Abstract
Psychrobacter arcticus strain 273-4, an isolate from a Siberian permafrost core, is capable of forming biofilms when grown in minimal medium under laboratory conditions. Biofilms form at 4 to 22°C when acetate is supplied as the lone carbon source and with 1 to 7% sea salt. P. arcticus is also capable of colonizing quartz sand. Transposon mutagenesis identified a gene important for biofilm formation by P. arcticus. Four transposon mutants were mapped to a 20.1-kbp gene, which is predicted to encode a protein of 6,715 amino acids (Psyc_1601). We refer to this open reading frame as cat1, for cold attachment gene 1. The cat1 mutants are unable to form biofilms at levels equivalent to that of the wild type, and there is no impact on the planktonic growth characteristics of the strains, indicating a specific role in biofilm formation. Through time course studies of the static microtiter plate assay, we determined that cat1 mutants are unable to form biofilms equivalent to that of the wild type under all conditions tested. In flow cell experiments, cat1 mutants initially are unable to attach to the surface. Over time, however, they form microcolonies, an architecture very different from that produced by wild-type biofilms. Our results demonstrate that Cat1 is involved in the initial stages of bacterial attachment to surfaces.
INTRODUCTION
Psychrobacter arcticus 273-4 is an isolate from a Siberian permafrost core that has been dated at 20,000 to 40,000 years old (1). The Siberian permafrost is considered an extreme environment due to a constant temperature of approximately −10°C, low abundance of unfrozen water and nutrients, and prolonged exposure to radiation from soil minerals (2, 3). However, the permafrost is not considered a dormant environment, and it is filled with metabolically active microbes (4). Results from a transcriptome analysis of P. arcticus cultures, grown from −6 to 22°C, suggest that this bacterium employs a resource efficiency strategy when grown under permafrost-like conditions of limited nutrients, low temperature, and low water availability (55).
The genus Psychrobacter, a member of the class Gammaproteobacteria, is predominantly isolated from cold and/or saline environments, such as Arctic permafrost, Antarctic ice pack, estuaries, and marine fish, including Korean fermented seafood (4–10). The frequent isolation of Psychrobacter species from these environments suggests that members of the genus have adapted to low temperatures and high-salt conditions, both of which share low water activity. P. arcticus can grow in the laboratory from −10 to 28°C, with an optimum growth temperature of 22°C, and in salinity ranging from 10 mM to 1.3 M NaCl (5). Compared to mesophiles, the amino acid usage in P. arcticus has significantly shifted to include amino acids known to increase protein flexibility at low temperatures (11). When P. arcticus is grown at low temperatures, a decrease in fatty acid saturation occurs, which is predicted to allow membranes to remain fluid (12).
Our study focuses on the ability of P. arcticus to attach to surfaces and form a biofilm. Biofilm formation may facilitate hydration in P. arcticus's natural environment, the Siberian permafrost, where the only unfrozen water is found as thin films surrounding soil, organic, and mineral particles (13–15). Scanning microscopy of Siberian permafrost soils has revealed tight associations between bacteria and soil particles (16, 17). Based on these observations, we hypothesize that P. arcticus is able to fasten to surfaces near unfrozen water in the permafrost and that this attachment will provide the bacterium with access to unfrozen water, ultimately increasing survivability in the permafrost environment.
Bacteria in nature are most often found associated with surfaces in communities known as biofilms and are not in the planktonic state (18, 19). Biofilm formation is a developmental process initiated when bacteria attach to a surface, followed by clonal growth, attachment of free-swimming microorganisms, and the production of an extracellular matrix (18). A mature biofilm is characterized by the production of exopolysaccharides, and bacteria residing in biofilms have been shown to have increased antimicrobial resistance and protection from osmotic shock, desiccation, and UV radiation (20–22). Microbes inhabiting extreme environments, such as hot springs and sabkha systems of the Red Sea, can be found as biofilms; the ability of the microorganism to form biofilms is believed to aid their adaptation and ultimate survival in the environment (23, 24).
In this study, we investigated the ability of P. arcticus to form biofilms under a variety of environmental conditions and identified a gene necessary for attachment. P. arcticus is capable of attaching to plastic surfaces from 4 to 22°C when grown in a defined medium supplemented with acetate and sea salt, but not when it is grown in a more nutrient-rich medium (half-strength tryptic soy broth [TSB] and LB). A large adhesin, which we refer to as Cat1, for cold attachment protein 1, plays a key role in the ability of P. arcticus to attach to plastic and sand surfaces.
MATERIALS AND METHODS
Bacterial growth conditions.
P. arcticus was grown in LB (Fisher), marine broth (MB) (30 g sea salt, 5 g peptone, and 1 g yeast extract per liter), half-strength Bacto TSB (Becton, Dickinson, and Company), or mineral medium with 1, 3, or 5% sea salt (MM; 50 mM morpholinepropanesulfonic acid [MOPS], 20 mM acetate, 5 mM NH4Cl, 1 mM K2HPO4, 1× Wolfe's mineral solution [55, 56], and 1× vitamin supplement [ATCC]). Escherichia coli WM3064 {ΔdapA1341::[erm pir(Wt)]} and E. coli EC100D pir+ (Epicentre) were grown in LB with the amendment of 100 μg diaminopimelic acid (DAP) ml−1 as appropriate. When required, kanamycin (Kn) was added to cultures at a final concentration of 50 μg/ml. P. arcticus cultures were incubated at 0, 4, 10, or 22°C, as indicated per experiment, and E. coli cultures were grown at 37°C. For flow cell studies, MM was modified with 10 mM acetate and 0.5% sea salt.
Transposon mutagenesis.
Transposon mutants were generated by utilizing a Tn5 transposon carried on plasmid pRL27 (25), with the following modifications. The recipient (P. arcticus) and donor (E. coli WM3064/pRL27) were grown to late log phase in LB and then diluted 1:10 into fresh medium until an absorbance at 600 nm of 0.8 was reached. One milliliter of recipient and 0.5 ml of donor were pelleted, washed with 500 μl of LB, pelleted, and resuspended together in 30 μl of LB. The cell suspension was spotted onto an LB plate supplemented with DAP and incubated at 22°C for 48 h. Cells were scraped from the plate and resuspended in 1 ml of LB, 100 μl was plated onto LB plates with Kn, and then the plates were incubated at 22°C for 48 to 72 h. The transposon mutants were screened for biofilm formation in the microtiter dish assay described below.
Biofilm assays.
The microtiter dish assay (26, 27) was employed to measure biofilm formation by P. arcticus under static conditions with the following modifications. Cultures were grown in LB, MB, half-strength TSB, or MM to saturation before a 1:50 dilution into fresh medium with 150 μl dispensed per well into a 96-well polyvinyl chloride (PVC) plate or a tissue culture-treated polystyrene plate. Plates were incubated at specified temperatures for times ranging from 12 to 120 h, the absorbance of cells was determined at 600 nm, and the plate was rinsed three times to remove unattached cells. To visualize and quantify attached cells, 175 μl 0.1% crystal violet was added to each well for 10 min and rinsed six times. The stain was solubilized by utilizing a solution containing 20% methanol and 10% acetic acid, and the absorbance was determined at 550 nm. Three biological replicates, each with four technical replicates, were averaged for each assay, and error bars represent the standard deviations.
Biofilms were studied in a flow system utilizing Stovall three-channel flow cells (Stovall Life Sciences Incorporated) with modified MM, as described above, as the growth medium. Overnight P. arcticus cultures were subcultured to a 1:5 dilution with fresh medium, and 0.5 ml was used to inoculate each channel. The medium flow was stopped for 1 h following inoculation. Medium was then pumped through the flow cell at a rate of 0.2 mm/s, and flow cells were incubated at 21°C for 3 days.
Sand attachment assay.
P. arcticus cultures were grown to stationary phase in MM with 1% sea salt and subcultured at a 1:50 dilution into fresh medium. Approximately 0.5 g of fine-grain quartz sand (Fisher) was added to the bottom of the well in a 24-well plate, and 200 μl of bacterial suspension was added. The plates were placed on a shaker, on low speed, at 21°C for 24 h. To visualize attached cells, a sample of sand was washed three times with 500 μl MM, and 250 μl of a 1:1,000 dilution of 3.34 mM Syto 9 (Invitrogen) was applied to the sand for 15 min. The sand was washed three times with MM to remove excess stain, and the sand was observed with an inverted fluorescence microscope. To quantify bacteria attached to sand, 0.1 g of sand was washed three times with 500 μl of sterile medium to remove any unattached bacteria, and then 50 μl of MM was added. Bacteria were then dislodged by an alternating series of vortexing and sonication as previously described (28). The total CFU/g of bacteria removed from the sand was determined by dilution plating on LB or LB Kn medium.
Imaging.
Phase-contrast and epifluorescence microscopy was performed using an IX71 inverted microscope (Olympus), and images were collected utilizing a DP72 (Olympus) camera.
qRT-PCR analysis.
The wild-type strain was grown in MM with 1% sea salt or LB, subcultured, and grown to mid-exponential phase or stationary phase at 4 or 22°C. Two milliliters of culture was immediately mixed with 4 ml of RNAprotect bacterial reagent (Qiagen) and spun down by following the manufacturer's instructions. RNA was isolated using the Qiagen RNeasy minikit according to the manufacturer's instructions, with the modification that lysozyme digestion was performed for 20 min and then an on-column DNase digestion. A second DNase treatment was carried out with 5 U DNase I (Epicentre) for 20 min at 37°C. The SurePrepRNA kit (Fisher Scientific) was used to purify and concentrate the RNA according to the manufacturer's instructions. One microgram of RNA was used for each first-strand cDNA synthesis, utilizing random hexamer primers and the Superscript III first-strand synthesis system (Invitrogen). Primers were designed to the constitutively expressed rplU gene as a normalization control (F1, 5′-AAATCCGTATCGTCAAGCACAA; R1, 5′-GCGGTGACCTTGCTCTTTG). For the detection of cat1, forward and reverse primers were 5′-TGCTACAGAGACTGCTGGCTTT and 5′-CAAGTCCTACGCCTGCAAGAC, respectively. A series of dilutions of cDNA was used in quantitative RT-PCR (qRT-PCR) with the Fast SYBR green master mix (Life Technologies), and reactions were performed in triplicate and repeated three to five times. Reactions were carried out in the Applied Biosystems StepOnePlus real-time PCR system (Life Technologies) according to the manufacturer's instructions. The efficiencies of both the target and reference reactions are approximately equal, allowing for analysis of comparative cycle threshold (CT) results. Relative quantitation of gene expression was analyzed with the comparative CT method (Applied Biosystems) and with a two-sample t test or with a one-way analysis of variance (ANOVA) with a Tukey pairwise comparison (Minitab 16 Statistical Software).
Cloning of cat1 transposon insertions.
To determine the DNA sequence flanking the transposon, one-step cloning, plasmid isolation, and sequencing reactions were performed as described previously (25), with the following modifications. Genomic DNA was digested with EcoRV, which does not cut within the transposon, followed by ligation with T4 DNA ligase. The ligated DNA was electroporated into E. coli EC100D pir+ (Epicentre), and transformants were selected on LB plates supplemented with Kn. The transposon-carrying plasmids were isolated and sequenced with the primer tpnRL17-1 (5′-AACAAGCCAGGGATGTAACG) at Iowa State University's DNA sequencing facility (25). The DNA sequence obtained was compared to the P. arcticus genome using the BLASTN program (29).
RESULTS
Characterization of biofilm formation.
To investigate the ability of wild-type P. arcticus to attach to surfaces, we employed the static PVC microtiter plate assay. P. arcticus grew to high cell density in a variety of media; however, only growth in mineral medium (MM), with acetate as the carbon source and 1% sea salts, promoted cell attachment to surfaces (Fig. 1). Measurable biofilm formation was observed after 10 h of incubation, which increased significantly through the first 48 h, when incubated at 22°C in the acetate medium (Fig. 2). P. arcticus was able to form considerable biofilms at 4 and 10°C, although the initial increase in biofilm formation was delayed, occurring between 3 to 5 days and 1 to 3 days, respectively (Fig. 2). Biofilms were observed for wild-type cells grown in MM with up to 7% sea salts, consistent with the high salinity tolerance previously observed for this bacterium (data not shown) (5).
Fig 1.
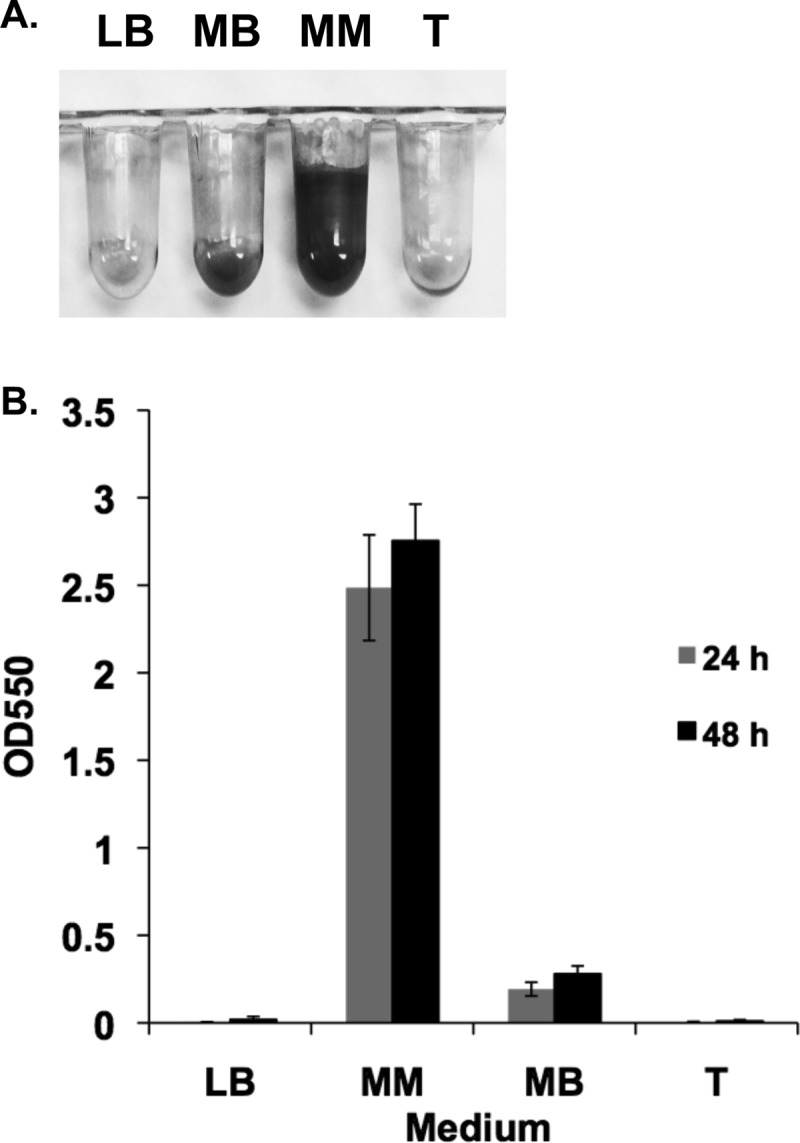
Biofilm formation by P. arcticus. Biofilm formation by wild-type P. arcticus was measured under static conditions in LB, half-strength tryptic soy broth (T), minimal defined medium (MM) supplemented with 20 mM acetate and 1% sea salt, and marine broth (MB). MM supports biofilm development. (A) Visualization of attached cells, which have been stained with crystal violet, following 48 h of incubation at 22°C. (B) Quantification of biofilm formation represented by crystal violet absorbance at 550 nm. Error bars represent standard deviations. n = 6.
Fig 2.
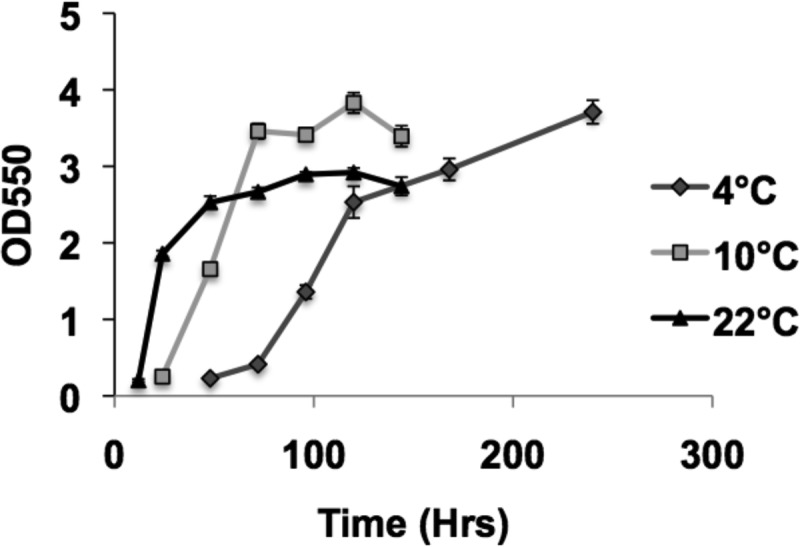
Kinetics of biofilm formation by wild-type P. arcticus grown in MM with 1% sea salt and acetate at three different temperatures. Quantification of biofilm formation is represented by crystal violet absorbance at 550 nm. Error bars represent standard deviations. n = 4.
To determine if P. arcticus is also able to attach to hydrophilic surfaces, we tested the ability of the wild-type strain to form biofilms when grown in MM and inoculated into tissue culture-treated polystyrene plates that were incubated up to 4 days at 22°C. P. arcticus only formed weak biofilms on this hydrophilic surface, with crystal violet absorbance readings at an optical density of 550 nm (OD550) reaching just 0.23 by day 4 (data not shown).
Isolation of biofilm formation mutants.
A transposon mutant library of P. arcticus containing 4,500 mutants was generated utilizing a Tn5 transposon, and it was screened in the microtiter plate assay to identify mutants that can grow as well as the wild-type strain but are unable to attach to the surface (27). Fifty-five mutants had growth rates similar to that of the wild-type strain but were unable to form a biofilm. Four of these mutants, with the most robust biofilm-deficient phenotypes, were further characterized in biofilm formation studies.
Sequence analysis of Cat1.
We cloned and sequenced the genomic region containing the transposon of the four mutants. All four transposons mapped to different locations within one 20,145-bp open reading frame, which encodes protein Psyc_1601, a predicted hypothetical protein (Fig. 3). This open reading frame had a distinctly high GC content, 56.9%, compared to the rest of the P. arcticus genome, which was 42.8% GC (11). We refer to this gene as cat1, for cold attachment gene 1, based on the role of the encoded protein in attachment to surfaces over a low temperature range. The first 10-bp stretches of cat1 sequence at each transposon junction, represented 5′ to 3′, are CACCAAGTGT, CTTTGCCGTG, CCAGTAGCTA, and TAGTAGGTGT. A gene predicted to encode an outer membrane protein (Psyc_1602) is located upstream of cat1, with 64 bp separating the two genes. Based on results of the sequence analysis programs FGENESB and BPROM (SoftBerry), the Psyc_1602-encoding gene and cat1 reside in an operon. Located 379 bp downstream of cat1 is a separate operon predicted to encode an ABC transporter (Psyc_1600, Psyc_1599, and Psyc_1598) (Fig. 3A).
Fig 3.
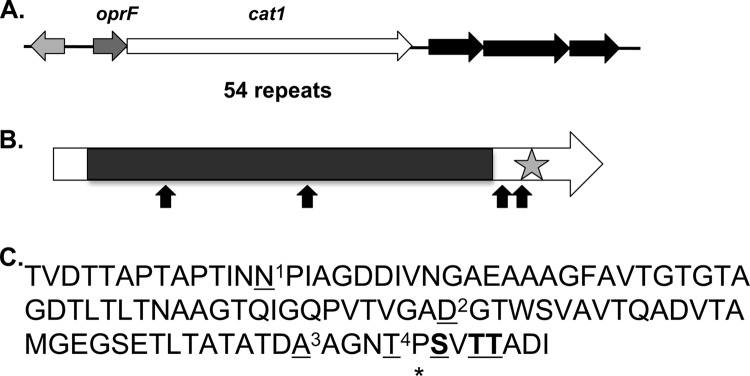
Analysis of the cat1 chromosomal region and the Cat1 protein sequence. (A) Organization of the chromosomal region surrounding cat1. The light gray gene encodes a hypothetical protein which is 799 bp upstream of a proposed two-gene operon containing an outer membrane protein-encoding gene, oprF, and cat1. The black arrows represent a predicted ABC transporter (Psyc_1600, Psyc_1599, and Psyc_1598). (B) Cat1 contains 54 repeats that constitute more than 80% of the protein. The star indicates a predicted calcium binding domain in the C terminus. The four transposon insertion sites are represented by arrows. (C) The repeat consensus sequence. The underlined amino acids differ in one or more of the repeats, and the boldface amino acids are conserved in 103-aa repeats but differ in 102-aa repeats. N1 is found in 52% of sequences, S is found in 48%, D2 is found in 98% of sequences, N is found in 2%, A3 is found in 94% of the sequences, T is found in 6%, T4 is found in 65% of sequences, A is found in 20%, and I is found in 15%. The asterisk indicates the amino acid found in the 103-aa repeats and is not present in the 102-aa repeats.
The Cat1 protein is 6,715 amino acids (aa) and contains 54 long repeats (11 repeats of 102 aa and 43 repeats of 103 aa), which constitute more than 80% of the total protein (Fig. 3B). The 102- and 103-aa repeats are more than 92% identical (Fig. 3C). Previous sequence analysis of Cat1 has placed it in a family of large adhesins, including Bap from Staphylococcus aureus, which are characterized by threonine-rich domains and either cadherin or calcium binding domains (30). A calcium binding domain is predicted to reside in the C terminus of the protein, based on the presence of two NodO calcium binding motifs flanking a cadherin 2 binding motif (5944-ELLGLTGNDTLT-5945; cadherin domain, 6660-GGVGTDLSD-6668) (http://www.ebi.ac.uk/Tools/printsscan/, http://toolkit.tuebingen.mpg.de/hhpred, and http://myhits.isb-sib.ch/cgi-bin/motif_scan) (31, 32). The conserved repeat domain of Cat1 is 61% identical to a sequence found in a 1,074-aa hypothetical protein from Psychrobacter sp. strain PAMC 21119 (PPAM21_07431) (BLASTP program; blast.ncbi.nlm.nih.gov/). The repeat region of Cat1 is predicted to form beta strands (http://smart.embl-heidelberg.de/) (33). Cat1 is predicted to localize to the outer membrane or be extracellular (PSortb program; http://www.psort.org/psortb/). Protein structural analysis has predicted a transmembrane helix region between amino acids 140 and 162 (http://smart.embl-heidelberg.de/ and http://www.sbg.bio.ic.ac.uk/∼phyre2/html/page.cgi?id=index) (33). Five of the nine large adhesins belonging to the Bap family (AAK38834, AAY28519, SC4140, SPA4077, and Psyc_1601) are predicted to contain a single internal helix domain, and all are predicted to localize to the outer membrane/cell wall or be extracellular. Similar to the other adhesins in this protein family coming from Gram-negative bacteria, Cat1 does not contain the predicted C-terminal secretion motif (GGXGXD) that is found in the LapA family of large adhesins (PSortb program) (28, 30).
Biofilm formation by cat1 mutants under static conditions.
The cat1 mutants are unable to form biofilms equivalent to those of the wild type in the static microtiter plate assay. All four mutants have a growth rate similar to that of the wild type in MM; however, the cells are unable to firmly attach to the surface. We have highlighted the phenotype of the cat1-E mutant strain in this paper, which is representative of phenotypes found for all four cat1 mutants (Fig. 4A). The cat1-E mutant strain is unable to form biofilms equivalent to those of the wild-type strain over the entire time course when grown at 22, 10, and 4°C (Fig. 4B). However, the cat1-E mutant strain is able to increase its surface attachment over time, indicating that although Cat1 is a major surface adhesin, it is not the only adhesin.
Fig 4.
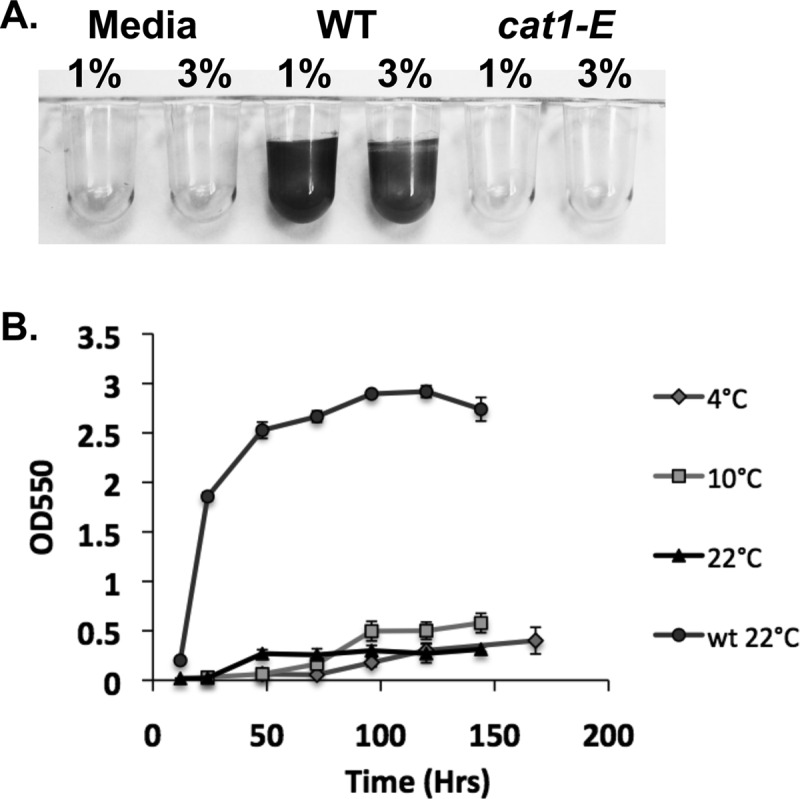
Kinetics of biofilm formation by cat1-E mutant cultures grown in MM at 4, 10, and 22°C. (A) Biofilm formation following 24 h of incubation at 22°C in MM supplemented with 1 or 3% sea salts. (B) Quantification of biofilm formation, represented by crystal violet absorbance at 550 nm. The cat1-E transposon mutant is unable to form a biofilm equivalent to that of the wild type over an extended time frame. Biofilm formation by wild-type P. arcticus at 22°C is represented by the top line with circles. Error bars represent standard deviations. n = 4.
Analysis of cat1 expression.
Based on the ability of the wild-type strain to form a biofilm when grown in MM, but not in LB, and the importance of Cat1 in the initiation of biofilm formation, we hypothesized that cat1 expression would be higher when cells are grown in MM than in LB. RNA was extracted from mid-exponential- and stationary-phase cultures grown at 22°C, because cell attachment to the surface is initiated during these stages of growth. There was no significant difference in cat1 expression relative to rplU expression in wild-type cells grown at 22°C in LB or MM at either mid-exponential or stationary phase (Table 1).
Table 1.
Relative cat1 expression under different growth conditions
| Growth medium, temperature, and phase | ΔCTa |
|---|---|
| MM | |
| Mid-exponential (22°C) | 2.4 ± 0.5A |
| Stationary (22°C) | −0.6 ± 0.2B |
| Stationary (4°C) | −1.5 ± 0.2C |
| LB | |
| Mid-exponential (22°C) | 3.0 ± 0.4A |
| Stationary (22°C) | −0.5 ± 1.3B |
The change in cycle threshold value (ΔCT) is generated by subtracting the rplU CT from the cat1 CT. The average ΔCT is shown, followed by the standard deviation. The experiments were repeated at least 3 times. Statistically different ΔCT values are represented by different letters (P < 0.05).
Due to the large size of the Cat1 protein and cellular demands for its synthesis, we hypothesized that there would be increased expression of cat1 relative to rplU expression when cells were grown to stationary phase rather than mid-exponential phase. In this case, we did find a significant difference in relative cat1 expression between cells grown in LB to mid-exponential versus stationary phase (Table 1). We expanded the studies to include cells grown to stationary phase in MM at 4°C and also found a significant difference in relative cat1 expression between cells grown in MM to mid-exponential phase at 22°C versus stationary phase at 22 or 4°C (Table 1). If the relative amount of cat1 mRNA is set to 1 during growth at 22°C to mid-exponential phase, on average, there would be 8 and 14.9 times more cat1 mRNA found in cells grown to stationary phase at 22 and 4°C, respectively, compared to rplU expression. Thus, growth phase and temperature impacted relative cat1 expression, while the medium conditions tested did not impact cat1 expression relative to rplU expression.
Attachment of P. arcticus to quartz sand.
To test the ability of P. arcticus to attach to an environmental surface, we investigated the ability of wild-type and cat1-E strains to attach to quartz sand. After 24 h of incubation on sand, unattached bacteria were washed away and attached bacteria were stained with Syto 9 for visualization by epifluorescence microscopy (Fig. 5A). Wild-type cells were observed attached on all sides of the sand particles, particularly colonizing cracks and crevices on the surface at high cell density. The attachment of cat1-E cells was more sporadic, with only a few cells observed attached to each sand particle surface and none of the clustering of cells that was observed with the wild type (Fig. 5A). We removed attached bacteria from the sand surface by sonication and vortexing and found 33-fold more wild-type cells attached than cat1-E cells (Fig. 5B).
Fig 5.
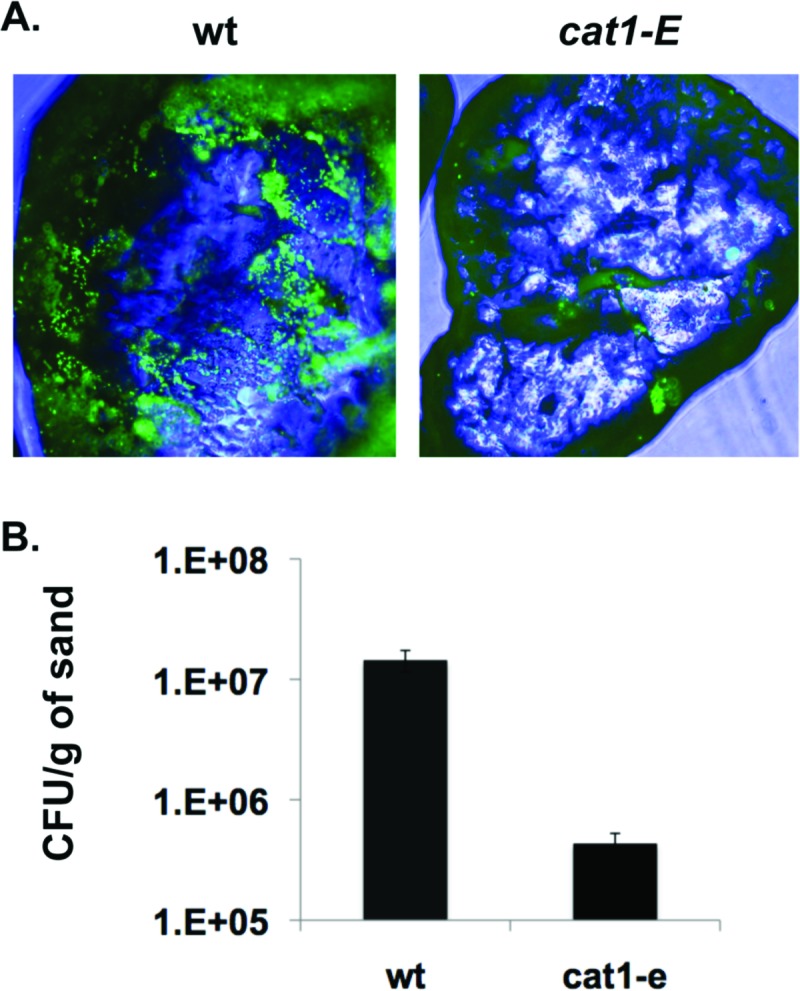
Cat1 is important for attachment to sand. (A) To visualize bacterial attachment to quartz sand at ×100 magnification, wild-type and cat1-E strains were incubated on sand for 24 h, and attached cells were stained with Syto 9. (B) Quantification of bacterial attachment to sand. Error bars represent standard deviations. n = 4.
Monitoring biofilm formation in a flow cell.
To observe mature biofilm structure, wild-type and cat1-E strains were inoculated into a flow cell system. Both the wild-type and cat1-E strains had difficulty colonizing the hydrophilic glass coverslip, but both were able to readily colonize the hydrophobic plastic surface on the back of the flow cell. These results are similar to what was observed in the static plate assay on hydrophobic and hydrophilic plates.
By 11 h, the wild-type cells were beginning to colonize the plastic flow cell surface, forming a monolayer, while few cat1-E cells attached to the surface (Fig. 6). After 1 day of development, wild-type cells formed a dense monolayer, while the cat1-E cells attached, forming small microcolonies on the surface. The wild-type strain maintained the dense layered architecture over the remainder of the experiment, while the cat1-E strain developed large patchy cell clusters throughout the flow cell chamber with increased surface coverage over time. In the flow cell assay, cat1-E cells are unable to initially form strong cell-surface interactions, but over time they form cell-cell interactions and some cell-surface interactions. Only single-cell-layer coverage was observed on the glass coverslip over the entire experimental period for cat1-E and wild-type strains.
Fig 6.
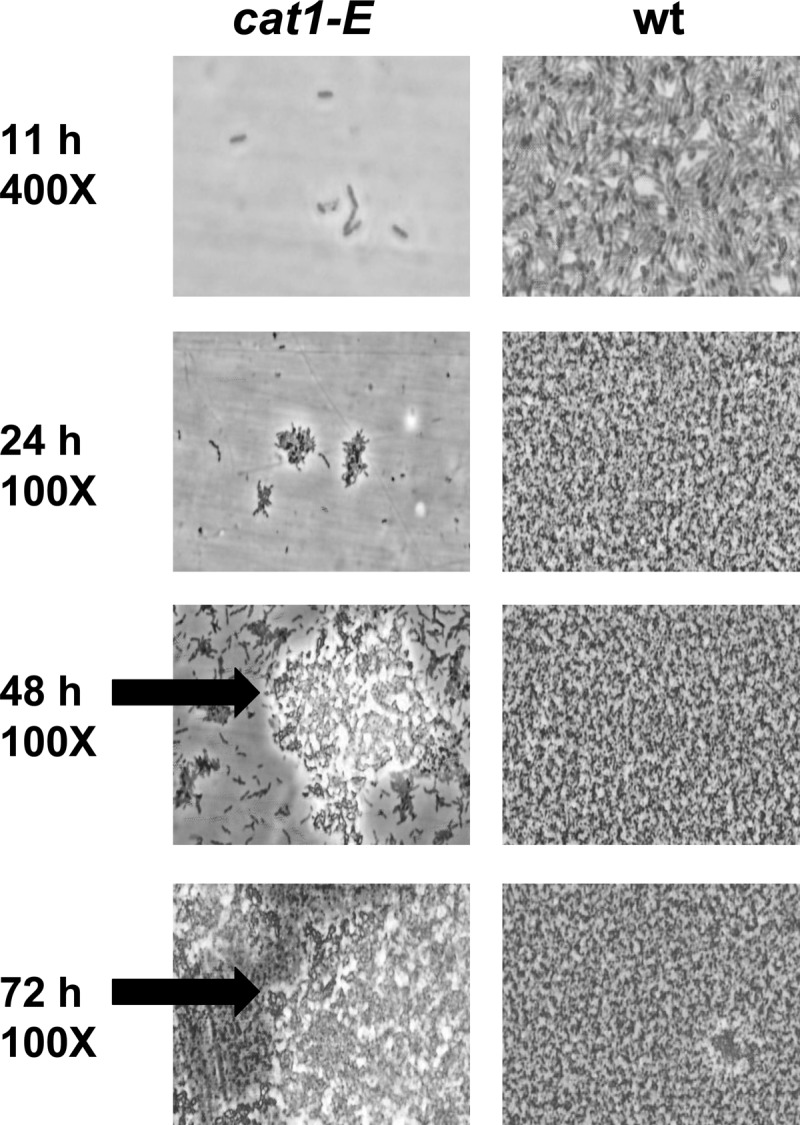
Biofilm development in a flow cell system. Wild-type and cat1-E strains were inoculated into flow cells and grown in modified MM (0.5% sea salt and 10 mM acetate) at room temperature. Biofilm formation was observed on the plastic surface of the flow cell over a 3-day time period. Although the cat1-E strain initially lagged in attachment, cells were able to attach over time and formed biofilms with a different architecture than that formed by wild-type cells. Arrows mark the large cell clusters observed only in cat1-E strain flow cells.
DISCUSSION
Wild-type P. arcticus 273-4 is capable of forming biofilms, a process we predict is advantageous for life in the permafrost. Psychrobacter sp. strain SW5H, a seawater isolate, was shown to attach to glass surfaces in parallel-plate and stagnation-point flow chambers (34, 35). We have found that a majority of Psychrobacter isolates are capable of attaching to surfaces, although to various degrees (unpublished observation). Together, these results suggest that the ability of Psychrobacter organisms to attach to surfaces is advantageous in their environments, which are generally cold and somewhat salty. However, further experimentation is required to prove a direct link between attachment and increased environmental survivorship. P. arcticus was found to attach in much higher densities to hydrophobic surfaces both in static assays and in flow cell experiments, indicating that hydrophobic interactions are important for attachment. Previous research has found the ability of bacterial cells to attach to hydrophobic and/or hydrophilic surfaces to be species or strain specific (27, 36).
Cat1 is a key adhesin involved in cell-surface interactions for P. arcticus. This extraordinarily large protein, containing 54 repeats that represent over 80% of the sequence, has similarities to other known adhesins found in Gram-positive and Gram-negative environmental and pathogenic bacterial strains (28, 30, 37–44). The hydrophilic amino acid repeats found in large adhesins, such as Cat1, are predicted to easily bind to water and other ions, keeping these molecules in close proximity to the cell (43). We hypothesize not only that attachment to environmental surfaces allows P. arcticus access to unfrozen water but also that Cat1 adhesin aids in retaining water close to the cell. Although Cat1 does not contain the secretion motif found in the LapA family of adhesins, we predict that Cat1 will be secreted to the cell surface via a type I secretion system, similar to what has been demonstrated for SiiE of Salmonella enterica (42). SiiE is a large adhesin belonging to the Bap family of adhesins, and it also lacks a secretion motif. We are currently investigating the localization and transport of Cat1.
Cat1 contains a predicted calcium binding domain, containing NodO and cadherin calcium binding motifs. Recent work by Martínez-Gil and colleagues has demonstrated calcium binding by the large adhesin LapF in Pseudomonas putida, which contains NodO calcium binding sites (45). Calcium was found to increase aggregate formation in P. putida (45).
Under the conditions tested in this study, P. arcticus formed robust biofilms only when grown in MM supplemented with acetate and sea salts. Previous studies have found that medium composition greatly impacts the ability of bacteria to attach to surfaces, and the optimal conditions may be unique for each bacterial species (27, 46–50). The MM employed in our study contained acetate as the sole carbon source. Acetate is an available carbon source in the permafrost environment that does not require a transport system for uptake. Growth studies and genomic analysis of P. arcticus have confirmed that key gluconeogenic enzymes are present, allowing this strain to use acetate as a carbon source (5, 11). Contrary to our hypothesis, we did not find a significant difference in cat1 expression between cells grown in LB and MM; however, differences in regulation of Cat1 export or protein stability could explain the differences in attachment.
We found increased relative expression of cat1 during times of slow growth and at low temperatures. This agrees with a prediction made by Reva and Tummler, that the production of large adhesins would be unlikely in cells growing in the mid-exponential phase (43). Further investigation is warranted to identify the regulatory molecule(s) controlling the transcription of cat1.
Based on the similar results from four independent cat1 transposon mutants, we propose that Cat1 is involved in the initial stages of bacterial attachment to surfaces. However, over time in both static and flow cell experiments, the cat1-E cells began to attach to the surface, although to a significantly lesser extent than wild-type cells in static assays. P. arcticus may express other adhesins which play roles in cell-cell interactions and are possibly masked by Cat1. One such adhesin could be the type IV pilus. The genes necessary for the formation of type IV pili are present in the P. arcticus genome, and type IV pili have been identified as adhesins for several different types of bacteria (51–54). P. arcticus does not have fimbriae or type I pilus genes. Another possible adhesin in P. arcticus is capsular-type exopolysaccharides (11). We are currently investigating the role of type IV pili and exopolysaccharides in biofilm formation by P. arcticus.
ACKNOWLEDGMENTS
We thank C. Montei, R. Cutler, R. Roewe. W. Aung, A. Weeks, J. H. Im, L. Imlay, A. Quinn, and L. Hinton for their technical contributions to the experiments presented.
This work was supported by Grinnell College and the NASA Astrobiology Institute at MSU. S. M. Hinsa-Leasure received funding as a NASA Astrobiology Fellow.
Footnotes
Published ahead of print 19 April 2013
REFERENCES
- 1. Vishnivetskaya T, Kathariou S, McGrath J, Gilichinsky D, Tiedje JM. 2000. Low-temperature recovery strategies for the isolation of bacteria from ancient permafrost sediments. Extremophiles 4:165–173 [DOI] [PubMed] [Google Scholar]
- 2. Gilichinsky D. 2002. Permafrost model of extraterrestrial habitat, p 125–142 In Horneck G, Baumstark-Khan C. (ed), Astrobiology: the quest for the conditions of life. Springer, Berlin, Germany [Google Scholar]
- 3. Gilichinsky DA. 2002. Permafrost as a microbial habitat, p 932–956 In Briton G. (ed), Encyclopedia of environmental microbiology. John Wiley & Sons, New York, NY [Google Scholar]
- 4. Azevedo JS, Correia A, Henriques I. 18 January 2013. Molecular analysis of the diversity of genus Psychrobacter present within a temperate estuary. FEMS Microbiol. Ecol. [Epub ahead of print.] 10.1111/1574-6941.12075 [DOI] [PubMed] [Google Scholar]
- 5. Bakermans C, Ayala-del-Rio HL, Ponder MA, Vishnivetskaya T, Gilichinsky D, Thomashow MF, Tiedje JM. 2006. Psychrobacter cryohalolentis sp. nov. and Psychrobacter arcticus sp. nov., isolated from Siberian permafrost. Int. J. Syst. Evol. Microbiol. 56:1285–1291 [DOI] [PubMed] [Google Scholar]
- 6. Borsodi AK, Kiss RI, Cech G, Vajna B, Toth EM, Marialigeti K. 2010. Diversity and activity of cultivable aerobic planktonic bacteria of a saline lake located in Sovata, Romania. Folia Microbiol. 55:461–466 [DOI] [PubMed] [Google Scholar]
- 7. Shivaji S, Reddy GS, Suresh K, Gupta P, Chintalapati S, Schumann P, Stackebrandt E, Matsumoto GI. 2005. Psychrobacter vallis sp. nov. and Psychrobacter aquaticus sp. nov., from Antarctica. Int. J. Syst. Evol. Microbiol. 55:757–762 [DOI] [PubMed] [Google Scholar]
- 8. Yoon JH, Kang KH, Park YH. 2003. Psychrobacter jeotgali sp. nov., isolated from jeotgal, a traditional Korean fermented seafood. Int. J. Syst. Evol. Microbiol. 53:449–454 [DOI] [PubMed] [Google Scholar]
- 9. Yoon JH, Lee CH, Kang SJ, Oh TK. 2005. Psychrobacter celer sp. nov., isolated from sea water of the South Sea in Korea. Int. J. Syst. Evol. Microbiol. 55:1885–1890 [DOI] [PubMed] [Google Scholar]
- 10. Yoon JH, Lee CH, Yeo SH, Oh TK. 2005. Psychrobacter aquimaris sp. nov. and Psychrobacter namhaensis sp. nov., isolated from sea water of the South Sea in Korea. Int. J. Syst. Evol. Microbiol. 55:1007–1013 [DOI] [PubMed] [Google Scholar]
- 11. Ayala-del-Rio HL, Chain P, Grzymski JJ, Ponder M, Ivanova N, Bergholz PW, Bartolo GD, Hauser L, Land M, Bakermans C, Rodrigues DF, Klappenback J, Zarka DG, Larimer F, Richardson P, Murray AE, Thomashow MF, Tiedje JM. 2010. The genome sequence of Psychrobacter arcticus 273-4, a psychroactive Siberian permafrost bacterium, reveals mechanisms for adaptation to low temperature. Appl. Environ. Microbiol. 76:2304–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ponder MA, Gilmour SJ, Bergholz PW, Mindock CA, Hollingsworth R, Thomashow MF, Tiedje JM. 2005. Characterization of potential stress responses in ancient Siberian permafrost psychroactive bacteria. FEMS Microbiol. Ecol. 53:103–115 [DOI] [PubMed] [Google Scholar]
- 13. Gilichinsky D, Wagener S, Vishnivetskaya T. 1995. Permafrost microbiology. Permafrost Periglac. Process. 6:281–291 [Google Scholar]
- 14. Rivkina EM, Friedmann EI, McKay CP, Gilichinsky DA. 2000. Metabolic activity of permafrost bacteria below the freezing point. Appl. Environ. Microbiol. 66:3230–3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Steven B, Leveille R, Pollard WH, Whyte LG. 2006. Microbial ecology and biodiversity in permafrost. Extremophiles 10:259–267 [DOI] [PubMed] [Google Scholar]
- 16. Ponder M, Vishnivetskaya T, McGrath J, Tiedje JM. 2004. Microbial life in permafrost: extended times in extreme conditions, p 151–170 In Fuller BJ, Lane N, Benson EE. (ed), Life in the frozen state. CRC Press, Boca Raton, FL [Google Scholar]
- 17. Soina VS, Mulyukin AL, Demkina EV, Vorobyova EA, El-Registan GI. 2004. The structure of resting bacterial populations in soil and subsoil permafrost. Astrobiology 4:345–358 [DOI] [PubMed] [Google Scholar]
- 18. Davey ME, O'Toole GA. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95–108 [DOI] [PubMed] [Google Scholar]
- 20. Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711–745 [DOI] [PubMed] [Google Scholar]
- 21. Flemming HC. 1993. Biofilms and environmental protection. Water Sci. Technol. 27:1–10 [Google Scholar]
- 22. Mah TF, O'Toole GA. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34–39 [DOI] [PubMed] [Google Scholar]
- 23. Allen CC, Albert FG, Chafetz HS, Combie J, Graham CR, Kieft TL, Kivett SJ, McKay DS, Steele A, Taunton AE, Taylor MR, Thomas-Keprta KL, Westall F. 2000. Microscopic physical biomarkers in carbonate hot springs: implications in the search for life on Mars. Icarus 147:49–67 [DOI] [PubMed] [Google Scholar]
- 24. Krumbein WE, Gorbushina AA, Holtkamp-Tacken E. 2004. Hypersaline microbial systems of sabkhas: examples of life's survival in “extreme” conditions. Astrobiology 4:450–459 [DOI] [PubMed] [Google Scholar]
- 25. Larsen RA, Wilson MM, Guss AM, Metcalf WW. 2002. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch. Microbiol. 178:193–201 [DOI] [PubMed] [Google Scholar]
- 26. O'Toole GA, Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295–304 [DOI] [PubMed] [Google Scholar]
- 27. O'Toole GA, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449–461 [DOI] [PubMed] [Google Scholar]
- 28. Hinsa SM, Espinosa-Urgel M, Ramos JL, O'Toole GA. 2003. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol. Microbiol. 49:905–918 [DOI] [PubMed] [Google Scholar]
- 29. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 30. Yousef F, Espinosa-Urgel M. 2007. In silico analysis of large microbial surface proteins. Res. Microbiol. 158:545–550 [DOI] [PubMed] [Google Scholar]
- 31. Economou A, Hamilton WD, Johnston AW, Downie JA. 1990. The Rhizobium nodulation gene nodO encodes a Ca2(+)-binding protein that is exported without N-terminal cleavage and is homologous to haemolysin and related proteins. EMBO J. 9:349–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fraiberg M, Borovok I, Weiner RM, Lamed R. 2010. Discovery and characterization of cadherin domains in Saccharophagus degradans 2-40. J. Bacteriol. 192:1066–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371 [DOI] [PubMed] [Google Scholar]
- 34. Bakker DP, Busscher HJ, van der Mei HC. 2002. Bacterial deposition in a parallel plate and a stagnation point flow chamber: microbial adhesion mechanisms depend on the mass transport conditions. Microbiology 148:597–603 [DOI] [PubMed] [Google Scholar]
- 35. Bakker DP, Klijnstra JW, Busscher HJ, van der Mei HC. 2003. The effect of dissolved organic carbon on bacterial adhesion to conditioning films adsorbed on glass from natural seawater collected during different seasons. Biofouling 19:391–397 [DOI] [PubMed] [Google Scholar]
- 36. Ohmura N, Kitamura K, Saiki H. 1993. Selective adhesion of Thiobacillus ferrooxidans to pyrite. Appl. Environ. Microbiol. 59:4044–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cucarella C, Solano C, Valle J, Amorena B, Lasa I, Penades JR. 2001. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183:2888–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Espinosa-Urgel M, Salido A, Ramos JL. 2000. Genetic analysis of functions involved in adhesion of Pseudomonas putida to seeds. J. Bacteriol. 182:2363–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lasa I, Penades JR. 2006. Bap: a family of surface proteins involved in biofilm formation. Res. Microbiol. 157:99–107 [DOI] [PubMed] [Google Scholar]
- 40. Latasa C, Roux A, Toledo-Arana A, Ghigo JM, Gamazo C, Penades JR, Lasa I. 2005. BapA, a large secreted protein required for biofilm formation and host colonization of Salmonella enterica serovar Enteritidis. Mol. Microbiol. 58:1322–1339 [DOI] [PubMed] [Google Scholar]
- 41. Martinez-Gil M, Yousef-Coronado F, Espinosa-Urgel M. 2010. LapF, the second largest Pseudomonas putida protein, contributes to plant root colonization and determines biofilm architecture. Mol. Microbiol. 77:549–561 [DOI] [PubMed] [Google Scholar]
- 42. Morgan E, Bowen AJ, Carnell SC, Wallis TS, Stevens MP. 2007. SiiE is secreted by the Salmonella enterica serovar Typhimurium pathogenicity island 4-encoded secretion system and contributes to intestinal colonization in cattle. Infect. Immun. 75:1524–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Reva O, Tummler B. 2008. Think big–giant genes in bacteria. Environ. Microbiol. 10:768–777 [DOI] [PubMed] [Google Scholar]
- 44. Toledo-Arana A, Valle J, Solano C, Arrizubieta MJ, Cucarella C, Lamata M, Amorena B, Leiva J, Penades JR, Lasa I. 2001. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl. Environ. Microbiol. 67:4538–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Martinez-Gil M, Romero D, Kolter R, Espinosa-Urgel M. 2012. Calcium causes multimerization of the large adhesin LapF and modulates biofilm formation by Pseudomonas putida. J. Bacteriol. 194:6782–6789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bernier SP, Ha DG, Khan W, Merritt JH, O'Toole GA. 2011. Modulation of Pseudomonas aeruginosa surface-associated group behaviors by individual amino acids through c-di-GMP signaling. Res. Microbiol. 162:680–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Donlan RM. 2002. Biofilms: microbial life on surfaces. Emerg. Infect. Dis. 8:881–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hancock V, Witso IL, Klemm P. 2011. Biofilm formation as a function of adhesin, growth medium, substratum and strain type. Int. J. Med. Microbiol. 301:570–576 [DOI] [PubMed] [Google Scholar]
- 49. Jamieson WD, Pehl MJ, Gregory GA, Orwin PM. 2009. Coordinated surface activities in Variovorax paradoxus EPS. BMC Microbiol. 9:124. 10.1186/1471-2180-9-124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wimpenny JW, Colasanti A. 1997. A unifying hypothesis for the structure of microbial biofilms based on cellular automaton models. FEMS Microbiol. Ecol. 22:1–16 [Google Scholar]
- 51. Frischkorn KR, Stojanovski A, Paranjpye R. 2013. Vibrio parahaemolyticus type IV pili mediate interactions with diatom-derived chitin and point to an unexplored mechanism of environmental persistence. Environ. Microbiol. 15:1416–1427 [DOI] [PubMed] [Google Scholar]
- 52. Wang S, Parsek MR, Wozniak DJ, Ma LZ. 28 January 2013. A spider web strategy of type IV pili-mediated migration to build a fibre-like Psl polysaccharide matrix in Pseudomonas aeruginosa biofilms. Environ. Microbiol. [Epub ahead of print.] 10.1111/1462-2920.12095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Conrad JC. 2012. Physics of bacterial near-surface motility using flagella and type IV pili: implications for biofilm formation. Res. Microbiol. 163:619–629 [DOI] [PubMed] [Google Scholar]
- 54. Richter LV, Sandler SJ, Weis RM. 2012. Two isoforms of Geobacter sulfurreducens PilA have distinct roles in pilus biogenesis, cytochrome localization, extracellular electron transfer, and biofilm formation. J. Bacteriol. 194:2551–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bergholz PW, Bakermans C, Tiedje JM. 2009. Psychrobacter arcticus 273-4 uses resource efficiency and molecular motion adaptations for subzero temperature growth. J. Bacteriol. 191:2340–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kostka J, Nealson KH. 1998. Isolation, cultivation and characterization of iron- and manganese-reducing bacteria, p 55–68 In Sayler G. (ed), Techniques in microbial ecology. Oxford University Press, New York, NY [Google Scholar]


