Abstract
Reactions associated with the geochemical process of serpentinization can generate copious quantities of hydrogen and low-molecular-weight organic carbon compounds, which may provide energy and nutrients to sustain subsurface microbial communities independently of the photosynthetically supported surface biosphere. Previous microbial ecology studies have tested this hypothesis in deep sea hydrothermal vents, such as the Lost City hydrothermal field. This study applied similar methods, including molecular fingerprinting and tag sequencing of the 16S rRNA gene, to ultrabasic continental springs emanating from serpentinizing ultramafic rocks. These molecular surveys were linked with geochemical measurements of the fluids in an interdisciplinary approach designed to distinguish potential subsurface organisms from those derived from surface habitats. The betaproteobacterial genus Hydrogenophaga was identified as a likely inhabitant of transition zones where hydrogen-enriched subsurface fluids mix with oxygenated surface water. The Firmicutes genus Erysipelothrix was most strongly correlated with geochemical factors indicative of subsurface fluids and was identified as the most likely inhabitant of a serpentinization-powered subsurface biosphere. Both of these taxa have been identified in multiple hydrogen-enriched subsurface habitats worldwide, and the results of this study contribute to an emerging biogeographic pattern in which Betaproteobacteria occur in near-surface mixing zones and Firmicutes are present in deeper, anoxic subsurface habitats.
INTRODUCTION
Subsurface habitats are estimated to host a large proportion of Earth's biomass (1–3), but the diversity, activity, and viability of subsurface organisms are largely unconstrained (4). Generalizations about subsurface habitats are further complicated by their wide range of geological, physical, chemical, and biological characteristics. Some subsurface habitats appear to host mostly dead or dormant organisms, while others provide sources of energy and nutrients that could potentially support active subsurface communities (5–7).
One potential source of biologically available energy for subsurface organisms is serpentinization: the aqueous alteration of iron minerals (from olivine to serpentine) that results in the release of hydrogen gas (H2) and organic carbon (particularly methane, with lesser amounts of larger hydrocarbons [8]). It occurs wherever groundwater interacts with peridotite (iron-rich rocks formed in the mantle), including sites of active serpentinization on all of the world's continents and throughout much of the seafloor. Nevertheless, microbiologists have only recently begun to investigate serpentinite habitats.
Because serpentinization represents a widespread and long-lasting subsurface source of geochemical energy and organic carbon, it has the potential to support vigorously active, self-sufficient subsurface ecosystems that may not require energy or nutrients from the photosynthetic surface biosphere. At the same time, serpentinization-associated processes create unusual and biologically challenging conditions (e.g., high pH, unavailable inorganic carbon, few electron acceptors) that are likely to constrain the genetic and metabolic diversity of serpentinite-associated microbial communities. Initial studies of microbial communities at the Lost City hydrothermal field are consistent with these hypotheses (9–12), but additional work is required to evaluate the global potential of serpentinite-powered subsurface life.
Ultrabasic continental serpentinite springs offer a distinct and complementary test for the potential of serpentinite-hosted ecosystems. Ultrabasic serpentinite springs exposed on land indicate the presence of underlying geochemical (and potential biogeochemical) activity in the subsurface and have the advantage of providing access to subsurface materials without need for drilling or other invasive or destructive sampling methods. Consequently, serpentinite springs can serve as natural windows into the continental subsurface in the same manner in which deep sea hydrothermal vents are used as windows into the marine subsurface (13, 14). As with deep sea hydrothermal fluids, however, the interpretation of microbiological data from serpentinite springs is complicated by the mixing of subsurface and surface fluids. The presence of genetic material in a serpentinite spring does not constitute convincing evidence that the genetic material represents a subsurface organism that is solely supported from serpentinization-associated reactions. An integrated biogeochemical approach designed to distinguish subsurface and surface biogeochemical signals is required to identify bona fide subsurface organisms.
Here we report on such a study of microbial communities associated with serpentinite springs of the Tablelands Ophiolite, Newfoundland, Canada (15). Ultramafic rocks of the Tablelands Ophiolite (also known as the Bay of Islands Ophiolite) were uplifted from the Iapetus Ocean and obducted onto the continental margin of Laurentia approximately 485 million years ago. Serpentinization remains evident today in the form of highly reducing, ultrabasic springs at several sites (15). This study examines the fluids discharged from these springs for evidence that they contain microorganisms flushed from subsurface habitats where they were likely to have been supported by the products of serpentinization-associated processes. Of the many taxa observed in the springs, we identify one lineage of Firmicutes, most closely related to the genus Erysipelothrix, as the most likely inhabitant of the serpentinite subsurface.
MATERIALS AND METHODS
Site description.
The Tablelands Ophiolite ultrabasic serpentinite springs are located within Gros Morne National Park, Newfoundland, Canada (N49°27′59″, W57°57′29″). Five springs (Winter House Canyon 1 [WHC1], Winter House Canyon 2 [WHC2], Winter House Canyon 75 [WHC75], Tablelands East [TLE], and Wallace Brook [WB]) and one “background” surface freshwater site (Winter House Brook [WHB]) were sampled for this study at three time points: June 2010, August 2010, and June 2011 (abbreviated in tables and figures as 2010J, 2010A, and 2011J). The WHC springs and WHB are located within ∼10 m of each other. WHC75 is located ∼75 m south of the WHC springs and WHB. TLE and WB are 2 km and 6 km west, respectively, of the WHC springs (see Szponar et al. [15] for a map and further descriptions). The WHC2 spring forms a shallow pool (∼40 cm deep and 126 cm wide) that includes three sampling sites (WHC2A, WHC2B, and WHC2C). WHC2A and WHC2B exhibited the most extreme pH and Eh measurements at every time point and represent putative sources of subsurface ultrabasic fluid. WHC2C receives surface runoff and therefore represents a likely mixing site between subsurface and surface fluids.
Aqueous geochemistry.
The general aqueous geochemistry of the springs discharging from the Tablelands Ophiolite was described by Szponar et al. (15). In short, ultrabasic (pH ≥11) and reducing groundwater springs (WHC1 and WHC2) discharge from hydrated peridotite along the base of Winterhouse Canyon. Moderately basic (pH ∼10.5) and oxidizing springs (TLE and WB) are located on the east facing slope of the Tablelands Massif and Wallace Brook. All of the spring waters are exposed to the atmosphere postdischarge and may mix with overland flow. The groundwater discharging from WHC1 could be isolated from overland flow and collected immediately upon its discharge due to its higher elevation and very small pool size. Thus, the water collected from WHC1 provided the best available proxy for the ultrabasic reducing groundwater end member. WHC1 consistently had the highest pH (12.2 ± 0.2), Cl−, Br−, and Ca2+ concentrations (479 ± 37 mg/liter, 1.1 ± 0.06 mg/liter, and 144 ± 0.5 mg/liter, respectively) (15). Using a conservative mixing model, Szponar et al. were able to estimate the relative fraction of ultrabasic groundwater (fUB) contributing to the water sample collected at each spring location. In short, the aqueous concentrations of conservative tracers Cl− and Br− in each fluid sample were positively correlated (r2 = 1), suggesting that within each pool there was conservative mixing between the two end member waters (i.e., ultrabasic groundwater and surface water). The relative fraction of ultrabasic groundwater (fUB) at each sampling site was calculated by using a 2-component mixing model: [Cl−]sample = fUB × [Cl−]UB + (1 − fUB) × [Cl−]brook, where [Cl−]sample, [Cl−]UB, and [Cl−]brook are the concentrations of chloride ions in the sampling locations of interest (WHC2A, WHC2B, WHC2C, etc.), the ultrabasic groundwater end member (WHC1), and the brook end member (WHB), respectively. The geochemistry suggested that the WHB surface water and WHC1 spring water were the best proxies attainable to represent the surface water and ultrabasic groundwater, respectively. Since WHB potentially receives a small amount of discharge from the ultrabasic springs, fUB represents the relative mixing between the freshwater and ultrabasic groundwater. This model can be used to predict the spring water concentrations of geochemical parameters solely due to physical mixing between the two water sources. Concentrations of geochemical parameters that are not well described by this model indicate that one or more processes, in addition to physical mixing (i.e., chemical and/or biological reactions) are occurring and affecting the parameter's concentrations.
Fluid sample collection.
Fluids were collected via sterile syringes or peristaltic pumping (Masterflex E/S portable sampler; Cole-Parmer, Vernon Hills, IL) through cleaned and flushed Masterflex C-Flex tubing. Samples for DNA analyses were filtered through 0.2-μm Sterivex filter cartridges (Millipore, Billerica, MA) in line with the peristaltic pumping and therefore simultaneous to sample collection. When practical, replicate filters were collected sequentially from the same site. At the WHC2 pool, chemical and biological measurements of collected samples were highly sensitive to minor variations in the placement of the sampling intake, due to strong chemical gradients between the bottom and the top of the pool. Therefore, on-site readings of pH (IQ Scientific Instruments GLP series IQ180G) and Eh (Oakton Testr10) were monitored before, during, and after sampling, and any changes were recorded so that each subsample for chemical and biological analyses could be correlated to the appropriate readings. Eh readings were corrected for the standard Ag/AgCl electrode (200 mV). DNA extracted from the August 2010 sample from WHC2B (i.e., WHC2B-2010A) and the June 2010 sample from TLE (i.e., TLE-2010J) were previously used in metagenomic analyses published in reference 16. WHC1 was sampled for chemical measurements, but collection of enough material for DNA analyses was not practical due to the extremely slow discharge rate.
Fluids (50 ml per replicate sample; numbers of replicates are noted in Table 1, below) were preserved in the field for cell abundance enumeration at a final concentration of 3.7% formaldehyde and stored at 4°C. In the laboratory, preserved fluids (5 to 20 ml each) were filtered through a 0.2 μm-filter, and cells stained with 4′,6-diamidino were counted with an Olympus BX61 spinning disk epifluorescence confocal microscope according to previously published protocols (17, 18). At least 30 fields containing between 10 and 30 cells/field were counted for each sample and used to calculate the average cell concentrations. The errors reported in Table 1 reflect the standard errors of the mean cell numbers for each sample.
Table 1.
Biogeochemical characteristics of each sample collected from Tablelands serpentinite springs and surface freshwater (WHB)
| Spring | Datea | fUB | pH | Eh (mV) | H2 (mg/liter) | CH4 (mg/liter) | TIC (mg/liter) | 13C-TIC (per ml) | DOC (mg/liter) | No. of cells/ml (×104)b | No. of TRFLP fragments | No. of CoDL OTUs | No. of JGI OTUs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WB | 2010J | 0.173 | 10.5 | 376 | 0 | 0 | 0.42 | ND | 0.1 | 0.4 ± 0.06 (2) | 24 | NDc | ND |
| WB | 2010A | ND | 10.91 | 328 | 0 | 0 | 1.07 | −16.5 | 0.32 | 2.2 | 30 | ND | ND |
| WB | 2011J | 0.153 | 10.76 | 250 | 0 | 0 | 0.9 | −20.7 | 0.15 | 1.3 ± 0.2 (2) | 21 | 1,108 | ND |
| TLE | 2010J | 0.166 | 10.5 | 224 | ND | 0 | 1.49 | −10 | 0.06 | 1.3 ± 0.4 (5) | 11 | ND | ND |
| TLE | 2011J | 0.000 | 10.81 | 253 | ND | 0 | 1.2 | −15.5 | 0.12 | 2.4 ± 1.1 (3) | 23 | 606 | ND |
| WHB | 2010J | 0.000 | 8.77 | 455 | 0 | 0 | 7.47 | −1.1 | 0.44 | ND | 40 | ND | ND |
| WHB | 2011J | 0.000 | 7.82 | 382 | 0 | 0 | 7.1 | −2.1 | 0.3 | 5.1 ± 1.3 (4) | 62 | 3,816 | 3,017 |
| WHC2A | 2010J | 0.796 | 12.57 | −647 | 0.75 | 0.25 | 0.39 | ND | 1.93 | 5.3 | 33 | ND | ND |
| WHC2A | 2010A | 0.931 | 12.4 | −642 | 0.69 | 0.31 | 1.17 | −19.2 | 2.67 | 8.3 ± 0.8 (4) | 23 | ND | ND |
| WHC2A | 2011J | 1.000 | 12.36 | −690 | 1.18 | 0.32 | 1.1 | −14.7 | 0.35 | 17.6 ± 4.5 (4) | 23 | ND | ND |
| WHC2B | 2010J | 0.615 | 12.63 | −642 | 0.73 | 0.25 | 1.73 | −13 | 0.44 | 17.4 ± 1.7 (3) | 26 | ND | ND |
| WHC2B | 2010A | 0.750 | 12.31 | −596 | 0.47 | 0.26 | 2.16 | −17.3 | 1.15 | 8.0 ± 1.1 (3) | 12 | 287 | 279 |
| WHC2B | 2011J | 0.529 | 12.28 | −618 | 1.04 | 0.32 | 1.2 | −17.6 | 0.29 | 22.1 ± 3.6 (7) | 22 | 464 | ND |
| WHC2C | 2010J | 0.318 | 11.5 | −468 | 0.57 | 0.03 | 11.75 | −11.5 | 1.93 | 5.6 ± 3.8 (2) | 18 | ND | ND |
| WHC2C | 2010A | 0.319 | 12.06 | −533 | ND | 0.01 | 14.78 | −12.6 | 1.37 | 40.2 | 24 | ND | ND |
| WHC2C | 2011J | 0.153 | 12.21 | −458 | 0.07 | 0.03 | 13.5 | −11.8 | 0.84 | 18.7 ± 1.5 (3) | 28 | ND | 1,899 |
| WHC75 | 2011J | ND | 11.2 | −603 | 0.32 | 0.04 | 0.76 | −11.7 | 1.54 | 7.3 ± 0.5 (3) | 25 | 815 | ND |
See Materials and Methods for an explanation of the sampling date format.
Values are means ± standard errors of the means; data in parentheses indicate the number of samples tested (n was 1 when a single value is reported without a standard error of the mean).
ND, not detected.
DNA extraction.
Sterivex filters were stored on wet ice in the field, frozen in liquid nitrogen as soon as possible (in many cases immediately and in others a few hours later), transported on dry ice, and stored at −80°C until DNA extraction, which was conducted according to the protocol described in references 19 and 20. DNA extracts were purified with QiaAmp columns (Qiagen, Hilden, Germany) according to the manufacturer's instructions for purification of genomic DNA.
16S rRNA tag sequencing.
Two separate data sets of 16S rRNA amplicon (tag) sequences were generated at two sequencing centers. Pyrosequencing (Roche Titanium platform) of bacterial 16S rRNA amplicons was conducted on six of the samples as part of the Census of Deep Life (CoDL), an initiative of the Deep Carbon Observatory (dco.gl.ciw.edu) performed at the Marine Biological Laboratory (Woods Hole, MA). This method produces ∼450-bp reads targeting the 16S rRNA V4 to V6 regions (Escherichia coli reference positions 518 to 1064). Additional CoDL amplification and sequencing methods have been published elsewhere (21) and are available at the VAMPS website (http://vamps.mbl.edu/resources/primers.php). Amplification, sequencing, data processing, chimera detection, and taxonomic classification procedures at the Josephine Bay Paul Center (Marine Biological Laboratory, Woods Hole, MA) have been previously described (20, 22, 23).
Three samples, two of which were also included in the six CoDL samples, were sequenced by the DOE Joint Genome Institute (JGI) with an Illumina MiSeq apparatus as described in reference 24. This method produces paired reads of ∼250 bp, each targeting the 16S rRNA V4 region (E. coli reference positions 533 to 786). After automated paired-end assembly, quality filtering, and chimera detection by the JGI, reads were further screened with Btrim (25) to remove all sequences less than 250 bp and to trim the remaining sequences so that their quality scores averaged ≥30 over a 15-bp sliding window.
For alpha diversity analyses and taxonomic classification, the CoDL and JGI data sets were processed separately but identically. In each case, sequences were aligned to a SILVA reference alignment (derived from SSURefv102, obtained November 2012 from http://www.mothur.org/wiki/Silva_reference_files) as described in reference 26. Taxonomic classification was performed using mothur with the SILVA taxonomy outline (27). Sequences were clustered into operational taxonomic units (OTUs) at a 3% distance threshold with the cluster.split command and using the average neighbor method in mothur (28). Numbers of OTUs reported in Table 1, below, reflect subsampling all CoDL and JGI data sets down to the size of the smallest data set (6,277 reads for CoDL; 6,305 reads for JGI). Prior to generating OTUs for beta diversity analyses (examining shared OTUs), all sequences from both data sets were merged and trimmed to the region where the CoDL (V4 to V6 regions) and JGI (V4 region) sequences overlap.
16S rRNA phylogeny.
Phylogenetic analyses were performed on the two most common CoDL tag sequences as an independent measure of their taxonomic affiliation (in addition to the automated classification described above). Reference sequences were obtained from the Ribosomal Database Project (29). References and tag sequences were aligned with the SINA webserver (30). A maximum-likelihood reference phylogeny of full-length 16S rRNA sequences (i.e., without CoDL tag sequences) was computed using RAxML (31). The short tag sequences were then placed next to their most likely neighbor in the reference phylogeny by using the evolutionary placement algorithm (32) implemented in RAxML version 7.4.2. This approach to identify the phylogenetic context of a short sequence is more conservative than inferring a new phylogeny that treats the new short sequence on the same basis as the previously published full-length sequences.
Molecular fingerprinting via TRFLP.
TRFLP (terminal restriction fragment length polymorphism) analysis utilized the same V4 to V6 primers as used for the CoDL tag sequencing: 518F, CCAGCAGCYGCGGTAAN; 1046R, 6-carboxyfluorescein–CGACARCCATGCANCACCT. PCR amplification reactions for TRFLP were performed with 0.05 to 2 ng of template DNA, 0.25 μM each primer, 0.2 mM each deoxynucleoside triphosphate, 1× GoTaq reaction buffer, and 1 U of GoTaq DNA polymerase (Promega, Madison, WI). We empirically determined that 34 PCR cycles was optimal for TRFLP, because it increased detection of rare fragments without significantly altering the relative abundance of common fragments (data not shown), and this is consistent with our previous work (33). At least 4 replicate reaction mixtures for each sample were pooled and purified with QiaQuick columns (Qiagen, Hilden, Germany). For each sample, 100 to 200 ng of purified amplicon was digested separately with 5 U HaeIII, MspI, or RsaI (New England BioLabs, Ipswich, MA) overnight at 37°C. Digests were precipitated in ethanol with ammonium acetate and resuspended in 5 μl formamide. Fragment sizing was performed with a 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA), BigDyeTerminator v3.1 chemistry, and the GeneScan 600 LIZ size standard at the East Carolina University Genomics Core Facility.
GeneMapper (Applied Biosystems) was used to generate data files in ABI format of fragment sizes by using the minimal peak detection threshold (1 RFU), as recommended in the documentation for fragment clustering software by Abdo et al. (34). The scripts AutomaticProgR.pl and FilteringandBinning.r were downloaded from the authors' website (http://www.ibest.uidaho.edu/tools/trflp_stats) and used to identify peaks from noise and cluster peak sizes according to the methods described in reference 34, with the only modification being that we analyzed peak height, not area. After clustering, all peaks of <60 bp were deleted from all samples, because many fragments of this size appeared to be PCR or digestion artifacts. We tested for correlation between the final number of terminal restriction fragment clusters for each sample and the total peak height for each sample in order to assess whether variability in loading and detection of DNA influenced the apparent fragment diversity, as recommended in reference 35. No correlation was detected, and outliers were removed from analysis or repeated until data quality was comparable to that for other samples.
Because the TRFLP data were generated with the same 16S rRNA primers as used for the CoDL tag sequences, we were able to assign taxonomy to fragment sizes by linking them with the CoDL sequences. If the predicted fragment sizes of a CoDL sequence for each of the three restriction enzymes matched the corresponding fragment sizes in the observed TRFLP data, then the taxonomic assignment for that CoDL tag sequence was applied to the matching set of fragment sizes. Only two matching digests were required if the predicted fragment size was outside the TRFLP analysis window (60 to 600 bp). Predicted and observed sizes often differed by 1 to 5 bp due to inherent variability in the electrophoretic mobility of DNA fragments, but the difference was consistent for each restriction enzyme in a matching set.
Statistical calculations.
The Sorensen index of community similarity was calculated within Primer 6 (Primer-E) for each pair of samples according to their shared presence/absence of TRFLP fragment clusters. Sorensen similarities were calculated separately for each of the three restriction enzyme digests and then averaged. The average Sorensen similarity for each pair of samples was imported into Primer 6 and subjected to the cluster function (36) to generate the final dendrogram of averaged community similarities. To obtain a measure of the overall goodness of fit of the dendrogram, the cophenetic correlation was calculated between the similarity values at which samples were clustered and the underlying similarity matrix. The significance of each similarity cluster in the resulting dendrogram was tested with the SIMPROF (similarity profile) test within Primer 6, which is a permutation test of the null hypothesis that a set of samples do not differ from each other in multivariate structure (36).
Our network association analysis was based on that described in reference 37), although our study did not attempt detection of time-lagged correlations by local similarity analysis, as used in that study. Pairwise Pearson correlations were computed with the rcor.test function in the R package ltm (38) from a matrix containing environmental data for each sample and the relative abundance of each fragment cluster in each sample. To adjust for multiple tests, the false-discovery rate (q value) was also computed for the distribution of Pearson P values. Only those pairwise correlations with a P level of <0.05 and q of <0.05 were included in subsequent analyses. Networks of significant correlations were visualized using Cytoscape v2.8.3 (39).
Nucleotide sequence accession number.
The CoDL nucleotide sequence data are available in the VAMPS database (http://vamps.mbl.edu) under project code DCO_ BRZ and NCBI SRA database under project accession number SRP021893.
RESULTS
Fluid chemistry.
Fluid chemistry data from Tablelands springs were previously reported (15); this report includes data from additional samples and provides time point-specific measurements. This study focused on the fraction of ultrabasic end member (fUB) as the most important variable for assessing the amount of mixing between subsurface and surface fluids, as described by Szponar et al. (15). The extremely high pH (10.5 to 12.6) of the springs in this study is consistent with active serpentinization, which is known to generate ultrabasic fluids. However, pH values cannot be used to quantitatively determine the contribution of ultrabasic groundwater to each sampling site, as hydrogen ions are highly reactive and are not conservative tracers for mixing. Therefore, Szponar et al. determined fUB values for each sample with a 2-component mixing model, using chlorine as the conservative tracer in which the surface freshwater stream WHB was defined as the freshwater end member (fUB = 0) and the spring WHC1 was defined as the ultrabasic end member (fUB = 1). Unfortunately, the extremely slow discharge rate of WHC1 did not allow collection of sufficient material for DNA extraction. WHC2A consistently had the highest fraction of ultrabasic water (fUB, 0.80 to 1). WHC2B was the next most ultrabasic (fUB, 0.53 to 0.75), and samples from the mixing site WHC2C were moderately ultrabasic (fUB, 0.15 to 0.32). Samples from the TLE and WB springs were much closer to the freshwater end member (0 to 0.17), but it must also be noted that they were not well represented by the mixing model (15). These springs are located 2 to 6 km from the other sampling sites (i.e., WHC1, WHC2, and WHB), so they may represent mixing of ultrabasic and/or freshwater end members with slightly different chemistries compared to WHB and WHC1.
The chemical characteristics of each sample are provided in Table 1. As expected, water samples with high pH values had the greatest fUB values, but the fUB more clearly distinguished the high pH springs from each other. A sample from TLE, for example, had a pH of 10.8 but an fUB of 0, indicating that this fluid maintained its high pH despite little or no contribution from subsurface fluids. This result is consistent with the lack of any detectable H2 or methane and with the measurement of highly oxidized reduction potentials (Eh, 253 mV) at this spring. In general, the samples with the highest fUB values also had the highest H2 and methane concentrations. WHC2A-2011J (fUB, 1) had the most H2 (1.18 mg/liter) and methane (0.32 mg/liter) concentrations, while the mixing site WHC2C (fUB, 0.15 to 0.32) exhibited much lower values (H2, 0.07 to 0.57 mg/liter; methane, 0.01 to 0.03 mg/liter). The Eh also appeared to track the fUB, with the highest-reducing fluid (−642 to −690 mV) at WHC2A and somewhat-lower reducing fluids (−468 to −533 mV) in the mixing site WHC2C.
As expected, total inorganic carbon (TIC) and dissolved organic carbon (DOC) were not tightly linked with fUB, because these compounds are affected by additional processes (physical, biological, and chemical) other than physical mixing of the two end member waters. For example, total inorganic carbon (TIC) was approximately 1 to 2 mg/liter for all springs except for WHC2C (11.8 to 14.8 mg/liter) and the surface freshwater end member WHB (7.1 to 7.5 mg/liter). The carbon-13 isotopic signatures of TIC (Table 1) at WHC2C were intermediate between the freshwater end member (WHB) and the most ultrabasic springs (WHC2A and WHC2B), indicating that WHC2C represents a mixing site between ultrabasic and freshwater end members. Therefore, the high TICs at WHC2C may indicate a stimulation of biological respiration and consequent production of inorganic carbon. Concentrations of DOC were also relatively high at WHC2C compared to WHB, consistent with the interpretation of stimulated biological production at this site. DOC measurements at WHC2A and WHC2B were highly variable between time points, and these fluctuations did not seem to track with other parameters.
Microbial cell abundance.
The most ultrabasic springs contained the highest cell densities (up to 2.2 × 105 cells ml−1) measured in this study (Table 1). The 2011 samples (n = 17) from WHC2A, WHC2B, WHC2C, and WHC75 contained significantly greater (P < 10−7, Student's t test) cell densities than the 2011 freshwater end member (WHB; 5.1 ± 1.3 × 104 cells ml−1), suggesting that the ultrabasic fluids stimulated microbial growth above background levels. Interestingly, the moderate springs WB and TLE had significantly fewer cells than WHB (as few as 4 × 103 cells ml−1). This result may reflect the biological challenges at the high pH of these moderate springs (pH 10.5 to 10.9) without the benefit of highly reducing compounds such as H2 and methane, which were present in the most ultrabasic and more densely populated springs.
16S rRNA diversity.
Bacterial diversity was estimated by molecular fingerprinting (TRFLP) and next-generation tag sequencing of the 16S rRNA gene. The freshwater end member (WHB) had more TRFLP fragment clusters than any of the ultrabasic springs (Table 1), but TRFLP did not resolve any fUB-correlated differences among ultrabasic springs (Fig. 1). Numbers of TRFLP fragment clusters is a rough approximation of species-level diversity of the 16S rRNA gene. Numbers of 16S rRNA OTUs (operational taxonomic units) generated by next-generation tag sequencing is a more robust and exhaustive measure of species-level diversity, but these data are available for fewer samples. Both the CoDL and JGI tag sequence data sets were consistent with the TRFLP results (Fig. 1). The freshwater end member WHB yielded the most OTUs, and the fluid sample in each data set with the highest (most ultrabasic) fUB value (WHC2B) had the fewest OTUs (Table 1). Rarefaction analysis of these data also pointed to WHB as the most diverse and the high-fUB samples as the least diverse (see Fig. S1 in the supplemental material).
Fig 1.
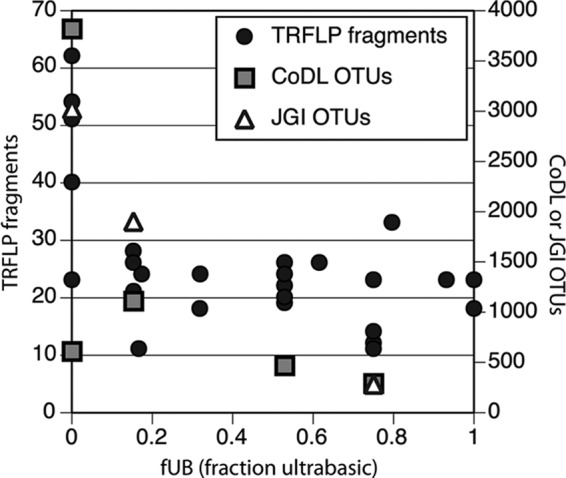
The bacterial diversity of Tablelands serpentinite springs decreases as the fUB increases. The freshwater end member (fUB, 0) has the highest number of TRFLP fragment clusters and OTUs produced by the CoDL and JGI tag sequencing projects. The most ultrabasic spring for which tag sequences were available yielded the fewest OTUs.
Community composition.
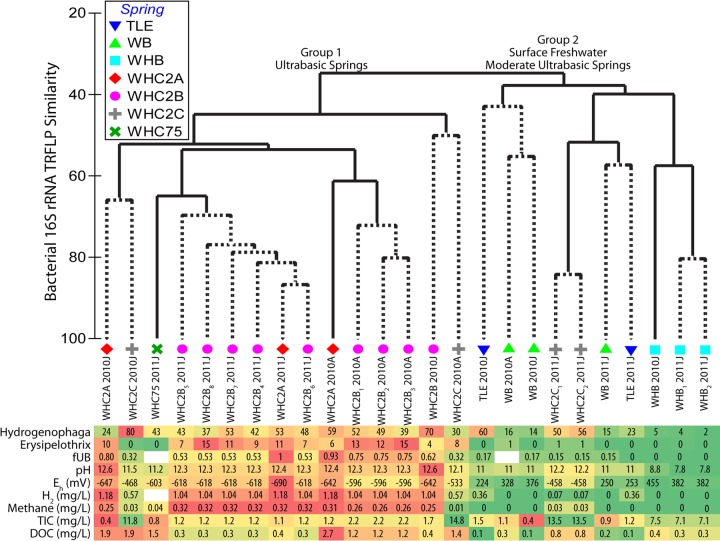
Community compositions of the fluids (measured by the presence/absence of TRFLP fragment clusters) generally reflect their geochemical characteristics. The community similarity dendrogram in Fig. 2 illustrates two main sample groupings: one group of the most ultrabasic springs (WHC2A and WHC2B) and another group with the freshwater end member (WHB) and moderately ultrabasic springs (TLE and WB). The cophenetic correlation was 0.88, indicating a good fit between the dendrogram clusters and the underlying similarity values between each sample. When replicate samples from the same site were available (indicated by subscript numbers in the sample labels of Fig. 2), the replicates showed high similarity (70 to 84%) to each other and supported the significance of the sample groupings.
Fig 2.
Dendrogram illustrating bacterial community similarity (as measured by the presence/absence of TRFLP fragment clusters) among samples collected from Tablelands serpentinite springs and surface freshwater. Dotted lines represent samples that could not be distinguished by a SIMPROF test. Subscript numbers indicate replicate samples from the same spring. The abundance levels of Hydrogenophaga and Erysipelothrix in the underlying table reflect the percent relative abundance of the corresponding TRFLP fragment cluster in each sample. Color coding of the table is an arbitrary visual aid to identify the highest and lowest numbers for each variable. Sample names and associated geochemical characteristics correspond to data provided in Table 1.
Exceptions and outliers to this overall pattern were also evident. The three samples from the mixing site WHC2C were found in both main groups and exhibited little similarity to each other or to any of the other samples. Such high variability in the bacterial community may reflect a highly variable mixing site. This variability, however, was not reflected in the geochemical characteristics of this site: fUB, pH, Eh, methane, and TIC all appeared to be remarkably consistent over time at WHC2C (Table 1; Fig. 2). Although the lower fUB of WHC2C-2011J compared to that of WHC2C-2010A and WHC2C-2010J may partially explain its anomalous bacterial community composition, the latter two samples had almost identical fUB values but very little similarity between their bacterial communities.
The June 2010 samples from WHC2A and WHC2B also appeare to have anomalous community compositions compared to other samples from the same springs. The fUB, pH, Eh, H2, and methane values for these samples were indistinguishable from values for other WHC2A and WHC2B samples, but their community compositions were most similar to samples from the WHC2C mixing site (Fig. 2). Therefore, these sites may have experienced more heterogeneity in their bacterial community compositions during the time of sampling than would be expected from their geochemical characteristics. Furthermore, it is always possible that these differences reflect environmental parameters that were not measured in this study.
Taxonomic diversity.
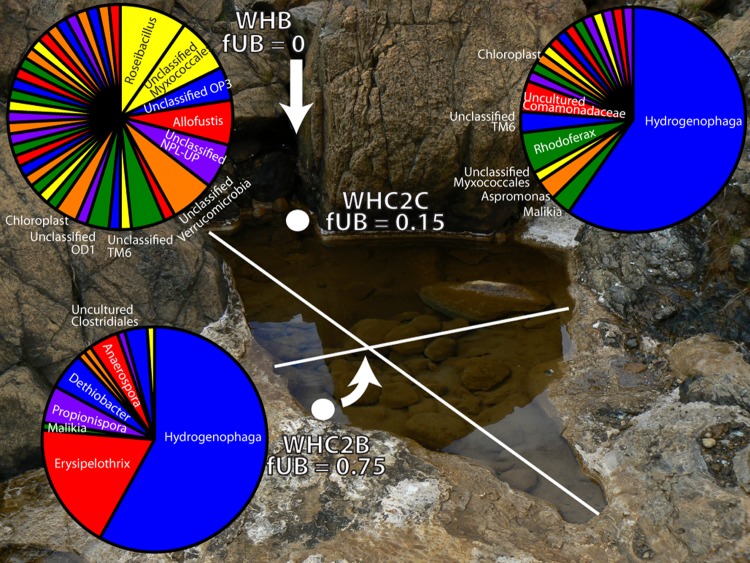
The TRFLP-based community composition data were further informed by taxonomic classification of 16S rRNA tag sequences from a few of the samples. The pie charts in Fig. 3 display the proportion of tag sequences assigned to each genus for three of the samples representing one example of an ultrabasic spring, the freshwater end member, and a mixing site. The most ultrabasic spring sample from which we were able to generate tag sequences (WHC2B-2010A; fUB, 0.75) was dominated by two genera, Hydrogenophaga (class Betaproteobacteria, order Burkholderiales, family Comamonadaceae) and Erysipelothrix (class Erysipelotrichi, order Erysipelotrichales, family Erysipelotrichaceae). Only 134 genera were assigned to the 19,930 sequences in WHC2B-2010, and 6 genera accounted for 89% of the sequences.
Fig 3.
Proportions of each genus in representative samples of an ultrabasic spring (WHC2B), a mixing site (WHC2C), and surface freshwater (WHB).White dots indicate sampling locations for WHC2B and WHC2C. The sampling location for WHB is located approximately 10 m upstream from the pool depicted in this photograph. Arrows indicate subsurface fluid exiting from WHC2B and surface freshwater entering the pool at WHC2C from upstream. The white lines indicate the width (∼1 m) and length (∼3 m) of the pool. Results in this figure are derived from JGI 16S rRNA tag sequencing. A full taxonomic summary is provided in Table S1 of the supplemental material.
Hydrogenophaga also appeared relatively abundant in the mixing site (WHC2C-2011J), but sequences assigned to genus Erysipelothrix comprised only 0.15% of this sample. Hydrogenophaga comprised only 0.09% of the freshwater end member WHB-2011J (pH 7.8; fUB, 0), and Erysipelothrix was completely absent. WHB exhibited much more evenness than the ultrabasic spring (Fig. 3), and its most abundant taxa were nearly completely absent in WHC2B-2010A. In the moderate spring WB-2011J (pH 10.8; fUB, 0.15), Hydrogenophaga represented 6% of all sequences, and Erysipelothrix was represented by a single sequencing read (0.02% of the total). In another moderate spring, TLE-2011J (pH 10.8; fUB, 0), Hydrogenophaga represented 11% of total sequences, and Erysipelothrix was completely absent (see Table S1 in supplemental material).
Bootstrap support for the automated classification of the most common Hydrogenophaga sequence was very strong (97%). Independent phylogenetic analysis was consistent with this classification, although the closest neighbors identified by evolutionary placement were members of the genus Malikia (Fig. 4A). In our phylogenetic analysis, however, this genus was not distinct from genus Hydrogenophaga. To our knowledge, Spring et al. (40) published the only phylogeny of Malikia, and those authors noted that Malikia formed a distinct lineage only in phylogenetic reconstructions that included all known members of the Comamonadaceae family. For convenience and clarity in this report, we will refer to these tag sequences as Hydrogenophaga. To our knowledge, no characterized Hydrogenophaga or Malikia isolates are capable of growth above pH 9, but growth of an uncharacterized Hydrogenophaga strain has been observed in pH 12 groundwater (41).
Fig 4.
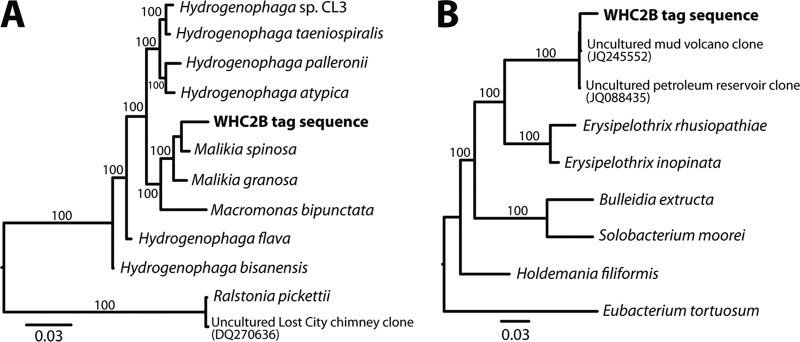
Maximum-likelihood phylogenetic trees of Hydrogenophaga-related (A) and Erysipelothrix-related (B) 16S rRNA sequences. For each tree, the representative tag sequence was placed next to its most likely neighbor in the reference phylogeny of nearly full-length sequences by using the evolutionary placement algorithm (32). Each tree represents the best topology after 20 maximum-likelihood references, and all nodes in the reference phylogeny received 100% bootstrap support.
Classification of tag sequences as Erysipelothrix was very weak (<50%), and >60% bootstrap support was only available for these sequences at the phylum level. Evolutionary placement of the most common Erysipelothrix CoDL tag sequence onto a reference phylogeny indicated two environmental clones, one from a pH 8.4 mud volcano (42) and one from a pH 8 to 9 oil well (43), as its closest neighbors (Fig. 4B). The most similar cultivated isolates to these sequences belong to genus Erysipelothrix, so both automated taxonomic classification and our phylogenetic analysis indicate this genus as the best taxonomic assignment for these tag sequences. It is possible, however, that future work with these organisms may reveal that they form a novel genus.
In addition to comparing taxonomic assignments, we also directly examined the distribution of tag sequence OTUs among samples independent of taxonomic classification. These results (see Table S1 and Fig. S2 in the supplemental material) highlighted the composite nature of the mixing site (WHC2C-2011J), which shared 28% of its sequences with the most ultrabasic spring (WHC2B-2010A) and 15% of its sequences with the freshwater end member (WHB). In contrast, WHC2B-2010A shared very few OTUs with WHB-2011J. The subset of sequences shared by all three samples (12 OTUs comprising 65%, 47%, and 1% of sequences in WHC2B-2010A, WHC2C-2011J, and WHB-2011J, respectively) was surprisingly large, and this was due to the dominance of Hydrogenophaga in the two ultrabasic samples and its minor representation in WHB-2011J.
The TRFLP and tag sequencing analyses employed in this study used Bacteria-specific primers, and neither analysis was expected to detect Archaea. Attempts to amplify archaeal 16S rRNA genes have been unsuccessful in our laboratory, and very few archaeal sequences were identified in the previously published metagenomic study of Tablelands springs (16).
Biogeochemical correlations.
We tested for Pearson correlations among geochemical factors and the distribution of individual bacterial types as measured by the relative abundance of their corresponding TRFLP fragment clusters in each sample. The tag sequence data were not informative for this correlation analysis due to the small number of samples for which those data were available. The TRFLP data were available for more samples (and replicate samples), and the relative abundance of the Hydrogenophaga and Erysipelothrix TRFLP fragment clusters were strongly correlated to the relative abundance of the corresponding tag sequence OTUs in the same samples (Pearson r > 0.9, P < 0.002). Therefore, biogeochemical correlations were tested with relative abundances of TRFLP fragment clusters, consistent with the practice of other researchers with similar data sets (e.g., reference 37).
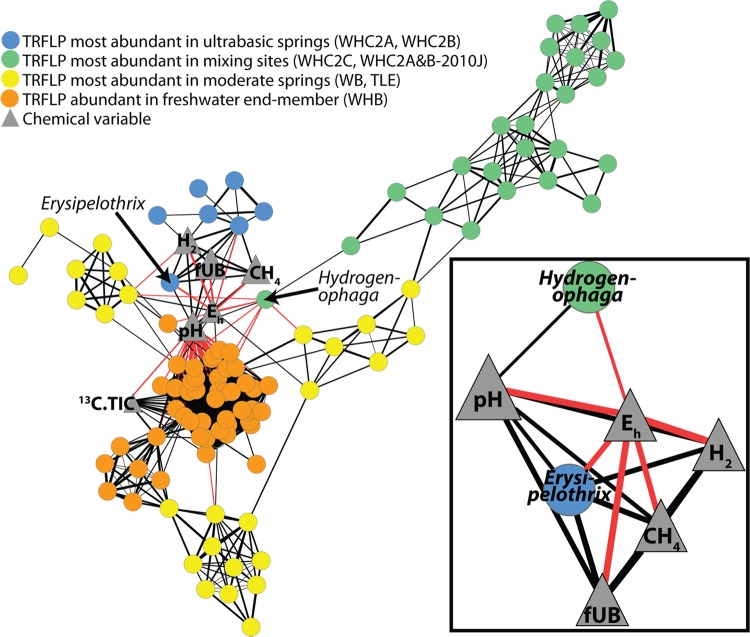
Statistically significant (P < 0.05 and q < 0.05) correlations are visualized as a network in Fig. 5. By linking predicted restriction fragment sizes of the tag sequences with the observed restriction fragment sizes in the TRFLP profiles, we were able to assign taxonomies to many of the TRFLP fragment clusters, including those corresponding to Hydrogenophaga and Erysipelothrix.
Fig 5.
Association network of the relative abundance of TRFLP fragment clusters and geochemical variables. Each circle is a TRFLP fragment cluster, and its color corresponds to the type of sample in which its relative abundance was greatest (see figure legend). Positive correlations are black lines; negative correlations are red lines; the width of each line is proportional to the r value. All correlations with P values of <0.05 and q levels of <0.05 are shown. The inset shows geochemical correlations only for Hydrogenophaga and Erysipelothrix, which were identified by linking observed TRFLP fragment sizes to predicted restriction sizes of tag sequences. For clarity, taxonomic assignments for other TRFLP fragment clusters are not shown. The six blue circles other than Erysipelothrix represent unidentified TRFLP fragment clusters, which did not constitute more than 3% of the total TRFLP signal in any sample.
The overview of the network highlights cooccurrence patterns distinguishing the few ultrabasic-enriched bacteria from those mostly present in mixing sites, moderate springs, or the freshwater end member. Interestingly, bacteria that were most abundant in the mixing sites or the moderate springs were not strongly correlated with any of the geochemical variables. Most of these bacteria formed cooccurrence subnetworks that were only indirectly linked to Eh and pH via their cooccurrence with Hydrogenophaga.
Interestingly, the fragment cluster corresponding to Erysipelothrix was the only one with significant correlations to all of the geochemical variables expected to indicate subsurface fluid (Fig. 5, inset): pH (r = 0.70, P = 0.005, q = 0.01); Eh (r = 0.81, P = 0.0004, q = 0.002); fUB (r = 0.83, P = 0.0002, q = 0.001); methane (r = 0.81, P = 0.004, q = 0.002), and hydrogen (r = 0.78, P = 0.001, q = 0.004). Six additional fragment clusters were, strictly speaking, most abundant in the ultrabasic springs, but even so, they were only present in very low abundance in those samples. Corresponding tag sequences could not be identified for these rare fragment sizes, and so taxonomic assignments are not available. These results (Fig. 5) provide further statistical support for the initial, qualitative interpretation from the community composition data (Fig. 2 and 3) that of all the taxa identified in these samples, Erysipelothrix is most strongly linked to ultrabasic (and potentially subsurface) fluids.
DISCUSSION
Serpentinite springs are potential windows into the subsurface biosphere and offer the advantages of convenient sampling (e.g., no deep sea submersibles required) and pristine environmental conditions (e.g., no drilling required). Because photosynthetic processes at the Earth's surface can swamp any signal from the subsurface biosphere, it is necessary to tightly integrate microbiological and geochemical analyses and to carefully interpret the presence of genetic material in the context of environmental measurements. Our analysis of the correlations among eight geochemical variables, >5,000 16S rRNA OTUs, and molecular fingerprinting (TRFLP) profiles of 17 serpentinite spring samples identified two main taxa as potentially important indicators of a serpentinization-influenced subsurface biosphere: Hydrogenophaga and Erysipelothrix.
The Hydrogenophaga 16S rRNA sequence that dominated the Tablelands ultrabasic springs matched that found in previously published metagenomic data from the same site (16). The Hydrogenophaga-associated metagenomic data included genes involved in carbon fixation via RuBisCO and in the aerobic oxidation of hydrogen and carbon monoxide. Characterized Hydrogenophaga species are known to be facultatively autotrophic, i.e., they oxidize hydrogen to power carbon fixation only when organic carbon is unavailable (44, 45). They are also aerobes or facultative anaerobes (45).
This prior knowledge is consistent with the data from the present study that indicate a role for Hydrogenophaga in oxic-anoxic transition zones in Tablelands springs. Their relative abundance was correlated with high pH and low Eh (conditions which can be achieved in near-surface environments), but they were not correlated with indicators of subsurface sources: H2, methane, or the fraction of ultrabasic water (fUB). Furthermore, they did not have significant cooccurrence patterns with taxa that were enriched in the most ultrabasic springs, but they did cooccur with taxa that were most abundant in mixing sites, moderate springs, and the freshwater end member (Fig. 5). In future studies, more quantitative analyses should be employed to estimate the absolute numbers of Hydrogenophaga cells and their activities under various environmental conditions, but all currently available information points to these organisms occupying transition zones where hydrogen-enriched, ultrabasic subsurface fluids mix with oxygenated surface freshwater. Therefore, Hydrogenophaga in the Tablelands springs appears to be a surface indicator of underlying subsurface geochemical processes that produce hydrogen-enriched, ultrabasic fluids.
In contrast to Hydrogenophaga, the Erysipelothrix sequences identified in the most ultrabasic Tablelands springs were strongly correlated with all of the available geochemical factors that are indicative of ultrabasic, anoxic subsurface fluids (Fig. 5, inset). Previously published metagenomic data from this same site featured many diverse hydrogenase-encoding sequences affiliated with Clostridia and Erysipelotrichi (16). The Clostridia and Erysipelotrichi are sister classes within the phylum Firmicutes, and the taxonomic binning and phylogenetic analyses in the previous metagenomics study could not distinguish them. Therefore, many of the Clostridia-related metagenomics sequences in reference 16, including some of the hydrogenases, probably correspond to the Erysipelothrix 16S rRNA sequences presented here.
The most common Erysipelothrix tag sequence was 98 to 99% similar over its length (450 bp) to environmental sequences from a terrestrial mud volcano (42) and a terrestrial oil well (43) in China. Interestingly, both of these studies of subsurface habitats also detected Hydrogenophaga. In the mud volcano study, Erysipelothrix was only detected in sediments 23 cm below the surface of the bubbling pool, where it formed 1% of the clone library (see the supplementary information for reference 42). Hydrogenophaga, in contrast, was absent at 23 cm, but it was the second-most-common taxon (11% of the clone library) in shallower sediments, 7 cm below the surface (see Fig. 4 in reference 42). These Hydrogenophaga clones are ∼95% similar to the Hydrogenophaga tag sequence that dominates the Tablelands springs.
Hydrogenophaga is also found within hydrogen-enriched deep boreholes in Finland (46) and South Africa (47). Erysipelothrix-related sequences have also been detected in the Finland borehole, but Clostridia are much more abundant in that environment. Clostridia also dominate deep fluids in the South Africa boreholes, where Hydrogenophaga is also more common in shallower fluids (47). Therefore, Hydrogenophaga appears to be a cosmopolitan inhabitant of hydrogen-enriched shallow subsurface environments, supporting the interpretation of this study that Hydrogenophaga is a near-surface indicators of hydrogen-producing reactions in the underlying subsurface. Furthermore, all of these hydrogen-enriched environments also featured abundant Clostridia and/or Erysipelotrichi in deeper, more anoxic zones than where Hydrogenophaga appears to thrive in the same locations.
Clostridia 16S rRNA sequences, including the same Desulfotomaculum genus that dominates deep borehole fluids in South Africa (47), are also abundant in the best-characterized marine site of active serpentinization, the Lost City hydrothermal field (33, 48). Hydrogenophaga appears to be absent in Lost City, but Ralstonia, a sister genus within the family Comamonadaceae, is present. Lost City chimneys also contain abundant hydrogenases that are similar to those found in both Hydrogenophaga- and Clostridia/Erysipelothrix-related metagenomic sequences from the Tablelands (16). Therefore, organisms related to the Comamonadaceae family and the Clostridia/Erysipelotrichi classes appear to be cosmopolitan in hydrogen-enriched subsurface environments in both marine and continental settings.
Although the in situ activities of Hydrogenophaga in these environments remain to be demonstrated experimentally, all of the available data indicate that they are most likely aerobic (or facultatively anaerobic) hydrogen-oxidizing chemolithoautotrophs. The biogeochemical roles of the Clostridia/Erysipelotrichi, however, are less clear. The most common Clostridia organisms in the South Africa deep boreholes are expected to be hydrogen-oxidizing sulfate reducers (49), but the Tablelands metagenomic data do not support this interpretation for the Erysipelothrix in Tablelands springs (16). Instead, the presence of many fermentation-associated hydrogenases in that study suggested the possibility that these organisms could be utilizing organic compounds and some of these organic compounds may be generated de novo by subsurface serpentinization-associated reactions (8, 10, 50).
The interdisciplinary approach of the present study provides additional support for Erysipelothrix as a bona fide denizen of the subsurface biosphere, but its physiology remains unclear. One possibility that should be addressed in future studies is that Clostridia/Erysipelotrichi sequences dominate low-diversity fluids because of their ability to tolerate extreme conditions (perhaps aided by spore formation) but not necessarily thrive in them. This possibility is somewhat contradictory to the observation that the Erysipelothrix phylotype is most abundant in springs that have greater cell densities than found in surface waters (Table 1), suggesting that serpentinization-associated subsurface processes are indeed stimulating its growth. Of course, this hypothesis should be tested in future studies exploring the physiology of these organisms.
All other bacteria detected in these springs are probably derived from the freshwater end member or, like Hydrogenophaga, inhabit transition zones where they may utilize serpentinization-associated products but are dependent on surface sources of energy or nutrients. Identification of the few subsurface-associated organisms from the many bacterial taxa detected in these springs required the biogeochemical correlation analyses made possible by the integrated, interdisciplinary approach of this study.
In summary, this study contributes to the emerging biogeographic trend that organisms related to the Comamonadaceae family cooccur with the Clostridia/Erysipelotrichi classes and that the hydrogen-oxidizing, chemolithoautrophic Comamonadaceae inhabit shallower zones than those inhabited by the most likely anaerobic Clostridia/Erysipelotrichi. Further insight into the biogeochemical roles of these organisms will require careful coordination and integration of a variety of biogeochemical measurements to distinguish subsurface signals from the photosynthetic surface biosphere. Our application of this approach in the present study was intended to demonstrate the advantages and challenges of applying this approach to natural, serpentinite springs.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge tremendous support and teamwork in the field from Mark Wilson, Candice Ratley, Chris Earle, Amanda Rietze, and Heidi Kavanagh. We also thank Katrina Twing for technical assistance and helpful discussions. The CoDL data were made possible by the Deep Carbon Observatory's Census of Deep Life, which is supported by the Alfred P. Sloan Foundation. Pyrosequencing was performed at the Marine Biological Laboratory (Woods Hole, MA), and we are grateful for the assistance of Mitch Sogin, Susan Huse, Joseph Vineis, Andrew Voorhis, Sharon Grim, and Hilary Morrison at the Marine Biological Laboratory. The JGI data were generated as part of the DOE JGI Community Sequencing Program (CSP-671; supported by the Office of Science of the U.S. Department of Energy under contract DE-AC02-05CH11231).
Funding for this study was provided by the NASA Astrobiology Institute through the Carnegie Institution for Science (CAN-5) to M.O.S. and a NASA Postdoctoral Fellowship to W.J.B. The geochemical component of this study was supported in part by grants from the Canadian Space Agency (CSA) Canadian Analogue Research Network (CARN) and CSA's Field Investigations, a Natural Sciences and Engineering Research Council (NSERC) Discovery Grant awarded to P.L.M., and an NSERC Alexander Graham Bell Canada Graduate Scholarship awarded to N.S.
ADDENDUM IN PROOF
During production of this paper, we became aware of a report on a similar ultrabasic ecosystem featuring Hydrogenophaga and Clostridia (I. Tiago and A. Veríssimo, Environ. Microbiol., 29 November 2012, doi:10.1111/1462-2920.12034).
Footnotes
Published ahead of print 12 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00330-13.
REFERENCES
- 1. Whitman WB, Coleman DC, Wiebe WJ. 1998. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. U. S. A. 95:6578–6583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Edwards KJ, Becker K, Colwell F. 2012. The deep, dark energy biosphere: intraterrestrial life on earth. Annu. Rev. Earth Planet. Sci. 40:551–568 [Google Scholar]
- 3. Kallmeyer J, Pockalny R, Adhikari RR, Smith DC, D'Hondt S. 2012. Global distribution of microbial abundance and biomass in subseafloor sediment. Proc. Natl. Acad. Sci. U. S. A. 109:16213–16216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Colwell FS, D'Hondt S. 2013. Nature and extent of the deep biosphere. Rev. Mineral. Geochem. 75:547–574 [Google Scholar]
- 5. Edwards KJ, Fisher AT, Wheat CG. 2012. The deep subsurface biosphere in igneous ocean crust: frontier habitats for microbiological exploration. Front. Microbiol. 3:8. 10.3389/fmicb.2012.00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nealson KH, Inagaki F, Takai K. 2005. Hydrogen-driven subsurface lithoautotrophic microbial ecosystems (SLiMEs): do they exist and why should we care? Trends Microbiol. 13:9. 10.1016/j.tim.2005.07.010 [DOI] [PubMed] [Google Scholar]
- 7. Orcutt BN, Sylvan JB, Knab NJ, Edwards KJ. 2011. Microbial ecology of the dark ocean above, at, and below the seafloor. Microbiol. Mol. Biol. Rev. 75:361–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Proskurowski G, Lilley MD, Seewald JS, Fruh-Green GL, Olson EJ, Lupton JE, Sylva SP, Kelley DS. 2008. Abiogenic hydrocarbon production at Lost City hydrothermal field. Science 319:604–607 [DOI] [PubMed] [Google Scholar]
- 9. Brazelton WJ, Mehta MP, Kelley DS, Baross JA. 2011. Physiological differentiation within a single-species biofilm fueled by serpentinization. mBio 2(4):e00127–11. 10.1128/mBio.00127-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lang SQ, Früh-Green GL, Bernasconi SM, Lilley MD, Proskurowski G, Méhay S, Butterfield DA. 2012. Microbial utilization of abiogenic carbon and hydrogen in a serpentinite-hosted system. Geochim. Cosmochim. Acta 92:82–99 [Google Scholar]
- 11. Schrenk MO, Kelley DS, Bolton SA, Baross JA. 2004. Low archaeal diversity linked to subseafloor geochemical processes at the Lost City hydrothermal field, mid-Atlantic ridge. Environ. Microbiol. 6:1086–1095 [DOI] [PubMed] [Google Scholar]
- 12. Kelley DS, Karson JA, Fru GL, Yoerger DR, Shank TM, Butterfield DA, Hayes JM, Schrenk MO, Olson EJ, Proskurowski G, Jakuba M, Bradley A, Larson B, Ludwig K, Glickson D, Buckman K, Bradley AS, Brazelton WJ, Roe K, Bernasconi SM, Elend MJ, Lilley MD, Baross JA, Summons RE, Sylva SP. 2005. A serpentinite-hosted ecosystem: the Lost City hydrothermal field. Science 307:1428–1434 [DOI] [PubMed] [Google Scholar]
- 13. Deming JW, Baross JA. 1993. Deep-sea smokers: windows to a subsurface biosphere? Geochim. Cosmochim. Acta 57:3219–3230 [DOI] [PubMed] [Google Scholar]
- 14. Schrenk MO, Huber JA, Edwards KJ. 2010. Microbial provinces in the subseafloor. Annu. Rev. Mar. Sci. 2:279–304 [DOI] [PubMed] [Google Scholar]
- 15. Szponar N, Brazelton WJ, Schrenk MO, Bower DM, Steele A, Morrill PL. Geochemistry of a continental site of serpentinization, the Tablelands Ophiolite, Gros Morne National Park: a Mars analogue. Icarus, in press. 10.1016/j.icarus.2012.07.004 [DOI] [Google Scholar]
- 16. Brazelton WJ, Nelson B, Schrenk MO. 2012. Metagenomic evidence for H2 oxidation and H2 production by serpentinite-hosted subsurface microbial communities. Front. Microbiol. 2:268. 10.3389/fmicb.2011.00268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schrenk MO, Kelley DS, Delaney JR, Baross JA. 2003. Incidence and diversity of microorganisms within the walls of an active deep-sea sulfide chimney. Appl. Environ. Microbiol. 69:3580–3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hobbie JE, Daley RJ, Jasper S. 1977. Use of nuclepore filters for counting bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 33:1225–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huber JA, Butterfield DA, Baross JA. 2002. Temporal changes in archaeal diversity and chemistry in a mid-ocean ridge subseafloor habitat. Appl. Environ. Microbiol. 68:1585–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, Arrieta JM, Herndl GJ. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc. Natl. Acad. Sci. U. S. A. 103:12115–12120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Marteinsson VT, Rúnarsson Á, Stefánsson A, Thorsteinsson T, Jóhannesson T, Magnússon SH, Reynisson E, Einarsson B, Wade N, Morrison HG, Gaidos E. 2013. Microbial communities in the subglacial waters of the Vatnajökull ice cap, Iceland. ISME J. 7:427–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huse SM, Dethlefsen L, Huber JA, Welch DM, Relman DA, Sogin ML. 2008. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 4:e1000255. 10.1371/journal.pgen.1000255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huse SM, Welch DM, Morrison HG, Sogin ML. 2010. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ. Microbiol. 12:1889–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Caporaso JG, Lauber CL, Walters WA, Berg-lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U. S. A. 108:4516–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kong Y. 2011. Btrim: a fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics 98:152–153 [DOI] [PubMed] [Google Scholar]
- 26. Schloss PD, Westcott SL. 2011. Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl. Environ. Microbiol. 77:3219–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pruesse E, Peplies J, Glöckner FO. 2012. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28:1823–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 [DOI] [PubMed] [Google Scholar]
- 32. Berger SA, Krompass D, Stamatakis A. 2011. Performance, accuracy, and web server for evolutionary placement of short sequence reads under maximum likelihood. Syst. Biol. 60:291–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brazelton WJ, Schrenk MO, Kelley DS, Baross JA. 2006. Methane- and sulfur-metabolizing microbial communities dominate the Lost City hydrothermal field ecosystem. Appl. Environ. Microbiol. 72:6257–6270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Abdo Z, Schüette UME, Bent SJ, Williams CJ, Forney LJ, Joyce P. 2006. Statistical methods for characterizing diversity of microbial communities by analysis of terminal restriction fragment length polymorphisms of 16S rRNA genes. Environ. Microbiol. 8:929–938 [DOI] [PubMed] [Google Scholar]
- 35. Osborne CA, Rees GN, Bernstein Y, Janssen PH. 2006. New threshold and confidence estimates for terminal restriction fragment length polymorphism analysis of complex bacterial communities. Appl. Environ. Microbiol. 72:1270–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Clarke KR. 1993. Non-parametric multivariate analyses of changes in community structure. Austral. Ecol. 18:117–143 [Google Scholar]
- 37. Fuhrman J, Steele J. 2008. Community structure of marine bacterioplankton: patterns, networks, and relationships to function. Aquat. Microb. Ecol. 53:69–81 [Google Scholar]
- 38. Rizopoulos D. 2006. ltm: an R package for latent variable modeling. J. Stat. Softw. 17:5 [Google Scholar]
- 39. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Spring S, Wagner M, Schumann P, Kämpfer P. 2005. Malikiagranosa gen. nov., sp. nov., a novel polyhydroxyalkanoate- and polyphosphate-accumulating bacterium isolated from activated sludge, and reclassification of Pseudomonas spinosa as Malikiaspinosa comb. nov. Int. J. Syst. Evol. Microbiol. 55:621–629 [DOI] [PubMed] [Google Scholar]
- 41. Roadcap GS, Sanford RA, Jin Q, Pardinas JR, Bethke CM. 2006. Extremely alkaline (pH >12) ground water hosts diverse microbial community. Ground Water 44:511–517 [DOI] [PubMed] [Google Scholar]
- 42. Cheng T-W, Chang Y-H, Tang S-L, Tseng C-H, Chiang P-W, Chang K-T, Sun C-H, Chen Y-G, Kuo H-C, Wang C-H, Chu P-H, Song S-R, Wang P-L, Lin L-H. 2012. Metabolic stratification driven by surface and subsurface interactions in a terrestrial mud volcano. ISME J. 6:2280–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tang Y-Q, Li Y, Zhao J-Y, Chi C-Q, Huang L-X, Dong H-P, Wu X-L. 2012. Microbial communities in long-term, water-flooded petroleum reservoirs with different in situ temperatures in the Huabei Oilfield, China. PLoS One 7:e33535. 10.1371/journal.pone.0033535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schwartz E, Voigt B, Zühlke D, Pohlmann A, Lenz O, Albrecht D, Schwarze A, Kohlmann Y, Krause C, Hecker M, Friedrich B. 2009. A proteomic view of the facultativelychemolithoautotrophic lifestyle of Ralstoniaeutropha H16. Proteomics 9:5132–5142 [DOI] [PubMed] [Google Scholar]
- 45. Willems A, Busse J, Goor M, Pot B, Falsen E, Jantzen E, Hoste B, Gillis M, Kersters K, Auling G, De Ley J. 1989. Hydrogenophaga, a new genus of hydrogen-oxidizing bacteria that includes Hydrogenophagaflava comb.nov. (formerlyPseudomonas flava), Hydrogenophagapalleronii (formerly Pseudomonas palleronii), Hydrogenophagapseudoflava (formerly Pseudomonas pseudoflava and “Pseudomonas carboxydoflava”), and Hydrogenophagataeniospiralis (formerly Pseudomonas taeniospiralis). Int. J. Syst. Bacteriol. 39:319–333 [Google Scholar]
- 46. Itävaara M, Nyyssönen M, Kapanen A, Nousiainen A, Ahonen L, Kukkonen I. 2011. Characterization of bacterial diversity to a depth of 1500 m in the Outokumpu deep borehole, Fennoscandian Shield. FEMS Microbiol. Ecol. 77:295–309 [DOI] [PubMed] [Google Scholar]
- 47. Moser DP, Gihring TM, Brockman FJ, Fredrickson JK, Balkwill DL, Dollhopf ME, Lollar BS, Pratt LM, Boice E, Southam G, Wanger G, Baker BJ, Pfiffner SM, Lin L, Onstott TC. 2005. Desulfotomaculum and Methanobacterium spp. dominate a 4- to 5-kilometer-deep fault. Appl. Environ. Microbiol. 71:8773–8783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Brazelton WJ, Ludwig KA, Sogin ML, Andreishcheva EN, Kelley DS, Shen C-C, Edwards RL, Baross JA. 2010. Archaea and bacteria with surprising microdiversity show shifts in dominance over 1000-year time scales in hydrothermal chimneys. Proc. Nat. Acad. Sci. U. S. A. 107:1612–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chivian D, Brodie EL, Alm EJ, Culley DE, Dehal PS, DeSantis TZ, Gihring TM, Lapidus A, Lin L-H, Lowry SR, Moser DP, Richardson PM, Southam G, Wanger G, Pratt LM, Andersen GL, Hazen TC, Brockman FJ, Arkin AP, Onstott TC. 2008. Environmental genomics reveals a single-species ecosystem deep within Earth. Science 322:275–278 [DOI] [PubMed] [Google Scholar]
- 50. Schrenk MO, Brazelton WJ, Lang SQ. 2013. Serpentinization, carbon and deep life. Rev. Mineral. Geochem. 75:575–606 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.