Abstract
The upstream region of antibiotic downregulatory wblA in Streptomyces coelicolor was found to contain AdpA binding motifs. A key morphological regulator, AdpA was shown to specifically bind these motifs by electrophoretic mobility shift assay. An adpA disruption mutant exhibited increased wblA transcription, suggesting that AdpA negatively regulates wblA transcription in S. coelicolor.
TEXT
Streptomyces is a Gram-positive, filamentous bacterial genus known for its ability to produce valuable antibiotics (and other secondary metabolites) and its complex life cycle, which involves significant changes in morphological differentiation (1, 2). An important member of this genus, Streptomyces coelicolor A3 (2), is a model species widely used in studies of morphological development and antibiotic regulation, and its genome was the first to be completely sequenced among the streptomycetes (3). The regulation of morphological differentiation in Streptomyces species is usually accompanied by the production of secondary metabolites and involves multiple regulatory networks (4–6). Among the complex Streptomyces regulatory networks, the A factor regulatory cascade is one of the best-characterized systems, working at both the onset of secondary metabolism and morphological differentiation (7, 8). A factor (2-isocapryloyl-3-R-hydroxymethyl-γ-butyrolactone), initially found in the streptomycin producer S. griseus (9, 10), derepresses the transcription of adpA (A-factor dependent protein A) by binding to ArpA (A-factor receptor protein A), which is bound to the adpA promoter (11). The S. griseus AdpA (designated AdpASG) plays a key role as a transcriptional regulator by binding to the promoter regions of various genes involved in the regulation of morphological differentiation and secondary metabolite biosynthesis (7, 12–14). Recently, chromatin immunoprecipitation studies found that AdpASG directly controls more than 500 genes in cooperation with other regulatory proteins, acting as a transcriptional repressor or activator in S. griseus (14). In S. coelicolor, however, an adpA ortholog gene (bldH) was found not to be under the control of an A factor-like γ-butyrolactone (3, 15, 16, 17), indicating that regulatory cascades are different between these two species. The S. coelicolor adpA mutation, however, causes a bald phenotype and a reduction of actinorhodin (15), suggesting that S. coelicolor AdpA (designated AdpASC) also has an important role in the regulation of both morphological differentiation and secondary metabolism (15, 16), and its functions are not identical to those of AdpASG (12, 17). Recently, AdpASG was identified as a transcriptional activator for its ability to influence the expression of STI (a protease inhibitor), RamR (an atypical response regulator required for aerial growth), and ClpP1 (an ATP-dependent protease) (18, 19). AdpASG contained a dual helix-turn-helix (HTH) motif in its C-terminal DNA binding domain that belongs to the AraC/XylS family of transcription regulators, and its consensus binding sequence is 5′-TGGCSNGWWY-3′ (S, G or C; W, A or T; Y, T or C; N, any nucleotide) (20, 21).
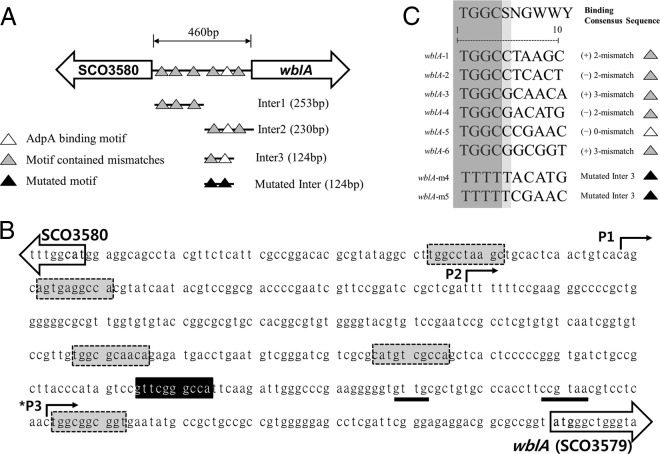
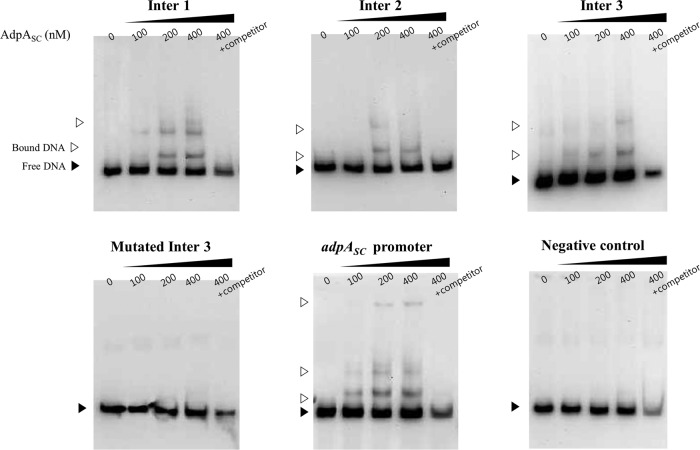
Another transcriptional regulatory gene, wblA (whiB-like gene A), identified in S. coelicolor as well as other Streptomyces species, encodes a pleiotropic downregulator of the biosynthesis of various antibiotics, including actinorhodin (22), doxorubicin (23), moenomycin (24), and tautomycetin (25). In addition, the S. coelicolor ΔwblA mutant showed sporulation-deficient aerial mycelium, thin hyphae, and nonsporulation pigments, suggesting that WblA has an important role in the formation of aerial hyphae (26). Recently, WblA was found to function in the response to oxidative stress among actinobacteria such as Corynebacteria glutamicum and S. coelicolor (27). Interestingly, we found six putative AdpA consensus binding sequences in the promoter region of the wblA gene in S. coelicolor. It has been demonstrated that the consensus binding sites for AdpASC are identical to those of AdpASG, and that the consensus binding sites for AdpASC exist in 157 intergenic regions in the genome of S. coelicolor (18). Thus, to test whether AdpASC directly binds to the wblA promoter, the EMSA (electrophoretic mobility shift assay) was performed to examine the ability of AdpASC binding to a promoter fragment of wblA. adpASC was amplified from the chromosome of S. coelicolor with primer pairs (SCO2792 exp-F, 5′-ACTATGAGCCACGACTCCACCG-3′; SCO2792 exp-R, 5′-CTCGAGTCACGGCGCGCTGC-3′), sequence verified, and cloned into pET21b (+) for expression as a C-terminal His-tagged protein (AdpASC-His) in the E. coli BL21 strain. Previously, it was confirmed that a C-terminal His-tagged AdpASG from S. griseus was functionally expressed and successfully bound to the AdpA binding motifs (14). AdpASC-His (predicted size, 45 kDa) was purified using nickel-nitrilotriacetic acid (Ni-NTA) columns (Qiagen) and analyzed by Western blotting as well as matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) (Genomine Inc., South Korea). Biotin-labeled PCR fragments of the wblA upstream region were prepared by PCR using the following primer pairs: inter1-F, 5′-GGCAGCCTACGTTCTCATTC-3′; inter1-R, 5′-GTCGGTTACCCAGCCCATAC-3′; inter2-R, 5′-TGTTGCGCCACAACGGACAC-3′; inter 3-F, 5′-GAGATGACCTGAATGTCGGG-3′; inter3-R, 5′-GCACAGCGCAACACCCCCTT-3′; mutated inter3-F, 5′-TCGCGCATGTAAAAAGCTCACTCCCCCGGGTGATCTGCCGCTTACCCATAGTCCGTTCGAAAAATTCAAG-3′; mutated inter3-R, 5′-CTTGAATTTTTCGAACGGACTATGGGTAAGCGGCAGATCCACCCGGGGGAGTGAGCTTTTTACATGCGCGA-3′ (mutated sequences are underlined); 2792 EMSA-F, 5′-GACAGGGTCTCGGGCCCCC-3′; 2792 EMSA-R, 5′-ACTGCTAAGCCCCCCTCGGT-3′; negative control-F, 5′-AGCGTGGAGGAGCGGAGCTCGT-3′; and negative control-R, 5′-CCAACCGCCGTGACTCCCAGGGGTACT-3′. As a negative control, a 244-bp DNA probe containing no AdpA binding motif was amplified from the chromosome of the intergenic region between SCO3581, encoding conserved hypothetical protein, and SCO3582, encoding putative secreted protein in S. coelicolor. Amplified probes containing the consensus binding sites were incubated with increasing amounts of purified AdpASC protein in the presence of a nonspecific competitor [poly(dI-dC)] with 2.5% glycerol, 0.05% NP-40, 1 mM EDTA, and 5 mg/ml bovine serum albumin (BSA) for 1 h at room temperature according to instructions of the LightShift chemiluminescent EMSA kit (Thermo Scientific). There are six binding motifs containing 0 to 3 mismatches (in the forward or reverse direction) in the upstream region of the wblA gene, and the consensus binding motifs are illustrated in Fig. 1. As a result, both the inter 1 fragment (253 bp) and inter 2 fragment (230 bp) were retarded by increasing the AdpASC concentration, although fragments possess mismatches, suggesting that AdpASC directly binds to the promoter of the wblA gene and controls the expression of wblA at the transcriptional level. The shifts caused by the binding of AdpASC to the short inter 3 fragment (124 bp), containing both perfect-match sequences and 2-mismatch sequences, were also detected. Perfect-match sequences for AdpA binding are commonly found in the company of mismatch sequences in both S. griseus and S. coelicolor strains, and these contiguous motifs are related to the function of AdpA (21, 28). Also, the TGGCS sequence is an important recognized motif for the N-terminal domain of AdpASG, with the C in the fourth position being essential for AdpA binding (12, 18). We also constructed a mutated inter fragment (Fig. 1 and 2) containing the TTTTT sequence in the consensus binding motifs of the inter 3 fragment by PCR and then performed EMSA with that fragment. No binding shift was detected (Fig. 2), indicating that the conserved TGGCS sequence was also essential as a binding motif for AdpASC. In an additional experiment, a clear shift in DNA-protein complexes was observed in EMSA performed with the adpA promoter fragment (Fig. 2), while shifts of similar strength were observed in EMSA performed with each inter fragment of the wblA upstream region. The binding strength of AdpASC to the wblA promoter was not as strong as the binding of AdpASC to the adpASC promoter (18). According to the reports (12, 14), however, the strength of the AdpA-DNA interaction is not related to the biological significance of AdpA binding sites. Promoter mapping of the wblA upstream region revealed that there are three putative transcriptional start points (26) (Fig. 1B). Among these three positions, P3 is the main transcriptional start point, and transcription begins before the formation of aerial mycelium at this position. Since putative AdpA binding sites are located throughout the three wblA promoter regions, it is not clear which promoter is the major target for the AdpASC binding in S. coelicolor. Thus, we suggest again that AdpASC directly binds to the upstream region of the wblA gene, regulating transcription of wblA in S. coelicolor. Based on these results, we wondered whether AdpASC regulates wblA gene expression as an activator or repressor. To test this, transcription analysis was conducted with strains of S. coelicolor M145, the S. coelicolor ΔwblA mutant (26, 29), and the S. coelicolor ΔadpA mutant (19). It was previously reported that the S. coelicolor ΔwblA mutant produces a much higher level of actinorhodin than S. coelicolor M145 (29, 30), whereas the adpA-deleted S. coelicolor strain does not produce actinorhodin at all in either liquid or plate cultures (15, 16). Each strain was grown in 100 ml of modified R5 (R5 without KH2PO4, CaCl2 · 2H2O, l-proline, and growth factor) liquid medium, and total RNAs were isolated (RNeasy miniprep kit; Qiagen) at 8 h (before any antibiotic production from all strains), 25 h (when there is a slight production of actinorhodin from just the ΔwblA mutant), and 35 h (when there is the production of actinorhodin from S. coelicolor M145 or the ΔwblA mutant and no production of actinorhodin from the ΔadpA mutant). cDNA synthesized from RNA isolated at each time point was analyzed for expression of the actinorhodin-specific activator gene actII-orf4 by normalizing results to the housekeeping gene, hrdB. To ensure that contaminating DNA was not present, we also confirmed that there was no real-time PCR amplification of adpA and wblA from the S. coelicolor ΔadpA and ΔwblA mutant strains, respectively (data not shown). As shown in Fig. 3, real-time PCR analyses demonstrated that the levels of actII-orf4 transcripts were higher in the ΔwblA mutant during the time course than in S. coelicolor M145 and the ΔadpA mutant. It is consistent that the ΔwblA mutant produced the highest level of actinorhodin and the ΔadpA mutant did not produce actinorhodin at all (15, 29, 30). The levels of adpA transcripts from S. coelicolor M145 were similar to those in the ΔwblA mutant, suggesting that the expression of adpA was not influenced by the wblA gene, and suggesting that the transcription of adpA occurs upstream of wblA transcription. The transcription of adpA was maintained at relatively high levels compared to those for wblA until 35 h. This result is similar to the facts that adpA promoter activity reached the peak during substrate mycelium growth in S. coelicolor (from 30 to 45 h) (15) and the maximal production of AdpASC protein was reached during the early stage of aerial mycelium formation in S. coelicolor (from 36 to 60 h) (18). In contrast to adpA, the level of wblA transcripts in the ΔadpA mutant was about 2-fold higher than those in the S. coelicolor M145 strain at 25 h, implying that adpA negatively controls wblA gene transcription. Interestingly, however, the regulation of wblA gene transcription by AdpASC seems to be transient (with apparently no significant differences at both 8 and 35 h), and its mechanism needs to be further characterized. This report shows that AdpASC regulates the transcription of wblA negatively in S. coelicolor, despite the fact that several genes related to the formation of aerial mycelium are regulated positively by AdpASC in S. coelicolor (6, 18, 31, 32). In addition, since the transcription of an actinorhodin-specific regulatory gene was reduced in the adpA disruption mutant, actinorhodin downregulation by WblA could be dependent on the AdpA regulatory pathway in S. coelicolor. By analyzing DNA microarray data from S. griseus, it was reported that the promoter region of wblA (SGR3340) also has an AdpA binding motif and that AdpASG binds to the promoter region of wblA (33), acting as a transcriptional activator for the wblA gene (14). Interestingly, this S. griseus result is the opposite of our current S. coelicolor result, indicating that the AdpA-WblA regulatory cascade mechanism for secondary metabolism and morphological differentiation is different between these two species. In conclusion, we report for the first time the direct influence of AdpASC, a key regulator of morphological differentiation and secondary metabolism in Streptomyces, on the wblA gene in S. coelicolor via EMSA analysis. We also show that wblA gene transcription was regulated by adpASC, suggesting that AdpASC not only acts as a central transcriptional regulator of several genes but also regulates wblA, which is known as a negative regulator in the antibiotic production and the formation of aerial mycelium.
Fig 1.
Putative AdpA binding motifs (A) and intergenic sequences (SCO3579 and SCO3580) in the wblA gene region of the S. coelicolor chromosome (B). Bent arrows, putative promoter designations (P1, P2, and *P3) presented in reference 26; *P3, major transcription start point; underlined sequences, close matches to the consensus sequence for promoters recognized by σF in M. tuberculosis (34); boxes, putative AdpA binding motifs (gray box, contains 2 to 3 mismatches; black box, contains a perfect match). (C) Alignment of AdpA binding consensus sequences in the upstream region of wblA represented with positions and directions.
Fig 2.
EMSA results. Increasing amounts of AdpASC protein were incubated with various regions of the wblA upstream fragment. PCR-amplified adpASC promoter was used as a positive control, and PCR-amplified intergenic sequence without a binding motif was used as a negative control.
Fig 3.
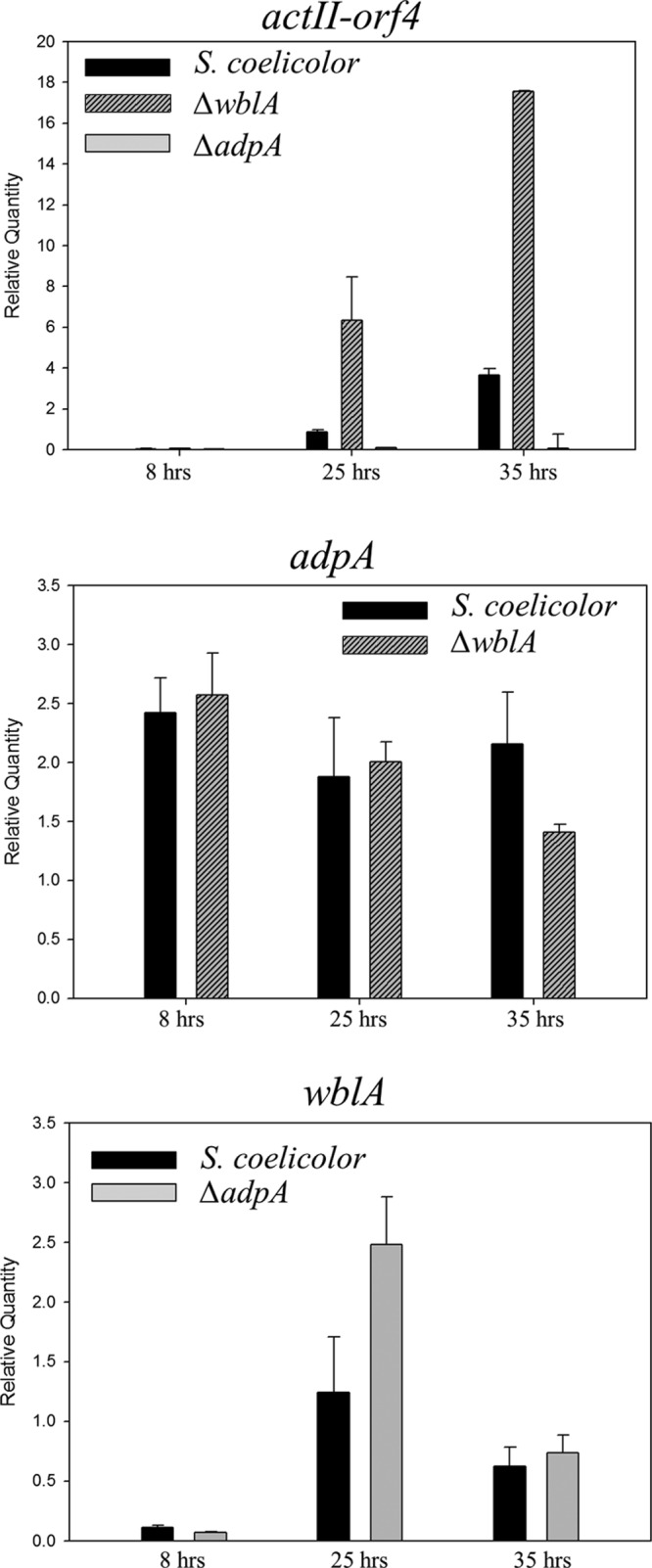
Transcriptional analysis of target genes from S. coelicolor M145, the S. coelicolor ΔadpASC mutant, and the S. coelicolor ΔwblA mutant at different time points (8, 25, and 35 h). All cultures were performed in triplicate, and the averages are shown. The y axis scale represents the expression value relative to that of hrdB, a housekeeping sigma factor (which was set to 1).
ACKNOWLEDGMENTS
The S. coelicolor ΔwblA mutant strain and S. coelicolor ΔadpA mutant strain were kindly provided by Keith Chater at John Innes Centre and Stanley N. Cohen at Stanford University, respectively.
This work was supported by the Korean Ministry of Education, Science, and Technology (MEST 2011-0027683, Program of the NRF).
Footnotes
Published ahead of print 19 April 2013
REFERENCES
- 1. Chater KF. 2006. Streptomyces inside-out: a new perspective on the bacteria that provide us with antibiotics. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:761–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Flardh K, Buttner MJ. 2009. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 7:36–49 [DOI] [PubMed] [Google Scholar]
- 3. Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O'Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147 [DOI] [PubMed] [Google Scholar]
- 4. Bibb MJ. 2005. Regulation of secondary metabolism in streptomycetes. Curr. Opin. Microbiol. 8:208–215 [DOI] [PubMed] [Google Scholar]
- 5. Strauch E, Takano E, Baylis HA, Bibb MJ. 1991. The stringent response in Streptomyces coelicolor A3(2). Mol. Microbiol. 5:289–298 [DOI] [PubMed] [Google Scholar]
- 6. McCormick JR, Flardh K. 2012. Signals and regulators that govern Streptomyces development. FEMS Microbiol. Rev. 36:206–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ohnishi Y, Kameyama S, Onaka H, Horinouchi S. 1999. The A-factor regulatory cascade leading to streptomycin biosynthesis in Streptomyces griseus: identification of a target gene of the A-factor receptor. Mol. Microbiol. 34:102–111 [DOI] [PubMed] [Google Scholar]
- 8. Takano E. 2006. γ-Butyrolactones: Streptomyces signalling molecules regulating antibiotic production and differentiation. Curr. Opin. Microbiol. 9:287–294 [DOI] [PubMed] [Google Scholar]
- 9. Horinouchi S, Beppu T. 1993. A-factor and streptomycin biosynthesis in Streptomyces griseus. Antonie Van Leeuwenhoek 64:177–186 [DOI] [PubMed] [Google Scholar]
- 10. Horinouchi S, Beppu T. 1994. A-factor as a microbial hormone that controls cellular differentiation and secondary metabolism in Streptomyces griseus. Mol. Microbiol. 12:859–864 [DOI] [PubMed] [Google Scholar]
- 11. Horinouchi S. 2002. A microbial hormone, A-factor, as a master switch for morphological differentiation and secondary metabolism in Streptomyces griseus. Front. Biosci. 7:d2045–d2057 [DOI] [PubMed] [Google Scholar]
- 12. Ohnishi Y, Yamazaki H, Kato JY, Tomono A, Horinouchi S. 2005. AdpA, a central transcriptional regulator in the A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus. Biosci. Biotechnol. Biochem. 69:431–439 [DOI] [PubMed] [Google Scholar]
- 13. Akanuma G, Hara H, Ohnishi Y, Horinouchi S. 2009. Dynamic changes in the extracellular proteome caused by absence of a pleiotropic regulator AdpA in Streptomyces griseus. Mol. Microbiol. 73:898–912 [DOI] [PubMed] [Google Scholar]
- 14. Higo A, Hara H, Horinouchi S, Ohnishi Y. 2012. Genome-wide distribution of AdpA, a global regulator for secondary metabolism and morphological differentiation in Streptomyces, revealed the extent and complexity of the AdpA regulatory network. DNA Res. 19:259–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nguyen KT, Tenor J, Stettler H, Nguyen LT, Nguyen LD, Thompson CJ. 2003. Colonial differentiation in Streptomyces coelicolor depends on translation of a specific codon within the adpA gene. J. Bacteriol. 185:7291–7296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Takano E, Tao M, Long F, Bibb MJ, Wang L, Li W, Buttner MJ, Bibb MJ, Deng ZX, Chater KF. 2003. A rare leucine codon in adpA is implicated in the morphological defect of bldA mutants of Streptomyces coelicolor. Mol. Microbiol. 50:475–486 [DOI] [PubMed] [Google Scholar]
- 17. Chater KF, Horinouchi S. 2003. Signalling early developmental events in two highly diverged Streptomyces species. Mol. Microbiol. 48:9–15 [DOI] [PubMed] [Google Scholar]
- 18. Wolanski M, Donczew R, Kois-Ostrowska A, Masiewicz P, Jakimowicz D, Zakrzewska-Czerwinska J. 2011. The level of AdpA directly affects expression of developmental genes in Streptomyces coelicolor. J. Bacteriol. 193:6358–6365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu W, Huang J, Lin R, Shi J, Cohen SN. 2010. Regulation of morphological differentiation in S. coelicolor by RNase III (AbsB) cleavage of mRNA encoding the AdpA transcription factor. Mol. Microbiol. 75:781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Onaka H, Horinouchi S. 1997. DNA-binding activity of the A-factor receptor protein and its recognition DNA sequences. Mol. Microbiol. 24:991–1000 [DOI] [PubMed] [Google Scholar]
- 21. Yamazaki H, Tomono A, Ohnishi Y, Horinouchi S. 2004. DNA-binding specificity of AdpA, a transcriptional activator in the A-factor regulatory cascade in Streptomyces griseus. Mol. Microbiol. 53:555–572 [DOI] [PubMed] [Google Scholar]
- 22. Kang SH, Huang J, Lee HN, Hur YA, Cohen SN, Kim ES. 2007. Interspecies DNA microarray analysis identifies WblA as a pleiotropic down-regulator of antibiotic biosynthesis in Streptomyces. J. Bacteriol. 189:4315–4319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Noh JH, Kim SH, Lee HN, Lee SY, Kim ES. 2010. Isolation and genetic manipulation of the antibiotic down-regulatory gene, wblA ortholog for doxorubicin-producing Streptomyces strain improvement. Appl. Microbiol. Biotechnol. 86:1145–1153 [DOI] [PubMed] [Google Scholar]
- 24. Rabyk M, Ostash B, Rebets Y, Walker S, Fedorenko V. 2011. Streptomyces ghanaensis pleiotropic regulatory gene wblA(gh) influences morphogenesis and moenomycin production. Biotechnol. Lett. 33:2481–2486 [DOI] [PubMed] [Google Scholar]
- 25. Nah JH, Park SH, Yoon HM, Choi SS, Lee CH, Kim ES. 2012. Identification and characterization of wblA-dependent tmcT regulation during tautomycetin biosynthesis in Streptomyces sp. CK4412. Biotechnol. Adv. 30:202–209 [DOI] [PubMed] [Google Scholar]
- 26. Fowler-Goldsworthy K, Gust B, Mouz S, Chandra G, Findlay KC, Chater KF. 2011. The actinobacteria-specific gene wblA controls major developmental transitions in Streptomyces coelicolor A3(2). Microbiology 157:1312–1328 [DOI] [PubMed] [Google Scholar]
- 27. Kim JS, Lee HN, Kim P, Lee HS, Kim ES. 2012. Negative role of wblA in response to oxidative stress in Streptomyces coelicolor. J. Microbiol. Biotechnol. 22:736–741 [DOI] [PubMed] [Google Scholar]
- 28. Kato JY, Ohnishi Y, Horinouchi S. 2005. Autorepression of AdpA of the AraC/XylS family, a key transcriptional activator in the A-factor regulatory cascade in Streptomyces griseus. J. Mol. Biol. 350:12–26 [DOI] [PubMed] [Google Scholar]
- 29. Kim SH, Lee HN, Kim HJ, Kim ES. 2011. Transcriptome analysis of an antibiotic downregulator mutant and synergistic actinorhodin stimulation via disruption of a precursor flux regulator in Streptomyces coelicolor. Appl. Environ. Microbiol. 77:1872–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee HN, Huang J, Im JH, Kim SH, Noh JH, Cohen SN, Kim ES. 2010. Putative TetR family transcriptional regulator SCO1712 encodes an antibiotic downregulator in Streptomyces coelicolor. Appl. Environ. Microbiol. 76:3039–3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim DW, Chater K, Lee KJ, Hesketh A. 2005. Changes in the extracellular proteome caused by the absence of the bldA gene product, a developmentally significant tRNA, reveal a new target for the pleiotropic regulator AdpA in Streptomyces coelicolor. J. Bacteriol. 187:2957–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nguyen KT, Willey JM, Nguyen LD, Nguyen LT, Viollier PH, Thompson CJ. 2002. A central regulator of morphological differentiation in the multicellular bacterium Streptomyces coelicolor. Mol. Microbiol. 46:1223–1238 [DOI] [PubMed] [Google Scholar]
- 33. Hara H, Ohnishi Y, Horinouchi S. 2009. DNA microarray analysis of global gene regulation by A-factor in Streptomyces griseus. Microbiology 155:2197–2210 [DOI] [PubMed] [Google Scholar]
- 34. Humpel A, Gebhard S, Cook GM, Berney M. 2010. The SigF regulon in Mycobacterium smegmatis reveals roles in adaptation to stationary phase, heat, and oxidative stress. J. Bacteriol. 192:2491–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]