Abstract
A prospective cohort study was performed to evaluate the prevalences and loads of Salmonella and Campylobacter spp. in farm and processing plant samples collected from 55 commercial broiler chicken flocks. Environmental samples were collected from broiler houses within 48 h before slaughter, and carcass rinses were performed on birds from the same flocks at 4 different stages of processing. Salmonella was detected in farm samples of 50 (90.9%) flocks and in processing samples of 52 (94.5%) flocks. Campylobacter was detected in farm samples of 35 (63.6%) flocks and in processing samples of 48 (87.3%) flocks. There was a significant positive relationship between environmental farm samples and processing plant carcass rinses with respect to both Salmonella and Campylobacter prevalences and loads. Campylobacter loads were significantly higher than Salmonella loads, and the correlations between samples collected from the same flocks were higher for Campylobacter than they were for Salmonella. Boot socks were the most sensitive sample type for detection of Salmonella on the farm, whereas litter samples had the strongest association with Salmonella loads in pre- and postchill carcass rinses. Boot socks, drag swabs, and fecal samples all had similar sensitivities for detecting Campylobacter on the farm, and all were more strongly associated with Campylobacter loads in carcass rinses than were litter samples. Farm samples explained a greater proportion of the variability in carcass rinse prevalences and loads for Campylobacter than they did for Salmonella. Salmonella and Campylobacter prevalences and loads both decreased significantly as birds progressed through the processing plant.
INTRODUCTION
Salmonella and Campylobacter cause an estimated 1.9 million food-borne illnesses in the United States each year (1), and poultry has been identified as a common source of the two pathogens (2–4). In a study ranking the importance of 104 different pathogen-food combinations with respect to their combined impact on the total cost of illness and loss of quality-adjusted life years, Campylobacter and Salmonella infections from poultry ranked first and fourth, respectively (5).
The regulatory approach to food-borne pathogen control in the U.S. broiler chicken industry is focused primarily on processing plants. A Salmonella performance standard was introduced in 1996, along with a requirement for slaughter establishments to implement a hazard analysis and critical control points (HACCP) program for pathogen reduction (6). While the percentage of Salmonella-positive broiler carcasses identified by regulatory testing decreased from an initial baseline prevalence of 20% in 1996 to 6.5% in 2011, the incidence of human Salmonella infections remained essentially unchanged over the same period of time (7–9). Consequently, a more stringent Salmonella performance standard was introduced in 2011, along with a new performance standard for Campylobacter (10).
Processing plant interventions such as the use of chlorinated water during immersion chilling are effective at reducing microbial contamination of broiler carcasses, although the magnitude of the reductions that can be achieved with these methods is limited (11, 12). Preharvest management practices with the potential to reduce pathogen contamination on the farm have been recommended as a way to provide a more integrated approach to pathogen control (13–15). Specific practices recommended by the U.S. Department of Agriculture's Food Safety Inspection Service for preharvest pathogen control in broiler chickens include having stringent biosecurity measures and sanitation practices, controlling litter moisture, using well-timed feed withdrawal before slaughter, using acids in drinking water during feed withdrawal, using vaccination programs, and screening flocks for pathogens before processing (16).
Although it seems reasonable that pathogen contamination on the farm would be associated with pathogen contamination at processing, there is currently little quantitative information available with respect to the relationships between these two environments. For Salmonella in particular, traditional enumeration methods are laborious and infrequently utilized in poultry processing studies (17). The lack of available information on pathogen concentrations has been identified as an important data gap with respect to evaluating the effectiveness of intervention and control measures in primary poultry production and processing (18). The primary objectives of this study were to obtain comparative information on the distributions of Salmonella and Campylobacter prevalences and loads in commercial broiler chicken flocks and to quantify the relationships between pathogen prevalences and loads in farm and processing plant samples.
MATERIALS AND METHODS
Study design.
A prospective cohort study was performed in cooperation with a commercial broiler production company in north Georgia. Environmental farm samples were collected from broiler houses within 48 h before processing, and carcass rinses were performed on birds from the same houses after they arrived at the processing plant. A flock was defined as a group of birds from the same broiler house. Only houses that were processed as the first flock immediately after a sanitation shift were included to minimize the risk of cross-contamination. The sanitation shift occurred at the end of each day and was a period of time when no birds were processed and all equipment was washed and sanitized. All flocks were harvested at a single processing plant. The study was approved by the University of Georgia Institutional Animal Care and Use Committee (AUP number A2007-10101-0).
Farm sample collection.
Two sets of environmental samples were collected simultaneously from each broiler house, with one set being cultured for Salmonella and the other for Campylobacter. Each sample set consisted of two drag swabs, two boot socks, four individual fecal samples, and two composite litter samples. Houses were subdivided for sampling so that half of the samples from each set were collected from each half of the house. Drag swabs were commercially prepared (catalog number DS-004; Solar Biologicals, Ogdensburg, NY). Boot socks consisted of a 10-cm section of tubular gauze bandage material (ConvaTec Tubigrip, size D, 3 inch, beige; Mölnlycke Health Care, Norcross, GA) that was worn over the foot while walking through the house. Drag swabs and boot socks were collected by walking around the interior perimeter of each half of the house. Boot socks were rotated on the foot after walking halfway around the house perimeter to ensure that both the top and bottom of the sock were exposed to the litter surface. Individual fecal samples were collected from the central area of the house by using a disposable wooden applicator to scoop samples into a sterile sampling bag (Whirl-Pak; Nasco, Fort Atkinson, WI). Composite litter samples were collected in sterile sampling bags by taking 20 grabs of litter at distributed locations in each half of the house for a total sample volume of approximately 25 g. One set of environmental samples from each house was placed on ice and sent by overnight shipment to the University of Minnesota for Salmonella culture, and the other set was placed on ice and sent by overnight shipment to the University of Arizona for Campylobacter culture.
Processing plant sample collection.
At the processing plant, whole-carcass rinses were performed on 30 birds from each flock: 12 birds outside the plant with feathers still attached; 6 birds at rehang, which is after defeathering but before evisceration; 6 birds after evisceration and inside-outside bird wash but immediately before entering the chill tank; and 6 birds immediately after exiting the chlorinated immersion chill tank. When birds arrived at the processing plant, forklift operators placed a transport container on the ground to enable access by study personnel. Live birds were manually removed from multiple compartments of the transport container and euthanized by cervical dislocation. For the outside plant carcass rinses, feathered birds were placed in individual plastic bags (Cryovac 14-by-20 bags, standard gauge; Sealed Air Corporation, Elmwood Park, NJ) with 450 ml of buffered peptone water (3M, St. Paul, MN). For the inside plant carcass rinses, defeathered birds were placed in individual plastic bags with 225 ml of buffered peptone water. Birds were shaken for 1 min in a mechanical carcass shaker (Simmons Engineering Company, Dallas, GA) to ensure consistent agitation (19). After removal from the carcass shaker, a bottom corner of the rinse bag was cut with scissors and 30 ml of the solution was decanted into each of two prelabeled sterile sample bags (Whirl-Pak; Nasco). Samples were kept on ice during transport to the laboratory. Salmonella cultures of carcass rinse samples were performed within 36 h at the University of Georgia's Poultry Diagnostic Research Center, and matching samples for Campylobacter culture were sent overnight on ice to the University of Arizona. Samples that were cultured at the University of Georgia were kept refrigerated overnight prior to processing to maintain comparability with shipped samples.
Salmonella enumeration.
Quantitative Salmonella cultures of environmental farm samples were performed at the University of Minnesota using a miniature three-tube most probable number (MPN) procedure. After arrival at the laboratory, tetrathionate (Hajna) broth (Difco, Division of Becton, Dickinson, and Co., Sparks, MD) was added to sample bags using the following volumes: 10 ml for drag swabs, 20 ml for boot socks, 9 ml for 1 g of feces, and 9 ml for 1 g of litter. Sample bags were mixed for 1 min in a stomacher, and then 1 ml of the mixed solution was transferred into each of 3 sterile 10-ml conical tubes. Next, a 96-well 2-ml deep-well plate (VWR International, West Chester, PA) was set up with 0.8 ml of tetrathionate broth in each well. An aliquot of 0.2 ml of the original mixed sample was added to the first well of each 8-well series, with each sample being run in triplicate. The first well of each series was mixed using a multichannel pipette before transferring 0.2 ml to the next well. This process was repeated, changing tips between each transfer, to create a serial 5-fold dilution series. The 96-well plate was incubated overnight at 41°C, and then 0.2-ml aliquots from each well were transferred to an identical plate containing 0.8 ml Rappaport-Vassiliadis R10 (RV10) broth (Difco, BD) per well. The three separate centrifuge tubes containing 1 ml of tetrathionate were treated similarly, so that the assay consisted of a total of 9 dilutions. After incubation overnight at 41°C, 1 μl of the solution from each well was transferred to a xylose lysine Tergitol 4 (XLT4; Difco, BD) agar plate in a single step using a pin-tool replicator (V & P Scientific, San Diego, CA). XLT4 plates were incubated overnight at 37°C and observed for 48 h to monitor for the development of black colonies. The number of wells with H2S-positive colonies was counted for each dilution, and MPN calculations were performed as previously described (20). Suspected colonies were confirmed as Salmonella using poly(O) antisera, and isolates were forwarded to the National Veterinary Services Laboratory (NVSL; Ames, IA) for serotyping. The smallest MPN values that could be obtained using this method were 2.7 CFU per drag swab, 2.7 CFU per gram of feces, 2.7 CFU per gram of litter, and 5.4 CFU per boot sock.
Quantitative Salmonella culture of processing plant carcass rinses was performed at the University of Georgia. The MPN procedure was similar to that described for the environmental samples with the exception that the initial dilution was made directly into the 96-well 2-ml deep-well plates rather than into a separate set of 3 individual tubes. For inside and outside plant carcass rinses, the 96-well plates were set up with 0.8 and 0.9 ml of tetrathionate (Hajna) broth in each well, respectively. For inside plant carcass rinses, an aliquot of 0.2 ml of the original rinse sample was added to the first well of each 8-well series, with subsequent 5-fold dilutions. For outside plant carcass rinses, an aliquot of 0.1 ml of the original rinse sample was added to the first well of each 8-well series, with subsequent 10-fold dilutions. Plates were incubated overnight at 41°C. For inside plant rinses, 0.2-ml aliquots from each well were then transferred to identical plates containing 0.8 ml RV10 broth per well. For outside plant rinses, 0.1-ml aliquots from each well were transferred to plates containing 0.9 ml RV10 per well. After overnight incubation at 41°C, 1 μl of the solution from each well was transferred to an XLT4 agar plate using a pin-tool replicator. XLT4 plates were incubated overnight at 37°C and observed for 48 h to monitor for the development of black colonies. The number of wells with H2S-positive colonies was counted for each dilution, and MPN calculations were performed as previously described (20). Suspected colonies were confirmed as Salmonella using poly(O) antisera, and isolates were forwarded to NVSL for serotyping. The smallest MPN values that could be obtained using this method were 300 CFU per carcass for inside plant rinses and 1,350 CFU per carcass for outside plant rinses.
In addition to the quantitative culture procedure, a separate qualitative Salmonella culture was performed for the processing plant carcass rinse samples to improve the detection sensitivity. For the inside plant rinses, 9 ml of the original rinse solution was added to a 50-ml conical tube containing 36 ml of tetrathionate broth. For the outside plant rinses, 4.5 ml of the original rinse solution was added to a 50-ml tube containing 40.5 ml of tetrathionate broth. Samples were incubated overnight at 41°C, with subsequent transfer of 1 μl to an XLT4 agar plate with overnight incubation at 37°C. Suspect colonies were confirmed as previously described for the quantitative cultures. The minimum detection limits for the qualitative cultures were 25 CFU per carcass for inside plant rinses and 100 CFU per carcass for outside plant rinses.
Campylobacter enumeration.
All Campylobacter cultures were performed at the University of Arizona. A miniature three-tube MPN procedure was used for Campylobacter enumeration to ensure comparability with the Salmonella enumeration methods. After arrival at the laboratory, Bolton broth (Hi Media, Mumbai, India) was added to environmental farm samples using the following volumes: 10 ml for drag swabs, 20 ml for boot socks, 9 ml for 1 g of feces, and 9 ml for 1 g of litter. For farm samples, 1 ml of the mixed solution was transferred to three adjacent wells on the first row of a 96-well 2-ml deep-well plate. An aliquot of 0.1 ml was then transferred to 0.9 ml of Bolton broth in the second row, and this process was repeated for the remaining rows to produce a total of 8 10-fold dilutions. For carcass rinse samples, 0.1 ml of the original rinse solution was transferred to 0.9 ml of Bolton broth in three adjacent wells of the first row of the plate, followed by subsequent 10-fold dilutions across the remaining rows of the plate. The 96-well plates were incubated at 42°C with 10% CO2 for 24 h, and then 10 μl of each dilution was plated on Campy-Cefex agar supplemented with 5% bovine blood citrate (Quad 5, Ryegate, MT) as described by Oyarzabal et al. (21), and incubated at 42°C for 48 to 72 h. The number of replicates with growth was counted for each dilution, and MPN calculations were performed as previously described (20). Suspected colonies were confirmed as Campylobacter and species typed using a multiplex PCR as described by Klena et al. (22). Each 25-μl reaction mixture included 15.65 μl sterile D2H2O, 2.5 μl reaction buffer, 2.0 μl magnesium chloride, 0.5 μl of a 10-mmol deoxynucleoside triphosphate (dNTP) mix, 0.25 μl Taq DNA polymerase, 2.5 μl DNA template, 0.5 μl of a 10-μmol solution of each forward primer for Campylobacter jejuni and Campylobacter coli (lpxACcoli-F and lpxACjejuni-F), and 0.6 μl of a 30 μM solution of the common reverse primer (lpxARkk2m). Polymerase, reaction buffer, magnesium, and dNTPs used were from Thermo Fisher Scientific, Rockford, IL. Primers were custom generated by Operon (Huntsville, AL). Reactions were run in an Eppendorf thermocycler under the following conditions: 94°C, 50°C, and 72°C for 1 min each, repeated 40 times, with a final extension at 72°C for 5 min. The PCR amplifies the lipid A gene, lpxA, resulting in a 391-bp product for C. coli and a 331-bp product for C. jejuni. Control strains C. jejuni NCTC 11168 and C. coli ATCC 33559 were used to determine the presence of Campylobacter in samples. The smallest MPN values that could be obtained using this method were 3.0 CFU per drag swab, 3.0 CFU per gram of feces, 3.0 CFU per gram of litter, 6.0 CFU per boot sock, 675 CFU per carcass for inside plant rinses, and 1,350 CFU per carcass for outside plant rinses.
An additional qualitative Campylobacter culture was performed for carcass rinse samples to improve the detection sensitivity. For both inside and outside plant carcass rinses, 10 ml of the rinse solution was added to 40 ml Bolton broth in a conical tube and incubated at 42°C with 10% CO2 for 24 h. A 0.1-ml aliquot of the enriched sample was then spread plated on Campy-Cefex agar and incubated at 42°C for 48 to 72 h. Suspected colonies were confirmed as Campylobacter and species typed using PCR. The minimum detection limit for the qualitative cultures was 23 CFU per carcass for inside plant rinses and 45 CFU per carcass for outside plant rinses.
Statistical analysis.
Salmonella and Campylobacter MPNs were reported on a per-sample basis for drag swabs and boot socks, on a per-gram basis for fecal and litter samples, and on a per-carcass basis for carcass rinses. Carcass rinse samples that were culture negative by the quantitative method but culture positive by the qualitative method were arbitrarily assigned an MPN value equal to one-half the minimum detection limit of the MPN procedure for statistical analyses. MPN values were log10 transformed prior to analysis to normalize the pathogen load distributions.
The correlations between mean pathogen loads of different sample types were estimated using the Spearman rank correlation coefficient. Logistic regression was used to determine what combination of farm samples best predicted Salmonella detection in postchill carcass rinses, and linear regression was used to determine what combination of farm samples best predicted postchill Campylobacter loads. Linear regression was used to quantify the relationship between the percentage of culture-positive farm samples obtained from each flock and the percentage of culture-positive processing plant carcass rinses. Linear regression was also used to quantify the relationship between the mean log-transformed pathogen loads of farm and processing plant samples.
Random-effects linear and logistic regression models were used to estimate the intracluster correlation (ICC) for quantitative and binary culture results, respectively, in samples collected from the same flock. ICCs for binary culture results were calculated assuming a latent-response model formulation (23, 24). The ICC is a measure of the degree of similarity between samples collected from the same flock. In theory, the ICC can take on any value between −1 and 1, although in practice most values range between 0 and 1. An ICC of 0 would correspond to independence between the samples, whereas an ICC of 1 would indicate that all samples yield the same result. The ICC is an important measure in determining the efficiency of cluster sampling designs and is a required input for sample size calculations in the design of cluster randomized trials (25, 26).
Mixed-effects linear and logistic regression models were used to evaluate the trend in pathogen loads and prevalences, respectively, as birds progressed through the processing plant. A mixed-effect linear model was also used to compare Salmonella and Campylobacter loads in culture-positive samples of each type. Flock was considered a random effect in all random- and mixed-effects models to account for the correlation between samples collected from the same flock.
All statistical testing assumed a two-sided alternative hypothesis, and P < 0.05 was considered statistically significant. Analyses were performed using commercially available software (Stata version 12.1; StataCorp LP, College Station, TX).
RESULTS
Fifty-five flock houses enrolled in the study were processed between May 2008 and July 2009. The number of birds per flock ranged from 16,700 to 40,000, with a mean of 28,590 birds (standard deviation [SD] = 5,704 birds). The age of birds at processing ranged from 36 to 43 days, with a mean of 38.2 days (SD = 1.6 days).
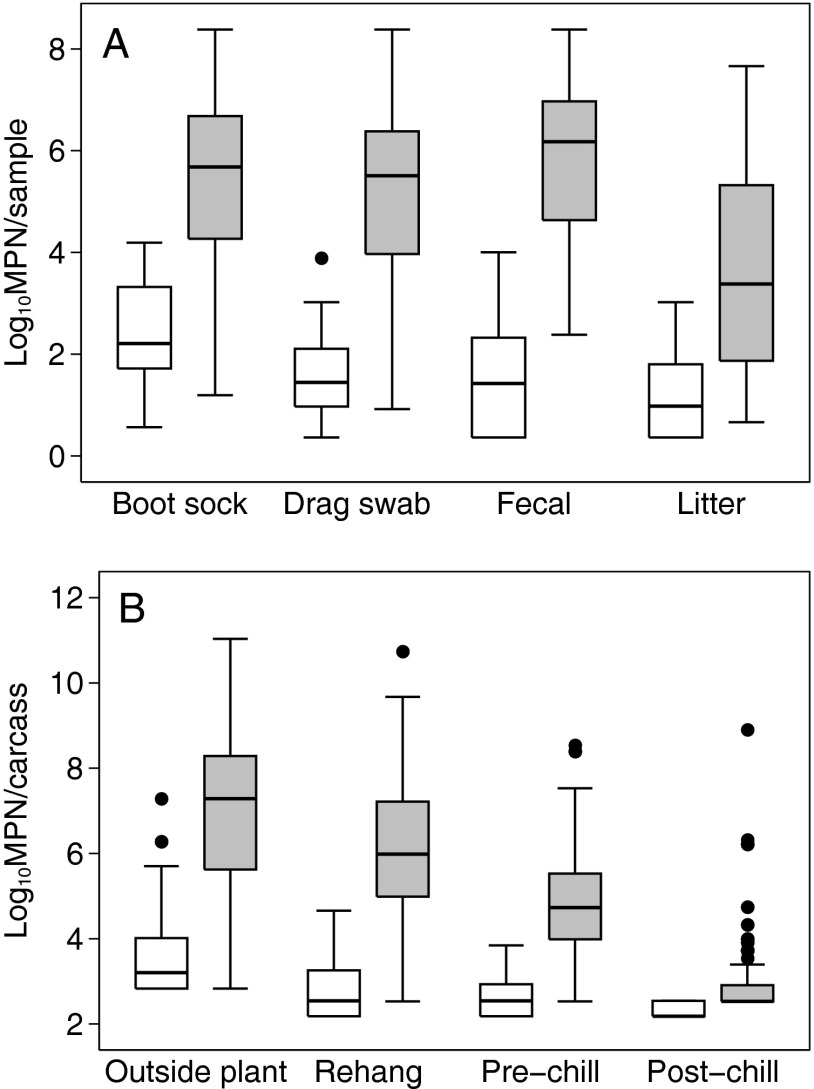
Salmonella was cultured from environmental farm samples of 50 (90.9%) flocks and from processing plant carcass rinses of 52 (94.5%) flocks. Campylobacter was cultured from environmental farm samples of 35 (63.6%) flocks and from processing plant carcass rinses of 48 (87.3%) flocks. Of the 52 flocks with one or more Salmonella-positive carcass rinses, 50 (96.2%) had at least one Salmonella-positive farm sample. Also, of the 48 flocks with one or more Campylobacter-positive carcass rinses, 34 (70.8%) had at least one Campylobacter-positive farm sample. A summary of Salmonella and Campylobacter prevalences and loads by sample type is shown in Table 1. With respect to farm samples, boot socks yielded the highest percentage of positive results for Salmonella and drag swabs yielded the highest percentage of positive results for Campylobacter. With respect to the processing samples, prevalences and loads of both Salmonella and Campylobacter were highest for the outside plant carcass rinses and decreased significantly (P < 0.001) as birds progressed through the processing plant. The distributions of Salmonella and Campylobacter loads in culture-positive samples are illustrated in Fig. 1. In culture-positive samples, Campylobacter loads were significantly higher than Salmonella loads for postchill carcass rinses (P = 0.020) and all other sample types (P < 0.001).
Table 1.
Salmonella and Campylobacter prevalences and loads in samples collected from 55 broiler chicken flocks
| Sample type | No. positive/total no. samples (%) |
Mean (SD) log10MPNa of culture-positive samples |
||
|---|---|---|---|---|
| Salmonella | Campylobacter | Salmonella | Campylobacter | |
| Environmental farm samples | ||||
| Boot sock | 75/110 (68.2) | 57/110 (51.8) | 2.34 (1.14) | 5.35 (1.64) |
| Drag swab | 47/109 (43.1) | 66/109 (60.6) | 1.53 (0.84) | 5.07 (1.89) |
| Fecal sample | 30/220 (13.6) | 113/220 (51.4) | 1.56 (1.13) | 5.89 (1.59) |
| Litter sample | 25/110 (22.7) | 26/110 (23.6) | 1.19 (0.83) | 3.63 (2.05) |
| Processing plant carcass rinses | ||||
| Outside plant | 303/660 (45.9) | 452/658 (68.7) | 3.44 (0.71) | 6.79 (1.93) |
| Rehang | 142/330 (43.0) | 205/330 (62.1) | 2.77 (0.59) | 6.08 (1.64) |
| Prechill | 60/330 (18.2) | 194/330 (58.8) | 2.57 (0.44) | 4.74 (1.29) |
| Postchill | 8/330 (2.4) | 144/330 (43.6) | 2.32 (0.19) | 2.96 (0.90) |
MPNs were reported on a per-sample basis for boot socks and drag swabs, per gram for fecal and litter samples, and per carcass for processing plant rinses.
Fig 1.
Box plots of Salmonella (white) and Campylobacter (gray) MPN distributions in culture-positive environmental farm samples (A) and processing plant carcass rinses (B) collected from 55 broiler chicken flocks. The number of culture-positive samples of each type is reported in Table 1.
The Spearman rank correlations between mean pathogen loads of individual sample types are summarized in Table 2. With respect to the predictive ability of the individual farm samples, only the Salmonella loads of boot socks and litter samples were significantly associated with Salmonella loads of postchill carcass rinses. The Campylobacter loads of all four farm sample types were significantly associated with Campylobacter loads of postchill carcass rinses. Because Salmonella loads in postchill carcass rinses were all near or below the minimum detection limit of the MPN assay, logistic regression was used to determine what combination of farm samples best predicted whether Salmonella would be detected in one or more postchill rinses. In this analysis, boot socks were the only farm sample type that significantly predicted the detection of Salmonella in postchill rinses, with the odds of detection increasing by a factor of 2.2 (95% confidence interval [CI], 1.0, 4.5; P = 0.038) for each 1 log10MPN increase in boot sock Salmonella loads. Linear regression was used to determine what combination of farm samples best predicted Campylobacter loads in postchill rinses. Boot socks (BS) and fecal samples (FS) were both significant predictors of postchill Campylobacter loads in the final multivariable model: y = 0.20 + 0.46(BS log10MPN) − 0.06(BS log10MPN)2 + 0.25(FS log10MPN), where y was the mean postchill log10MPN (R2 = 0.626; P < 0.001).
Table 2.
Spearman correlations for Salmonella and Campylobacter loadsa in different sample types collected from 55 broiler chicken flocks
| Sample type | Environmental farm samples |
Processing plant carcass rinses |
||||||
|---|---|---|---|---|---|---|---|---|
| Boot sock | Drag swab | Fecal sample | Litter sample | Outside plant | Rehang | Prechill | Postchill | |
| Environmental farm samples | ||||||||
| Boot sock | 0.33* | 0.36** | 0.49*** | 0.42** | 0.28* | 0.28* | 0.28* | |
| Drag swab | 0.87*** | 0.45*** | 0.38** | 0.11 | 0.22 | 0.28* | 0.19 | |
| Fecal sample | 0.78*** | 0.82*** | 0.54*** | 0.24 | 0.27* | 0.26 | 0.22 | |
| Litter sample | 0.59*** | 0.56*** | 0.59*** | 0.33* | 0.24 | 0.40** | 0.34* | |
| Processing plant carcass rinses | ||||||||
| Outside plant | 0.86*** | 0.83*** | 0.82*** | 0.58*** | 0.49*** | 0.35** | 0.13 | |
| Rehang | 0.87*** | 0.86*** | 0.85*** | 0.53*** | 0.91*** | 0.53*** | 0.01 | |
| Prechill | 0.85*** | 0.83*** | 0.84*** | 0.55*** | 0.89*** | 0.92*** | 0.05 | |
| Postchill | 0.62*** | 0.69*** | 0.74*** | 0.44*** | 0.75*** | 0.75*** | 0.81*** | |
Mean log10MPN of samples collected from the same flock. Salmonella values are shown in boldface, and Campylobacter values are shown in lightface. Statistical significance: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The overall relationships between farm and plant sample prevalences and loads are illustrated in Fig. 2. There was a direct linear relationship between the percentage of Salmonella-positive farm samples and the percentage of Salmonella-positive carcass rinses (Fig. 2A; P < 0.001, R2 = 0.191). There was also a direct linear relationship between the mean Salmonella load (log10MPN) of farm samples and the mean Salmonella load of carcass rinses (Fig. 2C; P < 0.001, R2 = 0.257). With respect to Campylobacter, there was a quadratic relationship between the percentage of Campylobacter-positive farm samples and the percentage of Campylobacter-positive carcass rinses (Fig. 2B; P < 0.001, R2 = 0.834). There was also a quadratic relationship between the mean Campylobacter load (log10MPN) of farm samples and the mean Campylobacter load of carcass rinses (Fig. 2D; P < 0.001, R2 = 0.908).
Fig 2.
Scatter plots of the combined environmental farm samples (n = 10 per flock) versus the combined processing plant carcass rinses (n = 30 per flock) for 55 broiler chicken flocks. (A) Salmonella prevalences. (B) Campylobacter prevalences. (C) Salmonella loads. (D) Campylobacter loads. Markers represent the number of flocks with each combination of coordinates.
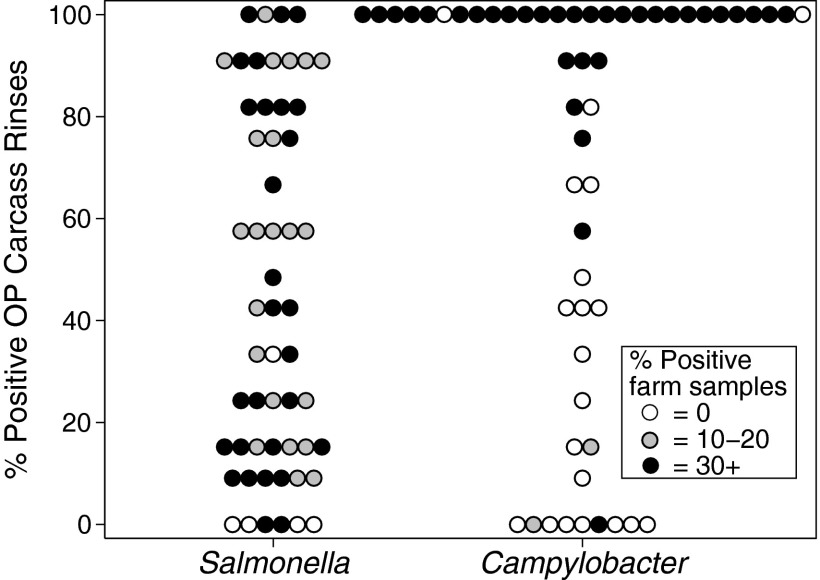
The ICCs for samples of the same type collected from the same flock are summarized for Salmonella and Campylobacter culture results in Table 3. ICCs were uniformly higher for Campylobacter than they were for Salmonella, reflecting greater similarity between samples collected from the same flock with respect to Campylobacter detection and enumeration. The distributions of Salmonella and Campylobacter prevalences in the outside plant carcass rinse samples are illustrated in Fig. 3. In comparison to the relatively uniform distribution of Salmonella prevalences, Campylobacter prevalences in the outside plant carcass rinses were more consistent with a bimodal distribution. There were only 4 flocks in which all 12 outside plant carcass rinses yielded a Salmonella-positive result, compared to 28 flocks in which all 12 outside plant carcass rinses yielded a Campylobacter-positive result.
Table 3.
Intracluster correlations (95% CI) for binary and quantitative Salmonella and Campylobacter culture results in samples collected from 55 broiler chicken flocks
| Sample type (samples/flock) | Intracluster correlation (95% CI) |
|||
|---|---|---|---|---|
|
Salmonella culture |
Campylobacter culture |
|||
| Binarya | Quantitativeb | Binarya | Quantitativeb | |
| Environmental farm samples | ||||
| Boot sock (n = 2) | 0.54 (0.22, 0.83) | 0.35 (0.16, 0.60) | 0.97 (0.91, 0.99) | 0.80 (0.69, 0.88) |
| Drag swab (n = 2) | 0.60 (0.29, 0.85) | 0.50 (0.31, 0.69) | 0.95 (0.89, 0.98) | 0.89 (0.82, 0.93) |
| Fecal sample (n = 4) | 0.12 (0.01, 0.68) | 0.02 (0.00, 0.94) | 0.98 (0.96, 0.99) | 0.86 (0.79, 0.90) |
| Litter sample (n = 2) | 0.36 (0.08, 0.78) | 0.24 (0.07, 0.55) | 0.71 (0.37, 0.91) | 0.58 (0.41, 0.74) |
| Processing plant carcass rinses | ||||
| Outside plant (n = 12) | 0.57 (0.44, 0.69) | 0.49 (0.38, 0.59) | 0.93 (0.85, 0.97) | 0.83 (0.77, 0.88) |
| Rehang (n = 6) | 0.65 (0.49, 0.79) | 0.52 (0.40, 0.63) | 0.97 (0.93, 0.99) | 0.88 (0.82, 0.91) |
| Prechill (n = 6) | 0.36 (0.19, 0.58) | 0.23 (0.14, 0.37) | 0.95 (0.86, 0.98) | 0.82 (0.75, 0.88) |
| Postchill (n = 6) | 0.00 (NEc) | 0.00 (NE) | 0.81 (0.66, 0.90) | 0.54 (0.42, 0.65) |
Presence or absence of the organism.
Enumeration of the organism (i.e., log10MPN).
NE, not estimable because Salmonella was detected in only 8 postchill carcass rinses.
Fig 3.
Distribution of Salmonella and Campylobacter prevalences in carcass rinses performed on birds outside the processing plant (OP; n = 12 per flock) for 55 broiler chicken flocks. The shading of markers indicates prevalences in the combined environmental farm samples (n = 10 per flock) collected from the same flocks within 48 h before processing.
The frequency distribution of Salmonella serotypes is summarized in Table 4. The most commonly identified serotypes were Kentucky and Enteritidis, accounting for a combined 77.9% of the isolates. With respect to Campylobacter species, 1,000 (79.6%) of the 1,257 Campylobacter-positive samples contained only C. jejuni, 153 (12.2%) contained only C. coli, and 104 (8.3%) contained both C. jejuni and C. coli.
Table 4.
Frequency distribution for Salmonella serotypes obtained from 55 broiler chicken flocks
| Serotype | No. of isolates (%)a |
|---|---|
| Kentucky | 448 (61.0) |
| Enteritidis | 124 (16.9) |
| Schwarzengrund | 44 (6.0) |
| Worthington | 35 (4.8) |
| Typhimurium var. 5− | 20 (2.7) |
| Kiambu | 15 (2.0) |
| Mbandaka | 12 (1.6) |
| 4,5,12:i:− | 12 (1.6) |
| Ohio | 8 (1.1) |
| Senftenberg | 5 (0.7) |
| Typhimurium | 4 (0.5) |
| Other | 7 (1.0) |
| Total | 734 (100) |
Two separate serotypes were isolated from 44 of the 690 Salmonella-positive samples.
DISCUSSION
Previous studies have identified significant relationships between farm and processing plant pathogen prevalences in broiler chicken flocks (27–29). Volkova et al. found that the prevalence of Salmonella in bird and environmental samples on the farm was associated with the prevalence of Salmonella in carcass rinses from the same flocks at processing (29). Fluckey et al. found significant associations between the prevalence of Salmonella in cecal samples collected from birds on the farm and the prevalence of Salmonella in carcass rinses at processing (27). Franz et al. reported significant associations between prevalences of both Salmonella and Campylobacter in fecal samples obtained on the farm, cecal swabs obtained at entry to the slaughterhouse, and skin samples at the end of processing (28). None of these previous studies reported the strength of associations between pathogen loads in farm and processing plant samples, although an enumeration of both Salmonella and Campylobacter was performed in the study by Fluckey et al. (27). In the current study, there were significant positive associations between farm and processing plant samples with respect to both Salmonella and Campylobacter prevalences and loads.
Salmonella was detected in farm samples from 96% of the flocks that had Salmonella identified at processing, while Campylobacter was detected in farm samples from 71% of the flocks that had Campylobacter identified at processing. The apparently lower sensitivity of farm sampling for the identification of flocks that had Campylobacter at processing may be a result of either late-term colonization events or cross-contamination of birds during transport to slaughter. Campylobacter colonization of broiler flocks can occur rapidly, achieving a within-flock prevalence of 95% within 4 to 7 days after colonization of the first bird (30). The stress associated with catching and transportation has also previously been shown to increase Campylobacter prevalences and loads (31). In addition, contamination of noncolonized birds exposed to contaminated transportation crates has previously been described (32, 33). Most flocks that had Campylobacter identified at the processing plant but not on the farm had lower prevalences and loads in carcass rinses than flocks that had Campylobacter detected on the farm, which would be consistent with either sampling shortly after a colonization event or limited surface contamination of birds during transport (Fig. 2 and 3).
Salmonella and Campylobacter prevalences and loads both decreased significantly as birds progressed through the processing plant. In comparing carcass rinses of birds at arrival to those of birds exiting the chill tank, the Salmonella prevalence decreased from 45.9% to 2.4%, and the Campylobacter prevalence decreased from 68.7% to 43.6%. Although there was a greater reduction in prevalence for Salmonella, there was a greater reduction in loads for Campylobacter (Table 1). Mean Campylobacter loads in culture-positive rinses decreased by 3.83 log10MPN during processing compared to a reduction of 1.12 log10MPN for Salmonella. The difference in the magnitude of reductions was likely due in part to the difference in initial concentrations; mean Campylobacter loads in the outside plant rinses were 3.35 log10MPN higher than those for Salmonella. Previous studies have also demonstrated significant reductions in prevalences and loads of Salmonella and Campylobacter during processing. Berrang et al. observed reductions of 52% in Salmonella prevalences and 2.23 log10 in Campylobacter loads between rehang and postchill carcass rinses (11, 34). Northcutt et al. observed overall reductions of 1.3 log10 for Campylobacter and 0.5 log10 for Salmonella in carcass rinses before and after chilling (35).
Several different types of environmental farm samples were evaluated in this study. Boot socks, drag swabs, and litter were all flock-level samples, while individual fecal droppings were bird-level samples. Salmonella and Campylobacter prevalences were both relatively high in boot socks and drag swabs and relatively low in composite litter samples. Individual fecal samples yielded a higher prevalence for Campylobacter than Salmonella, reflecting the tendency of Campylobacter to inhabit nearly all birds in a colonized flock (30).
The different farm sample types were all significantly correlated with respect to pathogen loads. Loads in carcass rinses at different stages of processing were also correlated with one another, with the exception that Salmonella loads in the postchill rinses were not significantly associated with Salmonella loads at the previous processing stages. This was likely because Salmonella was detected in only 8/330 (2.4%) postchill rinses, five of which had concentrations that were below the minimum detection limit of the MPN assay. This finding is similar to the results of the study by Volkova et al., which reported a lack of association between Salmonella prevalences in pre- and postchill carcass rinses due to the reductions in Salmonella that were achieved during the chlorinated immersion chilling process (29).
Of the different farm sample types, litter samples had the strongest correlation with Salmonella loads in postchill carcass rinses, and boot socks were the best predictors of whether Salmonella would be detected in one or more postchill samples. Boot socks were not evaluated in the study by Volkova et al., but that study did find that Salmonella prevalence in litter samples was significantly associated with Salmonella prevalence in postchill carcass rinses (29). Both boot socks and fecal samples were significant predictors of postchill Campylobacter loads in the multivariable analysis. In contrast to Salmonella, litter samples had the weakest correlation with Campylobacter loads in postchill carcass rinses. Correlations between all sample types were stronger with respect to Campylobacter loads than they were with respect to Salmonella loads, which is likely due in part to the larger magnitude and wider range of Campylobacter MPNs than of Salmonella MPNs (Fig. 1).
The combined farm samples explained a greater proportion of the variability in processing plant carcass rinses for Campylobacter than they did for Salmonella (Fig. 2). In addition to the narrower range of MPN values for Salmonella cultures, a higher level of sampling variability may help to account for the apparent difference in predictive ability. When flocks were colonized with Campylobacter, there was a tendency for most samples to have a positive culture result (Fig. 3). This was not the case for Salmonella, which had a more uniform prevalence distribution and lower ICCs for all sample types than did Campylobacter (Table 3). The ICC is a measure of the correlation between samples collected from the same flock. An ICC of 0 would indicate that there is no relationship between the samples (i.e., they are independent), whereas an ICC of 1 would indicate that all samples yield the same result. Collecting a larger number of samples per flock may help to improve the precision of estimation for Salmonella and reduce the unexplained variation in Salmonella results but would not be expected to meaningfully improve the precision of estimation for Campylobacter because of the higher level of correlation between Campylobacter cultures performed on the same flock.
Setting the processing order of broiler flocks based on their colonization status (i.e., logistic slaughter) has been suggested as one way to potentially reduce cross-contamination between flocks in the processing plant (36). Using such an approach, colonized flocks would be identified by farm sampling and scheduled for slaughter at the end of the processing shift. A modeling study by Evers indicated that such an approach was likely to yield little benefit for Salmonella (37), however, and a study of 62 Dutch broiler flocks by Nauta et al. found it impractical as a control strategy for Campylobacter (38). Logistic slaughter was not evaluated in the current study; all of the flocks enrolled were processed immediately after a sanitation shift. Nonetheless, the results suggest that logistic slaughter may not have been effective for reducing cross-contamination with Salmonella in the study population. More than 90% of flocks were colonized with Salmonella, and although one could attempt to order flocks based on the relative Salmonella loads in their farm samples, the ability to accurately predict processing loads for an individual flock was limited as evidenced by the relatively low coefficient of determination for this relationship (e.g., Fig. 2C; R2 = 0.26). Logistic slaughter may have had greater potential for reducing cross-contamination with Campylobacter in the study population. Although Campylobacter was detected in the processing samples of 13 (65%) of 20 flocks that did not have any Campylobacter detected on the farm, the Campylobacter loads that were observed in those flocks at processing were typically lower than those in flocks that did have Campylobacter detected on the farm (Fig. 2D). In addition, the coefficient of determination was higher for the relationship between farm and processing plant Campylobacter loads (i.e., R2 = 0.91), indicating that the accuracy of prediction for individual flocks was higher for Campylobacter than it was for Salmonella. Campylobacter seemed to exhibit a threshold effect, however, with the detection of any Campylobacter on the farm typically corresponding to relatively high loads at processing. In practice, additional scheduling constraints and the availability of accurate and timely testing results may limit the feasibility of logistic slaughter implementation in a commercial production setting.
Salmonella enterica serotype Kentucky (S. Kentucky) and serotype Enteritidis (S. Enteritidis) were the most frequently isolated Salmonella serotypes in the current study, and they were also the most frequently isolated serotypes identified during regulatory testing of all U.S. broiler processing plants during 2008 to 2010 (39). All Salmonella serotypes are considered potential pathogens, although S. Kentucky is infrequently associated with human illness, accounting for only 0.2% of all laboratory-confirmed human Salmonella isolates reported to the CDC between 1999 and 2009 (40). In contrast, S. Enteritidis accounted for 16.3% of all laboratory-confirmed human isolates reported to the CDC during the same time period. Human infection with S. Enteritidis has historically been associated with the consumption of undercooked eggs but increasingly has been associated with the consumption of chicken meat in recent years (41, 42). C. jejuni was the most frequently isolated Campylobacter species and is also the most common cause of human campylobacteriosis (43).
The current study has several limitations. Sampling was performed in cooperation with a single broiler production complex in north Georgia. Therefore, the associations that were observed may not be representative of the entire industry, although the management practices of the sampled complex were typical of most U.S. broiler companies. A relatively small number of samples were collected from each flock, considering that the mean flock size was greater than 28,000 birds. Collecting a larger number of samples might have reduced sampling variability, especially with respect to the estimation of Salmonella prevalences and loads. Shipment of samples likely also introduced a source of variability but was necessary to distribute the workload across laboratories.
In conclusion, Salmonella and Campylobacter prevalences and loads on the farm were significantly associated with prevalences and loads of the same pathogens at processing. Consequently, management practices that reduce pathogens on the farm would be expected to reduce contamination at processing. Vaccination of breeder hens, competitive exclusion products, and the use of acidified water during feed withdrawal have all been reported to reduce Salmonella colonization in commercial broiler flocks (14, 44, 45). Unfortunately, aside from the implementation of strict biosecurity protocols to reduce the likelihood of Campylobacter introduction, reliable approaches to reduce Campylobacter colonization are currently unavailable (15, 46, 47). As a postprocessing intervention, freezing has been shown to reduce Campylobacter counts of broiler carcasses by 0.65 to 2.87 log10 (48). Additional research is needed to develop and quantify the effectiveness of on-farm pathogen control methods in commercial production settings.
ACKNOWLEDGMENTS
This project was supported by U.S. Department of Agriculture-National Research Initiative, Food Safety Research and Response Network award no. 2004-1578-20.
We thank Jeannette Muñoz-Aguayo and Cristian Flores for technical assistance.
Footnotes
Published ahead of print 26 April 2013
REFERENCES
- 1. Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17:7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Domingues AR, Pires SM, Halasa T, Hald T. 2012. Source attribution of human campylobacteriosis using a meta-analysis of case-control studies of sporadic infections. Epidemiol. Infect. 140:970–981 [DOI] [PubMed] [Google Scholar]
- 3. Greig JD, Ravel A. 2009. Analysis of foodborne outbreak data reported internationally for source attribution. Int. J. Food Microbiol. 130:77–87 [DOI] [PubMed] [Google Scholar]
- 4. Guo C, Hoekstra RM, Schroeder CM, Pires SM, Ong KL, Hartnett E, Naugle A, Harman J, Bennett P, Cieslak P, Scallan E, Rose B, Holt KG, Kissler B, Mbandi E, Roodsari R, Angulo FJ, Cole D. 2011. Application of Bayesian techniques to model the burden of human salmonellosis attributable to U.S. food commodities at the point of processing: adaptation of a Danish model. Foodborne Pathog. Dis. 8:509–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Batz MB, Hoffmann S, Morris JG., Jr 2012. Ranking the disease burden of 14 pathogens in food sources in the United States using attribution data from outbreak investigations and expert elicitation. J. Food Prot. 75:1278–1291 [DOI] [PubMed] [Google Scholar]
- 6. US Department of Agriculture 1996. Pathogen reduction; hazard analysis and critical control point (HACCP) systems. Fed. Regist. 61:38806–38989 [Google Scholar]
- 7. Centers for Disease Control and Prevention 2011. Vital signs: incidence and trends of infection with pathogens transmitted commonly through food—foodborne diseases active surveillance network, 10 U.S. sites, 1996–2010. MMWR Morb. Mortal. Wkly. Rep. 60:749–755 [PubMed] [Google Scholar]
- 8. US Department of Agriculture 1996. Nationwide broiler chicken microbiological baseline data collection program: July 1994–June 1995. Food Safety and Inspection Service, US Department of Agriculture, Washington, DC: http://www.fsis.usda.gov/OPHS/baseline/broiler1.pdf Accessed 8 March 2013 [Google Scholar]
- 9. US Department of Agriculture 2012. Progress report on Salmonella and Campylobacter testing of raw meat and poultry products, 1998–2011. Food Safety and Inspection Service, US Department of Agriculture, Washington, DC: http://www.fsis.usda.gov/PDF/Progress_Report_Salmonella_Testing_1998–2011.pdf Accessed 8 March 2013 [Google Scholar]
- 10. US Department of Agriculture 2011. New performance standards for Salmonella and Campylobacter in young chicken and turkey slaughter establishments: response to comments and announcement of implementation schedule. Fed. Regist. 76:15282–15290 [Google Scholar]
- 11. Berrang ME, Bailey JS, Altekruse SF, Patel B, Shaw WK, Jr, Meinersmann RJ, Fedorka-Cray PJ. 2007. Prevalence and numbers of Campylobacter on broiler carcasses collected at rehang and postchill in 20 U.S. processing plants. J. Food Prot. 70:1556–1560 [DOI] [PubMed] [Google Scholar]
- 12. Northcutt JK, Smith D, Huezo RI, Ingram KD. 2008. Microbiology of broiler carcasses and chemistry of chiller water as affected by water reuse. Poult. Sci. 87:1458–1463 [DOI] [PubMed] [Google Scholar]
- 13. Arsenault J, Letellier A, Quessy S, Boulianne M. 2007. Prevalence and risk factors for Salmonella and Campylobacter spp. carcass contamination in broiler chickens slaughtered in Quebec, Canada. J. Food Prot. 70:1820–1828 [DOI] [PubMed] [Google Scholar]
- 14. Gast RK. 2007. Serotype-specific and serotype-independent strategies for preharvest control of food-borne Salmonella in poultry. Avian Dis. 51:817–828 [DOI] [PubMed] [Google Scholar]
- 15. Newell DG, Elvers KT, Dopfer D, Hansson I, Jones P, James S, Gittins J, Stern NJ, Davies R, Connerton I, Pearson D, Salvat G, Allen VM. 2011. Biosecurity-based interventions and strategies to reduce Campylobacter spp. on poultry farms. Appl. Environ. Microbiol. 77:8605–8614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. US Department of Agriculture 2010. Compliance guideline for controlling Salmonella and Campylobacter in poultry, 3rd ed Food Safety and Inspection Service, US Department of Agriculture, Washington, DC: http://www.fsis.usda.gov/PDF/Compliance_Guide_Controlling_Salmonella_Campylobacter_Poultry_0510.pdf Accessed 8 March 2013 [Google Scholar]
- 17. Brichta-Harhay DM, Arthur TM, Koohmaraie M. 2008. Enumeration of Salmonella from poultry carcass rinses via direct plating methods. Lett. Appl. Microbiol. 46:186–191 [DOI] [PubMed] [Google Scholar]
- 18. Food and Agriculture Organization/World Health Organization 2009. Salmonella and Campylobacter in chicken meat, meeting report. Food and Agriculture Organization/World Health Organization, Rome, Italy: http://www.fao.org/docrep/012/i1133e/i1133e00.htm Accessed 8 March 2013 [Google Scholar]
- 19. Dickens JA, Cox NA, Bailey JS, Thomson JE. 1985. Automated microbiological sampling of broiler carcasses. Poult. Sci. 64:1116–1120 [Google Scholar]
- 20. Blodgett RJ. October 2010, posting date Appendix 2: most probable number from serial dilutions. Bacteriological analytical manual. Food and Drug Administration, Silver Spring, MD: http://www.fda.gov/Food/ScienceResearch/LaboratoryMethods/BacteriologicalAnalyticalManualBAM/ucm109656.htm Accessed 8 March 2013 [Google Scholar]
- 21. Oyarzabal OA, Macklin KS, Barbaree JM, Miller RS. 2005. Evaluation of agar plates for direct enumeration of Campylobacter spp. from poultry carcass rinses. Appl. Environ. Microbiol. 71:3351–3354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Klena JD, Parker CT, Knibb K, Ibbitt JC, Devane PM, Horn ST, Miller WG, Konkel ME. 2004. Differentiation of Campylobacter coli, Campylobacter jejuni, Campylobacter lari, and Campylobacter upsaliensis by a multiplex PCR developed from the nucleotide sequence of the lipid A gene lpxA. J. Clin. Microbiol. 42:5549–5557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dohoo IR, Martin SW, Stryhn H. 2009. Veterinary epidemiologic research, 2nd ed, p 582–584 AVC Inc, Charlottetown, Prince Edward Island, Canada [Google Scholar]
- 24. Rabe-Hesketh S, Skrondal A. 2008. Multilevel and longitudinal modelling using Stata, 2nd ed, p 238–240 Stata Press, College Station, TX [Google Scholar]
- 25. Donner A, Klar N. 2000. Design and analysis of cluster randomization trials in health research, p 53–56 Arnold, London, United Kingdom [Google Scholar]
- 26. Lohr SL. 1999. Sampling: design and analysis, p 139–141 Duxbury Press, Pacific Grove, CA [Google Scholar]
- 27. Fluckey WM, Sanchez MX, McKee SR, Smith D, Pendleton E, Brashears MM. 2003. Establishment of a microbiological profile for an air-chilling poultry operation in the United States. J. Food Prot. 66:272–279 [DOI] [PubMed] [Google Scholar]
- 28. Franz E, van der Fels-Klerx HJ, Thissen J, van Asselt ED. 2012. Farm and slaughterhouse characteristics affecting the occurrence of Salmonella and Campylobacter in the broiler supply chain. Poult. Sci. 91:2376–2381 [DOI] [PubMed] [Google Scholar]
- 29. Volkova VV, Bailey RH, Rybolt ML, Dazo-Galarneau K, Hubbard SA, Magee D, Byrd JA, Wills RW. 2010. Inter-relationships of Salmonella status of flock and grow-out environment at sequential segments in broiler production and processing. Zoonoses Public Health 57:463–475 [DOI] [PubMed] [Google Scholar]
- 30. van Gerwe T, Miflin JK, Templeton JM, Bouma A, Wagenaar JA, Jacobs-Reitsma WF, Stegeman A, Klinkenberg D. 2009. Quantifying transmission of Campylobacter jejuni in commercial broiler flocks. Appl. Environ. Microbiol. 75:625–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stern NJ, Clavero MR, Bailey JS, Cox NA, Robach MC. 1995. Campylobacter spp. in broilers on the farm and after transport. Poult. Sci. 74:937–941 [DOI] [PubMed] [Google Scholar]
- 32. Berrang ME, Northcutt JK, Fletcher DL, Cox NA. 2003. Role of dump cage fecal contamination in the transfer of Campylobacter to carcasses of previously negative broilers. J. Appl. Poult. Res. 12:190–195 [Google Scholar]
- 33. Hansson I, Ederoth M, Andersson L, Vagsholm I, Olsson Engvall E. 2005. Transmission of Campylobacter spp. to chickens during transport to slaughter. J. Appl. Microbiol. 99:1149–1157 [DOI] [PubMed] [Google Scholar]
- 34. Berrang ME, Bailey JS, Altekruse SF, Shaw WK, Jr, Patel BL, Meinersmann RJ, Fedorka-Cray PJ. 2009. Prevalence, serotype, and antimicrobial resistance of Salmonella on broiler carcasses postpick and postchill in 20 U.S. processing plants. J. Food Prot. 72:1610–1615 [DOI] [PubMed] [Google Scholar]
- 35. Northcutt JK, Berrang ME, Dickens JA, Fletcher DL, Cox NA. 2003. Effect of broiler age, feed withdrawal, and transportation on levels of coliforms, Campylobacter, Escherichia coli and Salmonella on carcasses before and after immersion chilling. Poult. Sci. 82:169–173 [DOI] [PubMed] [Google Scholar]
- 36. Nauta MJ, Havelaar AH. 2008. Risk-based standards for Campylobacter in the broiler meat chain. Food Control 19:372–381 [Google Scholar]
- 37. Evers EG. 2004. Predicted quantitative effect of logistic slaughter on microbial prevalence. Prev. Vet. Med. 65:31–46 [DOI] [PubMed] [Google Scholar]
- 38. Nauta MJ, van der Wal FJ, Putirulan FF, Post J, van de Kassteele J, Bolder NM. 2009. Evaluation of the “testing and scheduling” strategy for control of Campylobacter in broiler meat in The Netherlands. Int. J. Food Microbiol. 134:216–222 [DOI] [PubMed] [Google Scholar]
- 39. US Department of Agriculture 2011. Serotypes profile of Salmonella isolates from meat and poultry products (January 1998 through December 2010). Food Safety and Inspection Service, US Department of Agriculture, Washington, DC: http://www.fsis.usda.gov/PDF/Serotypes_Profile_Salmonella_2010.pdf Accessed 8 March 2013 [Google Scholar]
- 40. Centers for Disease Control and Prevention 2010. Salmonella annual summary tables 2009. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/ncidod/dbmd/phlisdata/salmonella.htm Accessed 8 March 2013 [Google Scholar]
- 41. Kimura AC, Reddy V, Marcus R, Cieslak PR, Mohle-Boetani JC, Kassenborg HD, Segler SD, Hardnett FP, Barrett T, Swerdlow DL. 2004. Chicken consumption is a newly identified risk factor for sporadic Salmonella enterica serotype Enteritidis infections in the United States: a case-control study in FoodNet sites. Clin. Infect. Dis. 38(Suppl 3):S244–S252 [DOI] [PubMed] [Google Scholar]
- 42. Voetsch AC, Poole C, Hedberg CW, Hoekstra RM, Ryder RW, Weber DJ, Angulo FJ. 2009. Analysis of the FoodNet case-control study of sporadic Salmonella serotype Enteritidis infections using persons infected with other Salmonella serotypes as the comparison group. Epidemiol. Infect. 137:408–416 [DOI] [PubMed] [Google Scholar]
- 43. Hermans D, Pasmans F, Messens W, Martel A, Van Immerseel F, Rasschaert G, Heyndrickx M, Van Deun K, Haesebrouck F. 2012. Poultry as a host for the zoonotic pathogen Campylobacter jejuni. Vector Borne Zoonotic Dis. 12:89–98 [DOI] [PubMed] [Google Scholar]
- 44. Berghaus RD, Thayer SG, Maurer JJ, Hofacre CL. 2011. Effect of vaccinating breeder chickens with a killed Salmonella vaccine on Salmonella prevalences and loads in breeder and broiler chicken flocks. J. Food Prot. 74:727–734 [DOI] [PubMed] [Google Scholar]
- 45. Dorea FC, Cole DJ, Hofacre C, Zamperini K, Mathis D, Doyle MP, Lee MD, Maurer JJ. 2010. Effect of Salmonella vaccination of chicken breeders on reducing carcass contamination of broiler chickens in integrated poultry operations. Appl. Environ. Microbiol. 76:7820–7825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hermans D, Van Deun K, Messens W, Martel A, Van Immerseel F, Haesebrouck F, Rasschaert G, Heyndrickx M, Pasmans F. 2011. Campylobacter control in poultry by current intervention measures ineffective: urgent need for intensified fundamental research. Vet. Microbiol. 152:219–228 [DOI] [PubMed] [Google Scholar]
- 47. Lin J. 2009. Novel approaches for Campylobacter control in poultry. Foodborne Pathog. Dis. 6:755–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Georgsson F, Thornorkelsson AE, Geirsdottir M, Reiersen J, Stern NJ. 2006. The influence of freezing and duration of storage on Campylobacter and indicator bacteria in broiler carcasses. Food Microbiol. 23:677–683 [DOI] [PubMed] [Google Scholar]