Abstract
Electrical current can be used to supply reducing power to microbial metabolism. This phenomenon is typically studied in pure cultures with added redox mediators to transfer charge. Here, we investigate the development of a current-fed mixed microbial community fermenting glycerol at the cathode of a bioelectrochemical system in the absence of added mediators and identify correlations between microbial diversity and the respective product outcomes. Within 1 week of inoculation, a Citrobacter population represented 95 to 99% of the community and the metabolite profiles were dominated by 1,3-propanediol and ethanol. Over time, the Citrobacter population decreased in abundance while that of a Pectinatus population and the formation of propionate increased. After 6 weeks, several Clostridium populations and the production of valerate increased, which suggests that chain elongation was being performed. Current supply was stopped after 9 weeks and was associated with a decrease in glycerol degradation and alcohol formation. This decrease was reversed by resuming current supply; however, when hydrogen gas was bubbled through the reactor during open-circuit operation (open-circuit potential) as an alternative source of reducing power, glycerol degradation and metabolite production were unaffected. Cyclic voltammetry revealed that the community appeared to catalyze the hydrogen evolution reaction, leading to a +400-mV shift in its onset potential. Our results clearly demonstrate that current supply can alter fermentation profiles; however, further work is needed to determine the mechanisms behind this effect. In addition, operational conditions must be refined to gain greater control over community composition and metabolic outcomes.
INTRODUCTION
Bioelectrochemical systems (BESs) exploit microorganisms to catalyze redox reactions in an electrochemical cell. To date, the most widely studied type of BES is the microbial fuel cell (MFC), which produces electrical power by exploiting the ability of certain microorganisms to link the oxidation of organic substrates to the reduction of an anodic electrode, i.e., to perform (oxidative) microbial bioelectrocatalysis (1). A more recent application of BESs is to stimulate bioelectrochemical catalysis of reduction reactions by providing current to microorganisms associated with cathode compartments (2, 3). Examples of this process include (i) bioelectrocatalytic generation of hydrogen and acetate with pure cultures of Desulfovibrio (4) and Sporomusa ovata (5), respectively, and (ii) bioelectrocatalytic generation of hydrogen (6, 7, 8), methane (8, 9, 10, 11, 12), and acetate (8, 12) with mixed microbial communities.
Decades of MFC research have revealed a core set of anode-associated microorganisms and led to an advanced level of understanding of the metabolic niches occupied by different populations (i.e., pure fermentation or various modes of extracellular electron transfer that facilitate anaerobic respiration [1]). In contrast, little is known about the organisms that proliferate in cathodic environments. Current supply is believed to favor reductive processes by enhancing the ability of cells to regenerate NADH (2, 13, 14). Possible mechanisms include (i) the shuttling of electrons to microorganisms via hydrogen produced at the cathode, (ii) electron shuttling to organisms via another soluble mediator, and/or (iii) direct electron transfer (DET) from the cathode to microorganisms via membrane-bound redox active components (2, 8, 11). It has yet to be determined whether organisms are capable of accepting electrons directly from cathodic electrodes; however, this would eradicate the need to add mediators to exploit current supply.
The temporal dynamics of cathode-associated microbial communities and product outcomes is another poorly understood aspect of this emerging technology. Stable production of desired compounds by a mixed microbial community would avoid the costs associated with maintaining pure cultures; however, it is not known whether cathodic electrode-associated microbial communities change in composition and diversity over time and whether this leads to differences in product outcomes. In this study, we used high-throughput 16S rRNA gene sequencing to characterize the diversity and composition of a current-fed mixed microbial community fermenting glycerol over time. After 9 weeks of continuous galvanostatic operation during which correlations between microbial diversity and the respective product outcomes were identified, we characterized the production of metabolites after current supply was stopped and later restarted, as well as during a period in which hydrogen gas was supplied to the microbial community at open-circuit potential (OCP). This choice of control was preferred over a separate open-circuit experiment, as in the latter case, the physicochemical conditions would differ because of changes in localized pH, ion balance, and gas partial pressures, including hydrogen and bicarbonate. In combination with turnover cyclic voltammetry performed on sterile and biofilm-coated electrodes, the metabolite data were used to gain insight into the electrode-microbe interactions and the influence of current on glycerol fermentation (15). We tested the hypothesis that changes in community composition could be related to changes in product outcomes.
MATERIALS AND METHODS
Reactor design.
A lamellar-type reactor consisting of two end plates and three paired anode-cathode compartments was constructed from acrylic sheeting (16) (Fig. 1). Each cathode frame (inner volume 0.56 L) housed three IGS-743 graphite electrodes (4 by 30 by 150 mm; Morgan Industrial Carbon, Revesby, Australia) that were mounted in acrylic blocks with rubber O rings. Because of the insertion of the electrodes into the acrylic blocks and the 20-mm wall thickness of the acrylic frames, only 120 mm of the electrode's length was exposed to the medium. As there were nine electrodes, the total electrode surface area was 745 cm2. The cathodic electrodes could be removed and replaced independently without touching the sides of the reactor by replacing the screws with rails along which the electrodes could be slid. This allowed the nine cathodic electrodes inserted at the start of the experiment to be sampled at independent time points. In a previous study (16), we demonstrated that when using this reactor design, no spatial variation in microbial diversity could be observed between electrodes within a time point. This finding indicates that the microbial diversity on each electrode can be considered representative of that associated with any other electrode within a time point. Three graphite plates, each measuring 290 by 150 by 5 mm (Morgan Industrial Carbon), were used as (sacrificial) anodes. The anode and cathode compartments were separated by a cation-exchange membrane (Ultrex CMI7000; Membranes International Inc.) sandwiched between two 2-mm-thick rubber frames with the same dimensions as the electrode frames. Anode-cathode pairs were separated by a sheet of impermeable rubber positioned between the anode and the next cathode frame. When all of the layers were correctly positioned, the end plates were fastened in place and the reactor was connected to the recirculation, buffer vessels and feed circuits (see Fig. S1 in the supplemental material). The total volumes of the anode and cathode circuits (i.e., the frames, recirculation bottles, and tubing) were approximately 2.09 and 2.00 liters, respectively.
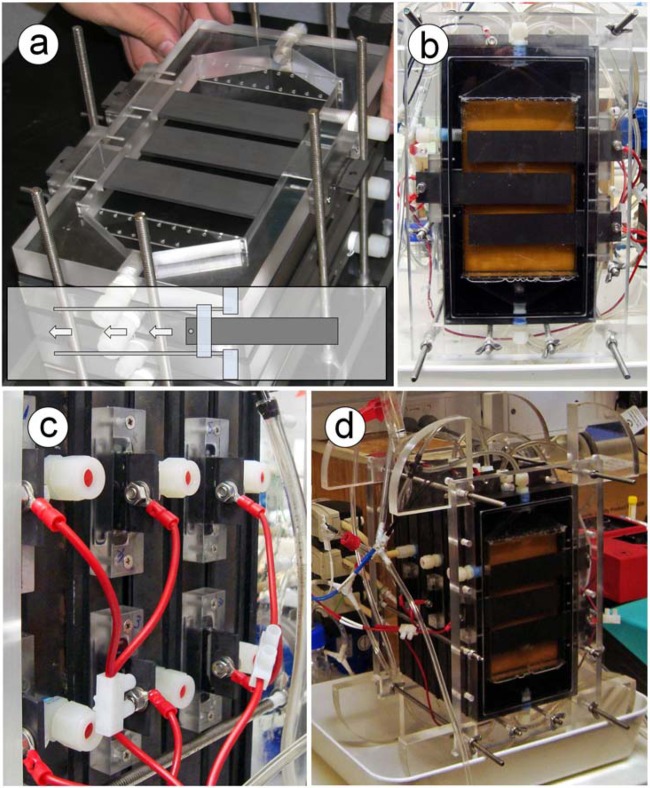
Fig 1.
Photographs of the reactor setup. Panels: a, a cathode frame fitted with three exchangeable cathodic electrodes; b, a cathode with a view of the electrodes; c, a closeup of the blocks used to attach the cathodic electrodes to the cathode frame; d, an overview of the assembled reactor.
The electrodes were connected in parallel to a modular five-channel potentiostat/galvanostat (Bio-Logic VSP). An Ag-AgCl reference electrode (RE-5b; Bioanalytical Systems Inc.; 0.197 V versus a standard hydrogen electrode) filled with 3 M KCl was fitted to a cathode compartment via a glass Luggin capillary. A pH electrode was fitted to the cathodic recirculation stream and connected to a pH logger and control unit (Endress+Hauser Liquisys M CPM223). The control unit maintained the cathodic pH at 5.5 by dosing 0.5 M NaOH.
Reactor operation and sample collection.
The cathodes were inoculated with a microbial community associated with a sewage sludge fermenter and supplied a total current of −240 mA, which equates to −0.32 mA cm−2 projected electrode surface in the galvanostat mode for a period of 9 weeks. The potential at each electrode was manually verified with a Fluke 179 multimeter. The catholyte contained 2 g liter−1 Na2HPO4, 1 g liter−1 KH2PO4, 1 g liter−1 NH4Cl, 0.0147 g liter−1 CaCl2 · 2H2O, 0.1 g liter−1 MgSO4 · 7H2O, and 8.25 g liter−1 glycerol and was autoclaved at 121°C for 60 min. After cooling, it was supplemented with 1 ml liter−1 of a mixed trace element solution (pH 7) and 1 ml liter−1 of a vitamin solution (17). The anolyte was 0.1 M H2SO4. The feed rate for both electrolytes was 3 liters day−1 (continuous-mode feeding), and the recirculation rate was 80 ml min−1; therefore, the hydraulic residence times were 16.7 and 16.0 h for the anolyte and catholyte, respectively.
During the initial 9-week period, cathode potential was monitored with the potentiostat/galvanostat and cathodic electrode- and electrolyte-associated microbial communities were sampled on a weekly basis. During sampling, all pumps and data loggers were briefly inactivated and a cathodic electrode was removed and replaced with a fresh sterile carbon electrode. Cathodic electrode-associated microbial communities were scraped from the surface of electrodes and placed in bead-beating tubes with sterile glass microscope slides. Microorganisms associated with 100 ml of cathodic electrolyte were recovered by centrifugation at 4,000 rpm for 25 min. Cell pellets were then resuspended in 0.5 ml of molecular biology grade water and transferred to bead-beating tubes for nucleic acid extraction. All samples were immediately frozen by immersion in liquid nitrogen and then stored at −80°C.
Additional electrolyte samples were filtered through 0.22-μm sterile filters and then used for chemical analyses. Glycerol and 1,3-propanediol concentrations were determined by high-performance liquid chromatography. Concentrations of ethanol, propanol, butanol, acetate, propionate, isobutyrate, butyrate, isovalerate, valerate, and hexanoate were determined by injecting a 0.9-ml sample mixed with 0.1 ml of 10% formic acid into a gas chromatography instrument fitted with a polar capillary column (DB-FFAP) at 140°C and a flame ionization detector at 250°C.
Characterization of microbe-electrode interactions.
To obtain metabolite profiles that were representative of the final week of galvanostatic operation (i.e., current supplied), three additional electrolyte samples were taken on subsequent days during week 8 for chemical analyses. As a comparison, after the sampling of the ninth electrode, the supply of current was stopped and electrolyte samples were then taken on 3 subsequent days. These samples were used as representative metabolite profiles for communities that were not supplied current (i.e., open-circuit controls). To investigate the effect of providing hydrogen gas (instead of electric current) on glycerol fermentation, the electrodes remained at OCP while a stainless steel mesh was introduced into the cathode compartment and used to generate hydrogen gas by electrolysis with an identical total current and thus charge flow as for electric current supply (calculated by using Faraday's law). Given that the mesh was freshly inserted and remained in the reactor for a short time only, biofilm formation would have been negligible. During the H2 fed period at OCP, electrolyte samples were taken on 3 consecutive days. To determine whether any observed effect of current on glycerol fermentation could be repeated, the supply of current was then resumed and samples were taken on 3 subsequent days. Data within these time periods were statistically compared to assess whether the supply of current or hydrogen gas influenced glycerol fermentation. Reactors held at OCP with and without the supply of hydrogen gas would not have represented valid controls because pH increases and changes in ionic content related to current supply could not be mimicked long term without considerable dosing of chemicals, which would lead to differences in salinity and alkalinity relative to the current-fed reactor.
Finally, turnover cyclic voltammetry (CV) was performed on independent triplicates of cathodes (15). CV was performed until stabilization, for at least three cycles, from −1.5 V to −0.2 V (versus an Ag-AgCl reference electrode) at a 1-mV s−1 scan rate. For comparison, identical CVs were recorded for sterile cathodes presoaked in catholyte for 12 h to mimic the chemical environment within the cathode compartment.
Characterization of cathodic microbial communities.
DNA was extracted with a MO-BIO PowerBiofilm DNA isolation kit according to the manufacturer's instructions. Extracted DNA concentrations were determined with a Qubit fluorometer with Quant-iT dsDNA BR assay kits (Invitrogen) and then normalized to 10 ng DNA μl−1. Eubacterial and archaeal 16S rRNA genes were amplified by PCR in 20-μl volumes containing 15 ng DNA, molecular biology grade water, 1× PCR buffer minus Mg (Invitrogen), 50 nM each deoxynucleoside triphosphate (Invitrogen), 1.5 mM MgCl2 (Invitrogen), 0.3 mg bovine serum albumin (New England BioLabs), 0.02 U Taq DNA polymerase (Invitrogen), 8 μM (each) primers 926F (5′-AAACTYAAAKGAATTGACGG-3′) and 1392R (18) (5′-ACGGGCGGTGTGTRC-3′) modified on the 5′ end to contain the 454 FLX Titanium Lib L adapters B and A, respectively. The reverse primers also contained a five- or six-base barcode sequence positioned between the primer sequence and the adapter. A unique barcode was used for each sample. The thermocycling conditions were as follows: 95°C for 10 min; 30 cycles of 95°C for 30 s, 55°C for 45 s, and 72°C for 90 s; and then 72°C for 10 min. Amplifications were performed with a Veriti 96-well thermocycler (Applied Biosystems). Amplicons were purified with a QIAquick PCR purification kit (Qiagen), quantified with a Qubit fluorometer with a Quant-iT dsDNA BR assay kit, normalized to 25 ng μl−1, and then pooled for 454 pyrosequencing. Sequencing was performed at the Australian Centre for Ecogenomics, The University of Queensland.
Data analyses.
Sequences were quality filtered and dereplicated with the QIIME script split_libraries.py with the homopolymer filter deactivated (19) and then checked for chimeras against the GreenGenes database with UCHIME ver. 3.0.617 (20) as previously described (21–24). Homopolymer errors were corrected with Acacia (25). Sequences were then subjected to the following procedures with QIIME scripts at the default settings. (i) Sequences were clustered at 97% similarity, (ii) cluster representatives were selected, (iii) GreenGenes taxonomy (26) was assigned to the cluster representatives by BLAST, and (iv) tables with the abundance of different operational taxonomic units (OTUs) and their taxonomic assignments in each sample were generated. The number of reads was then normalized to 1,050 per sample.
The mean number of OTUs (observed richness) and Simpson diversity index values (27) corresponding to 1,050 sequences per sample were calculated with QIIME. Generalized linear modeling (GLM) was used to assess whether variation in observed richness and Simpson diversity index values could be explained by time or location within the reactor (i.e., electrode- or electrolyte-associated communities). Rarefaction curves were generated with QIIME (see Fig. S2 in the supplemental material). Differences in the composition of electrode- and electrolyte-associated microbial communities were assessed by redundancy analysis (RDA) with Monte Carlo permutation tests (999 permutations). Metabolite profiles were expressed as percent chemical oxygen demand (COD) and were related to the community profiles by RDA. GLM and RDA were used to compare metabolite profiles associated with communities that were supplied current with those associated with communities held at (i) OCP and (ii) OCP with a supply of hydrogen gas. All analyses were implemented with R version 2.12.0.
Nucleotide sequence accession number.
Data from this study have been deposited in the Short Read Archive (SRA) under accession no. SRP021172.
RESULTS AND DISCUSSION
Glycerol degradation, product outcomes, and process insights.
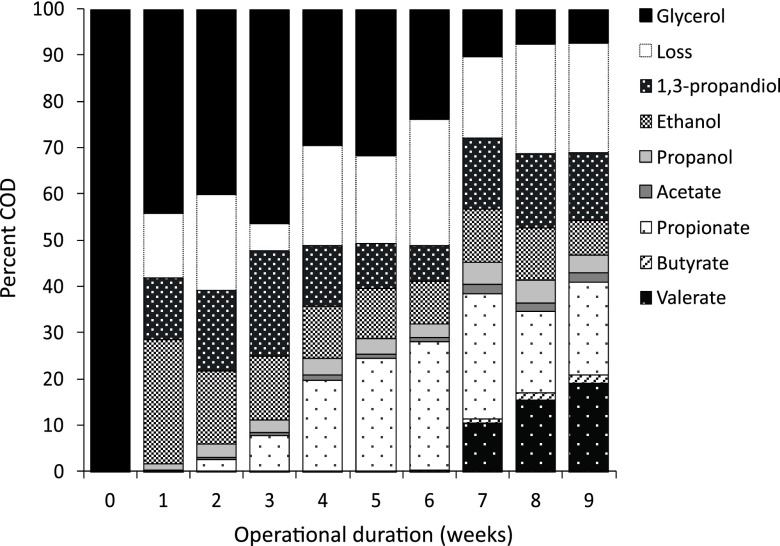
During the initial 9-week period in which current was supplied, glycerol degradation increased (P < 0.001, GLM) and the composition of metabolites changed (P < 0.001, RDA; Fig. 2). One week after inoculation, approximately 45% of the glycerol was converted to a mixture of 1,3-propanediol, ethanol, propanol, and trace quantities of volatile fatty acids (Fig. 2). From the second week of operation, glycerol was also converted to propionate. From week 7, 1 to 2% and 10 to 20% of the glycerol was converted to butyrate and valerate, respectively, and by week 8, the total glycerol degradation exceeded 90% (Fig. 2).
Fig 2.
Mean glycerol degradation and production of metabolites over time. The loss component is the discrepancy between the theoretical CODs in and out. The loss component represents COD associated with cells that were washed out of the system, compounds that were not characterized, and gaseous losses. Negligible concentrations of butanol, isobutyrate, isovalerate, and hexanoate are not shown.
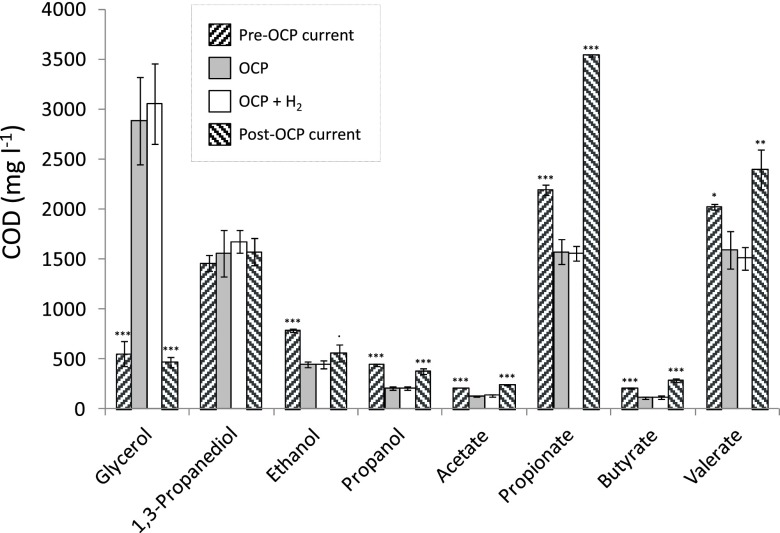
After 9 weeks of continuous galvanostatic operation, the current supply was stopped (i.e., set to OCP) and subsequent changes in metabolite profiles were characterized. Metabolite profiles associated with samples collected on 3 consecutive days prior to the cessation of current supply differed significantly from those associated with cathodes set to OCP (P < 0.001, RDA; Fig. 3). In comparison with cathodes at OCP, current supply was associated with enhanced glycerol degradation and the production of alcohols (ethanol and propanol) and C2-C5 carboxylates (acetate, propionate, butyrate, and valerate; Fig. 3). Subsequent supply of hydrogen gas to the cathodes at OCP did not lead to a further change in glycerol degradation or metabolite formation (P > 0.05, GLM, RDA); however, when current supply was resumed, glycerol and metabolite concentrations returned to values that closely resembled those measured during the initial current-fed period (Fig. 3). These results indicate that current supply significantly influenced fermentation product outcomes with an increase in more reduced compounds such as ethanol, propanol, and valerate (Fig. 3).
Fig 3.
Residual glycerol and concentrations of metabolites produced by microbial communities associated with cathodes that were (i) supplied −0.32 mA cm−2 (pre-OCP current), (ii) switched to OCP, (iii) maintained at OCP and then supplied hydrogen (OCP + H2), and finally (iv) resupplied −0.32 mA cm−2 (post-OCP current). Negligible concentrations of butanol, isobutyrate, isovalerate, and hexanoate are not shown. Significant differences (GLM) from OCP values are indicated for each compound as follows: P < 0.10, •; P < 0.05, *; P < 0.01, **; P < 0.001, ***.
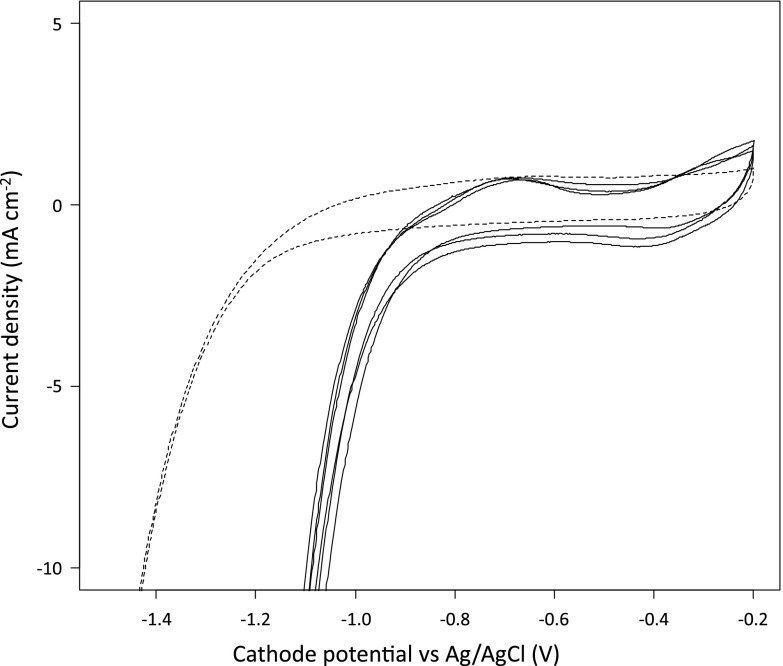
The mechanism(s) that mediates the effects of current supply on metabolic outcomes is unclear. One hypothesis is that electrons are transferred directly to cathode-associated microorganisms via membrane-bound redox-active complexes (8, 12); however, in our study, turnover CV of biofilm-coated and control electrodes (fresh electrodes with no biofilms) did not reveal any evidence of the presence of redox couples (Fig. 4). Voltammograms similar to ours have been attributed to DET (8). While we do not rule out this possibility, we cannot draw the same conclusion in the present study. All electrodes exhibited an electrocatalytic reduction curve for negative potentials greater than −1 V versus Ag-AgCl, which can be attributed to the hydrogen evolution reaction (HER; Fig. 4). Biofilm-coated electrodes were associated with a shift in the onset potential of the HER of approximately +0.4 V (Fig. 4). The onset potential for the HER associated with biofilm-coated electrodes was still more negative than the biological standard potential of the HER at pH 7 (E0′ = −0.612 V versus Ag-AgCl). Cathodic reduction locally leads to a pH increase that could have rendered the potential of the HER more negative, at about −0.059 V/pH unit above 7. We assume that this shift in the onset potential of the HER is a clear indication that microorganisms facilitated the HER (6); however, we cannot rule out the possibility that other factors (i.e., a change in the chemistry and/or real surface area of the electrodes) played a role. The latter was the very reason for not developing a separate control experiment in which a community would develop long term under different conditions. The potential of cathodic electrodes was similar throughout the galvanostatic operation (see Fig. S3 in the supplemental material), and the potentials of freshly inserted electrodes with no biofilms (−1,496 ± 20.7 mV versus Ag-AgCl; mean ± standard deviation) were similar to those with mature biofilms (−1,505 ± 2.3 mV). Given that a nonlimiting current density of −0.32 mA cm−2 was supplied, however, catalytic differences that could have been attributed to microorganisms would be difficult to detect because of other limitations such as diffusion.
Fig 4.
Cyclic voltammograms for a sterile electrode (dashed line) and cathodic electrodes with well-developed biofilms (solid lines) in the presence of glycerol (turnover conditions) with a scan rate of 1 mV s−1.
An alternative explanation for the observation that current induced changes in the metabolite profiles is that current supply led to very high partial pressures of hydrogen in the immediate vicinity of cathode surfaces compared with those achieved by simply pumping hydrogen through the electrolyte. Indeed, electrolysis leads to the formation of hydrogen in solution from water, and this hydrogen needs to come out of solution rather than dissolve. Low potentials such as those observed here for the cathode (see Fig. S3 in the supplemental material) would theoretically lead to extremely high hydrogen partial pressures. Thus, the observed changes in fermentation profiles upon current supply may have been induced by high partial pressures of hydrogen rather than DET.
Microbial diversity and links to the production of specific metabolites.
Relative to the starting inoculum, the observed richness (number of species) and equitability (Simpson diversity index) of cathode-associated microbial communities 1 week after inoculation decreased by 75 to 86% and 88 to 98%, respectively (Fig. 5). Within the cathode, no differences in richness, equitability, and composition were observed between electrode- and electrolyte-associated communities (P > 0.05, GLM, RDA). The inoculum sample was dominated by populations related to the Bacteroidales, Firmicutes, and Alcaligenaceae (Fig. 5). After 1 week of operation, populations that were abundant in the inoculum were not detected in the cathode (Fig. 5). Within less than 2 weeks, the cathode was dominated by an OTU that was most closely related to Citrobacter freundii and represented more than 95% of the electrode- and electrolyte-associated communities but was not detected among the total of 9,814 sequences obtained from the inoculum sample (Fig. 5). Over time, the richness and equitability of cathode-associated microbial communities increased, indicating that conditions became favorable for a broader range of populations (observed richness, t = 5.62, P < 0.001; equitability, t = 10.52, P < 0.001 [GLM]) (Fig. 5). The relative abundance of the Citrobacter population decreased with time until it represented approximately 50% of the community from weeks 7 to 9 (Fig. 5). From 2 weeks postinoculation, a Pectinatus-related population rapidly increased in relative abundance, and from 6 weeks postinoculation, several Firmicutes populations increased in relative abundance, albeit to a lesser extent than the Citrobacter and Pectinatus representatives, which remained the key cathode-associated populations (Fig. 5). These changes in composition over time were significant (P < 0.001, RDA).
Fig 5.
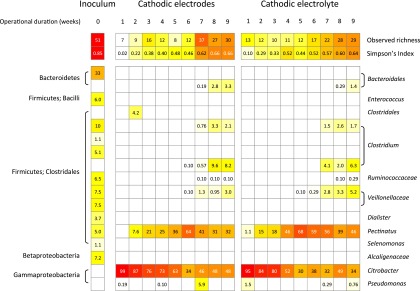
Heat map summarizing the observed richness and equitability (Simpson diversity index) of inoculum- and cathode-associated microbial communities over time, as well as the percent relative abundances of OTUs that were present at more than 1% in any sample (all normalized to 1,050 individuals). Richness and equitability values are rarefied means based on 50 resamplings of 1,050 individuals.
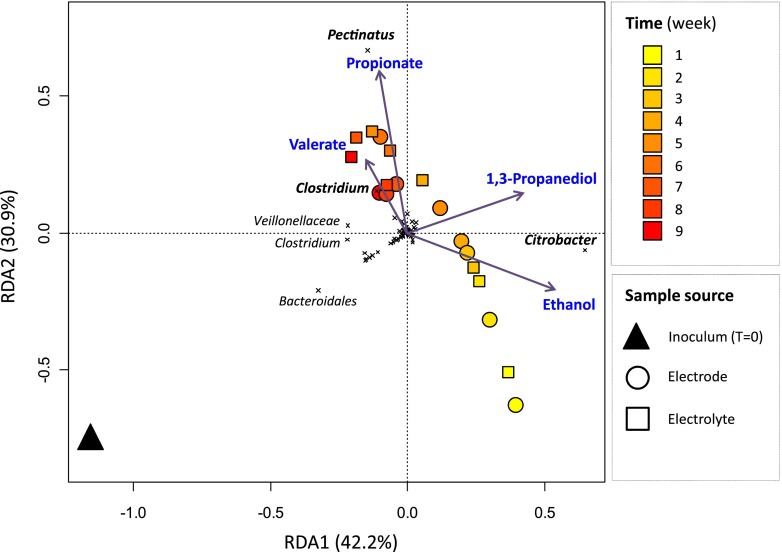
An RDA model was constructed that contained only metabolites that were significantly correlated with variation in the composition of microbial communities (Fig. 6). This model indicated that for current-fed glycerol-fermenting communities, (i) the Citrobacter population was positively correlated with the production of 1,3-propanediol and ethanol, (ii) the Pectinatus population was positively correlated with the production of propionate, and (iii) one of the Clostridium populations was positively correlated with the production of valerate (Fig. 6). Citrobacter species are known to produce 1,3-propanediol to regenerate NAD+ during glycerol fermentation (28, 29). In comparison with glycerol, 1,3-propanediol is a valuable product that can be used for the production of polymers, cosmetics, lubricants, and drugs (30). Given the dominance of Citrobacter in the reactor 1 week after inoculation, it is likely that it is also responsible for the production of propanol. Propanol is a valuable fuel and represented approximately 5% of the glycerol degraded in the latter third of the current-fed period (Fig. 2). Propionic acid is known to be produced by most members of the genus Pectinatus and is a valuable food preservative and intermediate in the production of polymers. Clostridium species are often associated with chain elongation of fatty acids (31). The association between Clostridium populations and valerate production in this study is consistent with previous studies and may relate to the reverse beta oxidation pathway used by organisms such as Clostridium kluyveri (32), although typically this is investigated for even-numbered carboxylates.
Fig 6.
RDA containing metabolites that were significantly correlated with variation in the composition of microbial communities. Metabolites that were not significantly correlated with variation in microbial community composition are not shown. The length of the arrows is proportional to the variance in microbial community composition that can be attributed to the constraints. The angle of an arrow reflects its correlation with the axes. The colors of the symbols are proportional to time, and the types of samples are indicated by different shapes as follows: triangle, inoculum; circle, cathodic electrode; square, cathodic electrolyte. Crosses represent bacterial OTUs. For clarity, taxonomic affiliations are shown for the most discriminating OTUs only.
To date, the majority of the studies involving microbial electrosynthesis have used pure cultures. While this approach is well suited for fundamental research, industrial application of BESs can benefit from mixed microbial communities, as avoiding contamination represents an additional cost. A key issue with mixed communities is a lack of specificity in terms of the end products obtained. In our study, we did not achieve product specificity either. We obtained not only 1,3-propanediol but also carboxylates, including chain-elongated products such as valerate, and alcohols such as propanol, which likely serve as precursors for chain elongation. From our study, it appears that hydrogen gas provided at 1 atm (and in identical stoichiometry) does not affect the metabolism of microbial communities in the same way as electrical current. We did not observe evidence of DET, but the biofilm appeared to catalyze the HER. It is likely, therefore, that high partial pressures of H2 are formed in the immediate vicinity of the cathode surface during current supply, and this may explain the observed change in fermentation outcomes. Overall, our results show that current can impact mixed-population fermentations and lead to attractive products. The challenge for future studies is to identify ways to reduce temporal variation in community composition to obtain more stable product outcomes.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Commonwealth Scientific and Industrial Research Organization (CSIRO) Flagship cluster Biotechnological Solutions to Australia's Transport Energy and Greenhouse Gas Challenges. K.R. acknowledges support by the Australian Research Council (ARC DP0879245) and the Multidisciplinary Research Partnership Ghent Bio-Economy. F.H. gratefully acknowledges the support by the DAAD (postdoctoral scholarship), the receipt of an Australia Award Endeavor Research Fellowship, and the Fonds der Chemischen Industrie.
Footnotes
Published ahead of print 19 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00569-13.
REFERENCES
- 1. Lovley DR. 2008. Extracellular electron transfer: wires, capacitors, iron lungs, and more. Geobiology 6:225–231 [DOI] [PubMed] [Google Scholar]
- 2. Rabaey K, Rozendal RA. 2010. Microbial electrosynthesis—revisiting the electrical route for microbial production. Nat. Rev. Microbiol. 8:706–716 [DOI] [PubMed] [Google Scholar]
- 3. Rosenbaum M, Aulenta F, Villano M, Angenent LT. 2011. Cathodes as electron donors for microbial metabolism: which extracellular electron transfer mechanisms are involved? Bioresour. Technol. 102:324–333 [DOI] [PubMed] [Google Scholar]
- 4. Aulenta F, Catapano L, Snip L, Villano M, Majone M. 2012. Linking bacterial metabolism to graphite cathodes: electrochemical insights into the H2-producing capability of Desulfovibrio sp. ChemSusChem 5:1080–1085 [DOI] [PubMed] [Google Scholar]
- 5. Nevin KP, Woodard TL, Franks AE, Summers ZM, Lovley DR. 2010. Microbial electrosynthesis: feeding microbes electricity to convert carbon dioxide and water to multicarbon extracellular organic compounds. mBio 1(2):e00103-10. 10.1128/mBio.00103-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rozendal RA, Hamelers HMV, Euverink GJW, Metz SJ, Buisman CJN. 2006. Principle and perspectives of hydrogen production through biocatalyzed electrolysis. Int. J. Hydrogen Energy 31:1632–1640 [Google Scholar]
- 7. Liu H, Grot S, Logan BE. 2005. Electrochemically assisted microbial production of hydrogen from acetate. Environ. Sci. Technol. 39:4317–4320 [DOI] [PubMed] [Google Scholar]
- 8. Marshall CW, Ross DE, Fichot EB, Norman RS, May HD. 2012. Electrosynthesis of commodity chemicals by an autotrophic microbial community. Appl. Environ. Microbiol. 78:8412–8420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheng S, Xing D, Call DF, Logan BE. 2009. Direct biological conversion of electrical current into methane by electromethanogenesis. Environ. Sci. Technol. 43:3953–3958 [DOI] [PubMed] [Google Scholar]
- 10. Villano M, Aulenta F, Ciucci C, Ferri T, Giuliano A, Majone M. 2010. Bioelectrochemical reduction of CO2 to CH4 via direct and indirect extracellular electron transfer by a hydrogenophilic methanogenic culture. Bioresour. Technol. 101:3085–3090 [DOI] [PubMed] [Google Scholar]
- 11. Pisciotta JM, Zaybak Z, Call DF, Nam JY, Logan BE. 2012. Enrichment of microbial electrolysis cell (MEC) biocathodes from sediment microbial fuel cell (sMFC) bioanodes. Appl. Environ. Microbiol. 78:5212–5219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nevin KP, Hensley SA, Franks AE, Summers ZM, Ou J, Woodard TL, Snoeyenbos-West OL, Lovley DR. 2011. Electrosynthesis of organic compounds from carbon dioxide is catalyzed by a diversity of acetogenic microorganisms. Appl. Environ. Microbiol. 77:2882–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zheng ZM, Xu YZ, Liu HJ, Guo NN, Cai ZZ, Liu DH. 2008. Physiologic mechanisms of sequential products synthesis in 1,3-propanediol fed-batch fermentation by Klebsiella pneumoniae. Biotechnol. Bioeng. 100:923–932 [DOI] [PubMed] [Google Scholar]
- 14. Thrash JC, Coates JD. 2008. Review: direct and indirect electrical stimulation of microbial metabolism. Environ. Sci. Technol. 42:3921–3931 [DOI] [PubMed] [Google Scholar]
- 15. Harnisch F, Freguia S. 2012. A basic tutorial on cyclic voltammetry for the investigation of electroactive microbial biofilms. Chem. Asian J. 7:466–475 [DOI] [PubMed] [Google Scholar]
- 16. Dennis PG, Guo K, Imelfort M, Jensen P, Tyson GW, Rabaey KR. 2013. Spatial uniformity of microbial diversity in a continuous bioelectrochemical system. Bioresour. Technol. 129:599–605 [DOI] [PubMed] [Google Scholar]
- 17. Rabaey K, Ossieur W, Verhaege M, Verstraete W. 2005. Continuous microbial fuel cells convert carbohydrates to electricity. Water Sci. Technol. 52:515–523 [PubMed] [Google Scholar]
- 18. Engelbrektson A, Kunin V, Wrighton KC, Zvenigorodsky N, Chen F, Ochman H, Hugenholtz P. 2010. Experimental factors affecting PCR-based estimates of microbial species richness and evenness. ISME J. 4:642–647 [DOI] [PubMed] [Google Scholar]
- 19. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Tumbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bourne DG, Dennis PG, Uthicke S, Soo R, Tyson G, Webster N. 10 January 2013. Coral reef invertebrate microbiomes correlate with the presence of photosymbionts. ISME J. (Epub ahead of print.) 10.1038/ismej.2012.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Carvalhais LC, Dennis PG, Badri DV, Tyson GW, Vivanco JM, Schenk PM. 2013. Activation of the jasmonic acid plant defence pathway alters the composition of rhizosphere bacterial communities. PLoS One 8(2):e56457. 10.1371/journal.pone.0056457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dennis PG, Seymour J, Kumbun K, Tyson G. 21 March 2013. Diverse populations of lake water bacteria exhibit chemotaxis towards inorganic nutrients. ISME J. (Epub ahead of print.) 10.1038/ismej.2013.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cayford BI, Dennis PG, Tyson GW, Keller J, Bond PL. 2012. High-throughput amplicon sequencing reveals distinct communities within a corroding concrete sewer system. Appl. Environ. Microbiol. 78:7160–7162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bragg L, Stone G, Imelfort M, Hugenholtz P, Tyson GW. 2012. Fast, accurate error-correction of amplicon pyrosequences using Acacia. Nat. Methods 9:425–426 [DOI] [PubMed] [Google Scholar]
- 26. DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. GreenGenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Simpson EH. 1949. Measurement of diversity. Nature 163:688 [Google Scholar]
- 28. Homann T, Tag C, Biebl H, Deckwer W-D, Schink B. 1991. Fermentation of glycerol to 1,3-propanediol by Klebsiella and Citrobacter strains. Appl. Microbiol. Biotechnol. 33:121–126 [Google Scholar]
- 29. Boenigk R, Bowien S, Gottschalk G. 1993. Fermentation of glycerol to 1,3-propanediol in continuous cultures of Citrobacter freundii. Appl. Microbiol. Biotechnol. 38:453–457 [Google Scholar]
- 30. Celińska E. 2010. Debottlenecking the 1,3-propanediol pathway by metabolic engineering. Biotechnol. Adv. 28:519–530 [DOI] [PubMed] [Google Scholar]
- 31. Lee SY, Mabee MS, Jangaard NO. 1978. Pectinatus, a new genus of the family Bacteroidaceae. Int. J. Syst. Bacteriol. 28:582–594 [Google Scholar]
- 32. Agler MT, Spirito CM, Usack JG, Werner JJ, Angenent LT. 2012. Chain elongation with reactor microbiomes: upgrading dilute ethanol to medium-chain carboxylates. Energy Environ. Sci. 5:8189–8192 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.