Abstract
Deep characterization, even by next-generation sequencing, of the vaginal microbiota in healthy women or posttreatment bacterial vaginosis patients is limited by the dominance of lactobacilli. To improve detection, we offer two approaches: quantitative PCR (qPCR) using phylogenetic branch-inclusive primers and sequencing of broad-spectrum amplicons generated with oligomers that block amplification of lactobacilli.
TEXT
Bacterial vaginosis (BV) is the most common vaginal infection worldwide, affecting 23% of premenopausal women (1, 2) and exhibiting high recurrence rates (3). Decades of microbiological and molecular analyses have not established an etiology (4). These difficulties result in part from the complex microbiota of the vagina, composed of hundreds of bacterial species, with titers ranging from billions to fewer than 100 cells, many of which are fastidious, unculturable, or difficult to identify (5–7).
PCR-based strategies include sequencing amplicons of the 16S rRNA gene generated by broad-spectrum primers after either cloning in Escherichia coli or physical separation of molecules in next-generation sequencing (NGS) (8–15). NGS has now been applied in at least 5 large-scale studies of bacterial populations in the vaginal mucosa of healthy women and asymptomatic and symptomatic BV patients (16–20). Limitations of this approach include the possibilities that broad-spectrum primers may miss whole phyla or fail to amplify some targets for unknown reasons (6, 7) and that rare species may be missed entirely in the presence of an overwhelmingly dominant species.
Blocking Lactobacillus amplification during broad-spectrum PCR.
Our first strategy was to block the amplification of Lactobacillus, using oligomers (LB-blockers) that partially overlap the broad-spectrum primers but complement only (and all) Lactobacillus species and are phosphorylated at their 3′ ends to prevent extension (Fig. 1). These primers and blockers and their PCR programs are described in Tables S1 and S2 in the supplemental material. Methods for their design, optimization, and initial testing are detailed in File S1 in the supplemental material. When applied to 8 species of Lactobacillus, LB blocking resulted in >24,000-fold inhibition; in contrast, no inhibition in 32 nontarget species and relatively modest inhibition of 7 species related to Lactobacillus were seen (Table S3).
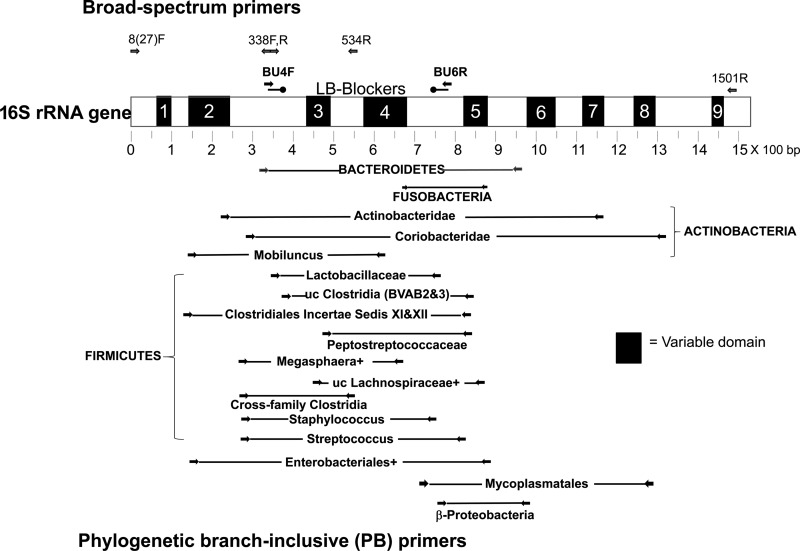
Fig 1.
Relative positions of primers and LB-blockers on 16S rRNA gene. The 16S rRNA gene is depicted with its variable domains based on E. coli. The positions of BU4F and BU6R used in this paper as broad-spectrum primers are shown above. Also included are other primers used for this purpose in published vaginal microbiome studies (see Table S1 in the supplemental material). LB-blockers overlap BU4F and BU6R primers and complement only Lactobacillus species. PB primers are positioned below the 16S rRNA gene (primers are detailed in Table S2). Primers overlapping 27F and 1501R were used as first-round primers in nested PCR to detect some targets at lower titers. uc, uncultured; +, other related genera in the target as described in Table S2.
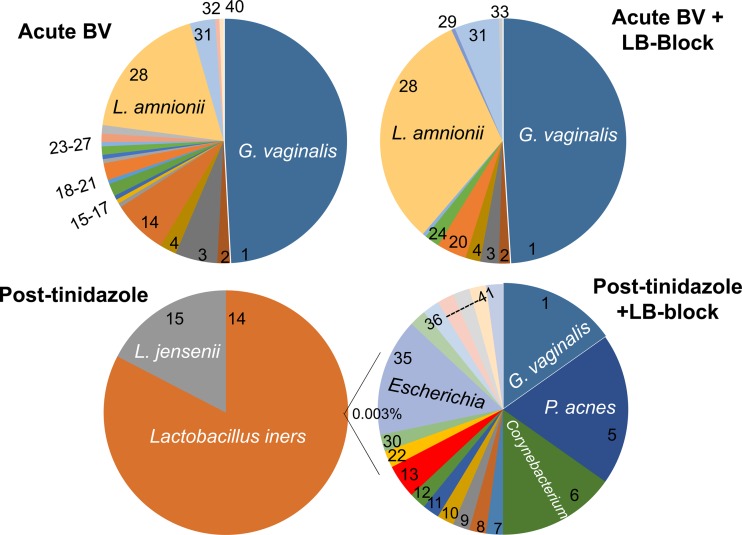
We used this approach on an acute BV sample (Fig. 2, top panels), comparing 204 LB-blocked versus 179 unblocked cloned sequences. LB blocking removed the Lactobacillales reads and 4 non-Lactobacillales reads seen only once without blocking, but other targets did not change significantly (Table 1). With or without blocking, the species detected are characteristic of those in acute BV samples in the literature. Chao1 prediction of actual diversity was 35 operational taxonomic units (OTUs) compared to 22 observed, with a Good's coverage of 88% and Shannon index of 1.8 (see Table S4 in the supplemental material).
Fig 2.
Compositions of vaginal microbiota from a patient with acute BV and after tinidazole treatment with and without Lactobacillus blocking. Amplicons from broad-spectrum primers, made with and without LB-blockers, were cloned and sequenced. The Post-tinidazole + LB-block chart represents the non-Lactobacillus component, collectively 0.003% of isolates. Numbers in the charts are defined in Table 1.
Table 1.
Species identified from sequences of cloned 16S rRNA genes amplified from vaginal bacteriaa
| Species no. | SeqMatch | Phylum |
|---|---|---|
| 1 | Gardnerella vaginalis | A |
| 2 | ∼Gardnerella | A |
| 3 | Atopobium vaginae | A |
| 4 | ∼Paraeggerthella | A |
| 5 | Propionbacteria acnes | A |
| 6 | Corynebacterium (uc) | A |
| 7 | Corynebacterium thomssenii | A |
| 8 | Corynebacterium amycolatum | A |
| 9 | Corynebacterium coyleae | A |
| 10 | Corynebacterium pyruviciproducens | A |
| 11 | ∼Corynebacterium ureicelerivorans | A |
| 12 | Brevibacterium | A |
| 13 | Actinomycetales (uc) | A |
| 14 | Lactobacillus iners | F |
| 15 | Lactobacillus jensenii | F |
| 16 | Lactobacillus 1 | F |
| 17 | Lactobacillus 2 | F |
| 18 | Aerococcus christensenii | F |
| 19 | ∼Aerococcus | F |
| 20 | ∼Roseburia | F |
| 21 | Lachnospiraceae | F |
| 22 | ∼Ruminococcus | F |
| 23 | Anaerococcus prevotii | F |
| 24 | Parvimonas | F |
| 25 | Peptoniphilus | F |
| 26 | Dialister | F |
| 27 | ∼Dialister | F |
| 28 | Leptotrichia amnionii | Fu |
| 29 | Leptotrichia | Fu |
| 30 | Bacteroides (uc) | B |
| 31 | Prevotella bivia | B |
| 32 | Prevotella disiens | B |
| 33 | Prevotella | B |
| 34 | Shingomonas aerolata | B |
| 35 | Escherichia coli | γ-P |
| 36 | Aggregatibacter | γ-P |
| 37 | Pseudomonas fluorescens | γ-P |
| 38 | Pseudomonas pseudoalcaligenes | γ-P |
| 39 | Methylobacterium aminovorans | α-P |
| 40 | Janthinobacterium lividum | β-P |
| 41 | Arcobacter cryaerophilus | ε-P |
Species or taxa were identified and tabulated from sequences as the nearest RDP hits, as described in File S1 in the supplemental material. Numbers refer to numbered sectors in the Fig. 2 charts. Phyla are abbreviated as follows: A, Actinobacteria; F, Firmicutes; Fu, Fusobacteria; B, Bacteroidetes; γ-P, Gammaproteobacteria; α-P, Alphaproteobacteria; β-P, Betaproteobacteria; ε-P, Epsilonproteobacteria. ∼, ≥3% divergent from the indicated, nearest RDP seqmatch hit. uc, uncultured.
The impact of application of LB-blockers to the posttreatment sample, which was dominated by Lactobacillus, was dramatic (Fig. 2, bottom panels). In the absence of Lactobacillus blocking, 100% of the reads represented Lactobacillus. In contrast, the presence of LB-blockers allowed detection of 21 other species in only 136 reads (Table 1), collectively representing only 0.003% of the total population. Chao1 prediction of actual diversity in the blocked sequencing was 135 OTUs compared to 21 observed, with a Good's coverage of 88.4% and Shannon index of 1.7 (see Table S4 in the supplemental material).
This pilot test of LB-blockers is limited to one pair of samples and small numbers of reads. Nevertheless, the data show that LB-blockers greatly enhance detection of non-Lactobacillus species without distorting the vaginal profile when Lactobacillus is subdominant. A more quantitative evaluation of the enhancement provided by LB blocking would require coupling it to NGS evaluation.
PB-qPCR.
Our second strategy employed a complementary approach of using quantitative PCR (qPCR) with 18 phylogenetic branch-inclusive (PB) primers. Group-specific primers have been used in other applications, especially in the gut (21–24), and in one vaginal study (25). These typically are focused on one or a few target genera, whereas our approach strives for inclusivity. Each PB primer targets its own phylogenetic branch, ranging from whole phyla to family or genus, and is far more inclusive than species-specific primers. Positions of PB primers are depicted in Fig. 1 and are described in detail in Table S2 in the supplemental material. The collection was initially validated with target and nontarget single species (listed in Table S3).
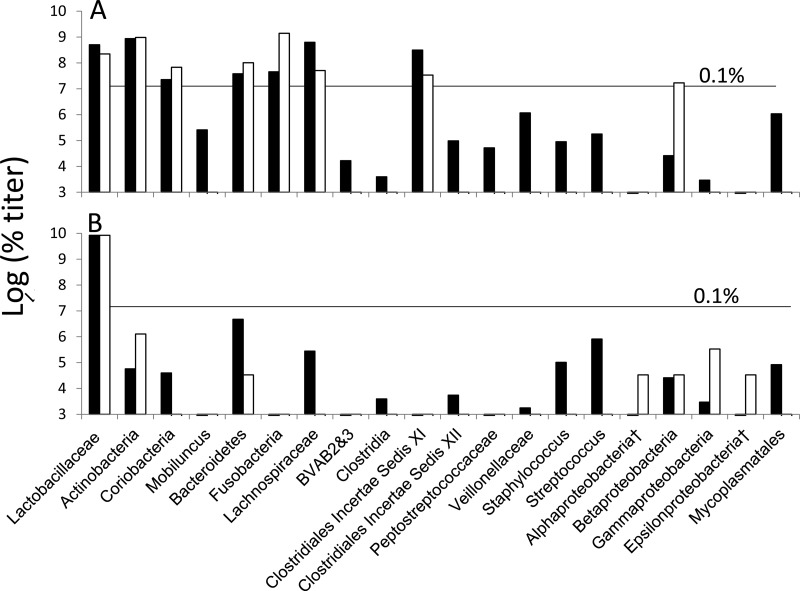
PB-qPCR performed on our pair of samples generated compositions (Fig. 3; see also Table S5 in the supplemental material) similar to those in acute BV and healthy samples in the literature (16–20). Titers of the more prevalent targets (Lactobacillaceae, Actinobacteria, Coriobacteridae, Bacteroidetes, Fusobacteria, and Clostridiales incertae sedis XI clostridia) were similar in proportion to the profiles generated by unblocked and LB-blocked sequencing; the less prevalent targets could not be compared due to low numbers of sequence reads. Sequencing of uncloned amplicons, when not mixed, all verified that the PB primers were measuring their intended targets. Some amplicons, those consisting of mixtures of codominant species, were resolved by sequencing of <20 clones (Table S5) from nested PCR, confirming that intended targets were being quantified and adding to the level of diversity. PB-qPCR detected 18 targets in the acute BV sample and 14 in the posttinidazole sample; these numbers were increased to 30 and 33, respectively, by sequencing mixed amplicons.
Fig 3.
Compositions of acute BV (A) versus posttinidazole (B) samples by PB-qPCR versus blocked and unblocked sequencing. Titers for PB-qPCR (black bars) were calculated as described in File S1 in the supplemental material. For 16S rRNA gene clone and sequence data (white bars), percent composition values (Fig. 2; see also Table S5 in the supplemental material) were converted to titers to facilitate comparison to the qPCR data, as described in File S1. †, no PB primer was designed. Species detected at titers below the 0.1% line would have been undetected or inaccurately counted in a 2,500-read NGS project.
Is there actually diversity in vaginal samples that warrants the use of PB primers instead of species-specific primers? An in silico comparison demonstrated that novel or atypical Clostridium species seen in this study had primer binding sites that had as many as 7 mismatches to the specific BVAB-1, -2, or -3 primers used in previous studies (12, 26). Consistently, these species-specific BVAB-1 primers failed to detect targets in our samples (data not shown). Furthermore, most of the variant species observed (see Table S5 in the supplemental material) have imperfect complementarity to primers specific for “expected” species in the target group and thus would be either missed or underestimated. These data support the premise that PB primers offer better profiling than species-specific primers and underscore the diversity of the vaginal microbiome. Combined with small-library sequencing, the PB-qPCR approach improves inclusiveness relative to species-specific qPCR and yet still allows rapid species identification.
Increased detection of diversity in vaginal samples.
Use of LB-blockers allowed the detection of 19 subdominant species present at 6 orders of magnitude below Lactobacillus. PB-qPCR detected 29 targets in the two samples with estimated titers < 1/1,000 of the total (Fig. 3) which would have not been detected in 2,500-read sets by NGS. Our observed diversity with just over a hundred reads exceeds that seen in recent pyrosequencing-based NGS studies of vaginal populations. For example, one NGS study analyzed 90 samples from among Amsel- and Nugent-based healthy samples, reading an average of 2,235 sequences per sample, and detected an average of only 3 non-Lactobacillus sequences per sample, with 18% of samples generating only Lactobacillus sequences (19). Another NGS study, averaging 2,236 reads per sample, found that, among 173 asymptomatic patients with a Nugent score of between 0 and 3 and pH < 4.5, an average of only 12 reads per sample represented species other than Lactobacillus (17). In a study using the Illumina platform, among 79 women with a Nugent score ≤ 3 and Amsel criteria > 2, with an average depth of 41,512 reads, an average of 51 OTUs, including Lactobacillus species, were seen (20). Our 21 OTUs from LB-blocked sequencing of only 136 clones, or 33 OTUs, including PB-qPCR, are similar in complexity, reflecting the power of the methods even on a small scale and supporting the idea that our observation of diversity in healthy women is not artifactual.
The tools described here, pending NGS validation, potentially allow feasible, in-depth characterization of large numbers of vaginal samples, documenting sequential changes in individual patients. It could be argued that there is little value in characterizing the less dominant species; however, the etiology and ecology of BV are complex and poorly understood (27). We do not know what initiates the changes in microbial populations as they transition from healthy to abnormal compositions, what causes refractory or recurrent responses to treatment, whether there is a sexual transmission component, or whether specific compositions pose higher risks for the complications associated with BV in general (28–33). It could also be argued that effective treatment of BV should have as an endpoint not merely of restoration of Lactobacillus species to dominance but of reduction of levels of BV-associated anaerobes below a threshold that is yet to be defined.
Supplementary Material
ACKNOWLEDGMENTS
This study was partly funded in part by a Focused Giving Grant from Johnson & Johnson, and by internal bridge funding from the Wayne State University Division of Research.
Footnotes
Published ahead of print 26 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01160-13.
REFERENCES
- 1. Sobel JD. 1997. Vaginitis. N. Engl. J. Med. 337:1896–1903 [DOI] [PubMed] [Google Scholar]
- 2. Allsworth JE, Peipert JF. 2007. Prevalence of bacterial vaginosis: 2001–2004 national health and nutrition examination survey data. Obstet. Gynecol. 109:114–120 [DOI] [PubMed] [Google Scholar]
- 3. Hay P. 2009. Recurrent bacterial vaginosis. Curr. Opin. Infect. Dis. 22:82–86 [DOI] [PubMed] [Google Scholar]
- 4. Bohbot JM, Lepargneur JP. 2012. Bacterial vaginosis in 2011: a lot of questions remain. Gynecol. Obstet. Fertil. 40:31–36 (In French.) [DOI] [PubMed] [Google Scholar]
- 5. Hillier SL. 2005. The complexity of microbial diversity in bacterial vaginosis. N. Engl. J. Med. 353:1886–1887 [DOI] [PubMed] [Google Scholar]
- 6. Kalra A, Palcu CT, Sobel JD, Akins RA. 2007. Bacterial vaginosis: culture- and PCR-based characterizations of a complex polymicrobial disease's pathobiology. Curr. Infect. Dis. Rep. 9:485–500 [DOI] [PubMed] [Google Scholar]
- 7. Srinivasan S, Fredricks DN. 2008. The human vaginal bacterial biota and bacterial vaginosis. Interdiscip. Perspect. Infect. Dis. 2008:e750479. 10.1155/2008/750479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hyman RW, Fukushima M, Diamond L, Kumm J, Giudice LC, Davis RW. 2005. Microbes on the human vaginal epithelium. Proc. Natl. Acad. Sci. U. S. A. 102:7952–7957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brown CJ, Wong M, Davis CC, Kanti A, Zhou X, Forney LJ. 2007. Preliminary characterization of the normal microbiota of the human vulva using cultivation-independent methods. J. Med. Microbiol. 56:271–276 [DOI] [PubMed] [Google Scholar]
- 10. Zhou X, Brown CJ, Abdo Z, Davis CC, Hansmann MA, Joyce P, Foster JA, Forney LJ. 2007. Differences in the composition of vaginal microbial communities found in healthy Caucasian and black women. ISME J. 1:121–133 [DOI] [PubMed] [Google Scholar]
- 11. Ferris MJ, Norori J, Zozaya-Hinchliffe M, Martin DH. 2007. Cultivation-independent analysis of changes in bacterial vaginosis flora following metronidazole treatment. J. Clin. Microbiol. 45:1016–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fredricks DN, Fiedler TL, Marrazzo JM. 2005. Molecular identification of bacteria associated with bacterial vaginosis. N. Engl. J. Med. 353:1899–1911 [DOI] [PubMed] [Google Scholar]
- 13. Verhelst R, Verstraelen H, Claeys G, Verschraegen G, Delanghe J, Van Simaey L, De Ganck C, Temmerman M, Vaneechoutte M. 2004. Cloning of 16S rRNA genes amplified from normal and disturbed vaginal microflora suggests a strong association between Atopobium vaginae, Gardnerella vaginalis and bacterial vaginosis. BMC Microbiol. 4:16. 10.1186/1471-2180-4-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhou X, Bent SJ, Schneider MG, Davis CC, Islam MR, Forney LJ. 2004. Characterization of vaginal microbial communities in adult healthy women using cultivation-independent methods. Microbiology 150:2565–2573 [DOI] [PubMed] [Google Scholar]
- 15. Oakley BB, Fiedler TL, Marrazzo JM, Fredricks DN. 2008. Diversity of human vaginal bacterial communities and associations with clinically defined bacterial vaginosis. Appl. Environ. Microbiol. 74:4898–4909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ling Z, Kong J, Liu F, Zhu H, Chen X, Wang Y, Li L, Nelson KE, Xia Y, Xiang C. 2010. Molecular analysis of the diversity of vaginal microbiota associated with bacterial vaginosis. BMC Genomics 11:488. 10.1186/1471-2164-11-488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, Brotman RM, Davis CC, Ault K, Peralta L, Forney LJ. 2011. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. U. S. A. 108(Suppl 1):4680–4687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gajer P, Brotman RM, Bai G, Sakamoto J, Schutte UM, Zhong X, Koenig SS, Fu L, Ma ZS, Zhou X, Abdo Z, Forney LJ, Ravel J. 2012. Temporal dynamics of the human vaginal microbiota. Sci. Transl. Med. 4:132ra52. 10.1126/scitranslmed.3003605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Srinivasan S, Hoffman NG, Morgan MT, Matsen FA, Fiedler TL, Hall RW, Ross FJ, McCoy CO, Bumgarner R, Marrazzo JM, Fredricks DN. 2012. Bacterial communities in women with bacterial vaginosis: high resolution phylogenetic analyses reveal relationships of microbiota to clinical criteria. PLoS One 7:e37818. 10.1371/journal.pone.0037818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hummelen R, Fernandes AD, Macklaim JM, Dickson RJ, Changalucha J, Gloor GB, Reid G. 2010. Deep sequencing of the vaginal microbiota of women with HIV. PLoS One 5:e12078. 10.1371/journal.pone.0012078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Di Cagno R, De Angelis M, De Pasquale I, Ndagijimana M, Vernocchi P, Ricciuti P, Gagliardi F, Laghi L, Crecchio C, Guerzoni ME, Gobbetti M, Francavilla R. 2011. Duodenal and faecal microbiota of celiac children: molecular, phenotype and metabolome characterization. BMC Microbiol. 11:219. 10.1186/1471-2180-11-219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mühling M, Woolven-Allen J, Murrell JC, Joint I. 2008. Improved group-specific PCR primers for denaturing gradient gel electrophoresis analysis of the genetic diversity of complex microbial communities. ISME J. 2:379–392 [DOI] [PubMed] [Google Scholar]
- 23. Parnell JA, Reimer RA. 2012. Prebiotic fibres dose-dependently increase satiety hormones and alter Bacteroidetes and Firmicutes in lean and obese JCR:LA-cp rats. Br. J. Nutr. 107:601–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schwab C, Ganzle M. 2011. Comparative analysis of fecal microbiota and intestinal microbial metabolic activity in captive polar bears. Can. J. Microbiol. 57:177–185 [DOI] [PubMed] [Google Scholar]
- 25. Vitali B, Biagi E, Brigidi P. 2012. Protocol for the use of PCR-denaturing gradient gel electrophoresis and quantitative PCR to determine vaginal microflora constitution and pathogens in bacterial vaginosis. Methods Mol. Biol. 903:177–193 [DOI] [PubMed] [Google Scholar]
- 26. Fredricks DN, Fiedler TL, Thomas KK, Oakley BB, Marrazzo JM. 2007. Targeted PCR for detection of vaginal bacteria associated with bacterial vaginosis. J. Clin. Microbiol. 45:3270–3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Turovskiy Y, Noll KS, Chikindas ML. 2011. The aetiology of bacterial vaginosis. J. Appl. Microbiol. 110:1105–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McDonald HM, O'Loughlin JA, Jolley PT, Vigneswaran R, McDonald PJ. 1994. Changes in vaginal flora during pregnancy and association with preterm birth. J. Infect. Dis. 170:724–728 [DOI] [PubMed] [Google Scholar]
- 29. Goyal R, Sharma P, Kaur I, Aggarwal N, Talwar V. 2004. Bacterial vaginosis and vaginal anaerobes in preterm labour. J. Indian. Med. Assoc. 102:548–550, 553 [PubMed] [Google Scholar]
- 30. Cohen CR, Lingappa JR, Baeten JM, Ngayo MO, Spiegel CA, Hong T, Donnell D, Celum C, Kapiga S, Delany S, Bukusi EA. 2012. Bacterial vaginosis associated with increased risk of female-to-male HIV-1 transmission: a prospective cohort analysis among African couples. PLoS Med. 9:e1001251. 10.1371/journal.pmed.1001251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hilber AM, Francis SC, Chersich M, Scott P, Redmond S, Bender N, Miotti P, Temmerman M, Low N. 2010. Intravaginal practices, vaginal infections and HIV acquisition: systematic review and meta-analysis. PLoS One 5:e9119. 10.1371/journal.pone.0009119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van de Wijgert JH, Morrison CS, Cornelisse PG, Munjoma M, Moncada J, Awio P, Wang J, Van der Pol B, Chipato T, Salata RA, Padian NS. 2008. Bacterial vaginosis and vaginal yeast, but not vaginal cleansing, increase HIV-1 acquisition in African women. J. Acquir. Immune Defic. Syndr. 48:203–210 [DOI] [PubMed] [Google Scholar]
- 33. Atashili J, Poole C, Ndumbe PM, Adimora AA, Smith JS. 2008. Bacterial vaginosis and HIV acquisition: a meta-analysis of published studies. AIDS 22:1493–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.