Abstract
During growth under conditions of glucose and oxygen excess, Staphylococcus aureus predominantly accumulates acetate in the culture medium, suggesting that the phosphotransacetylase-acetate kinase (Pta-AckA) pathway plays a crucial role in bacterial fitness. Previous studies demonstrated that these conditions also induce the S. aureus CidR regulon involved in the control of cell death. Interestingly, the CidR regulon is comprised of only two operons, both encoding pyruvate catabolic enzymes, suggesting an intimate relationship between pyruvate metabolism and cell death. To examine this relationship, we introduced ackA and pta mutations in S. aureus and tested their effects on bacterial growth, carbon and energy metabolism, cid expression, and cell death. Inactivation of the Pta-AckA pathway showed a drastic inhibitory effect on growth and caused accumulation of dead cells in both pta and ackA mutants. Surprisingly, inactivation of the Pta-AckA pathway did not lead to a decrease in the energy status of bacteria, as the intracellular concentrations of ATP, NAD+, and NADH were higher in the mutants. However, inactivation of this pathway increased the rate of glucose consumption, led to a metabolic block at the pyruvate node, and enhanced carbon flux through both glycolysis and the tricarboxylic acid (TCA) cycle. Intriguingly, disruption of the Pta-AckA pathway also induced the CidR regulon, suggesting that activation of alternative pyruvate catabolic pathways could be an important survival strategy for the mutants. Collectively, the results of this study demonstrate the indispensable role of the Pta-AckA pathway in S. aureus for maintaining energy and metabolic homeostasis during overflow metabolism.
INTRODUCTION
Staphylococcus aureus is a dangerous human pathogen, asymptomatically colonizing over 30% of healthy individuals worldwide yet capable of causing invasive opportunistic infections (1). It is a leading cause of both nosocomial and community-associated infections, ranging from mild skin infections to life-threatening diseases, such as severe sepsis, necrotizing pneumonia, endocarditis, and bacteremia (2, 3). S. aureus infections represent an enormous challenge to physicians, due to the ability of the bacterial pathogen to survive in diverse host environments and the emergence of multidrug-resistant strains (1, 3). The versatility of the bacterium is dependent on its proficiency in sensing and utilizing nutrients from different sources and responding appropriately to rapid environmental changes. This is achieved by maximizing virulence factor and energy production via modulation of the expression of genes involved in different metabolic pathways (4–9).
During growth, staphylococci catabolize glucose and other carbohydrates primarily through glycolysis (the Embden-Meyerhof-Parnas pathway) and the pentose phosphate pathway (10). Carbon flux through glycolysis produces two molecules of pyruvate per molecule of glucose consumed, generates two molecules of ATP, and reduces two molecules of NAD+ to NADH (8). During anaerobic growth, pyruvate undergoes mixed-acid fermentation, leading to excretion of lactate, formate, acetate, and ethanol, with the concomitant oxidation of reducing equivalents during lactate and ethanol fermentation (10–12). Under aerobic conditions, however, pyruvate is decarboxylated to acetyl-coenzyme A (Ac-CoA) and CO2 by the pyruvate dehydrogenase complex (PDHC), with reduction of one molecule of NAD+ to NADH (8, 13). Depending on the growth conditions, acetyl-CoA is then oxidized by the tricarboxylic acid (TCA) cycle and/or is hydrolyzed to acetate by the phosphotransacetylase-acetate kinase (Pta-AckA) pathway, which generates one molecule of ATP through substrate level phosphorylation. For example, it was shown that, under conditions of excess glucose and oxygen (overflow metabolism), when carbon flow into the TCA cycle is limited by carbon catabolite repression (6, 14–16), S. aureus predominantly excreted acetate into the culture medium (10, 17). This implies a critical role for the Pta-AckA pathway during overflow metabolism, and its activity may be essential for the growth and fitness of S. aureus, as has been reported for other bacteria (18–22). To date, however, the Pta-AckA pathway in staphylococci has not been characterized.
Interestingly, the LysR-type transcriptional regulator CidR, which is involved in control of the molecular components mediating cell death in S. aureus, exerts its control during aerobic growth under carbon excess (23). Inactivation of the cidR gene reduces transcription of the cid operon, encoding a putative holin that affects murein hydrolase activity and cell viability (23). Intriguingly, CidR function during overflow metabolism also affects the transcription of genes involved in diverse pathways of pyruvate catabolism (i.e., cidC and alsSD operons), leading to the generation of acetate and acetoin (24). It has been speculated that activation of the CidR regulon during overflow metabolism may result from the accumulation of intracellular pyruvate (25, 26).
In the present study, we highlight the contribution of the Pta-AckA pathway to staphylococcal metabolism. We demonstrate that disruption of either pta or ackA in S. aureus has a drastic inhibitory effect on growth during overflow metabolism and leads to significant accumulation of dead cells during the exponential phase of growth. Our results show that the detrimental effects on growth were not caused by the decrease in the energy status of bacteria, as intracellular concentrations of ATP, NAD+, and NADH were found to be higher in the mutants but appear to be associated with a metabolic block at the pyruvate node that globally alters the intracellular metabolic status and activates CidR-dependent pathways.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were grown in LB broth (Novagen, EMD) or on LB agar. S. aureus strains were grown in tryptic soy broth (TSB) without dextrose (BD Biosciences) supplemented with 0.25% glucose (Sigma-Aldrich) or on TSB containing agar. S. aureus cultures were inoculated to 0.06 optical density at 600 nm (OD600) units from overnight cultures, incubated at 37°C, and aerated at 250 rpm with a flask-to-medium ratio of 10:1. Bacterial growth was assessed by measuring the optical density at 600 nm or by determining the number of CFU ml−1. Antibiotics were purchased from Fisher Scientific and were used at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 10 μg/ml; and erythromycin, 10 μg/ml.
Table 1.
Strains and plasmids used in this study
| Bacterial strain or plasmid | Relevant properties | Source |
|---|---|---|
| Strains | ||
| DH5α | E. coli cloning host | Invitrogen |
| RN4220 | S. aureus restriction-deficient mutant of strain 8325-4 | 31 |
| UAMS-1 | S. aureus clinical isolate | 61 |
| KB1058 | UAMS-1-cidC::ermC | 62 |
| KB8000 | UAMS-1-ΔackA::ermB; Emr | This study |
| KB8001 | UAMS-1-Δpta | This study |
| KB8005 | UAMS-1-Δpta; cidC::ermC; Emr | This study |
| SA564 | S. aureus clinical isolate | 63 |
| KB8006 | SA564-ΔackA::ermB; Emr | This study |
| JE2 | Derivative of USA300 LAC | 64 |
| KB8007 | JE2-ΔackA::ermB; Emr | This study |
| Plasmids | ||
| pTS1-d | Shuttle vector; pE194orits; ColE1; Apr Cmr | 29 |
| pMRS78 | Derivative of pTS1-d containing 3.7-kb ackA::ermB fragment | This study |
| pWS2 | Derivative of pTS1-d containing 2-kb Δpta fragment | This study |
| pBK123 | Shuttle vector; Apr Cmr | 32 |
| pWS5 | Derivative of pBK123 containing wild-type allele of ackA | This study |
| pLI50 | Shuttle vector; Apr Cmr | 33 |
| pMRS102 | Derivative of pLI50 containing wild-type allele of pta | This study |
Construction of the ackA and pta mutants in S. aureus.
All primers (see Table S1 in the supplemental material) used for construction and confirmation of the ackA and pta mutations were generated based on the sequence of S. aureus strain MRSA252 (NC_002952.2). The ackA mutant was constructed by replacing a 1.2-kb region, which included the ackA gene, with an erythromycin resistance gene (ermB), using the gene splicing by overlap extension (SOE) technique (27). The ermB antibiotic resistance cassette was amplified from pEC4 (28), using ackA-ermB-f and ackA-ermB-r primers, which contain sequences homologous to the ackA gene. The primers SAR1790-f and ermB-ackA-r were used for amplification of a 1.4-kb region upstream of the ackA gene, while a 1.3-kb region downstream of the ackA gene was amplified using ermB-ackA-f and SacI-ald1-r primers. All three amplified fragments were mixed in equimolar ratios (1:1:1) and amplified using SAR1790-f and SacI-ald1-r primers. The resulting 3.7-kb PCR product consisted of the 1-kb ermB gene flanked by sequences upstream and downstream of the ackA gene. Following digestion with the restriction endonucleases SacI and SmaI, the 3.7-kb product was cloned into pTS1-d (29). The resulting plasmid (pMRS78) was used to construct the UAMS-1-ackA::ermB mutant through standard allelic-exchange methodology described in an earlier study (30). Replacement of the internal region of the ackA gene by the ermB cassette was verified by PCR using the primers ermB-f, ermB-r, ald1-r, and SAR1790-f1.
The pta mutant was constructed by in-frame deletion of the pta gene, using the gene SOE technique (27). Primers SacI-SAR0593-r and lplA-eutD-r were used for amplification of a 0.9-kb region upstream of the pta gene. Primers eutD-lplA-f and lplA-r were used to amplify a 1.1-kb region downstream of the pta gene containing the lplA gene. Importantly, primers lplA-eutD-r and eutD-lplA-f contain a ribosome binding site (RBS) of the pta (eutD) gene combined with the start codon (ATG) of the lplA gene. The amplified fragments were mixed in equimolar ratios (1:1) and amplified using SacI-SAR0593-r and lplA-r primers. The resulting 2.0-kb PCR product containing a single SacI site was cloned into pTS1-d (29) digested with SacI and SmaI to generate the plasmid pWS2. Plasmid pWS2 was used to construct the UAMS-1-Δpta mutant by allelic-exchange methodology (30), and the in-frame deletion of the pta gene in the KB8001 strain was verified by PCR and DNA sequencing, using primers SAR0592-f, SAR0596-r, lplA-r1, and SalI-P-eutD-f.
The pta-cidC double mutant was generated by bacteriophage Φ11-mediated transduction (31) of the cidC::ermC allele from KB1058 and confirmed by PCR using sets of primers, SAR0592-f and SAR0596-r, and cidA-f and cidC DN. The UAMS-1 background in all mutants was confirmed by pulsed-field gel electrophoresis (PFGE) and PCR using primers cna-f and cna-r.
Complementation of the ackA and pta mutants.
For complementation of the ackA mutation, a 1.2-kb PCR product containing the promoterless wild-type ackA gene was amplified using primers SalI-RBS-ackA-f and SacI-ackA-r (see Table S1 in the supplemental material). Following digestion with the restriction endonucleases SalI and SacI, the PCR product was ligated into the plasmid pBK123 under the control of the cadmium-inducible promoter from the cadC gene (32). The resulting recombinant plasmid was designated pWS5. For complementation of the pta mutation, a 1.2-kb PCR product containing the wild-type pta allele with its promoter region was amplified using primers SalI-P-eutD-f and SacI-eutD-r (see Table S1 in the supplemental material). The PCR product containing SalI and SacI sites was cloned into pLI50 (33) digested with SalI and SacI to generate the plasmid pMRS102. The plasmids pWS5 and pMRS102 were transformed into strain RN4220 by electroporation and then introduced into UAMS-1-ackA::ermB and UAMS-1-Δpta strains, respectively, by phage Φ11-mediated transduction (31).
Measurement of glucose, acetic acid, lactate, pyruvate, and acetoin concentrations in the culture medium.
Aliquots of bacterial cultures (1 ml) were centrifuged for 3 min at 14,000 rpm at 4°C. The supernatants were removed and stored at −20°C until use. Acetate, glucose, and d- and l-lactate concentrations were determined using kits purchased from R-Biopharm, according to the manufacturer's protocol. Pyruvate concentrations were determined using the Pyruvate Assay Kit (MBL) according to the manufacturer's protocol. Acetoin concentrations were determined at 560 nm as described previously (34).
Determination of intracellular ATP, NAD+, NADH, Ac-CoA, and pyruvate concentrations.
Intracellular ATP concentrations were determined using the BacTiter-Glo kit (Promega) according to the manufacturer's protocol and normalized to the corresponding viable-cell counts at the time of harvest.
Intracellular NAD+ and NADH concentrations were determined using a Fluoro NAD/NADH kit (Cell Technology). Aliquots of bacterial cultures (10 ml) were harvested by centrifugation at 4°C for 10 min at 4,000 rpm. The bacterial pellets were washed twice with 1 ml of phosphate-buffered saline, pH 7.4 (PBS). Then, the bacterial pellets were resuspended in 0.2 ml of the NAD/NADH extraction buffer and 0.2 ml of the lysis buffer. The cells were lysed using lysing matrix B tubes in a FastPrep instrument (Qbiogene), and then the lysates were incubated at 60°C for 15 min. The lysates were centrifuged at 4°C for 3 min at 14,000 rpm. NAD+ and NADH concentrations in the lysates were determined according to the manufacturer's protocol and normalized to the corresponding viable-cell counts determined at the time of harvest.
Intracellular Ac-CoA concentrations were determined using a PicoProbe Acetyl-CoA Assay Kit (BioVision). Aliquots of bacterial cultures (25 ml) were harvested by centrifugation at 4°C for 10 min at 4,000 rpm. The bacterial pellets were washed twice with 1 ml of PBS and resuspended in 0.5 ml of PBS, followed by the addition of 0.1 ml of ice-cold 3 M perchloric acid. The cells were lysed using lysing matrix B tubes in a FastPrep instrument (Qbiogene). The lysates were centrifuged at 4°C for 3 min at 14,000 rpm. Subsequently, 300 μl of supernatants was neutralized with 75 μl of saturated solution of potassium bicarbonate and centrifuged at 4°C for 3 min at 14,000 rpm. Ac-CoA concentrations were determined according to the manufacturer's protocol and normalized to the corresponding colony counts.
Intracellular pyruvate concentrations were determined using the Pyruvate Assay Kit (MBL). Aliquots of bacterial cultures (10 ml) were harvested by centrifugation at 4°C for 10 min at 4,000 rpm. The bacterial pellets were washed twice with 1 ml of PBS and then resuspended in 0.35 ml of pyruvate assay buffer, incubated for 20 min at 80°C, and lysed using lysing matrix B tubes in a FastPrep instrument (Qbiogene). The lysates were centrifuged at 4°C for 5 min at 14,000 rpm. Pyruvate concentrations were determined according to the manufacturer's protocol and normalized to the corresponding viable-cell counts at the time of harvest. All assays were performed in duplicate for three independent experiments.
Measurement of oxygen consumption.
Cells were cultured at 37°C in TSB supplemented with 0.25% glucose and aerated at 250 rpm with a flask-to-medium ratio of 10:1. After 3 h of growth, samples were collected and diluted in air-saturated TSB supplemented with 0.25% glucose to an OD600 of 0.1 to 0.8. Relative oxygen consumption rates were determined for up to 120 min at 37°C by using a MitoXpress oxygen-sensitive probe (Luxcel Biosciences) according to the manufacturer's instructions. The data were normalized to the corresponding viable-cell counts at the time of harvest. The results were recorded in duplicate for three independent experiments.
Flow cytometry.
Bacterial cell viability was measured by using a LIVE/DEAD BacLight Viability kit (Invitrogen), according to the manufacturer's protocol, on a FACSAria flow cytometer (BD Biosciences). Aliquots of bacterial cultures (1 ml) were harvested by centrifugation at 4°C for 10 min at 4,000 rpm. The bacterial pellets were washed twice with 1 ml of PBS. The bacteria (1 × 107 to 5 × 107 cells per ml) were then stained for 15 min with SYTO 9 and a propidium iodide dye mixture. Bacteria were distinguished from the background using a combination of forward-scatter and side-scatter parameters. In total, 50,000 events were collected for each sample, and fluorescent signals were measured using logarithmic amplifications. Flow cytometric measurements were performed with 488-nm excitation from a blue solid-state laser at 50 mW, and fluorescent emissions in the green and red spectra were detected using 530- and 610-nm band-pass filters, respectively. Data were analyzed with FlowJo software (Tree Star, Inc.). The results are representative of duplicates for three independent experiments.
NMR sample preparation.
Samples for two-dimensional (2D) 1H-13C heteronuclear single quantum coherence (HSQC) spectra were prepared from three independent 50-ml cultures of S. aureus strains UAMS-1, UAMS-1-ackA, and UAMS-1-pta at the exponential growth phase (3 h) on TSB containing 0.25% [13C6]glucose (Sigma-Aldrich). Bacteria (10 OD600 units) were harvested by centrifugation, suspended in 700 μl of quenching solution (60% ethanol, 40% D2O [Isotec], 50 mM phosphate buffer, pH 7.1), and lysed using lysing matrix B tubes in a FastPrep instrument (Qbiogene). To remove the cell debris, the lysates were centrifuged at 4°C for 5 min at 14,000 rpm. The samples were then lyophilized and suspended in a 99.8% D2O (Isotec), 50 mM phosphate buffer and used for nuclear magnetic resonance (NMR) analysis.
NMR data collection and analysis.
All cell lysates were stored in a −80°C freezer until the samples were utilized for NMR experiments. Each sample was thawed completely and allowed to equilibrate at 298.15 K for approximately 1 h. Then, 600 μl of deuterated 50 mM potassium phosphate buffer at pH 7.4 (uncorrected) with 500 μM 3(trimethylsilyl)propionic-2,2,3,3-d4 acid sodium salt (TMSP) was added to the cell lysates and transferred to an NMR tube. A Bruker Avance DRX 500-MHz spectrometer equipped with a 5-mm triple-resonance cryoprobe (1H, 13C, and 15N) with a z-axis gradient was utilized for all 2D 1H-13C HSQC NMR experiments. NMR spectra were acquired in random order, which included alternating between the three S. aureus strains. Also, a BACS-120 sample changer with Bruker Icon software was used to automate the NMR data collection and minimize instrument variations. The 2D 1H-13C HSQC spectra were collected at 298.15 K with 128 scans, 16 dummy scans, and a relaxation delay of 1.5 s. The spectra were acquired with a spectrum width of 5,000 Hz and 2,048 data points in the direct dimension and a spectrum width of 17,605.6 Hz and 64 data points in the indirect dimension. The NMR spectra were processed and analyzed using NMRPipe (35) and NMRViewJ (36) and following the protocol described previously (37). Briefly, peaks were assigned to a metabolite by matching 1H and 13C chemical shifts from the Human Metabolome Database (HMDB) (38), using error tolerances of 0.08 ppm and 0.25 ppm, respectively. The presence of metabolites and metabolic pathways within S. aureus was verified with the KEGG (39) and Metacyc (40) databases.
Peak intensities were normalized by setting the maximal peak intensity to 100. Peak intensities for a specific metabolite were then averaged for each individual 2D 1H-13C HSQC spectrum and subsequently across the set of triplicate spectra. To account for differences in the numbers of viable cells between the S. aureus strains, the peak intensities were scaled by 2.5 for the UAMS-1-ackA and UAMS-1-pta strains. This corresponds to the ratio of CFU to that of the UAMS-1 wild-type strain. The peak intensities from the S. aureus UAMS-1-ackA and UAMS-1-pta mutant strains were then compared to the peak intensities from the UAMS-1 wild-type strain, where metabolite concentration changes were reported as a percentage change relative to the wild type.
Quantification of the mRNA transcripts.
RNA isolation was carried out as described previously (29). Quantitative real-time PCR was performed using the sigA-, pfkA-, citZ-, cidA-, ldh1-, ddh-, and alsS-specific primers listed in Table S1 in the supplemental material. Total RNA (500 ng) was converted to cDNA using the Quantitect Reverse Transcription Kit (Qiagen). The samples were then diluted 1:50, and the cDNA products were amplified using the LightCycler DNA Master SYBR green I kit (Roche Applied Science) following the manufacturer's protocol. The relative transcript levels were calculated using the comparative threshold cycle (CT) method (41) with normalization to the amount of sigA transcripts present in the RNA samples. The results were recorded in duplicate and are representative of three independent experiments. The statistical significance of changes between wild-type and mutant strains was assessed using several statistical tests. The Shapiro-Wilk test was used to check the normality of the data. Levene's test was used to test the equality of variances among groups in comparison. Since the values were normally distributed, based on the Shapiro-Wilk test, and some groups in comparison have unequal variances, analysis of variance (ANOVA) with heterogeneous variance was used to compare the values between the ackA or pta mutants and the wild-type strain for experiments with certain primers, separately. Dunnett's method was used to account for multiple comparisons. A P value of ≤0.05 was considered statistically significant.
RESULTS
Inactivation of the Pta-AckA pathway inhibits growth of S. aureus.
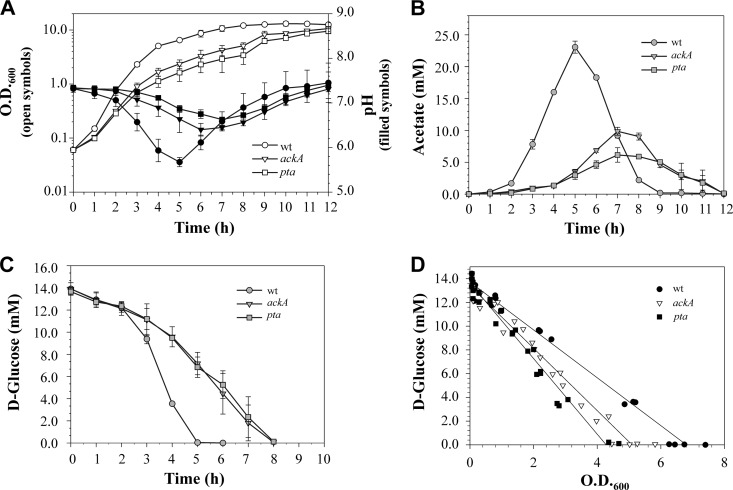
To examine the impact of the Pta-AckA pathway on S. aureus growth and carbon and energy metabolism, as well as CidR-mediated regulation, we constructed ΔackA::ermB and Δpta mutations in the S. aureus strain UAMS-1 (see Materials and Methods). Inactivation of the Pta-AckA pathway by disrupting either the ackA or pta gene had a drastic inhibitory effect on growth rates during the exponential phase in both mutants compared to the isogenic wild-type strain (Fig. 1A). The observed growth defects were accompanied by lower acidification of the culture media (Fig. 1A) and a marked decrease in the rate and concentrations of acetic acid accumulation in both mutants (Fig. 1B). Additionally, the decreased growth rates of the ackA and pta mutants were also reflected in the reduced temporal depletion of glucose (Fig. 1C). However, when the concentrations of glucose in the culture media were plotted as a function of growth, the rates of glucose consumption during the exponential growth phase were found to be higher in the ackA and pta mutants (Fig. 1D; see Fig. S1 in the supplemental material), suggesting carbon is redirected from cell growth to other metabolic pathways and cellular processes. To exclude second-site mutations as the cause of these phenotypes, we performed a complementation study using plasmids pWS5 and pMRS102, containing the wild-type alleles of the ackA and pta genes, respectively, and observed restoration of the growth characteristics in the mutants to wild-type levels (see Fig. S2 in the supplemental material).
Fig 1.
Inactivation of the Pta-AckA pathway has drastic effects on the growth characteristics of S. aureus. (A) Growth curves of the wild-type (wt) strain UAMS-1 and mutant strains UAMS-1-ackA and UAMS-1-pta grown aerobically in TSB containing 0.25% glucose. The OD600 and the pH of the culture medium were determined at the indicated times. (B) Temporal accumulation and depletion of acetic acid in the culture media of strains UAMS-1, UAMS-1-ackA, and UAMS-1-pta. (C) Temporal depletion of glucose from the culture media of strains UAMS-1, UAMS-1-ackA, and UAMS-1-pta. (D) Concentrations of glucose in the culture media of UAMS-1, UAMS-1-ackA, and UAMS-1-pta plotted as a function of growth. The results are presented as the means ± standard errors of the mean for at least three independent experiments.
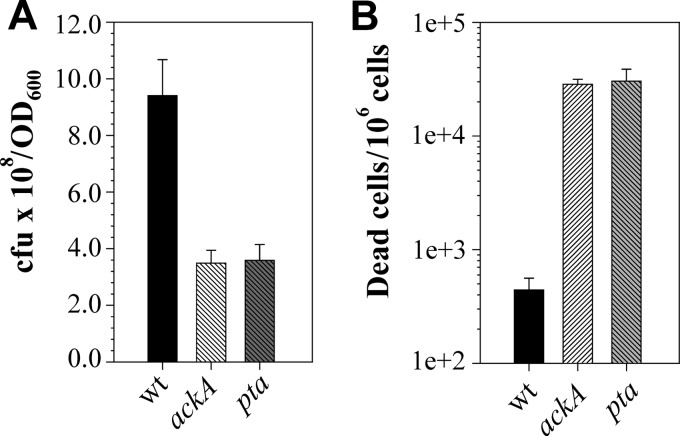
Disruption of the Pta-AckA pathway causes cell death.
During growth experiments, we noticed that the viable-cell count differences observed for the wild-type and mutant strains seemed to be disproportionately high (unpublished data) compared to the differences observed for their optical densities (Fig. 1A). As the optical-density values reflect the total number of cells, including both live and dead cells, we reasoned that during growth the mutant strains may undergo increased cell death. Indeed, the number of viable cells (CFU) per OD600 unit for the wild-type strain at the exponential phase of growth was approximately 2.5 times higher than that for the mutants (Fig. 2A). The observed accumulation of dead cells was not specific to the UAMS-1 strain, as inactivation of the ackA gene in strains JE2 and SA564 caused a similar decrease in the number of viable cells per OD600 unit (see Fig. S3 in the supplemental material). To estimate the number of dead cells in the wild-type and mutant strains during exponential growth, we performed flow cytometry analysis of cells treated with the LIVE/DEAD staining dyes. As anticipated, the numbers of accumulated dead cells in the mutants were nearly 2 orders of magnitude higher than in the wild-type strain (Fig. 2B). These results indicate that the activity of the Pta-AckA pathway in S. aureus is critical for cell viability during overflow metabolism.
Fig 2.
Inactivation of the Pta-AckA pathway causes cell death. (A) Number of viable cells per OD600 unit determined for strains UAMS-1, UAMS-1-ackA, and UAMS-1-pta after 3 h of growth. (B) Quantitation of dead cells in the strains UAMS-1, UAMS-1-ackA, and UAMS-1-pta after 3 h of growth by flow cytometry using the LIVE/DEAD BacLight viability kit. The results are presented as the means plus standard errors of the mean of duplicate determinations for at least three independent experiments.
Pta-AckA pathway inactivation does not decrease intracellular ATP pools.
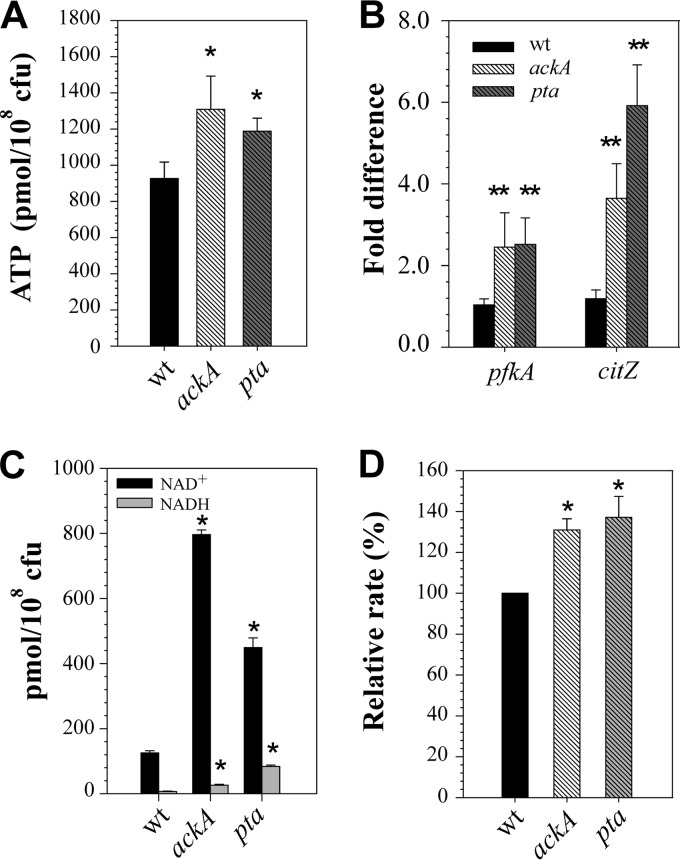
During aerobic growth in the presence of excess glucose or other rapidly metabolizable carbon sources, the TCA cycle in staphylococci is subject to carbon catabolite repression (6, 10, 14–16). To support growth under these conditions, ATP is generated through substrate level phosphorylation via glycolysis and the Pta-AckA pathway (8, 17). The net yield of ATP directly generated by these pathways is four molecules per molecule of glucose consumed, where two molecules of ATP are produced by the Pta-AckA pathway. Hence, we speculated that the observed negative effect on bacterial growth and viability following inactivation of the Pta-AckA pathway may potentially result from a depleted energy status of the cell. To test this hypothesis, we determined the intracellular ATP concentrations in the ackA and pta mutants at the exponential phase of growth. Surprisingly, the intracellular concentrations of ATP were significantly higher in the mutants than in the wild-type strain (Fig. 3A), suggesting that the bacteria efficiently meet their cellular energy requirements by relying on other metabolic pathways.
Fig 3.
Impact of Pta-AckA pathway inactivation on the energy status of bacteria. (A) Intracellular ATP concentrations determined for strains UAMS-1, UAMS-1-ackA, and UAMS-1-pta after 3 h of growth. (B) Relative transcript levels of pfkA and citZ genes in strains UAMS-1, UAMS-1-ackA, and UAMS-1-pta determined by quantitative RT-PCR after 3 h of growth. (C) Intracellular NAD+ and NADH concentrations determined for strains UAMS-1, UAMS-1-ackA, and UAMS-1-pta after 3 h of growth. (D) Relative oxygen consumption rates in strains UAMS-1, UAMS-1-ackA, and UAMS-1-pta were determined using an oxygen-sensitive probe after 3 h of growth. The results are presented as the means plus standard errors of the mean of duplicate determinations for at least two independent experiments. Statistical significance between the wild-type strain and pta and ackA mutants was determined by Student's t test (*, P ≤ 0.001) and by ANOVA with heterogeneous variances (**, P < 0.005) (see Materials and Methods).
Inactivation of the Pta-AckA pathway alters the metabolic status of S. aureus.
We considered two main possibilities that might explain the increased ATP pools in the ackA and pta mutants: (i) the mutants have increased glucose uptake and carbon flow through the glycolytic machinery and (ii) the mutants have a partial redirection of carbon into the TCA cycle. Enhanced carbon flow through these pathways could potentially compensate for the cellular ATP requirements of the mutants, as it increases the flow of electrons through the electron transport chain (ETC), resulting in increased oxidative phosphorylation. To determine whether inactivation of the ackA and pta genes altered the transcription of genes involved in the control of glycolysis and the TCA cycle, we performed a quantitative real-time reverse transcriptase PCR (RT-PCR) analysis using primers specific for pfkA, encoding the key glycolytic enzyme phosphofructokinase, and citZ, encoding the TCA cycle enzyme citrate synthase. Using this approach, we found that inactivation of the pta and ackA genes led to more than a 2-fold increase in the levels of pfkA transcripts and more than a 4-fold increase in the levels of citZ transcripts (Fig. 3B), indicating enhanced carbon flow through both glycolysis and the TCA cycle. In support of the RT-PCR results and consistent with the data for intracellular ATP levels, determination of the concentrations of intracellular NAD+ and NADH revealed a significant increase in the pools of these metabolites in the mutants (Fig. 3C). Additionally, enhanced respiration and carbon flow through the TCA cycle and glycolysis was also supported by higher oxygen consumption rates in the mutants (Fig. 3D).
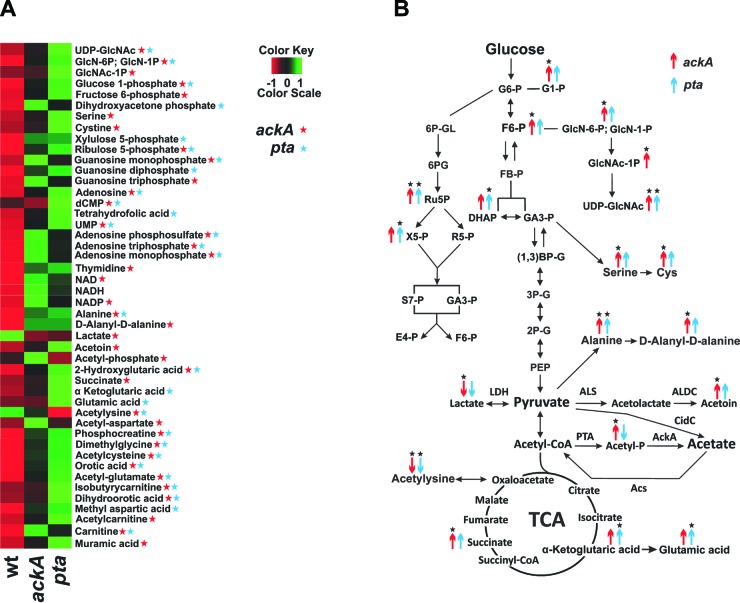
As a direct measure of the effect that disruption of the Pta-AckA pathway has on carbon flow through glycolysis and the TCA cycle, we performed an NMR-based metabolomic analysis of the wild-type and mutant strains at the exponential growth phase. Consistent with previous studies of the Pta-AckA pathway in other organisms (42, 43), this analysis revealed decreased and increased levels of acetyl phosphate in the pta and ackA mutants, respectively. In agreement with the results obtained by RT-PCR, indicating enhanced carbon flow through glycolysis in the mutants, the NMR metabolomics revealed increased levels of fructose 6-phosphate, dihydroxyacetone phosphate, and amino acids whose biosynthesis is linked to glycolysis (i.e., serine and cysteine) (Fig. 4A and B). In support of the RT-PCR results suggesting redirection of carbon into the TCA cycle, we observed an increase in the relative concentrations of the TCA cycle intermediates, as well as metabolites associated with its activity (succinate, α-ketoglutarate, and glutamate concentrations were higher in the mutants) (Fig. 4A and B). In addition, evidence for carbon overflow into the pentose phosphate pathway (PPP) and various biosynthetic pathways, including amino acid, nucleotide, peptidoglycan, and polysaccharide biosynthesis (Fig. 4A and B), was detected. Overall, these results showed that the metabolic block caused by inactivation of the Pta-AckA pathway leads to redirection of carbon flux into diverse metabolic pathways and thus alters the metabolic status of bacteria.
Fig 4.
Pta-AckA pathway inactivation alters the metabolic status of bacteria. (A) Heat map generated from the normalized mean peak intensities for each metabolite identified from the triplicate set of 2D 1H-13C HSQC NMR experiments. The normalized mean intensities are plotted on a color scale from −1 (red) to 1 (green). The red and blue asterisks denote statistical significance at the 90% confidence level (P < 0.10). (B) Metabolic pathway depicting the metabolites identified in the UAMS-1, UAMS-1-ackA, and UAMS-1-pta cellular metabolome by the 2D 1H-13C HSQC NMR experiments. The up arrows correspond to a relative increase in the metabolite concentration, and the down arrows correspond to a relative decrease in the metabolite concentration. Statistical significance at the 90% confidence level (P < 0.10) is denoted by asterisks above the arrows. Acetyl-P, acetyl-phosphate; (1,3)BP-G, 1,3-bisphosphoglycerate; F6-P, fructose 6-phosphate; FB-P, fructose 1,6-bisphosphate; DHAP, dihydroxyacetone phosphate; G6-P, glucose 6-phosphate; G1-P, glucose 1-phosphate; GlcN-6-P, glucosamine-6-phosphate; GlcN-1-P, glucosamine-1-phosphate; GA3-P, glyceraldehyde 3-phosphate; GlcNAc-1-P, N-acetyl-glucosamine-1-phosphate; 3P-G, 3-phosphoglycerate; 2P-G, 2-phosphoglycerate; PEP, phosphoenolpyruvate; Ru5P, ribulose 5-phosphate; S7-P, sedoheptulose 7-phosphate; UDP-GlcNAc, UDP N-acetylglucosamine; X5-P, xylulose 5-phosphate.
Disruption of the Pta-AckA pathway causes a metabolic block at the pyruvate node and activates the CidR regulon.
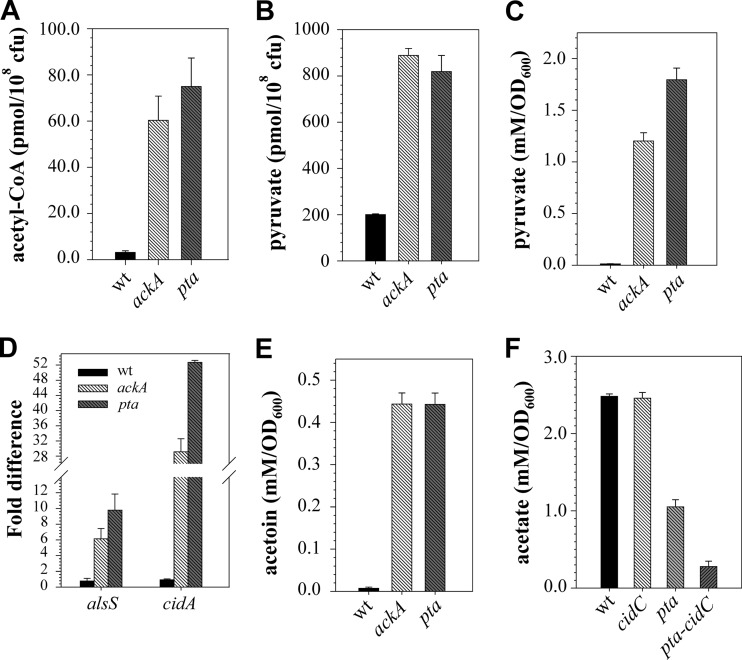
As described above, we observed significant perturbations in the catabolic fate of glucose in the mutants relative to the wild-type strain. In addition to the increased concentrations of glycolytic and TCA cycle intermediates in the mutants, NMR-based metabolomics revealed an abundance of metabolites linked to the pyruvate node. Specifically, intracellular concentrations of acetoin and alanine were higher in the mutants (Fig. 4A and B). Increased concentrations of these intermediates may result from an overflow of acetyl-CoA and pyruvate in the mutants. Since these metabolites were not detected in our NMR assays, we determined the intracellular acetyl-CoA and pyruvate concentrations in the wild-type and mutant strains during the exponential phase of growth using a spectrophotometric assay. As anticipated, inactivation of the Pta-AckA pathway led to accumulation of intracellular acetyl-CoA and pyruvate in the mutants (Fig. 5A and B). Moreover, similar to the results reported previously for Escherichia coli (19, 20, 44), measurements of the concentrations of extracellular pyruvate revealed a significant increase in the culture media for ackA and pta mutants (Fig. 5C). Pyruvate is an unusual by-product not normally excreted by S. aureus during overflow metabolism. Thus, its accumulation in the media may indicate that the surge in the levels of intracellular pyruvate exerts a negative effect on cell viability and the bacteria may need to redirect accumulated pyruvate into other metabolic pathways, or excrete it, in order to survive.
Fig 5.
Disruption of the Pta-AckA pathway alters carbon flow at the pyruvate node and activates the CidR regulon. (A) Intracellular Ac-CoA concentrations determined for strains UAMS-1, UAMS-1-ackA, and UAMS-1-pta after 3 h of growth. (B) Intracellular pyruvate concentrations determined for strains UAMS-1, UAMS-1-ackA, and UAMS-1-pta after 3 h of growth. (C) Concentrations of pyruvate in the culture medium determined for strains UAMS-1, UAMS-1-cidC, UAMS-1-pta, and UAMS-pta-cidC after 3 h of growth. (D) Relative transcript levels of the alsSD and cidABC operons in strains UAMS-1, UAMS-1-ackA, and UAMS-1-pta determined by quantitative RT-PCR after 3 h of growth. (E) Concentrations of acetoin in the culture medium determined for strains UAMS-1, UAMS-1-ackA, and UAMS-1-pta after 3 h of growth. (F) Concentrations of acetate in the culture medium determined for strains UAMS-1, UAMS-1-cidC, UAMS-1-pta, and UAMS-pta-cidC after 5 h of growth. All results are presented as the means plus standard errors of the mean of duplicate determinations for at least three independent experiments.
Unlike E. coli, where a pta mutation led to excretion of d-lactate (20), inactivation of the Pta-AckA pathway in S. aureus has only a minor effect on the generation of l- and d-lactate (Fig. 4; see Fig. S4A in the supplemental material). Moreover, quantitative real-time PCR analysis revealed a decrease in the relative ldh1 and ddh transcript levels (see Fig. S4B in the supplemental material), suggesting that inactivation of the Pta-AckA pathway in S. aureus did not redirect carbon into these fermentative pathways. An alternate route of pyruvate redirection is through the CidR-regulated AlsSD and CidC pathways (24). Given the observation that the S. aureus CidR regulator responds to growth under conditions of excess glucose, it is possible that the pta and ackA mutations affect the expression of the cid and als operons. Thus, to determine if inactivation of the Pta-AckA pathway has an effect on cidABC and alsSD expression, we performed quantitative RT-PCR analysis, using primers specific to cidA and alsS. Indeed inactivation of either pta or ackA resulted in a robust increase of both cidA- and alsS-specific transcripts (Fig. 5D). The transcriptional upregulation of the alsSD and cidABC operons in the pta and ackA mutants was also reflected by a significant increase in the levels of accumulated acetoin in the medium (Fig. 5E) and by increased generation of acetate via pyruvate oxidase, CidC (Fig. 5F).
DISCUSSION
Overflow metabolism in S. aureus occurs during aerobic growth in the presence of rapidly metabolizable carbon sources, when glycolytic flux exceeds the capacity of the TCA cycle and results in the production of acetate. It has been proposed that substrate level phosphorylation during acetate metabolism (catalyzed by AckA) is an important secondary energy-yielding pathway, since under carbon overflow conditions, TCA cycle activity is limited by carbon catabolite repression (the Crabtree effect) (8, 22, 45–48). Indeed, introduction of a metabolic block (mutation of either pta or ackA) that hinders substrate level phosphorylation during overflow metabolism gave rise to growth defects and increased the rate of cell death during exponential growth. However, these deleterious effects on growth and viability did not stem from depleted energy reserves of the cell, as both of the isogenic pta and ackA mutants had comparable or even elevated levels of ATP relative to that of the wild-type strain. Interestingly, similar results were also reported for E. coli, where inactivation of the pta gene resulted in a large growth defect but did not lead to a substantial loss in the amounts of generated ATP (90.5% of the total energy generated by the wild-type strain) (20).
How do the S. aureus pta and ackA mutants replenish their ATP pools during overflow metabolism? Several lines of evidence suggest that the energy derived from an increased rate of glycolysis combined with an increased flux through the TCA cycle may compensate for the loss of ATP following inactivation of the Pta-AckA pathway. First, the rate of glucose uptake was higher for both the pta and ackA mutants than for the wild-type strain, supporting a higher glycolytic flux in these mutants. Second, quantitative RT-PCR analyses confirmed the increased expression of both phosphofructokinase (a critical glycolytic enzyme) and citrate synthase (the first enzyme of the TCA cycle that catalyzes the production of citrate). Finally, metabolomic profiles of both the pta and ackA mutants not only demonstrated an abundance of multiple glycolytic and TCA cycle intermediates, but also indicated carbon overflow into peripheral pathways that are directly linked to central metabolism.
Despite achieving ATP homeostasis, the observation that the pta and ackA mutants still display a growth defect or undergo increased cell death remains a paradox. However, based on our analyses, several possibilities exist. Foremost among them is the drastic increase in the expression of CidR-regulated operons, i.e., cidABC and alsSD. Although CidC and AlsSD represent overflow metabolic enzymes that may redirect excess pyruvate, their transcriptional coupling to the CidAB-holin-like proteins that have been implicated in S. aureus cell death is noteworthy. Although the effects of CidAB overexpression in the pta and ackA mutants are yet to be characterized, their proposed role as effectors of cell death by oligomerizing and localizing to the S. aureus cytoplasmic membrane (49) makes this an intriguing possibility.
A second possibility that may account for the observed growth and viability defects of pta and ackA mutants involves intracellular metabolic toxicity that may arise from increased pyruvate and/or acetyl-CoA levels (Fig. 5A and B). Chang et al. proposed that the growth defect observed for the E. coli pta mutant resulted from the unbalanced flux of acetyl-CoA (20). Accumulation of acetyl-CoA caused inhibition of the pyruvate dehydrogenase complex, leading to increased concentrations of intracellular pyruvate and excretion of d-lactate and pyruvate into the medium (20). Recently, it was shown that triphenylbismuthdichloride (TPBC) suppresses growth and blocks pyruvate catabolism in S. aureus by specific inhibition of the PDHC (50). Using exometabolome analysis, Birkenstock et al. showed that the treatment of cells with TPBC led to an accumulation of pyruvate, reduced temporal depletion of glucose, and decreased acetate excretion (50). Although, the growth defect observed after TPBC treatment may result from decreased intracellular pools of acetyl-CoA and carbon flux into the Pta-AckA pathway and TCA cycle or associated effects on protein acetylation (51–53), a negative impact of the high levels of accumulated pyruvate on bacterial growth cannot be excluded from consideration.
Finally, fluctuations in the redox status or the generation of reactive oxygen species (ROS) as a result of enhanced oxidative phosphorylation in the pta and ackA mutants may yet constitute another reason for the observed deleterious growth characteristics. In support of this, determination of the oxygen consumption rates revealed increased levels of respiration in the mutants (Fig. 3D). Moreover, quantification of intracellular NAD+ and NADH demonstrated significantly higher concentrations of both species in the mutants (Fig. 3C and 4A), suggesting enhanced de novo biosynthesis of NAD+. Consistent with this observation, NMR-based metabolomics detected carbon redirection toward the nucleotide biosynthesis and pentose phosphate pathway that could generate precursors for NAD+ synthesis. Increased NADH levels could potentially result in the increased respiration observed for the mutants and generation of ROS (54–56).
Considering that inactivation of the Pta-AckA pathway alters the metabolic status of bacteria and has deleterious effects on bacterial growth and cell viability, our findings provide evidence that the activity of this pathway in S. aureus is critical under conditions of overflow metabolism. Although the exact mechanisms leading to cell death in the pta and ackA mutants still remain to be clarified, our data suggest that perturbations in the concentration of intracellular acetyl phosphate (Fig. 4A), which can affect two-component signal transduction systems and other cellular processes (22, 57–60), are not a primary cause of growth inhibition and cell death in the mutants, as disruption of either the pta or ackA gene had similar effects on bacterial growth and cell viability. Furthermore, inactivation of the Pta-AckA pathway did not decrease the energy status of bacteria; however, it did alter the metabolic state by causing a metabolic block at the pyruvate node, as well as an increase in carbon flux through both glycolysis and the TCA cycle.
Overall, the results of this study provide important insight into the Pta-AckA pathway in S. aureus, which is critical for acetate dissimilation (22). This pathway could also affect biofilm development, considering previous evidence suggesting acetyl phosphate functions as a global signal within a biofilm (59). Thus, further studies of this pathway will expand our understanding of the physiological processes that underlie the colonization and invasive mechanisms critical for staphylococcal infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank Victoria Smith of the UNMC Cell Analysis Facility for technical assistance with flow cytometry, Fang Yu of the UNMC Department of Biostatistics for statistical analysis, and Kari Nelson for editorial assistance. We thank Paul D. Fey and Jennifer L. Endres for comments on the manuscript.
This research was supported by grants P01-AI08321 (K.W.B.), R01-A1038901 (K.W.B.), P20 RR-17675 (R.P.), and P30-GM103335 (R.P.) from the National Institutes of Health and by Emergent BioSolutions, Inc. (Lansing, MI). Research was performed in facilities renovated with support from the NIH under grant RR015468-01.
Footnotes
Published ahead of print 26 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00042-13.
REFERENCES
- 1. Chambers HF, Deleo FR. 2009. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 7:629–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wertheim HF, Vos MC, Ott A, van Belkum A, Voss A, Kluytmans JA, van Keulen PH, Vandenbroucke-Grauls CM, Meester MH, Verbrugh HA. 2004. Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus non-carriers. Lancet 364:703–705 [DOI] [PubMed] [Google Scholar]
- 3. Lowy FD. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520–532 [DOI] [PubMed] [Google Scholar]
- 4. Seidl K, Stucki M, Ruegg M, Goerke C, Wolz C, Harris L, Berger-Bachi B, Bischoff M. 2006. Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob. Agents Chemother. 50:1183–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Majerczyk CD, Sadykov MR, Luong TT, Lee C, Somerville GA, Sonenshein AL. 2008. Staphylococcus aureus CodY negatively regulates virulence gene expression. J. Bacteriol. 190:2257–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Seidl K, Muller S, Francois P, Kriebitzsch C, Schrenzel J, Engelmann S, Bischoff M, Berger-Bachi B. 2009. Effect of a glucose impulse on the CcpA regulon in Staphylococcus aureus. BMC Microbiol. 9:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Majerczyk CD, Dunman PM, Luong TT, Lee CY, Sadykov MR, Somerville GA, Bodi K, Sonenshein AL. 2010. Direct targets of CodY in Staphylococcus aureus. J. Bacteriol. 192:2861–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Somerville GA, Proctor RA. 2009. At the crossroads of bacterial metabolism and virulence factor synthesis in staphylococci. Microbiol. Mol. Biol. Rev. 73:233–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chaffin DO, Taylor D, Skerrett SJ, Rubens CE. 2012. Changes in the Staphylococcus aureus transcriptome during early adaptation to the lung. PLoS One 7:e41329. 10.1371/journal.pone.0041329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Strasters KC, Winkler KC. 1963. Carbohydrate metabolism of Staphylococcus aureus. J. Gen. Microbiol. 33:213–229 [DOI] [PubMed] [Google Scholar]
- 11. Richardson AR, Libby SJ, Fang FC. 2008. A nitric oxide-inducible lactate dehydrogenase enables Staphylococcus aureus to resist innate immunity. Science 319:1672–1676 [DOI] [PubMed] [Google Scholar]
- 12. Leibig M, Liebeke M, Mader D, Lalk M, Peschel A, Gotz F. 2011. Pyruvate formate lyase acts as a formate supplier for metabolic processes during anaerobiosis in Staphylococcus aureus. J. Bacteriol. 193:952–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gardner JF, Lascelles J. 1962. The requirement for acetate of a streptomycin-resistant strain of Staphylococcus aureus. J. Gen. Microbiol. 29:157–164 [DOI] [PubMed] [Google Scholar]
- 14. Collins FM, Lascelles J. 1962. The effect of growth conditions on oxidative and dehydrogenase activity in Staphylococcus aureus. J. Gen. Microbiol. 29:531–535 [DOI] [PubMed] [Google Scholar]
- 15. Seidl K, Goerke C, Wolz C, Mack D, Berger-Bachi B, Bischoff M. 2008. Staphylococcus aureus CcpA affects biofilm formation. Infect. Immun. 76:2044–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sadykov MR, Hartmann T, Mattes TA, Hiatt M, Jann NJ, Zhu Y, Ledala N, Landmann R, Herrmann M, Rohde H, Bischoff M, Somerville GA. 2011. CcpA coordinates central metabolism and biofilm formation in Staphylococcus epidermidis. Microbiology 157:3458–3468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Somerville GA, Said-Salim B, Wickman JM, Raffel SJ, Kreiswirth BN, Musser JM. 2003. Correlation of acetate catabolism and growth yield in Staphylococcus aureus: implications for host-pathogen interactions. Infect. Immun. 71:4724–4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Grundy FJ, Waters DA, Allen SH, Henkin TM. 1993. Regulation of the Bacillus subtilis acetate kinase gene by CcpA. J. Bacteriol. 175:7348–7355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kakuda H, Shiroishi K, Hosono K, Ichihara S. 1994. Construction of Pta-Ack pathway deletion mutants of Escherichia coli and characteristic growth profiles of the mutants in a rich medium. Biosci. Biotechnol. Biochem. 58:2232–2235 [DOI] [PubMed] [Google Scholar]
- 20. Chang DE, Shin S, Rhee JS, Pan JG. 1999. Acetate metabolism in a pta mutant of Escherichia coli W3110: importance of maintaining acetyl coenzyme A flux for growth and survival. J. Bacteriol. 181:6656–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang YT, Aristidou AA, San KY, Bennett GN. 1999. Metabolic flux analysis of Escherichia coli deficient in the acetate production pathway and expressing the Bacillus subtilis acetolactate synthase. Metab. Eng. 1:26–34 [DOI] [PubMed] [Google Scholar]
- 22. Wolfe AJ. 2005. The acetate switch. Microbiol. Mol. Biol. Rev. 69:12–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang SJ, Rice KC, Brown RJ, Patton TG, Liou LE, Park YH, Bayles KW. 2005. A LysR-type regulator, CidR, is required for induction of the Staphylococcus aureus cidABC operon. J. Bacteriol. 187:5893–5900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang SJ, Dunman PM, Projan SJ, Bayles KW. 2006. Characterization of the Staphylococcus aureus CidR regulon: elucidation of a novel role for acetoin metabolism in cell death and lysis. Mol. Microbiol. 60:458–468 [DOI] [PubMed] [Google Scholar]
- 25. Rice KC, Bayles KW. 2008. Molecular control of bacterial death and lysis. Microbiol. Mol. Biol. Rev. 72:85–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sadykov MR, Bayles KW. 2012. The control of death and lysis in staphylococcal biofilms: a coordination of physiological signals. Curr. Opin. Microbiol. 15:211–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Horton RM, Cai ZL, Ho SN, Pease LR. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques 8:528–535 [PubMed] [Google Scholar]
- 28. Bruckner R. 1997. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 151:1–8 [DOI] [PubMed] [Google Scholar]
- 29. Sadykov MR, Olson ME, Halouska S, Zhu Y, Fey PD, Powers R, Somerville GA. 2008. Tricarboxylic acid cycle-dependent regulation of Staphylococcus epidermidis polysaccharide intercellular adhesin synthesis. J. Bacteriol. 190:7621–7632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Foster TJ. 1998. Molecular genetic analysis of staphylococcal virulence. Methods Microbiol. 27:433–454 [Google Scholar]
- 31. Novick RP. 1991. Genetic systems in staphylococci. Methods Enzymol. 204:587–636 [DOI] [PubMed] [Google Scholar]
- 32. Sharma-Kuinkel BK, Mann EE, Ahn JS, Kuechenmeister LJ, Dunman PM, Bayles KW. 2009. The Staphylococcus aureus LytSR two-component regulatory system affects biofilm formation. J. Bacteriol. 191:4767–4775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee CY, Buranen SL, Ye ZH. 1991. Construction of single-copy integration vectors for Staphylococcus aureus. Gene 103:101–105 [DOI] [PubMed] [Google Scholar]
- 34. Nicholson WL. 2008. The Bacillus subtilis ydjL (bdhA) gene encodes acetoin reductase/2,3-butanediol dehydrogenase. Appl. Environ. Microbiol. 74:6832–6838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6:277–293 [DOI] [PubMed] [Google Scholar]
- 36. Johnson BA. 2004. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 278:313–352 [DOI] [PubMed] [Google Scholar]
- 37. Zhang B, Halouska S, Schiaffo CE, Sadykov MR, Somerville GA, Powers R. 2011. NMR analysis of a stress response metabolic signaling network. J. Proteome Res. 10:3743–3754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, Mandal R, Sinelnikov I, Xia J, Jia L, Cruz JA, Lim E, Sobsey CA, Shrivastava S, Huang P, Liu P, Fang L, Peng J, Fradette R, Cheng D, Tzur D, Clements M, Lewis A, De Souza A, Zuniga A, Dawe M, Xiong Y, Clive D, Greiner R, Nazyrova A, Shaykhutdinov R, Li L, Vogel HJ, Forsythe I. 2009. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 37:D603–D610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. 2008. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36:D480–D484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Caspi R, Altman T, Dale JM, Dreher K, Fulcher CA, Gilham F, Kaipa P, Karthikeyan AS, Kothari A, Krummenacker M, Latendresse M, Mueller LA, Paley S, Popescu L, Pujar A, Shearer AG, Zhang P, Karp PD. 2010. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 38:D473–D479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3:1101–1108 [DOI] [PubMed] [Google Scholar]
- 42. Klein AH, Shulla A, Reimann SA, Keating DH, Wolfe AJ. 2007. The intracellular concentration of acetyl phosphate in Escherichia coli is sufficient for direct phosphorylation of two-component response regulators. J. Bacteriol. 189:5574–5581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ramos-Montanez S, Kazmierczak KM, Hentchel KL, Winkler ME. 2010. Instability of ackA (acetate kinase) mutations and their effects on acetyl phosphate and ATP amounts in Streptococcus pneumoniae D39. J. Bacteriol. 192:6390–6400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dittrich CR, Vadali RV, Bennett GN, San KY. 2005. Redistribution of metabolic fluxes in the central aerobic metabolic pathway of E. coli mutant strains with deletion of the ackA-pta and poxB pathways for the synthesis of isoamyl acetate. Biotechnol. Prog. 21:627–631 [DOI] [PubMed] [Google Scholar]
- 45. Crabtree HG. 1929. Observations on the carbohydrate metabolism of tumours. Biochem. J. 23:536–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hanson RS, Cox DP. 1967. Effect of different nutritional conditions on the synthesis of tricarboxylic acid cycle enzymes. J. Bacteriol. 93:1777–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Andersen KB, von Meyenburg K. 1980. Are growth rates of Escherichia coli in batch cultures limited by respiration? J. Bacteriol. 144:114–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sonenshein AL. 2007. Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol. 5:917–927 [DOI] [PubMed] [Google Scholar]
- 49. Ranjit DK, Endres JL, Bayles KW. 2011. Staphylococcus aureus CidA and LrgA proteins exhibit holin-like properties. J. Bacteriol. 193:2468–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Birkenstock T, Liebeke M, Winstel V, Krismer B, Gekeler C, Niemiec MJ, Bisswanger H, Lalk M, Peschel A. 2012. Exometabolome analysis identifies pyruvate dehydrogenase as a target for the antibiotic triphenylbismuthdichloride in multiresistant bacterial pathogens. J. Biol. Chem. 287:2887–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hu LI, Lima BP, Wolfe AJ. 2010. Bacterial protein acetylation: the dawning of a new age. Mol. Microbiol. 77:15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, Ning ZB, Zeng R, Xiong Y, Guan KL, Zhao S, Zhao GP. 2010. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327:1004–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Thao S, Escalante-Semerena JC. 2011. Control of protein function by reversible Nε-lysine acetylation in bacteria. Curr. Opin. Microbiol. 14:200–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen Q, Vazquez EJ, Moghaddas S, Hoppel CL, Lesnefsky EJ. 2003. Production of reactive oxygen species by mitochondria: central role of complex III. J. Biol. Chem. 278:36027–36031 [DOI] [PubMed] [Google Scholar]
- 55. Lenaz G, Bovina C, D'Aurelio M, Fato R, Formiggini G, Genova ML, Giuliano G, Merlo Pich M, Paolucci U, Parenti Castelli G, Ventura B. 2002. Role of mitochondria in oxidative stress and aging. Ann. N. Y. Acad. Sci. 959:199–213 [DOI] [PubMed] [Google Scholar]
- 56. Ishii N. 2007. Role of oxidative stress from mitochondria on aging and cancer. Cornea 26:S3–S9 [DOI] [PubMed] [Google Scholar]
- 57. Hong JS, Hunt AG, Masters PS, Lieberman MA. 1979. Requirements of acetyl phosphate for the binding protein-dependent transport systems in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 76:1213–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wanner BL, Wilmes-Riesenberg MR. 1992. Involvement of phosphotransacetylase, acetate kinase, and acetyl phosphate synthesis in control of the phosphate regulon in Escherichia coli. J. Bacteriol. 174:2124–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wolfe AJ, Chang DE, Walker JD, Seitz-Partridge JE, Vidaurri MD, Lange CF, Pruss BM, Henk MC, Larkin JC, Conway T. 2003. Evidence that acetyl phosphate functions as a global signal during biofilm development. Mol. Microbiol. 48:977–988 [DOI] [PubMed] [Google Scholar]
- 60. Mizrahi I, Biran D, Ron EZ. 2006. Requirement for the acetyl phosphate pathway in Escherichia coli ATP-dependent proteolysis. Mol. Microbiol. 62:201–211 [DOI] [PubMed] [Google Scholar]
- 61. Gillaspy AF, Hickmon SG, Skinner RA, Thomas JR, Nelson CL, Smeltzer MS. 1995. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect. Immun. 63:3373–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Patton TG, Rice KC, Foster MK, Bayles KW. 2005. The Staphylococcus aureus cidC gene encodes a pyruvate oxidase that affects acetate metabolism and cell death in stationary phase. Mol. Microbiol. 56:1664–1674 [DOI] [PubMed] [Google Scholar]
- 63. Somerville GA, Chaussee MS, Morgan CI, Fitzgerald JR, Dorward DW, Reitzer LJ, Musser JM. 2002. Staphylococcus aureus aconitase inactivation unexpectedly inhibits post-exponential-phase growth and enhances stationary-phase survival. Infect. Immun. 70:6373–6382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. 2013. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4:e00537-12. 10.1128/mBio.00537-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.