop of arylalkylamine N-acetyltransferase (EC 2.3.1.87). J. Biol. Chem. 283:14552–14558 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.