Abstract
The ASM 6th Conference on Biofilms was held in Miami, Florida, 29 September to 4 October, 2012. The conference provided an opportunity for the exchange of new findings and ideas with regard to biofilm research. A wide range of findings, spanning applied biology, evolution, ecology, physiology, and molecular biology, were presented at the conference. This review summarizes the presentations with regard to emerging biofilm-related themes.
INTRODUCTION
The ASM 6th Conference on Biofilms was held in Miami, Florida, 29 September to 4 October 2012, and was attended by 447 participants. Of these conferees, 180 were international. The meeting covered an exciting range of topics across the breadth of biofilm research and comprised three keynote lectures and 14 thematically organized sessions. The meeting included two extensive poster sessions for an overall 269 posters, where investigators presented their latest research. Of these posters, 8 distinguished young researchers were recognized at the conference banquet with awards in memory of international leaders in the field of biofilm research: The Bill Characklis Poster Award for Excellence in Engineering in Biofilm Research, the Terry Beveridge Poster Award for Excellence in Biofilm Microscopy, the Peter Gilbert Poster Award for Excellence in Innovation and Biofilm Control, and the Bill Costerton Poster Award for Outstanding Interdisciplinary Biofilm Research. In this review, we summarize the Biofilms 2012 presentations in the individual sessions on emerging biofilm-related themes, including the three keynote talks and several selected posters. We intend for this to provide the attendees of the Biofilms 2012 conference with a synopsis of the scientific highlights presented and to update those who were unable to attend.
Biofilms are the products of microbial multicellular interactions, and they adopt physical structures that reflect the summation of complex interactions among their individual constituents. Biofilms form at many different surfaces, including air-solid, air-liquid, and liquid-liquid interfaces, and range from single species to highly complex, multispecies assemblies. More and more laboratories have investigated the genetic and physiological bases of biofilm formation and structure for a wide range of bacteria. Various bacterial activities, including cell growth and cell death, nutrient acquisition, waste product accumulation, secretion, motility mechanisms, and exopolysaccharide synthesis, can influence the structure and emergent attributes of biofilms.
BIOFILMS, COLONIES, AND WRINKLES
Not surprisingly, many of the same properties that are key to biofilms also impact the morphology of simple colonies on solid medium. Although it has long been debated in the biofilm community whether a bacterial colony should be considered a specialized type of biofilm, it is unquestionable that colony morphologies can effectively reflect some of the important attributes of biofilms. At the Biofilms 2012 conference, it was striking how prominently the analysis of bacterial colonies featured in many different research contexts, and several of the most exciting presentations were squarely focused on understanding the dynamic structures of these colonies. How do bacterial assemblies, in particular, colonies on solid growth medium, develop higher-order conformations such as complex wrinkling patterns (Fig. 1)? The detailed answers to this seemingly simple question are beginning to emerge through a combination of molecular genetics, advanced microscopy, and biophysics. Wrinkle formation includes localized cell death, exopolysaccharide production, extracellular DNA release, and fibers or amyloid proteins. These answers pertain to many aspects of current biofilm research but also harken back to an earlier time in microbiology when biofilms were undefined and colonies, wrinkled, rhizoid, rugose, or otherwise (Fig. 1), were the most visible and recognizable manifestation of the microbial world. In the summaries of oral presentations and posters that follow, the influence of the simple bacterial colony should not be underestimated nor overlooked.
Fig 1.

Wrinkled and otherwise complex colony morphologies. Left to right: B. subtilis colony (reprinted from Cold Spring Harbor Perspectives in Biology [73] with permission of the publisher), Agrobacterium tumefaciens on agar plus Congo red (image courtesy of Jing Xu and C. Fuqua), Vibrio fischeri colony (image courtesy of V. Ray and K. Visick), and Pseudomonas fluorescens wrinkly spreader colony (image courtesy of Andrew Spier).
BIOFILM MORPHOLOGY AND COLONY STRUCTURE
Studies of surface-adherent biofilms have revealed an even greater level of heterogeneity than that within colonies, with complex chemical gradients and physiologically distinct subpopulations. Furthermore, unlike colonies, natural biofilms are often composed of multiple bacterial species and can show dramatically more complex structure and segregation of activity. Even so, properties of colonies can often predict certain important attributes of biofilms. The formation of wrinkles in colonies is one such example, where mutant derivatives that either develop or abolish wrinkles also manifest qualitative differences in biofilm formation and structure. The analysis of wrinkle formation provides fundamental information on how structural patterns can develop. Gurol Suel and colleagues have investigated the spatiotemporal dynamics of cell death, movement, and mechanical processes during self-organization of Bacillus subtilis cells into wrinkled colonies. As presented at the Biofilms 2012 conference by Munehiro Asally, it was determined that localized patterns of cell death, combined with passive and active cell population expansion, are coincident with areas of the colony that will develop wrinkles (Fig. 2). Production of extracellular matrix components physically confines cells in space, which over time results in the buildup of compressive forces that contributes to the tendency of the entire colony structure to begin to buckle and wrinkle (1). This analysis led to a remarkably detailed view of wrinkle formation and provided striking snapshots of the buckling process in action. One of the outstanding remaining questions is what drives the cell death and whether it is a passive or active process.
Fig 2.
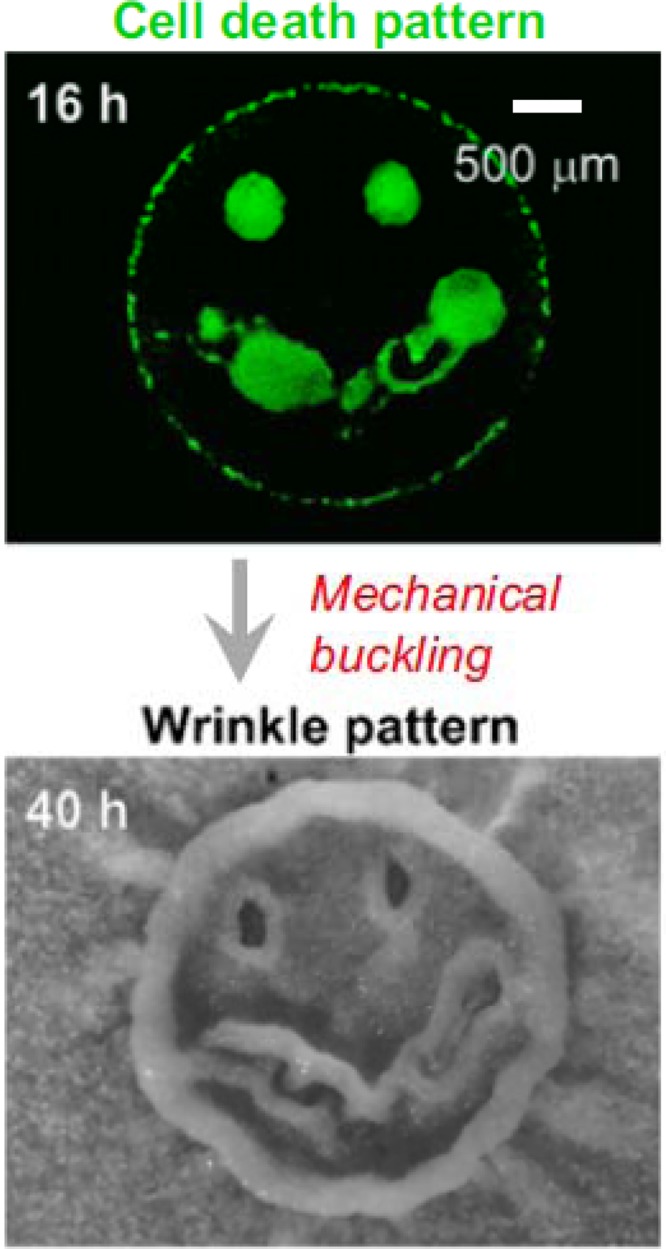
Cell death predicts wrinkle formation in B. subtilis colonies. (Top) Area of induced cell death; (bottom) wrinkle formation. Reprinted from Proceedings of the National Academy of Sciences of the United States of America (1) with permission of the publisher.
David Weitz has collaborated with Richard Losick using a rheological approach to examine the wrinkly colony phenotype exhibited by B. subtilis (Fig. 1). Matrix components, including the rigid TasA amyloid-type polymer and the more elastic exopolysaccharide, that impact biofilm formation also dictate the colony morphotypes observed (2). By injecting dyes into the wrinkles, Weitz et al. demonstrated the rapid transport of solutes through the open spaces underneath the wrinkles within the colony, and they suggest that similar structures act to propel more rapidly transported materials within biofilms. Evaporation from the surface is hypothesized to drive this rapid fluid movement. Josephine R. Chandler discussed the role of a quorum sensing-controlled exopolysaccharide (EPS) in rugose colonies of Burkholderia thailandensis (3). B. thailandensis EPS-producing colonies reach a higher growth yield than EPS mutant colonies, possibly because cells within wrinkles have better access to oxygen. EPS producers also have an advantage over nonproducers in coinoculated colonies. This suggests that for B. thailandensis EPS benefits only producing cells, instead of providing a freely shared “public” benefit to the population. EPS was not exploited by nonproducing individuals. The coregulation of EPS and public goods via quorum sensing may protect cooperators from exploitation and thereby stabilize cooperation. Doug B. Weibel presented a physical model that describes the spontaneous segregation of cells with different motilities in Proteus mirabilis communities. The formation of patterns involves the periodic differentiation of cells into highly flagellated, elongated, and flexible swarmers and consolidators unable to move though viscous fluids. Whereas the swarmers actively move, the consolidators replicate and increase the bacterial population, thus producing the characteristic colonial pattern of P. mirabilis swarms (4). One of the three Biofilms 2012 keynote talks was given by Regine Hengge. She presented striking high-resolution electron microscopy images of Escherichia coli colonies in which curli amyloid fibers are produced by the starving cells at the periphery, whereas closer to the agar surface, abundant entangled flagella are the dominating architectural element, indicating that spatial order in a colony reflects the physiology of different growth phases.
Two posters on the biophysical properties of bacterial biofilms were awarded the Bill Characklis Poster Award for Excellence in Engineering in Biofilm Research. The first went to Munehiro Asally, who presented additional insights into wrinkle formation in B. subtilis colonies and biofilms, combining experimental and modeling approaches to evaluate the roles of exopolysaccharide production and localized cell death. The other award was won by Leonid Pavlovsky for studies on the elasticity of Staphylococcus epidermidis biofilms and the impact of environmental stresses on their physical recalcitrance and rheology.
BIOFILM MATRIX
Within the immediate environment of biofilm bacteria is a self-produced matrix of hydrated extracellular polymeric substances (EPS) consisting mainly of polysaccharides, proteins, nucleic acids, and lipids. The matrix not only provides mechanical stability to biofilms but also mediates bacterial adhesion to surfaces, interconnecting and transiently immobilizing biofilm cells. The properties and formation of the matrix were addressed in several talks at the Biofilms 2012 meeting, with a notable focus on extracellular DNA and functional amyloids. Kai Thormann presented work on extracellular DNA as a structural matrix component during Shewanella oneidensis biofilm formation. The eDNA is released mainly due to prophage-mediated cell lysis and serves not only as a structural biofilm component but also as a source of nutrients. Three extracellular nucleases have been identified, with each of them exhibiting a distinct role with respect to establishment of biofilm structures, degradation of DNA, and control of DNA uptake (5). Matthew R. Chapman reported on molecular mechanisms that are important for the formation of pellicle biofilms, which consist of extracellular structures, including amyloids, flagella, and cellulose, in the uropathogenic strain Escherichia coli UTI89. CsgE was found to prohibit the self-assembly of CsgA into amyloid fibers by acting directly on the secretion substrate CsgA to prevent premature subunit assembly (6). Furthermore, genetic and biochemical analysis indicate that l-cysteine or downstream products in cysteine metabolism pathways prevent planktonic bacteria from forming pellicle biofilms. This knowledge might be exploited for the development of drugs that can inhibit biofilm formation or induce its disassembly as a treatment option for chronic urinary tract infections. A second talk on functional amyloids was given by Blaise Boles. He discussed the importance of amyloid-like fibers consisting of small peptides (phenol soluble modulins [PSMs]) in the formation of Staphylococcus aureus biofilms. Interestingly, whereas polymerized PSM peptides are an important biofilm-stabilizing component, the addition of soluble PSM peptides disperses biofilms (7). A poster presented by Will DePas from the M. Chapman lab, University of Michigan, reported microscopic analysis of the distribution of amyloid-fiber-expressing subpopulations of E. coli within rugose colony wrinkles induced by iron stress. High-resolution imaging revealed that bacteria on the surface of the wrinkle produce curli fibers and cellulose, whereas cells on the interior of the wrinkle do not. The dense matrix on the exterior of the wrinkle is coincident with greater resistance to oxidative stress (8). The striking images presented in this poster were awarded one of the two Terry Beveridge Poster Awards for Excellence in Biofilm Microscopy.
Cynthia B. Whitchurch reported on advanced microscopy techniques, including time-lapse movies, and presented spectacular images of cell lysis, membrane vesicle biogenesis, and extracellular DNA (eDNA) production in Pseudomonas aeruginosa moving on a surface. Exopolysaccharides are well-recognized matrix components, and Michael J. Franklin presented protein structural and bioinformatic information to outline the biosynthesis of the exopolysaccharides alginate, Psl, and Pel of P. aeruginosa (9). Microscopic analysis demonstrated that whereas alginate is more loosely associated with the cells, both Pel and Psl contribute to a mesh-like network that interconnects cells and attaches them to the surface. Furthermore, Gyanendra P. Dubey reported on intercellular nanotubes as a novel mode of interbacterial communication and molecular exchange within and between bacterial species (10). Intercellular and free-end scavenging tubes could be observed in living cells by the use of recently developed fluorescence microscopy imaging approaches.
REGULATION OF THE PLANKTONIC-TO-BIOFILM LIFESTYLE SWITCH
Whereas it is commonly articulated that the process of biofilm formation consists of several distinct developmental phases, the regulation of the transition between those phases—involving initial adherence of motile planktonic cells, followed by irreversible attachment onto the surface, microcolony growth through cell division, EPS synthesis, and biofilm maturation—remains a topic of intense study. For a range of bacteria, the intracellular secondary messenger cyclic diguanosine monophosphate (c-di-GMP) has been implicated in the regulation of transitions between the planktonic and sessile lifestyle. In her keynote address, Regine Hengge reported on the complex integrated c-di-GMP signaling network in E. coli that operates during the temporal succession from the post-exponential to the stationary growth phase. Whereas the housekeeping and the flagellar sigma factors dominate gene expression in growing cells, the need to optimize survival is orchestrated by the alternative sigma factor RpoS upon entry into stationary phase. In addition, c-di-GMP activates the RpoS-dependent expression of the biofilm regulator CsgD in a cascade involving several diguanylate cyclases and phosphodiesterases. Overall, the temporal succession of gene expression in different growth phases translates into a spatial pattern of heterogeneous gene expression that follows nutrient gradients within a colony biofilm. Tim Tolker-Nielsen presented examples of the diversity of c-di-GMP effectors in several bacteria. In Pseudomonas putida, c-di-GMP regulates biofilm formation by binding to the inner membrane protein LapD, which controls the activity of the periplasmic proteinase LapG, which in turn targets the large adhesive outer membrane protein LapA, essential for P. putida biofilm formation (11). In Burkholderia cenocepacia, c-di-GMP regulates biofilm formation by binding to the transcriptional regulator Bcam1349, which induces the expression of genes encoding enzymes for the synthesis of biofilm matrix exopolysaccharide and fimbriae (12). Evidence was also presented that overproduction of a c-di-GMP-hydrolyzing phosphodiesterase cleared P. aeruginosa biofilms on silicone tubes in the mouse peritoneum very effectively, indicating that targeting c-di-GMP signaling pathways might be a promising therapeutic option for clearing chronic biofilm infections. Clay Fuqua reported on recent work with the alphaproteobacterium Agrobacterium tumefaciens in which the motile-to-sessile switch, particularly the production of a unipolar polysaccharide adhesin, is highly dependent on c-di-GMP synthases, diguanylate cyclases with GGDEF domains. These enzymes are under the control of a pair of transcription factors known as VisN and VisR, which regulate motility and attachment in opposite directions and function as a major decision point for the transition from free-living to sessile growth modes. Ruchi Jain reported that FimX, a GGDEF/EAL protein suggested to bind and degrade c-di-GMP, is required for type IV pilus assembly in P. aeruginosa in low c-di-GMP concentrations. Thereby, binding of c-di-GMP to the EAL domain of FimX seems to be important for its localization to the cell poles, whereas release or hydrolysis of c-di-GMP may also be required for pilus assembly (13).
The response of bacteria to surface attachment can be complex, and Thomas K. Wood presented findings on the MqsR-MqsA toxin-antitoxin (TA) system that was first discovered to be upregulated during biofilm formation in E. coli and with increased levels of c-di-GMP. The Mqs system in turn regulates a newly discovered TA system called GhoT-GhoS, described as the first type V TA system, in which the antitoxin acts by cleaving the ghoT transcript, encoding the toxin. These systems can act as global regulatory circuits, which in these specific examples are integrated with the adaptation to surfaces. Alain Filloux presented data on the role of two-component systems on P. aeruginosa biofilm formation. The production of exopolysaccharides is controlled by a complex regulatory system involving the sensor kinases RetS, GacS, and LadS and small regulatory RNAs, as well as c-di-GMP (14). The production of Cup fimbriae, particularly CupD, is controlled by another two-component system, Rcs/Pvr, in which the activity of the response regulator RcsB is controlled by a kinase, PvrS, and a phosphatase, RcsB (15–17). Gerard Wong reported on how the production of the Psl exopolysaccharide impacts P. aeruginosa surface behavior. Since P. aeruginosa can both secrete and associate with Psl, the more Psl there is on a local surface region, the higher the probability of additional Psl deposition in that region (described as the “rich get richer” model). Wong et al. showed using an arabinose-controlled promoter mutant that they can continuously tune surface usage via different degrees of Psl production and predict where microcolonies will be formed (18). G. Wong also addressed physical aspects of social motility in myxobacteria, where a chaotic stick-slip motion (analogized as a process similar to earthquakes) is observed only in bacteria producing an EPS with lubricant-like rather than cellular glue-like properties.
The leadoff keynote presentation in the opening session of Biofilms 2012 was given by Richard Losick and focused on the processes underlying the transition from the colonial biofilm state to an independent lifestyle, biofilm disassembly. Whereas several environmental signals trigger biofilm formation of B. subtilis, self-produced factors are clearly important for biofilm disassembly. For two of those factors, d-amino acids and polyamines, Losick and coworkers have found that these compounds exhibit a biofilm disassembling activity not only in B. subtilis but also in the Gram-positive bacterial pathogen Staphylococcus aureus (19, 20). Ilana Kolodkin-Gal, a researcher in the Losick group, furthermore reported on self-produced factors that promote disassembly of aged S. aureus biofilms. Specific d-amino acids (distinct from those found to disassemble B. subtilis biofilms), a putative peptide, and possibly an aromatic compound were identified as exhibiting disassembly activity in aged S. aureus biofilms. The dispersal activity of self-produced compounds might in the future be exploited as leads to develop novel antibiofilm therapeutic drugs.
MECHANISMS OF BIOFILM FORMATION
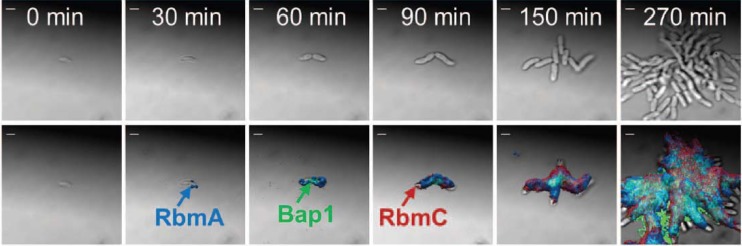
The formation of three-dimensional biofilm structures is dependent on complex regulatory processes which involve the fine-tuned expression of factors important for biofilm formation in response to distinct environmental cues. The molecular mechanisms of the underlying genetic processes are far from being understood and were addressed in the Biofilms 2012 meeting in several talks. Work on the role of the cell surface adhesin protein LapA in irreversible attachment of P. fluorescens was presented by George O′Toole. LapA is a cell surface adhesin (21) and the target of LapD, a c-di-GMP effector, which regulates the periplasmic protease LapG. LapG-LapD cocrystallization studies provided new insights into structural and functional aspects of differential LapA surface expression, including the formation of an unusual multimeric interface between the periplasmic portion of LapD and LapG. Veysel Berk described work performed in collaboration with Fitnat Yildiz on Vibrio cholerae biofilm formation using very-high-resolution time-lapse confocal laser scanning microscopy and tagged extracellular matrix constituents. The additional application of superresolution fluorescence microscopy provided incredible detail of the extracellular material in association with single cells and small cell clusters (22) (Fig. 3). Time-lapse analysis enabled assignment of the four matrix constituents, the RbmA, RbmC, and Bap1 proteins and the Vibrio polysaccharide, to discrete steps in the formation of multicellular biofilms, with the attached cells eventually being enmeshed within an extracellular envelope. In B. subtilis biofilm formation, there is a bimodal switch between biofilm-forming cells and those that are free-living, resulting in a bifurcated population. Victoria L. Marlow presented work using a combination of genetics and single cell analysis with fluorescent reporter genes for matrix components (exopolysaccharide and amyloid fibers) to examine how the phosphorylated form of the transcription factor DegU influences the frequency of cells activating transcription from the matrix operons, and that phosphorylated DegU does this by affecting the level of phosphorylated Spo0A, another key transcription factor in biofilm formation. Karen Visick presented findings regarding the ability of bioluminescent Vibrio fischeri to form biofilms on the surface of the light organ of the squid Euprymna scolopes. Biofilm formation is regulated by the response regulator SypE through the opposing activities of its serine kinase and serine phosphatase domains (23). SypE thereby controls the phosphorylation state of SypA, a downstream effector that, when unphosphorylated, promotes biofilm formation and thus colonization of the squid light organ. Intriguing work on the role of pyruvate fermentation on P. aeruginosa microcolony formation was presented by Karin Sauer (24). Expression profiling of the MifRS two-component regulatory pathway, required for microcolony formation within biofilms, led to the discovery of pyruvate metabolism as a central control point in the process. Remarkably, exogenous addition of pyruvate dehydrogenase removes external pyruvate and inhibits microcolony formation. In another example of the influence of central metabolism on biofilm formation, Vinai C. Thomas elucidated the role of programmed cell death for the formation of structured biofilms of S. aureus. Evidence was presented that glucose catabolism, and the resulting balance between acetoin and acetate formation, influences the intracellular pH and thus provides the mechanistic basis for bacterial programmed cell death possibly also in communities of other species.
Fig 3.
Temporal progression of matrix components during the early stages of V. cholerae biofilm formation. (Top) Phase-contrast microscopy; (bottom) same images merged with fluorescence channels for matrix components. From V. Berk et al., Science 337:236–239, 2012 (22). Reprinted with permission from AAAS.
In a departure from studies on gravity-restricted biofilm formation, Wooseong Kim presented a poster on comprehensive analysis of S. aureus biofilms formed during spaceflight on two separate space shuttle missions. S. aureus contamination of surfaces in manned space vehicles, as well as infections of personnel, are a major concern. This project provided detailed information on the structure of these biofilms and gene expression profiles, suggesting significant yet subtle differences between these microgravity biofilms and those formed in normal gravity. Fittingly, this presentation was awarded one of the Peter Gilbert Poster Awards for Excellence in Innovation and Biofilm Control.
BIOFILM COMMUNITIES—NATURAL AND HOST ASSOCIATED
Monospecies biofilms generated in vitro for a limited number of model bacteria have been a subject of intense research and have provided profound insights into dynamic structural and functional aspects of biofilm formation processes. However, technological advances primarily in the field of imaging and genomics are opening new perspectives on the natural microbial world, with novel scientific findings on the complex microbial communities that can drive geochemical cycles and are critical to human and environmental health (Fig. 4) (25, 26). The third keynote lecture of Biofilms 2012 was presented by Scott J. Hultgren and focused on multicellular behavior of uropathogenic E. coli (UPEC). UPEC strains not only form biofilms on abiotic surfaces but also form biofilm-like intracellular bacterial communities (IBC) within the bladder epithelial cells. These IBCs evolve very rapidly, facilitate bacterial survival, and promote spread to neighboring cells and bacterial persistence. By the use of a UPEC transposon mutant library screened under various in vitro biofilm conditions, several mutants with variable defects in type I pili expression were identified that exhibited both defects in biofilm formation and were unable to form IBCs in a murine model of urinary tract infection (27). These results are in line with the finding that mannosides, small-molecule inhibitors of type 1 pilus adhesion, provided significant protection against urinary tract infections and potentiated the activity of antibiotic treatment regimes. Fernando Andrade presented data on an in vitro anaerobic biofilm model for dental caries. In response to modulations in sucrose concentrations the population dynamics of four bacterial species within the cariogenic biofilm model was explored. High-resolution vertical scanning interferometry revealed the cariogenic potential of sucrose and confirmed the usefulness of the model for further studies on dental caries. Heidi B. Kaplan also focused on oral biofilms and discussed whether nitrate-reducing oral bacterial communities generate nitrite that can be used as a substrate for the generation of NO, an important human signal molecule. Higher nitrate-reducing activity in tongue biofilms correlated with a higher diversity, and a significantly higher abundance of efficient nitrate-reducing bacteria was detected by the use of 16S pyrosequencing. One of the major issues related to biofilm research is the paucity of model hosts for studying bacterial interactions within biofilms. Eleftherios Mylonakis presented work on the establishment of host models using nematodes (Caenorhabditis elegans) and insects (Galleria mellonella) (28). The latter served as a model for studying the biofilm-forming capability of oral and systemic fungal isolates and to distinguish biofilm-forming capabilities from virulence phenotypes. The C. elegans model was used to show that a cross-kingdom interaction between the pathogenic yeast Candida albicans and Salmonella enterica serovar Typhimurium attenuates the virulence of C. albicans. The C. elegans model can also be exploited for high-throughput compound screens and has led to the identification of novel natural compounds with antifungal and antibiofilm activity. Moving to a mammalian model host system, Irena Pastar reported on a porcine epithelial wound healing model for P. aeruginosa and methicillin-resistant S. aureus (MRSA) polymicrobial biofilms, which are a frequent cause of chronic wound infections in humans. Porcine skin is morphologically and physiologically similar to human skin and has been used for over 25 years to examine efficacy of various antimicrobials and presence of wound biofilms (29, 30). Although the presence of P. aeruginosa suppressed S. aureus growth in mixed-species wound infections, it also enhanced the production of S. aureus virulence factors, which resulted in inhibition of wound healing. This animal model might not only significantly contribute to a more detailed understanding of the effects of the wound environment on polymicrobial biofilm formation and bacterial virulence but might also serve as an in vivo model to assess the activity of various compounds on polymicrobial biofilms. Antibiotic perturbation of the intestinal microbiota, e.g., by the use of clindamycin is well known to facilitate the invasion of human pathogens, e.g., Clostridium difficile. By the use of a mouse model, Joao B. Xavier and colleagues demonstrated that microbial diversity in the gut dropped even after a single clindamycin dose and that there was evidence of selection for enterobacteria (31). The development of a minimal ecological model (32) served to explain catastrophic shifts in the microbiota upon antibiotic treatment and might help to estimate infection risks following antibiotic treatment, guiding clinicians in their choice of antibiotic treatment regimes.
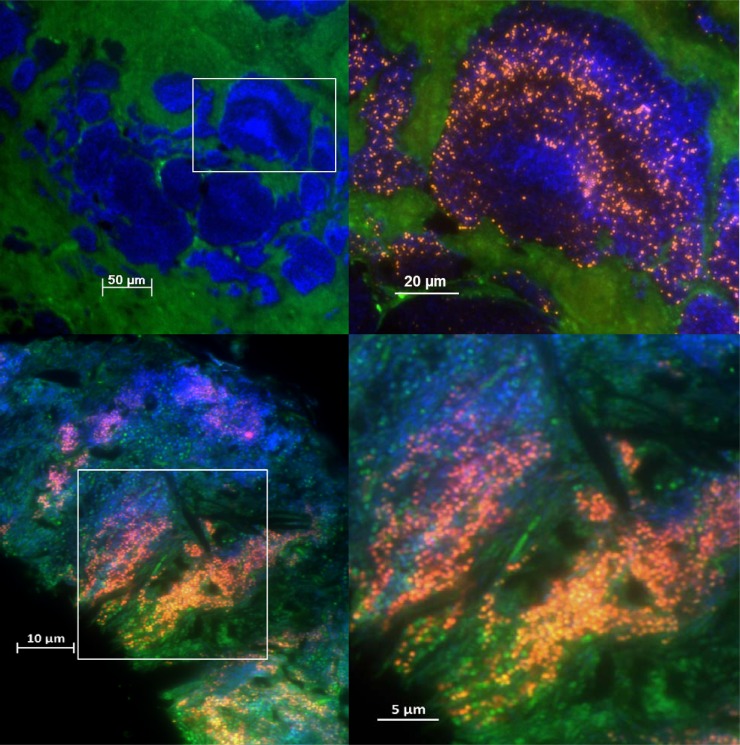
Fig 4.
(Top) Fluorescence in situ hybridization (FISH) of a human heart valve section, showing streptococci in a culture-negative case of infective endocarditis. The overview (left) shows a structured biofilm (blue) within the heart valve tissue (green background fluorescence). Nucleic acids were nonspecifically stained with DAPI (4′,6-diamidino-2-phenylindole dihydrochloride) (blue). (Inset, right) Layers of Streptococcus spp. (hybridized with a genus-specific FISH probe, in orange) show high ribosomal content within this monospecies biofilm. (Bottom) FISH of a subgingival biofilm from a periodontitis patient. The overview (left) shows the complex multispecies biofilm stained with a genus-specific probe for streptococci (orange) combined with EUB338 (green), which detects most bacteria, and the nucleic acid stain DAPI (blue). At higher resolution (inset, right), the streptococci appear to grow along the orientation of the rods. Images courtesy of Annette Moter.
In a poster presentation that was awarded one of the Bill Costerton Poster Awards for Outstanding Interdisciplinary Biofilm Research, Stephanie Roberfroid genetically examined the response of Salmonella Typhimurium to existence in a multispecies biofilm. Using a clever screening strategy described as differential fluorescence induction (DFI), in which random green fluorescent protein (GFP) fusions with Salmonella genes were compared for monospecies and multispecies biofilms, a number of candidate genes specifically induced in response to multispecies environments were identified.
Integrating concepts on host-associated communities and environmental assemblies, Alex Rickard articulated the hypothesis that species-specific coaggregation—mediated by protein adhesins or polysaccharide receptors—enhance the biofilm forming potential of freshwater bacteria (33). By the use of a genetic approach and imaging technology, evidence was provided that Sphingomonas natatoria is a highly promiscuous bridging microorganism that facilitates the recruitment of diverse species into freshwater biofilms. Representing a natural environment with huge human health impact, water distribution systems are engineered to carry, store, and remove massive amounts of water for cities and municipalities all over the world. Biofilms within these systems have been of significant interest for many years, as a potential source and reservoir of human pathogens. David Schwake, of Arizona State University, is specifically interested in identifying Legionella strains harbored in engineered water systems and evaluating the propensity for this pathogen colonizing biofilms. He reported the frequent but nonuniform detection of Legionella spp. from biofilms within distribution systems in central Arizona, using PCR-based detection methods. A separate experiment demonstrated the capability of Legionella to associate within biofilms formed on several drinking water pipe materials, including copper. In addition, inoculation of a model water distribution system revealed rapid colonization of biofilms by Legionella which was sustained for months. In an analysis of another important waterborne pathogen, Marzia Sultana discussed the impact of climate and seasonality on V. cholerae in the Bay of Bengal estuarine ecosystem. Whereas temperature and salinity influence the seasonal abundance of V. cholerae, biofilm formation promotes survival and act as reservoirs when conditions are unfavorable. This work suggests an important role for biofilms in the persistence of waterborne disease agents. Tom Battin reported on biodiversity patterns of complex bacterial communities from various stream ecosystems, ranging from glacier-fed streams to larger rivers. Up to several thousand bacterial operational taxonomic units (OTUs) in these benthic biofilms assemble from the stream water, obviously serving as a seed bank (34, 35). Species sorting governed by the local environment drives the community composition, whereas the biofilm architecture is largely dependent on hydrodynamic forces.
HOST RESPONSES TO BIOFILMS
Biofilms represent a protected mode of growth, and bacteria that reside within biofilms not only withstand diverse environmental insults but also are very effectively protected against the host immune response. Lauren O. Bakaletz presented evidence that in biofilms of a number of bacterial pathogens, not only extracellular DNA but also the DNA-binding protein IHF (integration host factor) is critical for the integrity of the matrix. Interestingly, anti-IHF antibodies disrupted biofilms in vitro but also eradicated Haemophilus influenzae biofilms in a chinchilla model of otitis media (ear infections) (36). Furthermore, anti-IHF antibodies were also effective on B. cepacia biofilms and dissolved microcolonies in sputum from cystic fibrosis (CF) patients (37). Mark E. Shirtliff discussed the role of the adaptive immune system and the detrimental effect it had in nonspecific inflammation and neutrophil recruitment that leads to host damage, thereby preventing the clearance of S. aureus infections (38). He also discussed an in vitro analysis of antigen expression in order to design a vaccine containing a combination of four S. aureus antigens. Vaccine testing in a rabbit osteomyelitis infection model revealed promising first results, especially when vaccination was combined with the administration of vancomycin and led to staphylococcal clearance following challenge. Furthermore, preliminary passive immunization studies in a mouse tibia implant model revealed up to 70% clearance of S. aureus. Çagla Tükel reported findings that curli amyloid fibrils found in biofilms of enteric bacteria are recognized by the Toll-like receptor TLR 2/TLR1 complex (39). New data indicate that recognition of curli fibrils of S. Typhimurium via TLR2 by epithelial cells helps reinforce the intestinal barrier function limiting bacterial translocation during infection. Nonetheless, S. Typhimurium overcomes this immune protection mechanism, translocating into the tissue (40). Across the intestinal barrier, T cells generate IL-17A and IL-22 in response to curli fibrils in a TLR2-dependent manner (41). TLRs also play a dominant role in the recognition of flagella for Gram-negative bacterial pathogens, and loss of flagella—as commonly observed in biofilm bacteria—seems to be an advantage with respect to immune evasion. Rustin Lovewell provided evidence that the loss of motility rather than the loss of the flagella per se is critical for the development of resistance to phagocytic clearance by P. aeruginosa, V. cholerae, and E. coli. Interestingly, phagocytic susceptibility is thereby proportional to bacterial motility (42). John G. Younger reported that a major biofilm matrix constituent, the polysaccharide intercellular adhesin (PIA) of Staphylococcus epidermidis, is important for the activation of the major complement anaphylatoxin C5a in human serum. However, detailed pharmacokinetic modeling revealed that the magnitude of C5a release would produce local concentrations far below the KD of human C5a receptors (43). Thus, even in the clinically frequent setting of a bloodstream catheter massively colonized by S. epidermidis, the rate at which C5a is produced would likely be too low to alert either passing phagocytes or nearby endothelial cells. This is consistent with clinical experience, in which S. epidermidis catheter contamination frequently produces very few clinical signs and symptoms. Avoidance of complement attack by biofilms of Streptococcus pneumoniae was also reported in a poster presentation by Elisa Ramos-Sevillano. Biofilms of S. pneumoniae impaired the deposition of complement components C3b and C1q and also contributed to resistance against phagocytosis by human neutrophils.
NEW INSIGHTS INTO CHRONIC BIOFILM DISEASES
Bacterial pathogens that cause acute infections are generally dispersed planktonic bacteria, whereas those that cause chronic infections and can persist for decades have evolved ways to join together into biofilm communities that resist antimicrobial therapy and immune defense mechanisms. Aspergillus fumigatus has been described as a key fungal pathogen in the cystic fibrosis lung. Gordon Ramage presented an in vitro model to monitor adaptive antifungal resistance mechanisms. A. fumigatus biofilms exhibit a phase-dependent resistance against antifungal agents (44), and transcriptional analysis revealed a higher expression of drug efflux pumps in more developed biofilms (45). Furthermore, eDNA, which is released in a phase-dependent manner, not only is important for the biofilm architecture but also contributes to resistance (46). Joe J. Harrison presented results of a nearly saturated transposon mutant screen in P. aeruginosa that aimed at the identification of genetic determinants underlying the evolution of rugose small-colony variants (RSCVs). A striking majority of the identified mutants had transposons disrupting genes encoding flagellar biogenesis. The same type of flagellum mutant was also abundant in biofilm populations from P. aeruginosa drip flow reactors. Interestingly, these flagellar mutants exhibited an increase in the surface contact-dependent production of the exopolysaccharides Pel and Psl. These findings suggest that the loss of motility provides the strains with a gain of function due to enhanced EPS synthesis, which was crucial for the increased fitness of flagellum mutants in biofilms. Chuanwu Xi hypothesized that extracellular ATP (eATP)—a eukaryotic stress signal—promotes Acinetobacter baumannii biofilm formation (47). In a mouse wound infection model it was demonstrated that treatment with the ATP-degrading enzyme apyrase reduced necrosis and the inflammatory response at wound sites, increasing the influx of macrophages and diminishing bacterial colonization. Bacterial colonization has emerged as a contributing factor in the development of the circulatory disorder arteriosclerosis (arterial hardening) in humans. Bernard B. Lanter presented a poster describing analysis of a collection of carotid artery samples in which bacteria were consistently detected by molecular probing, and microscopic analysis revealed the formation of aggregates and biofilms. A model was proposed in which dispersal of these biofilms occurs in response to fluctuations of blood iron levels, potentially causing the arterial plaque to dislodge.
BIOFILM ANTIMICROBIAL TOLERANCE
Bacteria that reside within biofilm structures exhibit up to 1,000-fold-increased antibiotic resistance to a broad range of antimicrobial agents. Despite being a focus of intense research, the mechanisms underlying this recalcitrance of biofilm bacteria are only poorly understood. Pradeep K. Singh provided evidence that the sensing of nutrient limitation and the subsequent activation of the general stress responses induce antibiotic tolerance in P. aeruginosa biofilms. Tolerance was shown to be dependent on an intact stringent response and the cell's ability to modulate the production of pro-oxidant quinolones (48). This work indicates that antibiotic tolerance of nutrient-limited and biofilm Pseudomonas aeruginosa is mediated by active responses to starvation rather than by the passive effects of growth arrest. It also adds to the emerging concepts that antibiotics often kill bacteria by oxidative stress and that mechanisms which increase stress may enhance antimicrobial activity whereas protecting against oxidative stress increases tolerance (49, 50).
Heleen Van Acker reported on toxin-antitoxin (TA) modules that play a role in the tolerance of Burkholderia cenocepacia biofilms against antibiotics and provided evidence for complex differential expression of 17 TA modules in biofilm-grown bacteria. It is clear that different components of the TA system network are active under different conditions and can contribute to antibiotic tolerance to different extents. Tom Coenye provided novel insight into changes of the B. cenocepacia biofilm transcriptome upon tobramycin treatment. A significant fraction of genes were found to be differentially regulated due to an increase in reactive oxygen species. In addition, small regulatory RNAs as well as several of the toxin-antitoxin systems were differentially regulated, indicating a role in biofilm tolerance. Interestingly, in a small tobramycin-resistant subpopulation, genes from the tricarboxylic acid cycle and genes involved in the electron transport chain were downregulated, whereas genes from the glyoxylate shunt were upregulated. These results are consistent with the finding that itaconate, which shuts down the glyoxylate shunt, increased tobramycin sensitivity in sessile cells. Phil Stewart presented a transcriptomic approach aimed at uncovering the physiological and genetic basis for the increased antibiotic tolerance of biofilm-grown bacteria, in which 293 genes were found to be expressed at higher levels in P. aeruginosa biofilm-grown bacteria than in planktonic bacteria. The finding that many of them are associated with oxygen limitation and stationary phase indicates that starvation/stress responses contribute to the tolerance of biofilm bacteria to ciprofloxacin and, by extension, other antibiotics (51).
Boo Shan Tseng was awarded one of the Bill Costerton Poster Awards for Outstanding Interdisciplinary Biofilm Research for presenting findings that the retardation of tobramycin transport into P. aeruginosa biofilms contributes to enhanced tolerance to this antibiotic. The penetration of fluorescent tobramycin was visualized in exposed biofilms and evidence was presented in support of ionic interactions impeding access of the antibiotic to the interior of the biofilm.
ANTIBIOFILM STRATEGIES
The recalcitrance of bacterial biofilms to classical antibiotic treatment regimens emphasizes the need to develop novel approaches to reduce bacterial attachment to abiotic surfaces and/or to eradicate existing biofilms. Ehud Banin presented exciting work on the use of sonochemistry to synthesize nanoparticles with antibiofilm properties (52). The synthesis of metal oxide and metal fluoride nanoparticles that inhibited biofilm development in a number of bacterial pathogens holds promise for a novel antibiofilm strategy that could be clinically useful. Interestingly, nano-antibiotics that were generated by the use of sonochemistry proved to be more active and more effective than classical antibiotics against drug-resistant pathogens. This effect might be due to improved permeability through the cell wall as well as override of bacterial antibiotic resistance mechanisms (e.g., efflux pumps or enzymes capable of degrading the antibiotics). A recognized problem in treating biofilms is the presence of small but significant persister cell populations that are physiologically tolerant to antibiotics (53). Jean-Marc Ghigo discussed whether the alleviation of persister cells within biofilms and the simultaneous administration of potent antibiotics could be an effective strategy to combat biofilm infections. Since the level of the proton motive force influences sensitivity to antibiotics (e.g., tobramycin due to an increased uptake of the antibiotic), elevation of the pH should lead to a decrease in antibiotic resistance. Indeed, the addition of alkaline amino acids such as arginine not only increased the pH but also significantly enhanced bacterial susceptibility. A promising combination therapy was demonstrated using an implant venous access catheter model in rats, in which gentamicin, ineffective on its own, becomes quite effective when combined with the basic amino acid arginine (54).
Tim H. Jakobsen highlighted the use of the anti-quorum-sensing activity of ajoene isolated from crude garlic extracts for the development of a novel antibiofilm strategy (55). Ajoene exhibited a clear synergistic effect with tobramycin on P. aeruginosa biofilms in vitro as well as in vivo and prevented killing of polymorphonuclear leukocytes in vitro. In order to expedite the identification of new antibiofilm compounds, approaches for large-scale screens are important. Roger Linington presented an image-based screening system that can be exploited for the discovery of novel biofilm-inhibiting compounds. This open-source screening tool, available through the University of California San Diego Chemical Screening Center, has already identified new natural products with activities against Pseudomonas and Vibrio biofilms (56). Biofilm inhibitors were reported that could both inhibit the formation of new biofilm structures and induce the detachment of preformed biofilms. Finally, it was shown that one of these inhibitors has the ability to sensitize preformed biofilms to the effect of antibiotics, including tetracycline and ciprofloxacin. Ronn S. Friedlander presented data using a defined in vitro assay to demonstrate that mucin biopolymers prevent surface colonization of pathogenic bacteria. The mucins promote bacterial segregation and inhibit attachment; however, these effects were shown to be overcome by nonmotile alginate- and Psl-producing P. aeruginosa bacteria, which form suspended aggregates. In a departure from chemical approaches to antibiofilm strategies, Paul Stoodley presented the work of his Ph.D. student Amir Rmaile, who employed high-speed imaging technology to provide a detailed analysis of the forceful dislocation of a dental biofilm from the interproximal space between teeth by an AirFloss device, which creates bursts of water with an initial velocity of 60 m/s. The shear stress and its spatial distribution as generated by the AirFloss device played the key role in efficient biofilm removal.
In a twist on the theme of enhanced antibiotic tolerance in biofilms, Anna Fàbrega Santamaria reported that development of ciprofloxacin resistance in Salmonella Typhimurium compromised its ability to form biofilms, for which she was named recipient of the one of the Peter Gilbert Poster Awards for Excellence in Innovation and Biofilm Control.
ECOLOGY AND EVOLUTION IN MICROBIAL COMMUNITIES
The effects of biodiversity on ecosystem function in the face of various degrees of environmental selective pressures have been intensively studied in microbial communities. The prominence of bacterial colonies as biofilm models was revisited in this session. Kevin Foster concentrated on the question of the degree to which cells cooperate within biofilms. By combining simulation modeling (57) and experiments with Pseudomonas colonies (58), spatio-genetic structure was shown to be critical for the emergence of cooperative phenotypes. In colonies, mucoid variants arose frequently due to mutations in the regulator rsmE, whereas when colonies were regularly dispersed, the mutant's advantage over the wild type was removed. In contrast, when multispecies bacterial consortia were examined, competition appeared to be more important than cooperation (59). Søren Molin presented work on the adaptive processes of P. aeruginosa in association with establishing chronic infections in cystic fibrosis airways, which involve generation of genetic variants that come to dominate these clinical populations (60). P. aeruginosa morphotypes that are commonly recovered from the chronically infected CF lung are mucoid isolates producing excessive amounts of the exopolysaccharide alginate, most arising due to mutations in mucA, an anti-sigma factor that inhibits the alternative sigma factor AlgT, which positively regulates alginate production (61). However, some of the dominant CF isolates are stably nonmucoid, and the genetic basis for this is also mutation of the MucA-AlgT pathways. In such variants, mucoidy can occur due to the emergence of loss-of-function mutations in the sigma factor RpoD. This analysis reveals the strong selective pressures on the mucoid phenotype in the CF lung and how mutations can shift the balance between AlgT and RpoD in the competition for RNA polymerase and control of the alginate genes. Zeinab Hosseinidoust reported on phenotypic diversification of P. aeruginosa strains that have been infected with two bacteriophages. Significant changes in fitness and more importantly virulence were observed, indicating that phenotypic changes of bacteria under phage selective pressure might be clinically relevant (62). David Bruce Borenstein focused on the question of whether cooperators that produce a diffusible public good can coexist with cheating neighbors. Using a computational model of public good dispersal in a dense biofilm environment, it was found that local competition prevents coexistence. The introduction of quorum sensing moderately enhanced cooperator fitness but did not create a coexistence regime. However, for a growing population, interactions at the front led to a series of distinct growth regimes, creating the possibility of coexistence for populations cycling through periods of growth and dispersal. Joshua A. Granek provided new insight into the processes that regulate the formation of natural Saccharomyces cerevisiae biofilms. Using next-generation sequencing of fungal isolates exhibiting substantial variation in colony architecture and a subsequent genome-wide expression analysis of biofilm and nonbiofilm segregants, candidate genes responsible for biofilm variation were identified. The results led to the development of a mechanistic model that relates genetic variation to gene network function and phenotypic outcomes (63).
BIOFILM APPLICATIONS
As our understanding of biofilms continues to grow, a greater number of potential practical uses for this knowledge, outside clinical issues, also emerges. An exciting application for microbial biofilms was described by Birthe Kjellerup, who is exploring the use of biofilms fostered on activated carbon particles for the remediation of polychlorinated biphenyls (PCBs) in the environment. Bacterial biofilms associated with granular activated carbon significantly enhanced PCB degradation, most likely due to better access to the PCBs sequestered onto the biofilm-covered carbon particles. Biofilm-coated granular carbon holds substantial promise to accelerate the rate of PCB degradation, and this approach may find application at contaminated sites in the environment. Jay M. Regan described research on biofilms that form in microbial fuel cells and their impact on power generation from these cells (64). Biofilms can form on either cathodes or anodes, and these adherent communities can exhibit very different properties. The efficiency of power generation from these microbial fuel cells can be strongly impacted by the composition and structure of this exoelectrogenic biofilm community. The fuel cells can be fed simple, inexpensive starting materials such as wastewater with the appropriate biofilm community constituents, driving electrical power generation while potentially generating useful products. In her presentation, Gemma Reguera reported that biofilms of cellulose degraders regulate carbon and nitrogen cycling in terrestrial environments and that, conversely, biofilm formation is fine-tuned to the carbon/nitrogen ratio (65). Nitrogen-limiting conditions induce biofilm formation and enable a more efficient degradation of the cellulosic substrate, leading to carbon sequestration and storage in the biofilm matrix. Conversely, nitrogen input leads to the use of the biofilm matrix as a carbon and energy source for biofilm dispersal, thus linking nitrogen inputs to carbon sequestration and remobilization in terrestrial environments.
In a poster presentation, Karl Anderson reported on the design and use of fluidized bed biofilms comprised of natural isolates from contaminated water sources to remove contaminating bromate from water. Similarly, industrial wastewater contaminated with copper is a pollution hazard. Aaron Mosier reported results of studies on copper removal from copper-polluted wastewater using a mixed-species biofilm reactor and the impact of this water on the structural integrity of these biofilms. Both of these studies suggest that biofilm treatment of wastewater contamination may emerge as a useful water remediation approach.
EMERGING TECHNOLOGIES IN BIOFILM RESEARCH
New imaging technologies have had a profound impact on the evolution of biofilm research. The advent and use of scanning laser confocal microscopy and optical sectioning revolutionized our view of biofilms, driving their conceptual transition from one-dimensional slime layers to complex, three-dimensional structures. Development and implementation of novel imaging approaches continue to propel new findings and expand our understanding of biofilm structure, formation, and activity, and several presentations at Biofilms 2012 highlighted this trend. Yves Dufrene summarized a complement of techniques developed by his laboratory and others, using atomic force microscopy (AFM) to analyze cell and biofilm structure (66, 67). Initially restricted to tracing the external form of cells and extracellular structures, new approaches allow much finer-scale analysis of attached cells and the biofilms they form. For example, specific conjugation of recognition molecules such as antibodies or other ligands to the AFM probe tip allows single-molecule recognition imaging. Once the tip-borne molecule has bound the target molecule(s), the AFM probe can be used to detect and to stretch the associated component, providing a measure of the localization and binding strength. Multiple steps above single molecule interactions are afforded through this specialized type of AFM. Eva Potthoff introduced fluidic force microscopy, a modification of atomic force microscopy in which the cantilever is hollow and through which fluid suction and expulsion can be controlled. Fluidic force microscopy can be used not only to quantify the adhesion force of a single cell onto a surface (68) but also for injections into cells, spatial manipulations, and the release of defined volumes of liquid (69). The data presented were largely related to manipulation of cells of the yeast C. albicans; future work will aim at extending the technology to bacterial systems. Similar goals of characterizing the architecture of biofilms were the target of Eduard Torrents, who presented data generated using mechanical profilometry and interferometry to physically and optically determine the surface topography of biofilms. These techniques utilize relatively simple instrumentation and data acquisition and can still yield important information regarding the nanoscale surface properties of biofilms. Gee Chong Ling introduced a microfabricated polydimethylsiloxane (PDMS) surface and demonstrated that an altered surface topography delayed biofilm formation even in a field study. The surface microtopography also selected for distinct bacterial communities with unique succession over an extended period of time and might be further explored as an effective tool for biofilm control. Aaron Packman addressed the internal heterogeneity of biofilms as a result of transport limitation that leads to nutrient and substrate gradients. Flow cells with imposed gradients simulate flow and influx of substrates and nutrients. Furthermore, by the use of confocal video microscopy and particle tracking velocity, changes in fluid flow around microcolonies, spatial patterns of growth, evolution of patterns of biofilm morphologies, and antimicrobial killing patterns can be observed (70). He also described development of a new generation of biofilm analysis software based within MATLAB, which will soon be available for public distribution and will provide many enhanced capabilities for biofilm image analysis. Complex natural biofilms were the focus of Jessica L. Mark Welch, who presented findings utilizing CLASI-FISH (combinatorial labeling and spectral imaging fluorescence in situ hybridization). The use of 8 different fluorophores in combinations of two per cell resulted in a large number of separable signatures for specific microbial taxa that can be distinguished using ribosomal and spectral imaging (71, 72). The presentation included several beautiful multicolor images of bacteria analyzed by CLASI-FISH applied to semithin sections of dental plaque and provided information on their relative organization within the biofilm. CLASI-FISH can potentially be applied to a wide range of complex microbial communities.
Metal corrosion is a consistent problem in industrial systems and is highly accelerated by certain microbial biofilms. Danielle France from the National Institute of Standards and Technology described the use of a quartz crystal microbalance (QCM) with an iron surface to monitor biofilm formation in flowing systems. Using a metal-corroding bacterial isolate as a model, early colonization events could be effectively followed using the QCM technique, and a correlation between the density of bacteria on the surface and degree of corrosion was readily demonstrated. QCM adds a potentially powerful approach for monitoring corrosion in situ. The chemical and physical modification of abiotic surfaces is a promising approach to prevent biofilm formation. A powerful technique for evaluating the propensity of bacterial biofilms to interact with redox-active metal compounds was presented in a poster by Sara Belchik, who was awarded one of the Terry Beveridge Poster Awards for Excellence in Biofilm Microscopy. This project utilized a novel synchrotron-based Fourier transform infrared (FTIR) microimaging technology operated at the Infrared Environmental Imaging (IRENI) beamline to analyze cryosections of S. oneidensis biofilms. Signatures for biofilm components such as nucleic acids, lipids, and flavins within biofilms and the interactions of specific metals with exopolysaccharides were spatially localized and correlated to high-resolution electron micrographs to provide novel insights into the metal reduction process.
CONCLUSIONS AND FUTURE DIRECTIONS
Reflective of the broad implications of biofilms in industrial and clinical settings and the highly interdisciplinary nature of biofilm research, the Biofilms 2012 meeting in Miami, FL, attracted a large international scientific participant group. The conference provided a framework for the exchange of exciting ideas which stimulate research and development on biofilms. A wide range of findings regarding biofilms, spanning applied biology, evolution, ecology, physiology, and molecular biology, were presented at the conference. Studies that either focused on colony structure or utilized colony morphology as a proxy for biofilms were featured prominently at the meeting. Major advances in our understanding of the physiological and structural processes that underlie formation of wrinkles and buckles in bacteriological colonies can be extrapolated to the formation of complex structures in other types of biofilms. In current biofilm research, new microscopic approaches also continue to shed light on the physical and structural properties of biofilms. In parallel, increasingly powerful, high-resolution genomic and proteomic approaches continue to uncover complex interactions of many genes and gene products important for the establishment of structured biofilms.
We are confident that a compelling, scientifically relevant, and diverse program such as that provided in Miami will continue to attract a wide range of researchers at different levels to the areas of biofilms and catalyze new ideas. Future conferees can look forward to participating in an international and multidisciplinary platform for the exchange of new scientific concepts and the opportunity to learn about and develop novel experimental approaches, cutting-edge technologies, and innovative thinking.
ACKNOWLEDGMENTS
We thank Matthew R. Parsek and Fitnat Yildiz for compiling a highly attractive program and the perfect organization of the Biofilms 2012 meeting.
This meeting was supported by the NIH. Research on biofilms in the lab of C.F. is supported through the National Institutes of Health (GM080546) and in the lab of S.H. by the EU (starter grant 260276-RESISTOME), the German Research Organization (SFB 900, A2), and the Helmholtz Association.
Footnotes
Published ahead of print 26 April 2013
REFERENCES
- 1. Asally M, Kittisopikul M, Rue P, Du Y, Hu Z, Cagatay T, Robinson AB, Lu H, Garcia-Ojalvo J, Suel GM. 2012. Localized cell death focuses mechanical forces during 3D patterning in a biofilm. Proc. Natl. Acad. Sci. U. S. A. 109:18891–18896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Seminara A, Angelini TE, Wilking JN, Vlamakis H, Ebrahim S, Kolter R, Weitz DA, Brenner MP. 2012. Osmotic spreading of Bacillus subtilis biofilms driven by an extracellular matrix. Proc. Natl. Acad. Sci. U. S. A. 109:1116–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chandler JR, Duerkop BA, Hinz A, West TE, Herman JP, Churchill ME, Skerrett SJ, Greenberg EP. 2009. Mutational analysis of Burkholderia thailandensis quorum sensing and self-aggregation. J. Bacteriol. 191:5901–5909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tuson HH, Copeland MF, Carey S, Sacotte R, Weibel DB. 2012. Flagella density regulates Proteus mirabilis swarm cell motility in viscous environments. J. Bacteriol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Heun M, Binnenkade L, Kreienbaum M, Thormann KM. 2012. Functional specificity of extracellular nucleases of Shewanella oneidensis MR-1. Appl. Environ. Microbiol. 78:4400–4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nenninger AA, Robinson LS, Hammer ND, Epstein EA, Badtke MP, Hultgren SJ, Chapman MR. 2011. CsgE is a curli secretion specificity factor that prevents amyloid fibre aggregation. Mol. Microbiol. 81:486–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schwartz K, Syed AK, Stephenson RE, Rickard AH, Boles BR. 2012. Functional amyloids composed of phenol soluble modulins stabilize Staphylococcus aureus biofilms. PLoS Pathog. 8:e1002744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Depas WH, Hufnagel DA, Lee JS, Blanco LP, Bernstein HC, Fisher ST, James GA, Stewart PS, Chapman MR. 2013. Iron induces bimodal population development by Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Franklin MJ, Nivens DE, Weadge JT, Howell PL. 2011. Biosynthesis of the Pseudomonas aeruginosa Extracellular Polysaccharides, Alginate, Pel, and Psl. Front. Microbiol. 2:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dubey GP, Ben-Yehuda S. 2011. Intercellular nanotubes mediate bacterial communication. Cell 144:590–600 [DOI] [PubMed] [Google Scholar]
- 11. Gjermansen M, Nilsson M, Yang L, Tolker-Nielsen T. 2010. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms: genetic elements and molecular mechanisms. Mol. Microbiol. 75:815–826 [DOI] [PubMed] [Google Scholar]
- 12. Fazli M, O'Connell A, Nilsson M, Niehaus K, Dow JM, Givskov M, Ryan RP, Tolker-Nielsen T. The CRP/FNR family protein Bcam1349 is a c-di-GMP effector that regulates biofilm formation in the respiratory pathogen Burkholderia cenocepacia. Mol. Microbiol. 82:327–341 [DOI] [PubMed] [Google Scholar]
- 13. Jain R, Behrens AJ, Kaever V, Kazmierczak BI. 2012. Type IV pilus assembly in Pseudomonas aeruginosa over a broad range of cyclic di-GMP concentrations. J. Bacteriol. 194:4285–4294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Moscoso JA, Mikkelsen H, Heeb S, Williams P, Filloux A. 2011. The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ. Microbiol. 13:3128–3138 [DOI] [PubMed] [Google Scholar]
- 15. Mikkelsen H, Ball G, Giraud C, Filloux A. 2009. Expression of Pseudomonas aeruginosa CupD fimbrial genes is antagonistically controlled by RcsB and the EAL-containing PvrR response regulators. PLoS One 4:e6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bordi C, Lamy MC, Ventre I, Termine E, Hachani A, Fillet S, Roche B, Bleves S, Mejean V, Lazdunski A, Filloux A. 2010. Regulatory RNAs and the HptB/RetS signalling pathways fine-tune Pseudomonas aeruginosa pathogenesis. Mol. Microbiol. 76:1427–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mikkelsen H, Sivaneson M, Filloux A. 2011. Key two-component regulatory systems that control biofilm formation in Pseudomonas aeruginosa. Environ. Microbiol. 13:1666–1681 [DOI] [PubMed] [Google Scholar]
- 18. Colvin KM, Gordon VD, Murakami K, Borlee BR, Wozniak DJ, Wong GC, Parsek MR. 2011. The pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 7:e1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kolodkin-Gal I, Romero D, Cao S, Clardy J, Kolter R, Losick R. 2010. D-amino acids trigger biofilm disassembly. Science 328:627–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kolodkin-Gal I, Cao S, Chai L, Bottcher T, Kolter R, Clardy J, Losick R. 2012. A self-produced trigger for biofilm disassembly that targets exopolysaccharide. Cell 149:684–692 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21. Ivanov IE, Boyd CD, Newell PD, Schwartz ME, Turnbull L, Johnson MS, Whitchurch CB, O'Toole GA, Camesano TA. 2012. Atomic force and super-resolution microscopy support a role for LapA as a cell-surface biofilm adhesin of Pseudomonas fluorescens. Res. Microbiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Berk V, Fong JC, Dempsey GT, Develioglu ON, Zhuang X, Liphardt J, Yildiz FH, Chu S. 2012. Molecular architecture and assembly principles of Vibrio cholerae biofilms. Science 337:236–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Morris AR, Visick KL. 2012. The response regulator SypE controls biofilm formation and colonization through phosphorylation of the syp-encoded regulator SypA in Vibrio fischeri. Mol. Microbiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Petrova OE, Schurr JR, Schurr MJ, Sauer K. 2012. Microcolony formation by the opportunistic pathogen Pseudomonas aeruginosa requires pyruvate and pyruvate fermentation. Mol. Microbiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marsh PD, Moter A, Devine DA. 2011. Dental plaque biofilms: communities, conflict and control. Periodontol 2000 55:16–35 [DOI] [PubMed] [Google Scholar]
- 26. Mallmann C, Siemoneit S, Schmiedel D, Petrich A, Gescher DM, Halle E, Musci M, Hetzer R, Gobel UB, Moter A. 2010. Fluorescence in situ hybridization to improve the diagnosis of endocarditis: a pilot study. Clin. Microbiol. Infect. 16:767–773 [DOI] [PubMed] [Google Scholar]
- 27. Hadjifrangiskou M, Gu AP, Pinkner JS, Kostakioti M, Zhang EW, Greene SE, Hultgren SJ. 2012. Transposon Mutagenesis Identifies Uropathogenic Escherichia coli Biofilm Factors. J. Bacteriol. 194:6195–6205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Desalermos A, Fuchs BB, Mylonakis E. 2012. Selecting an invertebrate model host for the study of fungal pathogenesis. PLoS Pathog. 8:e1002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Davis SC, Ricotti C, Cazzaniga A, Welsh E, Eaglstein WH, Mertz PM. 2008. Microscopic and physiologic evidence for biofilm-associated wound colonization in vivo. Wound Repair Regen. 16:23–29 [DOI] [PubMed] [Google Scholar]
- 30. Mertz PM, Alvarez OM, Smerbeck RV, Eaglstein WH. 1984. A new in vivo model for the evaluation of topical antiseptics on superficial wounds. The effect of 70% alcohol and povidone-iodine solution. Arch. Dermatol. 120:58–62 [PubMed] [Google Scholar]
- 31. Buffie CG, Jarchum I, Equinda M, Lipuma L, Gobourne A, Viale A, Ubeda C, Xavier J, Pamer EG. 2012. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect. Immun. 80:62–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bucci V, Bradde S, Biroli G, Xavier JB. 2012. Social interaction, noise and antibiotic-mediated switches in the intestinal microbiota. PLoS Comput. Biol. 8:e1002497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Min KR, Zimmer MN, Rickard AH. 2010. Physicochemical parameters influencing coaggregation between the freshwater bacteria Sphingomonas natatoria 2.1 and Micrococcus luteus 2.13. Biofouling 26:931–940 [DOI] [PubMed] [Google Scholar]
- 34. Besemer K, Peter H, Logue JB, Langenheder S, Lindstrom ES, Tranvik LJ, Battin TJ. 2012. Unraveling assembly of stream biofilm communities. ISME J. 6:1459–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Augspurger C, Karwautz C, Mussmann M, Daims H, Battin TJ. 2010. Drivers of bacterial colonization patterns in stream biofilms. FEMS Microbiol. Ecol. [DOI] [PubMed] [Google Scholar]
- 36. Goodman SD, Obergfell KP, Jurcisek JA, Novotny LA, Downey JS, Ayala EA, Tjokro N, Li B, Justice SS, Bakaletz LO. 2011. Biofilms can be dispersed by focusing the immune system on a common family of bacterial nucleoid-associated proteins. Mucosal Immunol. 4:625–637 [DOI] [PubMed] [Google Scholar]
- 37. Gustave JE, Jurcisek JA, McCoy KS, Goodman SD, Bakaletz LO. 2012. Targeting bacterial integration host factor to disrupt biofilms associated with cystic fibrosis. J. Cyst. Fibros. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Prabhakara R, Harro JM, Leid JG, Harris M, Shirtliff ME. 2011. Murine immune response to a chronic Staphylococcus aureus biofilm infection. Infect. Immun. 79:1789–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tukel C, Nishimori JH, Wilson RP, Winter MG, Keestra AM, van Putten JP, Baumler AJ. 2010. Toll-like receptors 1 and 2 cooperatively mediate immune responses to curli, a common amyloid from enterobacterial biofilms. Cell Microbiol. 12:1495–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Oppong GO, Rapsinski GJ, Newman TN, Nishimori JH, Biesecker SG, Tukel C. 2013. Epithelial Cells Augment Barrier Function via Activation of the Toll-Like Receptor 2/Phosphatidylinositol 3-Kinase Pathway upon Recognition of Salmonella enterica Serovar Typhimurium Curli Fibrils in the Gut Infect. Immun. 81:478–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nishimori JH, Newman TN, Oppong GO, Rapsinski GJ, Yen JH, Biesecker SG, Wilson RP, Butler BP, Winter MG, Tsolis RM, Ganea D, Tukel C. 2012. Microbial Amyloids Induce Interleukin 17A (IL-17A) and IL-22 Responses via Toll-Like Receptor 2 Activation in the Intestinal Mucosa. Infect. Immun. 80:4398–4408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lovewell RR, Collins RM, Acker JL, O'Toole GA, Wargo MJ, Berwin B. 2011. Step-wise loss of bacterial flagellar torsion confers progressive phagocytic evasion. PLoS Pathog. 7:e1002253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Conrad EC, Hsu YY, Bortz DM, Younger JG. 2013. Spatiotemporal Dynamics of Complement C5a Production within Bacterial Extracellular Polymeric Substance. J. Innate Immun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mowat E, Lang S, Williams C, McCulloch E, Jones B, Ramage G. 2008. Phase-dependent antifungal activity against Aspergillus fumigatus developing multicellular filamentous biofilms. J. Antimicrob. Chemother. 62:1281–1284 [DOI] [PubMed] [Google Scholar]
- 45. Rajendran R, Mowat E, McCulloch E, Lappin DF, Jones B, Lang S, Majithiya JB, Warn P, Williams C, Ramage G. 2011. Azole resistance of Aspergillus fumigatus biofilms is partly associated with efflux pump activity. Antimicrob. Agents Chemother. 55:2092–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rajendran R, Williams C, Lappin DF, Millington O, Martins M, Ramage G. 2013. Extracellular DNA release acts as an antifungal resistance mechanism in mature Aspergillus fumigatus biofilms. Eukaryot. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xi C, Wu J. 2010. dATP/ATP, a multifunctional nucleotide, stimulates bacterial cell lysis, extracellular DNA release and biofilm development. PLoS One 5:e13355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK. 2011. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334:982–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810 [DOI] [PubMed] [Google Scholar]
- 50. Haussler S, Becker T. 2008. The pseudomonas quinolone signal (PQS) balances life and death in Pseudomonas aeruginosa populations. PLoS Pathog. 4:e1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Folsom JP, Richards L, Pitts B, Roe F, Ehrlich GD, Parker A, Mazurie A, Stewart PS. 2010. Physiology of Pseudomonas aeruginosa in biofilms as revealed by transcriptome analysis. BMC Microbiol. 10:294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lellouche J, Friedman A, Gedanken A, Banin E. 2012. Antibacterial and antibiofilm properties of yttrium fluoride nanoparticles. Int. J. Nanomedicine. 7:5611–5624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Allison KR, Brynildsen MP, Collins JJ. 2011. Heterogeneous bacterial persisters and engineering approaches to eliminate them. Curr. Opin. Microbiol. 14:593–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chauhan A, Lebeaux D, Decante B, Kriegel I, Escande MC, Ghigo JM, Beloin C. 2012. A rat model of central venous catheter to study establishment of long-term bacterial biofilm and related acute and chronic infections. PLoS One 7:e37281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jakobsen TH, van Gennip M, Phipps RK, Shanmugham MS, Christensen LD, Alhede M, Skindersoe ME, Rasmussen TB, Friedrich K, Uthe F, Jensen PO, Moser C, Nielsen KF, Eberl L, Larsen TO, Tanner D, Hoiby N, Bjarnsholt T, Givskov M. 2012. Ajoene, a sulfur-rich molecule from garlic, inhibits genes controlled by quorum sensing. Antimicrob. Agents Chemother. 56:2314–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Peach KC, Bray WM, Shikuma NJ, Gassner NC, Lokey RS, Yildiz FH, Linington RG. 2011. An image-based 384-well high-throughput screening method for the discovery of biofilm inhibitors in Vibrio cholerae. Mol. Biosyst. 7:1176–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nadell CD, Foster KR, Xavier JB. 2010. Emergence of spatial structure in cell groups and the evolution of cooperation. PLoS Comput. Biol. 6:e1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Korolev KS, Xavier JB, Nelson DR, Foster KR. 2011. A quantitative test of population genetics using spatiogenetic patterns in bacterial colonies. Am. Nat. 178:538–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Foster KR, Bell T. 2012. Competition, no cooperation, dominates interactions among culturable microbial species. Curr. Biol. 22:1845–1850 [DOI] [PubMed] [Google Scholar]
- 60. Folkesson A, Jelsbak L, Yang L, Johansen HK, Ciofu O, Hoiby N, Molin S. 2012. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat. Rev. Microbiol. 10:841–851 [DOI] [PubMed] [Google Scholar]
- 61. Sautter R, Ramos D, Schneper L, Ciofu O, Wassermann T, Koh CL, Heydorn A, Hentzer M, Hoiby N, Kharazmi A, Molin S, Devries CA, Ohman DE, Mathee K. 2012. A complex multilevel attack on Pseudomonas aeruginosa algT/U expression and algT/U activity results in the loss of alginate production. Gene 498:242–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hosseinidoust Z, Van de Ven TG, Tufenkji N. 2011. Bacterial capture efficiency and antimicrobial activity of phage-functionalized model surfaces. Langmuir 27:5472–5480 [DOI] [PubMed] [Google Scholar]
- 63. Granek JA, Murray D, Kayikci O, Magwene PM. 2012. The Genetic Architecture of Biofilm Formation in a Clinical Isolate of Saccharomyces cerevisiae. Genetics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Logan BE, Regan JM. 2006. Electricity-producing bacterial communities in microbial fuel cells. Trends Microbiol. 14:512–518 [DOI] [PubMed] [Google Scholar]
- 65. Young JM, Leschine SB, Reguera G. 2012. Reversible control of biofilm formation by Cellulomonas spp. in response to nitrogen availability. Environ. Microbiol. 14:594–604 [DOI] [PubMed] [Google Scholar]
- 66. Alsteens D, Dupres V, Mc Evoy K, Wildling L, Gruber HJ, Dufrene YF. 2008. Structure, cell wall elasticity and polysaccharide properties of living yeast cells, as probed by AFM. Nanotechnology. 19:384005. [DOI] [PubMed] [Google Scholar]
- 67. Dufrene YF. 2008. AFM for nanoscale microbe analysis. Analyst 133:297–301 [DOI] [PubMed] [Google Scholar]
- 68. Potthoff E, Guillaume-Gentil O, Ossola D, Polesel-Maris J, Leibundgut-Landmann S, Zambelli T, Vorholt JA. 2012. Rapid and serial quantification of adhesion forces of yeast and Mammalian cells. PLoS One 7:e52712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Stiefel P, Schmidt FI, Dorig P, Behr P, Zambelli T, Vorholt JA, Mercer J. 2012. Cooperative vaccinia infection demonstrated at the single-cell level using FluidFM. Nano Lett. 12:4219–4227 [DOI] [PubMed] [Google Scholar]
- 70. Zhang W, Sileika TS, Chen C, Liu Y, Lee J, Packman AI. 2011. A novel planar flow cell for studies of biofilm heterogeneity and flow-biofilm interactions. Biotechnol. Bioeng. 108:2571–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Valm AM, Mark Welch JL, Borisy GG. 2012. CLASI-FISH: principles of combinatorial labeling and spectral imaging. Syst. Appl. Microbiol. 35:496–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Valm AM, Mark Welch JL, Rieken CW, Hasegawa Y, Sogin ML, Oldenbourg R, Dewhirst FE, Borisy GG. 2011. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc. Natl. Acad. Sci. U. S. A. 108:4152–4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lopez D, Vlamakis H, Kolter R. 2010. Biofilms. Cold Spring Harbor Perspect. Biol. 2:a000398. [DOI] [PMC free article] [PubMed] [Google Scholar]