Abstract
Lyme disease spirochetes possess complex genomes, consisting of a main chromosome and 20 or more smaller replicons. Among those small DNAs are the cp32 elements, a family of prophages that replicate as circular episomes. All complete cp32s contain an erp locus, which encodes surface-exposed proteins. Sequences were compared for all 193 erp alleles carried by 22 different strains of Lyme disease-causing spirochete to investigate their natural diversity and evolutionary histories. These included multiple isolates from a focus where Lyme disease is endemic in the northeastern United States and isolates from across North America and Europe. Bacteria were derived from diseased humans and from vector ticks and included members of 5 different Borrelia genospecies. All erp operon 5′-noncoding regions were found to be highly conserved, as were the initial 70 to 80 bp of all erp open reading frames, traits indicative of a common evolutionary origin. However, the majority of the protein-coding regions are highly diverse, due to numerous intra- and intergenic recombination events. Most erp alleles are chimeras derived from sequences of closely related and distantly related erp sequences and from unknown origins. Since known functions of Erp surface proteins involve interactions with various host tissue components, this diversity may reflect both their multiple functions and the abilities of Lyme disease-causing spirochetes to successfully infect a wide variety of vertebrate host species.
INTRODUCTION
One striking aspect of the Lyme disease-causing spirochete, Borrelia burgdorferi sensu lato, is the abundance of naturally occurring DNA replicons within each bacterium. The type strain, B31, contains 25 distinct DNA entities: a 911-kb linear main chromosome, and a variety of 5- to 56-kb linear and circular elements, loosely described as plasmids (1, 2, 3, 4, 5). Among the smaller borrelial DNAs are a family of circular replicons of approximately 32 kb in size, known as cp32 elements, that appear to be episomal prophages (2, 6, 7, 8, 9, 10, 11). Individual bacteria have been identified that simultaneously harbor 9 or more distinct cp32s, which is possible due to each possessing a different segregation locus (2, 3, 7, 12, 13, 14, 15, 16).
In addition, cp32 elements exhibit considerable variability at two loci that encode outer surface lipoproteins (2, 3, 7, 12, 13, 15, 16, 17). One of the variable cp32 loci is designated erp (OspEF-related protein) (3, 6, 7, 15). An alternative nomenclature has been proposed, dividing erp genes into three groups, named ospE (outer surface protein E), ospF (outer surface protein F), and elp (OspEF-like leader peptide), under the assumption that each group represents a discrete evolutionary lineage (13). Closely related cp32s of different bacterial isolates often contain extremely diverse erp sequences.
The mono- or bicistronic erp operons encode surface-exposed lipoproteins that are produced throughout vertebrate infection but are largely repressed during colonization of vector ticks (18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28). Known functions of Erp proteins include binding of host plasmin (which could facilitate dissemination through host tissues), complement factor H and factor H-related proteins (which protect against killing by host complement), and laminin (which could enhance colonization of host extracellular matrices) (29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44). Sequences of erp alleles often vary dramatically between different strains. It has been hypothesized that such variations in the encoded host-interactive proteins contribute to the ability of Lyme disease-causing spirochetes (here, Lyme spirochetes) to efficiently infect a wide variety of vertebrate host species (32, 37). Notably, Borrelia species that cause relapsing fever also contain cp32 episomes, but none possesses erp loci (45).
Previous sequence analyses of cp32 elements from four strains of B. burgdorferi indicated that, in nature, cp32s move between bacteria, and erp sequences shuffle between different cp32 prophages (3, 12, 46). Recently, the entire erp sequence repertoires of 18 additional strains of Lyme spirochetes have been determined (this work and references 47, 48, 49, 50, 51, 52, and 53). These bacteria were isolated from humans and ticks throughout the world and include multiple isolates from an area where Lyme disease is endemic in the northeastern United States. Herein we report an analysis of the evolutionary histories of sequences encoded at the erp locus, with an emphasis on the implications of sequence conservation and diversity. These results indicate that all open reading frames at this locus contain a well-conserved 5′ end, indicative of a common ancestry, while the 3′ part of each allele is a unique chimera of DNAs from many diverse origins.
MATERIALS AND METHODS
Bacterial strains.
The strains analyzed in this study are listed in Table 1. The primary goal of the present study was to assess the variation among erp alleles both within individual bacteria and between different bacterial strains. For that reason, we analyzed all strains of Lyme disease spirochetes for which the entire repertoire of erp loci has been sequenced. The whole genomes for the majority of strains analyzed were sequenced as parts of unrelated projects by us or by other researchers (3, 47, 48, 49, 51, 52, 53). As a result, the analyzed strains included numerous bacteria isolated from a fairly small geographical area, the northeastern United States, and individual bacteria from many locations across North America and Europe. Genetic variations among Lyme disease borreliae have led to division of the original species B. burgdorferi into several genospecies, including B. burgdorferi sensu stricto, B. bissettii, B. garinii, B. afzelii, and B. valaisiana (54). Included in our analyses were at least one member of each of those named species. Thus, our panel of strains included both groups of closely related organisms and individual examples of genetically distant bacteria, permitting a broad view of erp diversity in nature.
Table 1.
The cp32 DNA elements and associated erp loci naturally contained in the analyzed Lyme disease-causing spirochetesa
| Genospecies and strain | cp32 segregation locus type |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cp32-1 | cp32-2/7 | cp32-3 | cp32-4 | cp32-5 | cp32-6 | cp32-8 | cp32-9 | cp32-10 | cp32-11 | cp32-12 | cp32-13 | |
| B. burgdorferi B31b | erpA1B2 | erpCD | erpG | erpHY | erpAB5 | erpK | erpAB8 | erpPQ | erpX | |||
| erpLM | ||||||||||||
| B. burgdorferi BL206b | erpLM | erpG | erpHY | erpAB5 | erpK | erpPQ | erpX | erpAB11 | ||||
| B. burgdorferi bu Bol26 | R10 | V38 | N41, N42 | Q34 | W10 | X41, X42 | ||||||
| B. burgdorferi 297c | ospE1, elpB1-1 | elpA2 | ospF | elpA1 | ospE5, elpB1-5 | bbk2.10 | p21, elpB2 | bbk2.11 | ospE12, elpB1-12 | |||
| B. burgdorferi Sh-2-82 | erp41-1, erp42-2 | erp43 | erp44 | erp45 | erp41-5, erp42-5 | erp46 | erp50, erp51 | erp47, erp48 | erp49 | erp41-12, erp42-12 | ||
| B. burgdorferi 156a | P15 | O27 | S39 | R40 | M38 | L38, L39 | W37 | X39, X40 | ||||
| B. burgdorferi ZS7d | P41 | AC26, AC64 | R38, R39 | N42, N43 | AC26, AC64 | X38 | ||||||
| B. burgdorferi 64be | P38 | O38 | SL81, SL82 | R39, R40 | V38, V39 | M38, M39 | SL81, SL82 | Truncated plasmid, no erp locus | W38 | |||
| B. burgdorferi WI91-23 | P39 | O42 | Truncated plasmid, no erp locus | V22 | M39, M40 | Q39 | W40, W41 | |||||
| B. burgdorferi 94a | O39 | V39 | M40, M41 | Q40, Q41 | W38, W39 | |||||||
| B. burgdorferi JD1f | PV38, PV80 | S38 | M38, M39 | L38, L39 | N44, N45 | Q40 | W44 | X44 | ||||
| B. burgdorferi CA-11.2A | P40 | S38 | V38 | Q39, Q40 | N38 | |||||||
| B. burgdorferi 118ag | ON39, ON83 | R38 | V25 | M39, M40 | L38, L39 | ON39, ON83 | Q40, Q41 | AB39 | ||||
| B. burgdorferi N40 | ospEF | erp23, erp24 | erp25 | p21, erp22 | erp26 | erp27 | ||||||
| B. burgdorferi 72a | O40 | Truncated plasmid, no erp locus | V24 | Q39, Q40 | X36 | N39 | ||||||
| B. burgdorferi 29805 | O38 | R39, R40 | V28 | N43, N44 | X36 | |||||||
| SV1h (unnamed species) | O39, O40 | S38, S39 | Truncated plasmid, no erp locus | M31 | X50, X51 | |||||||
| B. valaisiana VS116 | O43 (aka H460) | V36, V37 | Q36 | |||||||||
| B. bissettii DN127i | O40 | S39 | R38, R39 | V40 | M40 | (i) | (i) | (i) | ||||
| B. afzelii PKo | P38 | O31 | S39 | V39 | N36, N37 | W37 | X38, X39 | |||||
| B. afzelii ACA-1 | P41, P42 | S38, S39 | R38 | V40 | ||||||||
| B. garinii PBr | V40 | Q67 | ||||||||||
Plasmid nomenclature is standardized according to segregation locus type, such that all plasmids designated cp32-1, for example, contain a similar segregation locus. All cp32 segregation loci fell into the 12 previously described groups (see Fig. 1). Note that the first examined strain, B. burgdorferi B31, contains 2 distinct prophages that possess identical maintenance loci, cp32-2 and cp32-7, and define segregation locus type 32-2/7. As a result, the numbering scheme for the 12 groups goes up to cp32-13. For those erp genes that have been previously described, the published names are used. Various naming schemes were applied to these genes by their discoverers. Previously undescribed erp alleles found by mining GenBank are identified by the open reading frame designations of the entries.
The cp32-10 plasmids of strain B31 and BL206 are naturally integrated into an unrelated linear replicon, which created the ca. 56-kb linear plasmid lp56.
The cp32-7 and cp32-9 plasmids of strain 297 are naturally truncated and have been named cp18-a and cp18-2, respectively.
The open reading frames are designated with the two letters, AC.
The cp32-3 and cp32-8 plasmids of strain 64b are fused together into a single circular replicon, cp32-3-8. Its open reading frames are designated with the two letters, SL.
The cp32-1 and cp32-5 of strain JD1 are fused together into a single circular replicon, cp32-1-5. Its open reading frames are designated with the two letters, PV.
The cp32-2/7 and cp32-9 plasmids of strain 118a are fused into a single circular replicon, cp32-7-9. Its open reading frames are designated with the two letters, ON.
Strain SV1 also contains an additional legitimate erp locus (ORF A100) on a linear plasmid that contains an lp54-type replication locus. This strain falls within an as-yet-unnamed group of Lyme disease-causing Borrelia.
Strain DN127 contains a large circular plasmid that includes portions of four distinct cp32s and includes replication loci of types cp32-9, cp32-11, and cp32-12. Three erp loci are located on this chimera, two monocistronic and one bicistronic. For the purposes of this study, the erp genes are designated open reading frames Quad-A, Quad-B1, Quad-B2, and Quad-C.
Naming of cp32 elements follows the convention of grouping borrelial DNAs according to sequences of the replication/segregation locus (5, 12). For example, all cp32s that possess an episomal maintenance locus similar to the cp32-1 of type strain B31 are designated cp32-1. Some episomes are fusions of two or more distinct cp32s and contain multiple segregation loci. In such cases, the replicon is given a name indicative of all its ancestral prophages (e.g., cp32-1-5 of strain JD1 is a chimera of cp32-1 and cp32-5). Strain DN127 maintains a chimera of four different cp32 types and is referred to as DN127-Quad.
Sequence data.
Sequences of erp loci were previously determined for B. burgdorferi strains B31 (2, 3, 6), 297 (13), Sh-2-82 (46), and N40 (46). The erp and cp32 segregation loci of B. burgdorferi strain BL206 were cloned and sequenced as part of the current work, using the previously described method of PCR with oligonucleotide primers based on conserved erp locus sequences (12, 46). Strain BL206 was selected because it was originally isolated from the blood of a human Lyme disease patient and is highly virulent in experimental mouse models (55, 56).
To ensure that all alleles were identified, GenBank was searched using the 5′-noncoding regions of previously identified erp alleles and the predicted sequences of known proteins encoded at this locus, using BlastN or BlastP, respectively (http://blast.ncbi.nlm.nih.gov/Blast.cgi). As new alleles were identified from the searches, they were used in turn as queries for additional BLAST searches. This was continued until BLAST searches failed to uncover new alleles.
The sequences of a single or pair of erp alleles from many Lyme spirochetes are included in GenBank. Most were identified and sequenced because they are closely related to previously identified sequences (57, 58). Such orphan sequences were not included in these analyses, since the cp32 and erp diversity in those bacteria cannot be assessed. Only strains for which the entire cp32 repertoire had been sequenced were considered for this work.
A locus was operationally defined as an erp if it (i) encodes one or more predicted lipoproteins and (ii) has a 5′-noncoding region similar to those originally identified in B. burgdorferi strains N40, B31, ZS7, and 297 (6, 18, 19, 57, 59). Those open reading frames within identified erp operons that encode predicted lipoproteins were operationally defined as being erp alleles. This last definition is important, as some erp operons include a nonlipidated, unrelated gene named bapA (6, 19, 46, 58, 59). erp sequences can vary greatly in size but generally fall into three groups, described here as small-, medium-, and large-sized erps. These groups roughly correspond with the previously suggested ospE, ospF, and elp groups, respectively (13).
Sequence analyses.
Alignments of DNA sequences were performed using Clustal W2 (http://bips.u-strasbg.fr/fr/Documentation/ClustalX/) with a gap opening penalty of 35.0, gap extension penalty of 6.66, and DNA transition weight of 0. The amino acid sequences of protein-coding regions were aligned using MAFFT (60) to correctly conserve gaps and were converted into the corresponding codon alignment by using PAL2NAL (61). Similarities among cp32 ParA-like proteins were assessed by neighbor-joining analysis using Clustal X (62).
Divergence within and among the three size-grouped erp alleles was investigated using global alignment tests for all pairwise combinations of peptide sequences by using the Needleman-Wunsch algorithm (63). The Gonnet matrix (64) was used as the scoring matrix for the alignments, with a gap penalty set to 10 and gap extension penalty set to 0.1. For all alignments, the percent sequence identity was calculated using either the proportion of identical residues in the alignment or the proportion of identical residues in the alignment without gaps.
Recombination events among alleles within each group of erp sequences were detected using RDP3 program (65). The RDP3 program incorporates the recombination-detecting algorithms RDP (66), GENECONV (67), Chimaera (68), MaxChi (68, 69), Bootscan (70, 71), Siscan (72), and 3Seq (73), all of which mandate aligned sequences that share a recent common ancestry to detect recombination breakpoints and to identify the source of recombinant fragments. Thus, sequences in different-sized groups could not be analyzed together, limiting the detection of recombination events between sequences in different groups to manual investigation of sequences with high homology. The RDP3 analyses were conservative, in that only events in which at least three independent algorithms detected recombination sites within the same region of the alignment are described.
Nucleotide sequence accession numbers.
New strain BL206 sequences were deposited in GenBank with the following accession numbers (in parentheses): erpAB5 (DQ872880), erpAB11 (DQ872883), erpCD (DQ872878), erpG (AF475925), erpHY (DQ872879), erpK (DQ872881), erpX (DQ872882), the cp32-11 replication/segregation locus (DQ872884), and the left and right recombination sites within lp56 (DQ872885 and DQ872886, respectively).
RESULTS
cp32 elements in Lyme disease-causing spirochetes.
The number of cp32 episomes in individual sequenced genomes was highly variable (Table 1). Additional, apparently linear plasmids were identified that contained cp32-like replication/segregation loci but did not carry erp loci or any other genes characteristic of cp32s (Fig. 1); these latter plasmids are not discussed further. B. garinii strain Far04 contains a total of 7 small DNA elements, a number comparable to cultures of B. burgdorferi strain B31 that have been serially passaged in culture media for many years and have lost the majority of their small DNAs (2, 50, 74). The sequenced culture of B. garinii Far04 does not contain any cp32 elements (50), but whether this is also true of the bacterium in nature or is due to plasmid loss in culture is not known.
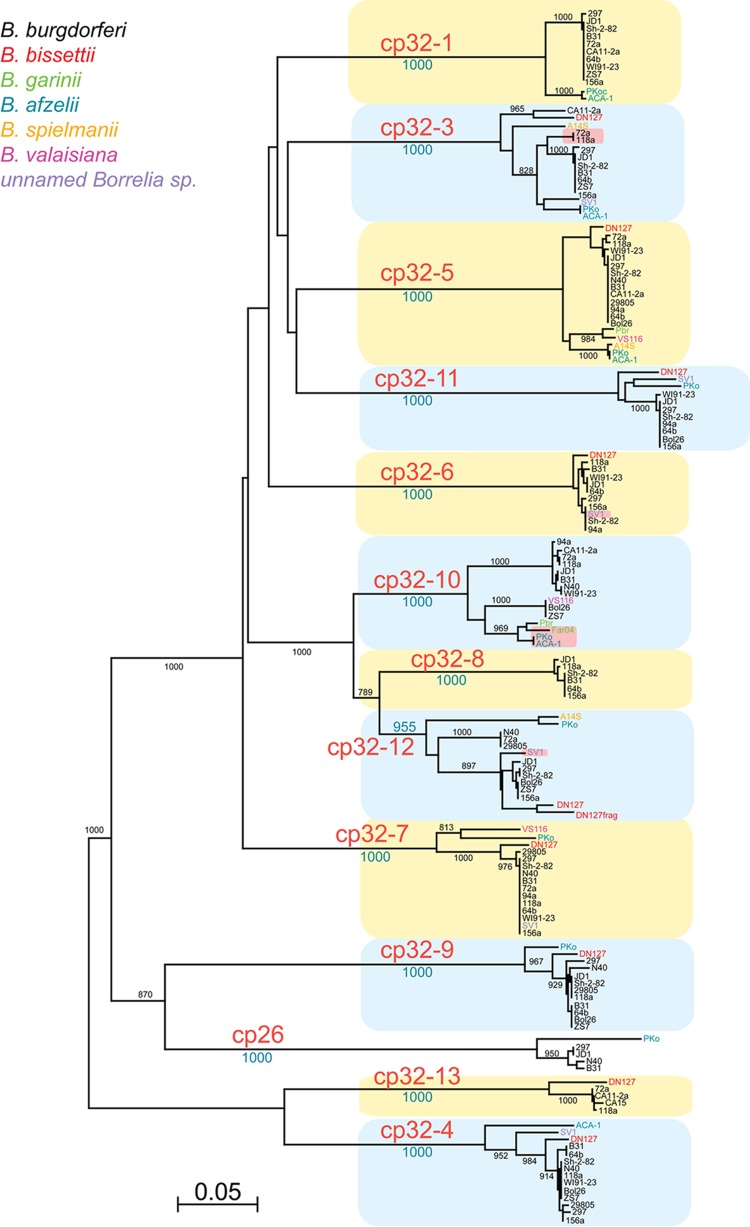
Fig 1.
Neighbor-joining tree of the ParA-like proteins encoded on cp32 elements. The tree was constructed from all the proteins encoded by the parA-like genes of the cp32s of the sequenced B. burgdorferi sensu lato genomes and those known from strains CA15, Sh-2-82, and Far04. Numbers on the tree branches are bootstrap support values from 1,000 trials. Blue and yellow boxes highlight the different sequence types (tree branches) for the cp32 ParA proteins. These types correlate with episomes named in large red type. The species which harbored each cp32 are indicated by the color of the strain name at the right branch tips (see color key in upper left of figure). The DN127 protein labeled DN127frag in the cp32-12 branch is a ParA fragment from the DN127 cp32-Quad element. All the replicons in the figure carry highly related complements of non-partition cluster genes, except for seven “lp32” plasmids, noted by a pink background in the figure, which encode cp32-like ParA proteins but do not carry erp sequences (these will be described elsewhere [S. Casjens, unpublished data]). Replicon cp26 is an unrelated, universally present circular “plasmid” and is shown only for reference. Bar, 0.05 fractional change.
For all newly analyzed cp32 plasmids, the cluster of genes involved with plasmid maintenance were similar to the 12 previously identified types (Fig. 1) (2, 3, 5, 7, 12, 16, 46, 58). The degree of conservation among cp32 maintenance loci in different species of Lyme spirochetes is striking. For example, the 257-amino-acid ParA-like protein encoded by the cp32-5 of B. garinii PBr differs by only 2 residues from that encoded by the cp32-5 of B. valaisiana VS116, by 12 from that of B. afzelii ACA-1, by 13 from that of B. spielmanii A14S, and by 14 from that of B. burgdorferi 64b. These results suggest that there exist a limited number of cp32 borreliaphage types in nature. We also note that the definitions of these 12 cp32 types are somewhat arbitrary. The 12 types indicated in Fig. 1 are presumed, based on the fact that they are found coexisting with other types in natural Borrelia isolates, to represent different plasmid compatibility types (5); however, in most cases this has not been proven experimentally. Thus, some of the relatively deep branching subclusters of ParA-like proteins, such as those represented by VS116 cp32-7, ZS7 cp32-10, and PKo cp32-12, could have different compatibility properties than others in those “types.”
Although most cp32 plasmids are syntenic and quite similar to each other in size and overall gene content, several cp32 elements exhibit evidence of naturally occurring gross rearrangements (Table 1). As examples, strains N40 and 297 both contain cp32s with large deletions that yield final sizes of approximately 18 kb (75, 76). Strain SV1 contains an 83-kb chimeric plasmid that is comprised of part of a cp32 integrated into a linear plasmid, and strains B31 and BL206 both contain a 56-kb linear replicon that consists of a cp32-10 integrated into a different linear plasmid (2, 3, 77). Strains ZS7, 64b, JD1, and 118a each contain unique replicons that are chimeras of two distinct cp32s (52).
erp 5′-noncoding sequences.
Analyses of 5′-noncoding DNA sequences from erp loci revealed extensive sequence conservation that is unique to the cp32 replicon family (Fig. 2A). Furthermore, every sequence in GenBank having a 5′-end homologous to a defined erp operon is located on a cp32-type replicon. High degrees of similarity were observed across all Borrelia genospecies from all geographic locations of origin.
Fig 2.
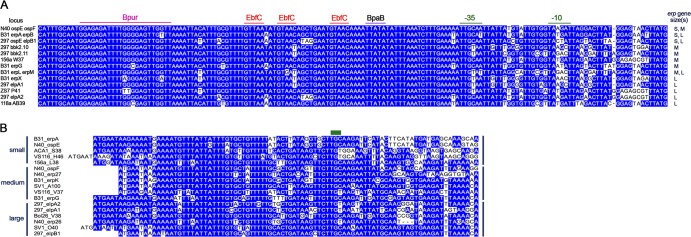
The 5′-noncoding and amino-terminal-coding sequences are conserved among erp operons. (A) Alignment of 5′-noncoding DNAs of 13 representative erp operons. Nucleotides occurring in >50% of all loci are boxed in blue. The ATG initiation codon of the proximal erp gene is on the right. On the far right are indications of whether each operon contains one or two erp open reading frames and the relative sizes of those sequences (S, small-sized erp; M, medium-sized erp; L, large-sized erp). The promoter −10 and −35 sites and the high-affinity binding sites of the regulatory factors BpaB, EbfC, and Bpur are indicated above the alignment (our unpublished results and references 79, 80, 81, and 82). (B) Alignment of 5′ ends of the erp alleles characterized below in Fig. 5A to C. Nucleotides occurring in >50% of all loci are boxed in blue. The green bar above the alignment indicates the codon for the lipid-modified cysteine of the mature lipoprotein. The leader polypeptide is encoded by nucleotides to the left of the cysteine codon, and localization/trafficking signals are encoded by the nucleotides to the right (85). The relative size of each gene is indicated to the left of the alignment.
The erp operator, located immediately 5′ of each erp operon promoter, is required for transcriptional regulation and contains high-affinity binding sites for three borrelial proteins (B. Jutras et al., submitted for publication, and references 11, 78, 79, 80, 81, 82, and 83). Although there is extensive sequence conservation among erp operator sequences, some variations were observed (Fig. 2A and data not shown). With four exceptions, all analyzed erp operators include two full binding sites and one half-site, for the antirepressor EbfC (80). Three erp loci of strain SV1 each contain one full and two half-sites, and one locus of strain JD1 contains three half-sites. Differences of 1 or 2 bp in the binding sites for the BpaB repressor and Bpur modulatory proteins were also observed for a small number of erp operons (Fig. 2A and data not shown). Such variations may affect the affinities of the proteins for each erp operon in a cell and may contribute to the reported differences in erp transcription levels (19, 59, 78, 84).
The transcriptional promoter, especially the −10 element, is the most variable part of the erp 5′ region (Fig. 2A), and this may contribute to different expression levels among loci. For example, B. burgdorferi B31 erpAB and erpG are transcribed at different levels due to differences in their promoter sequences (78). However, there are no patterns regarding linkage of various promoter −10 sequences with any particular of type of erp allele (Fig. 2A).
erp coding sequences.
All erp sequences contain highly conserved 5′ termini, encoding the leader polypeptide and cysteine lipidation signal (Fig. 2B). Also well conserved are the first 5 codons beyond the lipidated cysteine, which contain the membrane sorting code, consistent with the conserved cellular localization of Erp proteins in the outer membrane (85, 86, 87). Beyond these initial nucleotides, tremendous diversity is apparent. erp sequences can vary greatly in size, and robust DNA sequence alignments require division into three groups, described here as small-, medium-, and large-sized erps. These groups roughly correspond with the alternative nomenclature names ospE, ospF, and elp, respectively (13). Even after removing gaps from the DNA sequence alignments, sequence identities between pairs of sequences from different groups averaged in the range of 20% (Fig. 3).
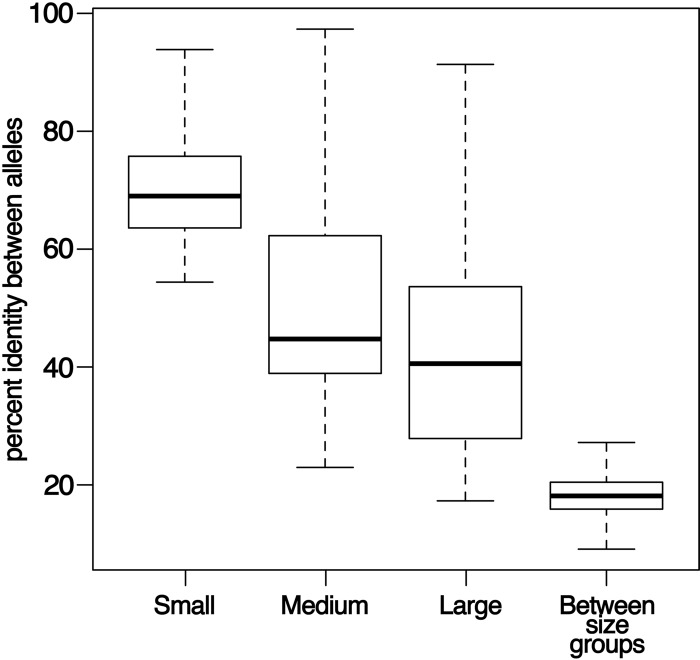
Fig 3.
Sequence similarities within and between erp sequence size classes are limited, except within the small-sized sequences. For each comparison, the horizontal line is the median, the box indicates the upper and lower quartiles, and whiskers indicate the maximum and minimum values. Average pairwise similarities among sequences within the large-sized sequences or within the medium-sized sequences are very low, with some pairs of sequences demonstrating as little as 17% identity. Small-sized sequences are a more homogeneous group, with an average of 69% sequence identity among pairs of sequences. Identity among pairs of sequences from different size groups was generally low, although some pairs of sequences between groups were more similar than pairs of sequences within groups. This result may indicate the presence of recombination events among sequences from different groups. Gap regions were not included in these analyses.
The small erp sequences (ospE) represent a relatively homogenous group with modest sequence and length diversities (69% average pairwise sequence identity) (Fig. 3, 4A, and 5A). In contrast, the medium- and large-sized erp sequences (ospF and elp, respectively) have undergone considerably greater numbers of insertion and deletion events (Fig. 4B and C and 5B and C). Further, pairwise identity is much lower within the medium- and large-sized erp groups, with 44 and 40% average pairwise identity, respectively (Fig. 3). In many cases, such as with the B. burgdorferi strain 297 large-sized elpA1 and elpB1 genes, similarly sized genes share almost no sequences in common (Fig. 5C). Some alleles that group as medium- or large-sized erp genes have less homology with other members of their own size group (as low as 17%) than with members of other groups (as high as 34%), suggesting that recombination has frequently occurred between alleles of different size groups (Fig. 3).
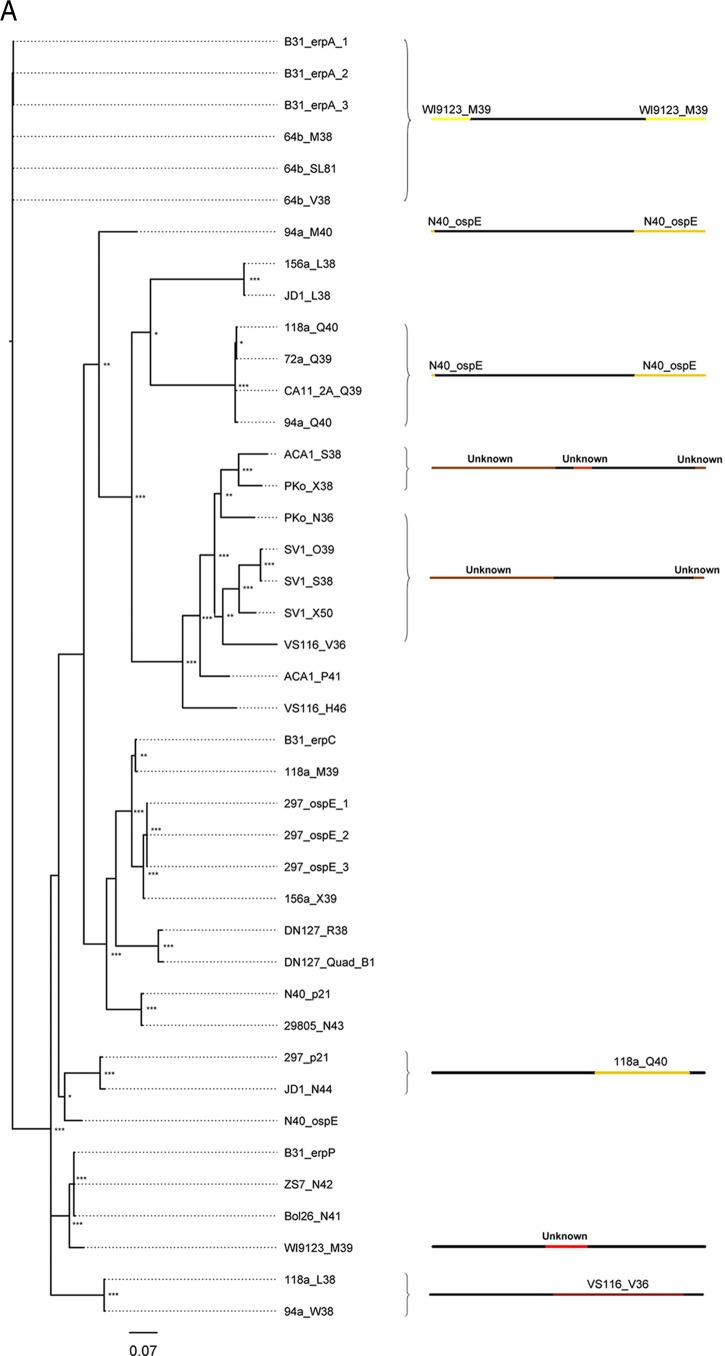
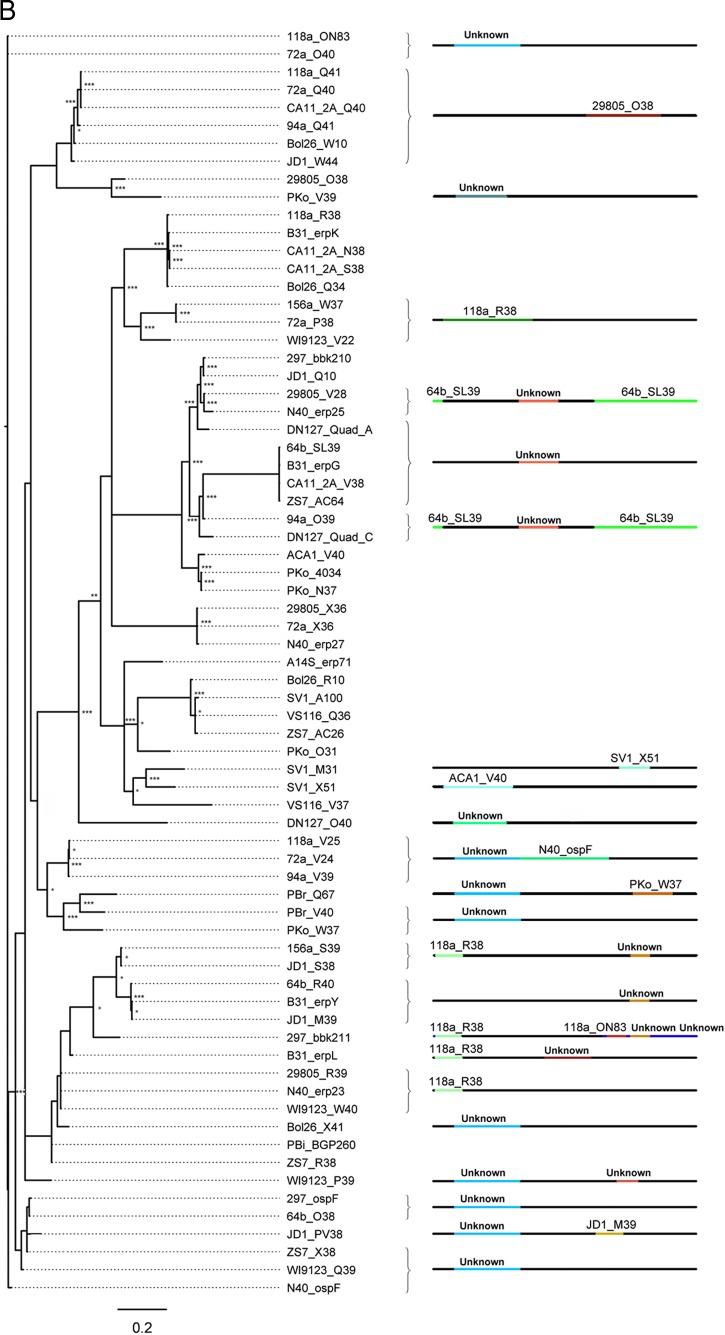
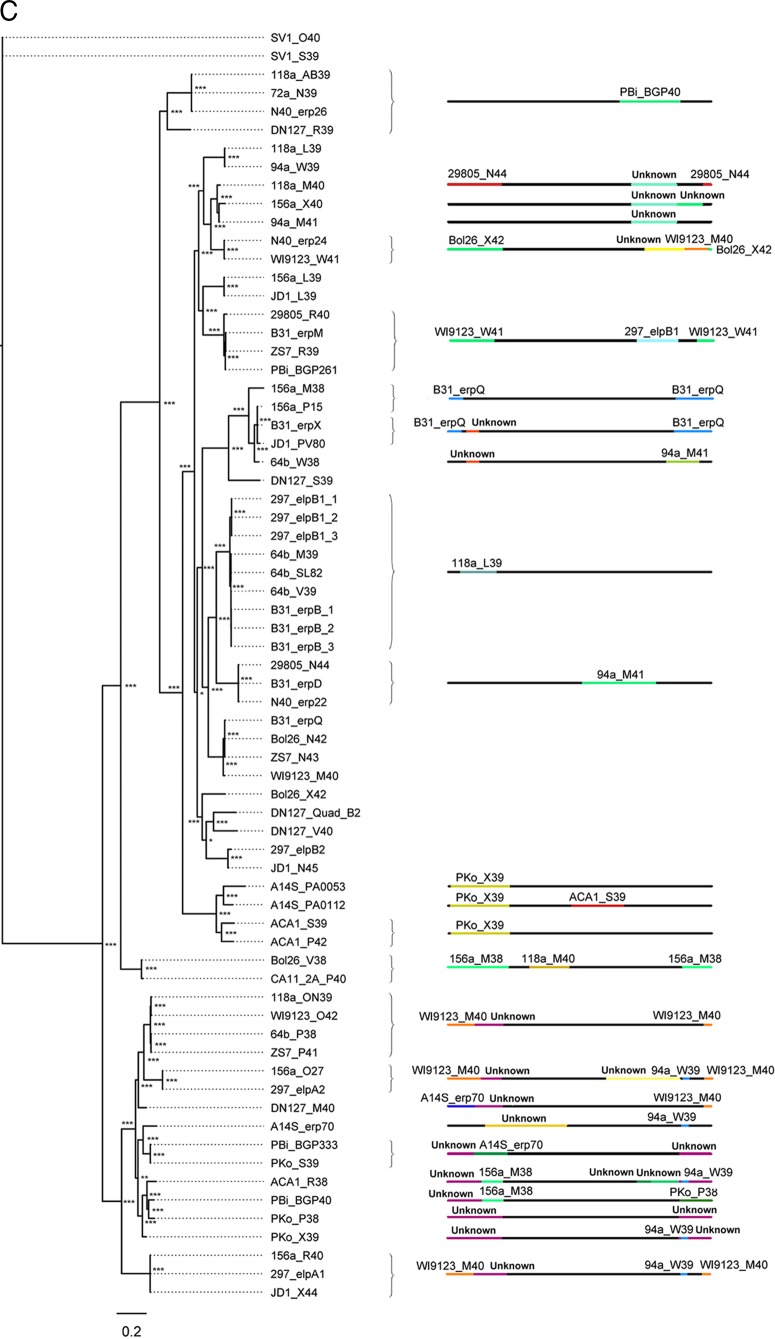
Fig 4.
Sequence diversity is generated by intragenic recombination. Evidence of intragenic recombination was apparent in the majority of analyzed alleles from small-, medium, and large-sized erp sequences (A, B, and C, respectively). The locations of recombination events are visualized by the changes in colors along the line representing the erp sequence; the identity of the donor erp allele is denoted above the colored line. For example, erp sequences 118a_AB39, 72a_N39, N40_erp26, and DN127_R39 (C, top) all contain a segment homologous to the erp sequence 94a_M41 near the 3′ end of the protein. Despite the diversity generated by recombination, identical alleles are present in multiple strains in all three size classes of erp sequences. All described recombination events were supported by at least three independent algorithms (see Table S1 in the supplemental material). Recombination events between alleles of different size classes could not be detected by the methods used here. The sequences of erp alleles carried by strains BL206 and Sh-2-82 are identical to sequences maintained by strains B31 and 297, respectively, and were omitted from these analyses since they did not provide unique information on genetic diversity (this work and reference 46). Asterisks at tree branches indicate bootstrap support values from 1,000 trials (***, >950; **, >850; *, >750; unmarked, ≤750).
Fig 5.
Alignments of representative erp sequences, illustrating sequence similarities and diversities within each size group. For all panels, bases that share >50% identity are boxed in blue and spaces introduced to maximize alignment are indicated by dashes. (A) Small-sized erp sequences; (B) medium-sized erp sequences; (C) large-sized erp sequences.
Evidence of recombination is common among alleles within each group (Fig. 4 and 5). Indeed, there is strong evidence that the majority of erp alleles are chimeras, comprised of erp DNA segments from multiple phylogenetic branches within each group of erp sizes. That is, a part of the protein-coding region of nearly every erp allele is more similar to a region of an otherwise distantly related allele than it is to alleles with which it is, overall, closely related. The locations of the recombination breakpoints, as well as the donors of the inserted fragments, were identified with high confidence by at least three independent statistical algorithms (Fig. 4; see also Table S1 in the supplemental material).
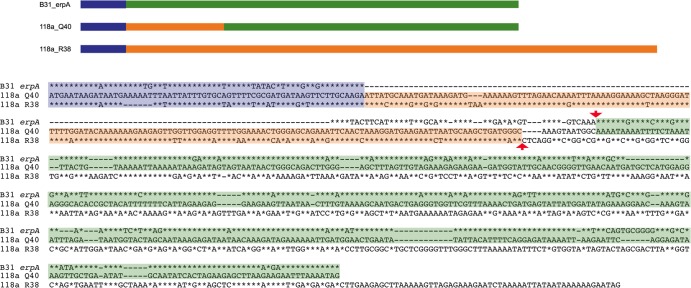
Recombination events between sequences from different erp size groups were also apparent after manual searching for homologous sequences. For example, four identical small erp alleles (strain 118a Q40, strain 72a Q39, CA-11.2A Q39, and 94a Q40) are chimeras of a medium-sized and a small-sized erp gene (Fig. 6).
Fig 6.
Strains 118a, 72a, CA-11.2A, and 94a each contain an identical erp gene that is a chimera of a small- and a medium-sized erp gene. Illustrated are a schematic and a sequence alignment comparing the strain 118a Q40 gene with the strain B31 erpA gene, which is a similar small-sized allele, and the strain 118a R38 gene, which is a similar medium-sized allele. All three open reading frames possess the characteristically conserved erp 5′ sequence, indicated in blue. Sequences of 118a Q40 that are similar to those of erpA or R38 are indicated by green or orange, respectively. In the sequence alignment, sequences of erpA and R38 that are identical to those of Q40 are indicated by asterisks. The approximate region of the recombination event that combined a small- and medium-sized erp to produce the chimera is bordered by red arrowheads.
DISCUSSION
The examined Lyme disease spirochetes naturally contain numerous, distinct cp32 prophages. Our previous analyses detected 12 types of cp32 replication/segregation loci (46, 58). The present analysis of 22 Lyme spirochete strains found only those 12 groups, suggesting that this may represent the total diversity of cp32 incompatibility groups in nature. Despite the limited number of cp32 plasmids in Lyme spirochetes, diversity at the erp loci appears to be almost endless. In addition to simple genetic drift, diversity among erp alleles has been increased by repeated recombination events among sequences of the same and different size groups and from many different, unidentified sources.
The operator regions of all erp loci are highly conserved. This is consistent with previous observations that different erp operons are similarly affected by culture conditions and are expressed during vertebrate infection (18, 19, 20, 21, 23, 24, 25, 26, 28, 59, 78, 82, 88, 89, 90, 91). The region of greatest diversity in erp 5′-noncoding DNAs is the promoter −10 sequence. erp transcription utilizes both σ70 and an alternative sigma, σS, with some operons being primarily dependent upon σS while others are not detectably affected by that RNA polymerase subunit (92, 93, 94). Some erp sequences are poorly transcribed in rpoS mutant B. burgdorferi, a phenomenon that has been linked to differences in promoter −10 sequences (92, 93). There is not an obvious pattern indicating whether an erp promoter contains a −10 element that strongly resembles the apparent consensus σ70-dependent sequence or a divergent −10 element. Identical −10 sequences are located in operons that include all different sizes of erp genes and in mono- and bicistronic arrangements.
All erp sequences share a highly conserved 70- to 80-bp sequence at the 5′ end, encoding the leader polypeptide, lipidation processing site, and the membrane sorting signal (3, 6, 13, 15, 40, 57, 88). Hence, it can be concluded that all erp open reading frames share a single ancestor for their initial 20 to 30 codons. Examined Erp proteins localize to the outer leaflet of the Lyme spirochete's outer membrane (18, 22, 25), and the extensive homology among erp gene 5′ termini suggests that all Erp proteins are surface-exposed outer membrane proteins.
Beyond the sorting signal codons, erp sequences exhibit enormous diversity. As was also noted in a previous study (13), erp sequences were more reliably compared if first separated into groupings of small-, medium-, and large-sized genes. However, the evolution of erp genes is far more complex than the previously proposed three lineages (13). Pairwise comparisons of many sequences within the large- and medium-sized erp groups indicate little to no sequence homology. Those data indicate that, beyond the processing and sorting codons, erp sequences have descended from a very large number of independent evolutionary origins.
The extraordinary diversity of sequences at the erp locus are, in large part, derived from rampant intragenic and intergenic recombination. Many erp sequences contain large segments of unique DNA of unknown origins. It is probable that even more recombination events occurred during the evolution of the analyzed 122 alleles, but since all available computational algorithms require alignments of stretches of similar sequences, we lack the ability to detect them. Thus, a phylogenetic map of erp evolution does not much resemble a tree(s) with linear branches but is more aptly described as a patchwork of criss-crossed paths. Nevertheless, all erp open reading frames include a 5′ end of singular origin.
The complex evolutionary histories of erp alleles, along with their conserved position, regulatory sequences, and 5′-coding sequences, argue against a simple three-named division of erp genes, since none of the proposed named groups includes only alleles with a single ancestor while also excluding unrelated sequences (13). Indeed, the same arguments used to justify splitting erp into three groups with distinct names are equally valid for further splitting of those groups into dozens of smaller groups (13). Given the complex and intertwined evolutionary histories of sequences encoded at the erp locus, along with their regulatory similarities, we suggest retaining a common term to identify sequences at this locus, with the understanding that these sequences evolved along many distinct lineages. Future studies investigating the molecular consequences of sequence mosaics from distinct lineages and the actions of specific DNA segments are essential to infer the natural selection forces acting on this locus.
In addition to a wide range of sequence diversity among erp sequences, there are also numerous examples of sequence conservation. This tendency toward sequence conservation has previously been noted in analyses of four strains from eastern North America (46). Identity among different isolates was again found in the current studies, with the erp loci present in B. burgdorferi strain BL206 being identical to those of type strain B31, each differing from the other by the presence or absence of three cp32s. Additionally, as is readily apparent from the phylogenies illustrated in Fig. 4, identical alleles were found in many divergent strains, such as erpK of New York isolate B31 and open reading frames N38 and S38 of the California isolate CA-11.2A. Identical sequences were also found in bacteria isolated from North America and Europe, such as the erpP allele of B31 and open reading frame N42 in German strain ZS7. Near identity was also detected in erp sequences of different Borrelia genospecies. For example, the medium-sized AC26 locus of B. burgdorferi ZS7 is 99.5% identical to the Q36 locus of B. valaisiana VS116 (4 differences over 801 bp) and 98.6% identical to the A100 locus of unnamed genospecies SV1 (11 bp differences). These latter data suggest that “genospecies” of Lyme Borrelia are not genetically isolated from each other. As also noted previously (46), the present study identified examples of erp sequences moving among different cp32s and probable horizontal transmission of cp32 elements.
The impacts of erp variation on bacterial phenotypes are largely unknown, as functions have yet to be defined for most Erp proteins. However, two examples of loss or gain of function have been identified. The Erp63 protein of B. spielmanii strain TIsar3 is nearly identical to the B. spielmanii strain TIsar2 Erp62 protein. Yet, a single-nucleotide difference encodes a histidine residue in Erp62 and an arginine residue in Erp63 (95). As a result, Erp62 binds both mammalian plasmin and factor H, while Erp63 can bind plasmin but not factor H (95). B. burgdorferi strain B31 ErpX protein binds mammalian laminin, while closely related proteins, such as the B. burgdorferi strain N40 Erp26, lack that activity (42).
The point(s) in the borrelial vertebrate-tick infection cycle at which interbacterial genetic exchange and erp locus mutations occur has yet to be precisely defined. Several studies demonstrated that erp gene sequences remain stable throughout mammalian infection (25, 89, 96, 97, 98). Although one study reported evidence of erp recombination during mammalian infection (98), a more detailed analysis indicated that the reported differences were PCR artifacts and that genetic variation had not occurred (97). Presumably, then, erp recombination takes place during colonization of the tick midgut, a time at which numerous genetically distinct bacteria are found in close proximity to each other. The resulting increased sequence diversity of Erp surface adhesins may thus enhance the probability that transmitted bacteria will form beneficial interactions with host components.
In conclusion, Lyme spirochetes generally contain multiple cp32 replicons. Every complete cp32 replicon carries an erp locus, and erp loci were found only on cp32-related replicons. All erp alleles share a highly conserved 5′ end, while the remainder of each gene has evolved through numerous mutational events, including acquisition of novel DNA sequences. Known functions of Erp proteins include adherence to vertebrate host proteins that may facilitate immune evasion and colonization of host tissues. Given that Erp proteins are exposed on the bacterial outer surface and are produced during vertebrate infection, it is reasonable to predict that all Erp proteins interact with host tissues. Presumably, these interactions enhance bacterial survival, thereby promoting persistence of cp32 elements in Lyme spirochetes.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by U.S. NIH grant R01-AI44254 and an exploratory grant from the University of Kentucky College of Medicine to B. Stevenson, and by U.S. NIH grant R01-AI074825 to S. Casjens.
We thank Darrin Akins and Justin Radolf for constructive discussions on this research, and Alyssa Antonicello, Amy Bowman, Catherine Brissette, Nicholas Brown, Logan Burns, Alicia Chenail, Marc Suchard, and Ashutosh Verma for assistance and helpful comments on the manuscript.
Footnotes
Published ahead of print 26 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00817-13.
REFERENCES
- 1. Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb J-F, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MD, Gocayne J, Weidmann J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fujii C, Cotton MD, Horst K, Roberts K, Hatch B, Smith HO, Venter JC. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580–586 [DOI] [PubMed] [Google Scholar]
- 2. Casjens S, van Vugt R, Tilly K, Rosa PA, Stevenson B. 1997. Homology throughout the multiple 32-kilobase circular plasmids present in Lyme disease spirochetes. J. Bacteriol. 179:217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Casjens S, Palmer N, van Vugt R, Huang WM, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson RJ, Haft D, Hickey E, Gwinn M, White O, Fraser C. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs of an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490–516 [DOI] [PubMed] [Google Scholar]
- 4. Miller JC, Bono JL, Babb K, El-Hage N, Casjens S, Stevenson B. 2000. A second allele of eppA in Borrelia burgdorferi strain B31 is located on the previously undetected circular plasmid cp9-2. J. Bacteriol. 182:6254–6258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Casjens SR, Eggers CH, Schwartz I. 2010. Borrelia genomics: chromosome, plasmids, bacteriophages and genetic variation, p 27–54 In Samuels DS, Radolf JD. (ed), Borrelia: molecular biology, host interaction and pathogenesis. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 6. Stevenson B, Tilly K, Rosa PA. 1996. A family of genes located on four separate 32-kilobase circular plasmids in Borrelia burgdorferi B31. J. Bacteriol. 178:3508–3516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stevenson B, Zückert WR, Akins DR. 2001. Repetition, conservation, and variation: the multiple cp32 plasmids of Borrelia species, p 87–100 In Saier MH, García-Lara J. (ed), The spirochetes: molecular cellular biology. Horizon Press, Oxford, England: [PubMed] [Google Scholar]
- 8. Eggers CH, Samuels DS. 1999. Molecular evidence for a new bacteriophage of Borrelia burgdorferi. J. Bacteriol. 181:7308–7313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eggers CH, Kimmel BJ, Bono JL, Elias AF, Rosa P, Samuels DS. 2001. Transduction by ϕBB-1, a bacteriophage of Borrelia burgdorferi. J. Bacteriol. 183:4771–4778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang H, Marconi RT. 2005. Demonstration of cotranscription and 1-methyl-3-nitroso-nitroguanidine induction of a 30-gene operon of Borrelia burgdorferi: evidence that the 32-kilobase circular plasmids are prophages. J. Bacteriol. 187:7985–7995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chenail AM, Jutras BL, Adams CA, Burns LH, Bowman A, Verma A, Stevenson B. 2012. Borrelia burgdorferi cp32 BpaB modulates expression of the prophage NucP nuclease and SsbP single-stranded DNA-binding protein. J. Bacteriol. 194:4570–4578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stevenson B, Casjens S, Rosa P. 1998. Evidence of past recombination events among the genes encoding the Erp antigens of Borrelia burgdorferi. Microbiology 144:1869–1879 [DOI] [PubMed] [Google Scholar]
- 13. Akins DR, Caimano MJ, Yang X, Cerna F, Norgard MV, Radolf JD. 1999. Molecular and evolutionary analysis of Borrelia burgdorferi 297 circular plasmid-encoded lipoproteins with OspE- and OspF-like leader peptides. Infect. Immun. 67:1526–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Eggers CH, Caimano MJ, Clawson ML, Miller WG, Samuels DS, Radolf JD. 2002. Identification of loci critical for replication and compatibility of a Borrelia burgdorferi cp32 plasmid and use of a cp32-based shuttle vector for the expression of fluorescent reporters in the Lyme disease spirochaete. Mol. Microbiol. 43:281–295 [DOI] [PubMed] [Google Scholar]
- 15. Stevenson B, Bykowski T, Cooley AE, Babb K, Miller JC, Woodman ME, von Lackum K, Riley SP. 2006. The Lyme disease spirochete Erp lipoprotein family: structure, function and regulation of expression, p 354–372 In Cabello FC, Godfrey HP, Hulinska D. (ed), Molecular biology of spirochetes. IOS Press, Amsterdam, Netherlands [Google Scholar]
- 16. Casjens SR, Huang WM, Gilcrease EB, Qiu W, McCaig WD, Luft BJ, Schutzer SE, Fraser CM. 2006. Comparative genomics of Borrelia burgdorferi, p 79–95 In Cabello FC, Godfrey HP, Hulinska D. (ed), Molecular biology of spirochetes. IOS Press, Amsterdam, Netherlands [Google Scholar]
- 17. Porcella SF, Popova TG, Akins DR, Li M, Radolf JD, Norgard MV. 1996. Borrelia burgdorferi supercoiled plasmids encode multi-copy tandem open reading frames and a lipoprotein gene family. J. Bacteriol. 178:3293–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lam TT, Nguyen T-PK, Montgomery RR, Kantor FS, Fikrig E, Flavell RA. 1994. Outer surface proteins E and F of Borrelia burgdorferi, the agent of Lyme disease. Infect. Immun. 62:290–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Akins DR, Porcella SF, Popova TG, Shevchenko D, Baker SI, Li M, Norgard MV, Radolf JD. 1995. Evidence for in vivo but not in vitro expression of a Borrelia burgdorferi outer surface protein F (OspF) homologue. Mol. Microbiol. 18:507–520 [DOI] [PubMed] [Google Scholar]
- 20. Stevenson B, Schwan TG, Rosa PA. 1995. Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 63:4535–4539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Akins DR, Bourell KW, Caimano MJ, Norgard MV, Radolf JD. 1998. A new animal model for studying Lyme disease spirochetes in a mammalian host-adapted state. J. Clin. Invest. 101:2240–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. El-Hage N, Babb K, Carroll JA, Lindstrom N, Fischer ER, Miller JC, Gilmore RD, Mbow ML, Stevenson B. 2001. Surface exposure and protease insensitivity of Borrelia burgdorferi Erp (OspEF-related) lipoproteins. Microbiology 147:821–830 [DOI] [PubMed] [Google Scholar]
- 23. Hefty PS, Jolliff SE, Caimano MJ, Wikel SK, Radolf JD, Akins DR. 2001. Regulation of OspE-related, OspF-related, and Elp lipoproteins of Borrelia burgdorferi strain 297 by mammalian host-specific signals. Infect. Immun. 69:3618–3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hefty PS, Brooks CS, Jett AM, White GL, Wikel SK, Kennedy RC, Akins DR. 2002. OspE-related, OspF-related, and Elp lipoproteins are immunogenic in baboons experimentally infected with Borrelia burgdorferi and in human Lyme disease patients. J. Clin. Microbiol. 40:4256–4265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hefty PS, Jolliff SE, Caimano MJ, Wikel SK, Akins DR. 2002. Changes in the temporal and spatial patterns of outer surface lipoprotein expression generate population heterogeneity and antigenic diversity in the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 70:3468–3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miller JC, von Lackum K, Babb K, McAlister JD, Stevenson B. 2003. Temporal analysis of Borrelia burgdorferi Erp protein expression throughout the mammal-tick infectious cycle. Infect. Immun. 71:6943–6952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miller JC, Narayan K, Stevenson B, Pachner AR. 2005. Expression of Borrelia burgdorferi erp genes during infection of non-human primates. Microb. Pathog. 39:27–33 [DOI] [PubMed] [Google Scholar]
- 28. Miller JC, von Lackum K, Woodman ME, Stevenson B. 2006. Detection of Borrelia burgdorferi gene expression during mammalian infection using transcriptional fusions that produce green fluorescent protein. Microb. Pathog. 41:43–47 [DOI] [PubMed] [Google Scholar]
- 29. Hellwage J, Meri T, Heikkilä T, Alitalo A, Panelius J, Lahdenne P, Seppälä IJT, Meri S. 2001. The complement regulatory factor H binds to the surface protein OspE of Borrelia burgdorferi. J. Biol. Chem. 276:8427–8435 [DOI] [PubMed] [Google Scholar]
- 30. Kraiczy P, Skerka C, Brade V, Zipfel PF. 2001. Further characterization of complement regulator-acquiring surface proteins of Borrelia burgdorferi. Infect. Immun. 69:7800–7809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Alitalo A, Meri T, Lankinen H, Seppälä I, Lahdenne P, Hefty PS, Akins D, Meri S. 2002. Complement inhibitor factor H binding to Lyme disease spirochetes is mediated by inducible expression of multiple plasmid-encoded outer surface protein E paralogs. J. Immunol. 169:3847–3853 [DOI] [PubMed] [Google Scholar]
- 32. Stevenson B, El-Hage N, Hines MA, Miller JC, Babb K. 2002. Differential binding of host complement inhibitor factor H by Borrelia burgdorferi Erp surface proteins: a possible mechanism underlying the expansive host range of Lyme disease spirochetes. Infect. Immun. 70:491–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kraiczy P, Hellwage J, Skerka C, Kirschfink M, Brade V, Zipfel PF, Wallich R. 2003. Immune evasion of Borrelia burgdorferi: mapping of a complement inhibitor factor H-binding site of BbCRASP-3, a novel member of the Erp protein family. Eur. J. Immunol. 33:697–707 [DOI] [PubMed] [Google Scholar]
- 34. Metts MS, McDowell JV, Theisen M, Hansen PR, Marconi RT. 2003. Analysis of the OspE determinants involved in binding of factor H and OspE-targeting antibodies elicited during Borrelia burgdorferi infection. Infect. Immun. 71:3587–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Alitalo A, Meri T, Chen T, Lankinen H, Cheng Z-Z, Jokiranta TS, Seppälä IJT, Lahdenne P, Hefty PS, Akins DR, Meri S. 2004. Lysine-dependent multipoint binding of the Borrelia burgdorferi virulence factor outer surface protein E to the C terminus of factor H. J. Immunol. 172:6195–6201 [DOI] [PubMed] [Google Scholar]
- 36. Kraiczy P, Hartmann K, Hellwage J, Skerka C, Brade V, Zipfel PF, Wallich R, Stevenson B. 2004. Immunological characterization of the complement regulator factor H-binding CRASP and Erp proteins of Borrelia burgdorferi. Int. J. Med. Microbiol. 293(Suppl 37):152–157 [DOI] [PubMed] [Google Scholar]
- 37. Hovis KM, Tran E, Sundy CM, Buckles E, McDowell JV, Marconi RT. 2006. Selective binding of Borrelia burgdorferi OspE paralogs to factor H and serum proteins from diverse animals: possible expansion of the role of OspE in Lyme disease pathogenesis. Infect. Immun. 74:1967–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bykowski T, Woodman ME, Cooley AE, Brissette CA, Brade V, Wallich R, Kraiczy P, Stevenson B. 2007. Coordinated expression of Borrelia burgdorferi complement regulator-acquiring surface proteins during the Lyme disease spirochete's mammal-tick infection cycle. Infect. Immun. 75:4227–4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Haupt K, Kraiczy P, Wallich R, Brade V, Skerka C, Zipfel PF. 2007. Binding of human FHR-1 to serum resistant Borrelia burgdorferi is mediated by borrelial complement regulator-acquiring surface proteins. J. Infect. Dis. 196:124–133 [DOI] [PubMed] [Google Scholar]
- 40. Brissette CA, Cooley AE, Burns LH, Riley SP, Verma A, Woodman ME, Bykowski T, Stevenson B. 2008. Lyme borreliosis spirochete Erp proteins, their known host ligands, and potential roles in mammalian infection. Int. J. Med. Microbiol. 298(Suppl 1):257–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brissette CA, Haupt K, Barthel D, Cooley AE, Bowman A, Skerka C, Wallich R, Zipfel PF, Kraiczy P, Stevenson B. 2009. Borrelia burgdorferi infection-associated surface proteins ErpP, ErpA, and ErpC bind human plasminogen. Infect. Immun. 77:300–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brissette CA, Verma A, Bowman A, Cooley AE, Stevenson B. 2009. The Borrelia burgdorferi outer-surface protein ErpX binds mammalian laminin. Microbiology 155:863–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Siegel C, Hallström T, Skerka C, Eberhardt H, Uzonyi B, Beckhaus T, Karas M, Wallich R, Stevenson B, Zipfel PF, Kraiczy P. 2010. Complement factor H-related proteins CFHR2 and CFHR5 represent novel ligands for the infection-associated CRASP proteins of Borrelia burgdorferi. PLoS One 5:e13519. 10.1371/journal.pone.0013519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kenedy MR, Akins DR. 2011. The OspE-related proteins inhibit complement deposition and enhance serum resistance of Borrelia burgdorferi, the Lyme disease agent. Infect. Immun. 79:1451–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stevenson B, Porcella SF, Oie KL, Fitzpatrick CA, Raffel SJ, Lubke L, Schrumpf ME, Schwan TG. 2000. The relapsing fever spirochete Borrelia hermsii contains multiple, antigen-encoding circular plasmids that are homologous to the cp32 plasmids of Lyme disease spirochetes. Infect. Immun. 68:3900–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stevenson B, Miller JC. 2003. Intra- and interbacterial genetic exchange of Lyme disease spirochete erp genes generates sequence identity amidst diversity. J. Mol. Evol. 57:309–324 [DOI] [PubMed] [Google Scholar]
- 47. Qiu WG, Schutzer SE, Bruno JF, Attie O, Xu Y, Dunn JJ, Casjens SR, Luft BJ. 2004. Genetic exchange and plasmid transfers in Borrelia burgdorferi sensu stricto revealed by three-way genome comparisons and multilocus sequence typing. Proc. Natl. Acad. Sci. U. S. A. 101:14150–14155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wywial E, Haven J, Casjens SR, Hernandez YA, Singh S, Mongodin EF, Fraser-Liggett CM, Luft BJ, Schutzer SE, Qiu WG. 2009. Fast, adaptive evolution at a bacterial host-resistance locus: the PFam54 gene array in Borrelia burgdorferi. Gene 445:26–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Casjens SR, Fraser-Liggett CM, Mongodin EF, Qiu WG, Dunn JJ, Luft BJ, Schutzer SE. 2011. Whole genome sequence of an unusual Borrelia burgdorferi sensu lato isolate. J. Bacteriol. 193:1489–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Casjens SR, Mongodin EF, Qiu WG, Dunn JJ, Luft BJ, Fraser-Liggett CM, Schutzer SE. 2011. Whole-genome sequences of two Borrelia afzelii and two Borrelia garinii Lyme disease agent isolates. J. Bacteriol. 193:6995–6996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schutzer SE, Fraser-Liggett CM, Casjens SR, Qiu WG, Dunn JJ, Mongodin EF, Luft BJ. 2011. Whole genome sequences of thirteen isolates of Borrelia burgdorferi. J. Bacteriol. 193:1018–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Casjens SR, Mongodin EF, Qiu WG, Luft BJ, Schutzer SE, Gilcrease EB, Huang WM, Vujadinovic M, Aron JK, Vargas LC, Freeman S, Radune D, Weidman JF, Dimitrov GI, Khouri HM, Sosa JE, Halpin RA, Dunn JJ, Fraser CM. 2012. Genome stability of Lyme disease spirochetes: comparative genomics of Borrelia burgdorferi plasmids. PLoS One 7:e33280. 10.1371/journal.pone.0033280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schutzer SE, Fraser-Liggett CM, Qiu WG, Kraiczy P, Mongodin EF, Dunn JJ, Luft BJ, Casjens SR. 2012. Whole-genome sequences of Borrelia bissettii, Borrelia valaisiana, and Borrelia spielmanii. J. Bacteriol. 194:545–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Morlon H, Kemps BD, Plotkin JB, Brisson D. 2012. Explosive radiation of a bacterial species group. Evolution 66:2577–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang G, Ojaimi C, Iyer R, Saksenberg V, McClain SA, Wormser GP, Schwartz IA. 2001. Impact of genotypic variation of Borrelia burgdorferi sensu stricto on kinetics of dissemination and severity of disease in C3H/HeJ mice. Infect. Immun. 69:4303–4312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ojaimi C, Mulay V, Liveris D, Iyer R, Schwartz I. 2005. Comparative transcriptional profiling of Borrelia burgdorferi clinical isolates differing in capacities for hematogenous dissemination. Infect. Immun. 73:6791–6802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Marconi RT, Sung SY, Hughes CAN, Carlyon JA. 1996. Molecular and evolutionary analyses of a variable series of genes in Borrelia burgdorferi that are related to ospE and ospF, constitute a gene family, and share a common upstream homology box. J. Bacteriol. 178:5615–5626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Miller JC, Stevenson B. 2003. Immunological and genetic characterization of Borrelia burgdorferi BapA and EppA proteins. Microbiology 149:1113–1125 [DOI] [PubMed] [Google Scholar]
- 59. Wallich R, Brenner C, Kramer MD, Simon MM. 1995. Molecular cloning and immunological characterization of a novel linear-plasmid-encoded gene, pG, of Borrelia burgdorferi expressed only in vivo. Infect. Immun. 63:3327–3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33:511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Suyama M, Torrents D, Bork P. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34:W609–W612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403–405 [DOI] [PubMed] [Google Scholar]
- 63. Needleman SB, Wunsch CD. 1970. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48:443–453 [DOI] [PubMed] [Google Scholar]
- 64. Gonnet GH, Cohen MA, Benner SA. 1992. Exhaustive matching of the entire protein sequence database. Science 256:1443–1445 [DOI] [PubMed] [Google Scholar]
- 65. Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. 2010. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26:2462–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Martin D, Rybicki E. 2000. RDP: detection of recombination amongst aligned sequences. Bioinformatics 16:562–563 [DOI] [PubMed] [Google Scholar]
- 67. Padidam M, Sawyer S, Fauquet CM. 1999. Possible emergence of new geminiviruses by frequent recombination. Virology 265:218–225 [DOI] [PubMed] [Google Scholar]
- 68. Posada D, Crandall KA. 2001. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc. Natl. Acad. Sci. U. S. A. 98:13757–13762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Smith JM. 1992. Analyzing the mosaic structure of genes. J. Mol. Evol. 35:126–129 [DOI] [PubMed] [Google Scholar]
- 70. Salminen M, Carr J, Burke D, McCutchan F. 1995. Identification of breakpoints in intergenotypic recombinants of HIV-1 by bootscanning. AIDS Res. Hum. Retroviruses 11:1423–1425 [DOI] [PubMed] [Google Scholar]
- 71. Martin DP, Posada D, Crandall KA, Williamson C. 2005. A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res. Hum. Retroviruses 21:98–102 [DOI] [PubMed] [Google Scholar]
- 72. Gibbs MJ, Armstrong JS, Gibbs AJ. 2000. Sister-scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 16:573–582 [DOI] [PubMed] [Google Scholar]
- 73. Boni MF, Posada D, Feldman MW. 2007. An exact nonparametric method for inferring mosaic structure in sequence triplets. Genetics 176:1035–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sadziene A, Wilske B, Ferdows MS, Barbour AG. 1993. The cryptic ospC gene of Borrelia burgdorferi B31 is located on a circular plasmid. Infect. Immun. 61:2192–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Stevenson B, Casjens S, van Vugt R, Porcella SF, Tilly K, Bono JL, Rosa P. 1997. Characterization of cp18, a naturally truncated member of the cp32 family of Borrelia burgdorferi plasmids. J. Bacteriol. 179:4285–4291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Caimano MJ, Yang X, Popova TG, Clawson ML, Akins DR, Norgard MV, Radolf JD. 2000. Molecular and evolutionary characterization of the cp32/18 family of supercoiled plasmids in Borrelia burgdorferi 297. Infect. Immun. 68:1574–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zückert WR, Meyer J. 1996. Circular and linear plasmids of Lyme disease spirochetes have extensive homology: characterization of a repeated DNA element. J. Bacteriol. 178:2287–2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Babb K, McAlister JD, Miller JC, Stevenson B. 2004. Molecular characterization of Borrelia burgdorferi erp promoter/operator elements. J. Bacteriol. 186:2745–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Babb K, Bykowski T, Riley SP, Miller MC, DeMoll E, Stevenson B. 2006. Borrelia burgdorferi EbfC, a novel, chromosomal encoded protein, binds specific DNA sequences adjacent to erp loci on the spirochete's resident cp32 prophages. J. Bacteriol. 188:4331–4339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Riley SP, Bykowski TT, Cooley AE, Burns LH, Babb K, Brissette CA, Bowman A, Rotondi M, Miller MC, DeMoll E, Lim K, Fried MG, Stevenson B. 2009. Borrelia burgdorferi EbfC defines a newly-identified, widespread family of bacterial DNA-binding proteins. Nucleic Acids Res. 37:1973–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Burns LH, Adams CA, Riley SP, Jutras BL, Bowman A, Chenail AM, Cooley AE, Haselhorst LA, Moore AM, Babb K, Fried MG, Stevenson B. 2010. BpaB, a novel protein encoded by the Lyme disease spirochete's cp32 prophages, binds to erp operator 2 DNA. Nucleic Acids Res. 38:5443–5455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Jutras BL, Verma A, Adams CA, Brissette CA, Burns LH, Whetstine CR, Bowman A, Chenail AM, Zückert WR, Stevenson B. 2012. BpaB and EbfC DNA-binding proteins regulate production of the Lyme disease spirochete's infection-associated Erp surface proteins. J. Bacteriol. 194:778–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Jutras BL, Bowman A, Brissette CA, Adams CA, Verma A, Chenail AM, Stevenson B. 2012. EbfC (YbaB) is a new type of bacterial nucleoid-associated protein and a global regulator of gene expression in the Lyme disease spirochete. J. Bacteriol. 194:3395–3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Das S, Barthold SW, Stocker Giles S, Montgomery RR, Telford SR, Fikrig E. 1997. Temporal pattern of Borrelia burgdorferi p21 expression in ticks and the mammalian host. J. Clin. Invest. 99:987–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Schulze RJ, Zückert WR. 2006. Borrelia burgdorferi lipoproteins are secreted to the outer surface by default. Mol. Microbiol. 59:1473–1484 [DOI] [PubMed] [Google Scholar]
- 86. Setubal JC, Reis M, Matsunaga J, Haake DA. 2006. Lipoprotein computational prediction in spirochaetal genomes. Microbiology 152:113–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kumru OS, Schulze RJ, Rodnin MV, Ladokhin AS, Zückert WR. 2011. Surface localization determinants of Borrelia OspC/Vsp family lipoproteins. J. Bacteriol. 193:2814–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Stevenson B, Bono JL, Schwan TG, Rosa P. 1998. Borrelia burgdorferi Erp proteins are immunogenic in mammals infected by tick bite, and their synthesis is inducible in cultured bacteria. Infect. Immun. 66:2648–2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. McDowell JV, Sung SY, Price G, Marconi RT. 2001. Demonstration of the genetic stability and temporal expression of select members of the Lyme disease spirochete OspF protein family during infection in mice. Infect. Immun. 69:4831–4838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. El-Hage N, Stevenson B. 2002. Simultaneous coexpression of Borrelia burgdorferi Erp proteins occurs through a specific, erp locus-directed regulatory mechanism. J. Bacteriol. 184:4536–4543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Jutras BL, Chenail AM, Stevenson B. 2013. Changes in bacterial growth rate govern expression of the Borrelia burgdorferi OspC and Erp infection-associated surface proteins. J. Bacteriol. 195:757–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Eggers CH, Caimano MJ, Radolf JD. 2006. Sigma factor selectivity in Borrelia burgdorferi: RpoS recognition of the ospE/ospF/elp promoters is dependent on the sequence of the −10 region. Mol. Microbiol. 59:1859–1875 [DOI] [PubMed] [Google Scholar]
- 93. Eggers CH, Caimano MJ, Radolf JD. 2004. Analysis of promoter elements involved in the transcription initiation of RpoS-dependent Borrelia burgdorferi genes. J. Bacteriol. 186:7390–7402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Caimano MJ, Iyer R, Eggers CH, Gonzalez C, Morton EA, Gilbert MA, Schwartz I, Radolf JD. 2007. Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Mol. Microbiol. 65:1193–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Seling A, Siegel C, Fingerle V, Jutras BL, Brissette CA, Skerka C, Wallich R, Zipfel PF, Stevenson B, Kraiczy P. 2010. Functional characterization of Borrelia spielmanii outer surface proteins that interact with distinct members of the human factor H protein family and with plasminogen. Infect. Immun. 78:39–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. El-Hage N, Lieto LD, Stevenson B. 1999. Stability of erp loci during Borrelia burgdorferi infection: recombination is not required for chronic infection of immunocompetent mice. Infect. Immun. 67:3146–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Stevenson B. 2002. Borrelia burgdorferi erp (ospE-related) gene sequences remain stable during mammalian infection. Infect. Immun. 70:5307–5311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Sung SY, McDowell JV, Carlyon JA, Marconi RT. 2000. Mutation and recombination in the upstream homology box-flanked ospE-related genes of the Lyme disease spirochetes result in the development of new antigenic variants during infection. Infect. Immun. 68:1319–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.