Abstract
Acinetobacter baumannii has become a major problem in the clinical setting with the prevalence of infections caused by multidrug-resistant strains on the increase. Nevertheless, only a limited number of molecular mechanisms involved in the success of A. baumannii as a human pathogen have been described. In this study, we examined the virulence features of a hypermotile derivative of A. baumannii strain ATCC 17978, which was found to display enhanced adherence to human pneumocytes and elevated levels of lethality toward Caenorhabditis elegans nematodes. Analysis of cellular lipids revealed modifications to the fatty acid composition, providing a possible explanation for the observed changes in hydrophobicity and subsequent alteration in adherence and motility. Comparison of the genome sequences of the hypermotile variant and parental strain revealed that an insertion sequence had disrupted an hns-like gene in the variant. This gene encodes a homologue of the histone-like nucleoid structuring (H-NS) protein, a known global transcriptional repressor. Transcriptome analysis identified the global effects of this mutation on gene expression, with major changes seen in the autotransporter Ata, a type VI secretion system, and a type I pilus cluster. Interestingly, isolation and analysis of a second independent hypermotile ATCC 17978 variant revealed a mutation to a residue within the DNA binding region of H-NS. Taken together, these mutants indicate that the phenotypic and transcriptomic differences seen are due to loss of regulatory control effected by H-NS.
INTRODUCTION
Acinetobacter baumannii is widely recognized as a highly multidrug-resistant pathogen that causes major problems in hospitals globally (1, 2). Antimicrobial resistance in A. baumannii has been extensively studied, and resistance determinants novel to Acinetobacter are being identified regularly. These resistance genes can often be found on horizontally acquired genetic material, increasing the risk of generating pan-resistant clones. As such, global dissemination of these multidrug-resistant clones poses a serious threat. Furthermore, biofilm formation has also previously been associated with increased resistance and survival as well as immune evasion and may therefore play a significant role in A. baumannii pathogenicity. Adherence characteristics, such as biofilm formation and binding to eukaryotic cells, have been studied in a large number of A. baumannii strains (3–9) and constitute but one of many potential factors involved in virulence. An additional factor that can influence virulence potential is motility; two distinct forms of surface migration have been described for this species (3, 7, 10–12). Expression of these phenotypes varies between strains and to a degree between clonal groups, such as the conserved expression of twitching motility by isolates in the international clone I group (3). Although motility has been associated with increased biofilm formation and virulence in other bacteria, such as Pseudomonas aeruginosa and Dichelobacter nodosus (13–18), the significance of motility in the virulence potential of A. baumannii remains to be confirmed.
Various molecular mechanisms contributing to the virulence potential of A. baumannii have been identified (19). For example, one of the most extensively studied virulence determinants in A. baumannii is OmpA, which not only facilitates adherence to eukaryotic cells and abiotic surfaces but also mediates invasion and promotes cell death of lung epithelial cells (20–24). The penicillin binding protein 7/8, phospholipase D, lipopolysaccharides, and production of K1 capsule in A. baumannii have also been shown to play important roles in survival in a host environment (25–28). Furthermore, phospholipase D facilitates bacterial crossing of the blood-lung barrier, as shown in a rat model system (26). More recently, siderophore-mediated iron acquisition mechanisms have been demonstrated to be essential for lethality in mice (29). Many virulence determinants found in other Gram-negative pathogens have been identified in the genome sequences of A. baumannii, such as the type VI secretion system (30, 31). However, to date, their role in virulence and persistence of A. baumannii has not been characterized.
The regulatory networks controlling expression of A. baumannii virulence determinants, such as iron acquisition, motility, attachment, and biofilm formation, remain largely unknown. Transcriptomic studies have indicated that the ferric uptake regulator (FUR) is the primary regulator of A. baumannii iron acquisition mechanisms (10, 32), and a number of studies have shown that quorum sensing influences both motility and biofilm formation (12, 33, 34). Additionally, a two-component regulatory system, encoded by bfmRS, plays a critical role in the regulation of the Csu type I pili, which may influence attachment and motility (35).
It is likely that A. baumannii has also acquired virulence factor genes via exogenous DNA uptake. In other bacterial genera, transcriptional regulation of horizontally acquired genetic material is in part controlled by the histone-like nucleoid structuring (H-NS) protein and provides a level of protection against expression of genes that encode products with detrimental effects to the host bacteria (36, 37). Although Acinetobacter spp. are known to possess large quantities of horizontally acquired genetic material, to date the function of H-NS in A. baumannii has not been described.
This article describes the comprehensive characterization of a hypermotile derivative of A. baumannii strain ATCC 17978. Whole-genome sequencing of this strain revealed the insertional inactivation of a gene encoding the global regulator H-NS. Beyond the dramatic changes observed with respect to motility, the mutant strain also displayed altered adherence phenotypes for biotic surfaces, as well as an increase in virulence using a Caenorhabditis elegans model system. In summary, this study describes the importance of H-NS in expression of A. baumannii persistence and virulence genes.
MATERIALS AND METHODS
Bacterial strains.
Acinetobacter strain ATCC 17978 (CP000521) was obtained from the American Type Culture Collection (ATCC), and Escherichia coli OP50 was kindly provided by Hannah Nicholas (The University of Sydney). Freeze-dried Acinetobacter cells were revived as recommended by the ATCC, and colony material from the overnight cultures grown on Luria-Bertani (LB) medium were transferred to a cryopreservation tube containing 20% glycerol in LB broth and placed at −80°C for long-term storage. The isolated variants, i.e., 17978hm and HNSmut88, were similarly stored. For subsequent culturing purposes, material was scraped from the top of the frozen stocks and streaked onto LB medium.
Biofilm assays.
The static biofilm formation assay was performed as described previously (3). In brief, overnight cultures were diluted 1:100 in fresh Mueller-Hinton (MH) broth in polystyrene microtiter trays and incubated overnight at 37°C. Adherent cells were washed once with phosphate-buffered saline (PBS), stained by incubation with 0.1% crystal violet for 30 min at 4°C, and washed three times with PBS. Dye was released from the cells using ethanol-acetone (4:1) and shaking at 200 rpm for 30 min at room temperature. Absorbance was measured at 595 nm by using a Fluostar Omega spectrometer (BMG Labtech, Offenburg, Germany).
Pellicle formation assays.
Pellicle formation assays were based on a method used previously (6). Overnight bacterial cultures in LB broth were diluted in fresh LB broth containing 100 mM NaCl. Pellicle formation assays were performed in polypropylene tubes with incubation at room temperature without shaking for 72 h. The pellicle film was separated from the tube by the addition of methanol. The pellicle biomass was measured (optical density at 600 nm [OD600]) after resuspending pelleted cells in 1 ml PBS. The experiments were performed at least three times.
Eukaryotic cell adherence assays.
The adherence of A. baumannii cells to A549 human pneumocytes was investigated as described previously (3). Cell lines were grown in Dulbecco's modified Eagle medium (Invitrogen, Australia) supplemented with 10% fetal bovine serum (Bovogen, Australia). Prior to use, the cell monolayer was examined microscopically to ensure >95% coverage. Washed A549 monolayers in 24-well tissue culture plates were subsequently infected with a bacterial inoculum containing ∼1 × 107 CFU. After incubation at 37°C for 4 h, culture medium was removed, and the monolayers were washed three times with 1 ml of PBS. Cell monolayers were detached from the plate by treatment with 100 μl of 0.25% trypsin and 0.02% EDTA in PBS. Eukaryotic cells were subsequently lysed by the addition of 400 μl 0.025% Triton X-100, and serial 10-fold dilutions thereof were plated on LB agar to determine the number of CFU of adherent bacteria per well. Collated data for adherence assays were obtained from at least three independent experiments and represent the data points for each experiment for quadruplicate wells. The CFU of the cell culture medium after 4 h of incubation was determined to ensure that strains did not display differences in their respective growth rates during the adherence assay.
Caenorhabditis elegans killing assays.
C. elegans N2 nematodes were synchronized in their development by initially placing nematode eggs on E. coli OP50 seeded nematode growth medium (NGM) (38). The larval L3/L4-stage nematodes were harvested and placed on NGM agar plates seeded with A. baumannii strains. Viability of at least 200 individual nematodes found in random fields of view was determined by microscopic examination. The viability was determined at 24-h intervals for up to 144 h and expressed as a percentage of live nematodes. The results (n = 4) represent duplicate experiments performed on two different days. Two independent researchers determined the viability of the nematodes in a “blind” experiment.
Cell surface hydrophobicity tests.
Cell surface hydrophobicity was examined as described previously (39). The OD of the cell suspensions before (ODinitial) and after (ODfinal) addition of xylene was measured at 600 nm. Experiments were performed three times. The hydrophobicity index (HI), expressed as a percentage, was calculated using the following formula: HI (%) = [(ODintial − ODfinal)/ODinitial] × 100. Experiments were performed at least three times.
Phenotype MicroArray analysis.
A. baumannii ATCC 17978 wild-type (WT) and 17978hm mutant strains were cultured on LB agar overnight. Cells were transferred to 1× IF-0 inoculation fluid (Biolog, Inc.) to 42% transmittance. Cells were then diluted 1:5 in 1× IF-0 containing dye A (Biolog, Inc.) with a final transmittance of 85%. One hundred microliters of the cell suspensions were transferred to each well of Biolog MicroPlate PM01 and PM02A plates. Plates were incubated in the OmniLog Phenotype MicroArray system (Biolog, Inc.) at 37°C for 48 h, with readings taken every 15 min. Data from the WT and mutant strains were overlaid using the OmniLog File Management/Kinetic Analysis software, v1.20.02, and analyzed using OmniLog Parametric Analysis software, v1.20.02 (Biolog, Inc.).
Fatty acid analyses.
The strains for fatty acid analysis were grown for 8 h on MH plates containing 0.25% agar. Cells were collected and washed with prechilled PBS. Total cellular lipids were extracted using chloroform-isopropanol and were subsequently methylated using a 1% sulfuric acid solution in methanol, as described previously (40). The fatty acid methyl esters were separated by gas chromatography at the School of Agriculture, Food and Wine, University of Adelaide. The abundance of each fatty acid was expressed as the percentage of the total of all fatty acids. All analyzed bacterial strains were prepared on two separate occasions, and the data represent the averages over those two experiments.
RNA isolation.
Cells used for measurement of transcription levels were harvested from semisolid MH medium (0.25% agar) using prechilled PBS. The cells were subsequently pelleted by centrifugation and lysed in TRIzol reagent (Invitrogen, Australia) and chloroform. Following phase separation by centrifugation, RNA was extracted from the aqueous phase using a RNA isolation kit (Bioline). DNase I (Promega) treatment was performed as per the manufacturer's recommendations.
Transcriptome analysis.
We used a custom genomic microarray for A. baumannii ATCC 17978, as previously described (10). The results represent data obtained from three biological replicates and a dye swap experiment. The array was designed to harbor at least four probes per gene.
qRT-PCR.
cDNA was synthesized using random hexamers (GeneWorks, Australia) and Moloney murine leukemia virus (M-MLV) reverse transcriptase (Promega) as per the manufacturer's recommendations. Oligonucleotides used in this study were designed using the software program Primer3 (41) as an integral part of UGENE v1.6.1 (Unipro) and are listed in Table S1 in the supplemental material. Quantitative reverse transcription-PCR (qPCR) was performed on a Rotor-Gene RG-3000 instrument (Corbett Life Science, Australia) using DyNAmo SYBR green qPCR kits (Finnzymes, Australia). Internal qPCR controls used primers designed to 16S rRNA (A1S_r01) to ensure that differences seen are due solely to alterations in the target gene expression and not to mRNA quality or quantity. Transcriptional differences were calculated using the ΔΔCT method (42), and the data represent experiments performed as biological triplicates.
Genome sequence analysis.
A. baumannii ATCC 17978 and 17978hm were sequenced using Illumina BeadArray technology, performed by the Ramaciotti Centre for Gene Function Analysis, University of New South Wales, Sydney, Australia. The whole-genome shotgun sequence reads were assembled using the software program Velvet 1.1 (43).
Genetic complementation of the hypermotile A. baumannii mutant strains.
The hns PCR product was cloned into BamHI-digested pWH1266 (44). Electro-competent A. baumannii cells freshly prepared on the day of use were incubated on ice with plasmid DNA for 5 min followed by electroporation using a MicroPulser instrument (Bio-Rad) at 2.5 kV, 200 Ω, 25 μF. After recovery in 1 ml of LB medium for at least 1 h at 37°C, cells were cultured overnight at 37°C on LB medium containing 200 μg/ml ampicillin.
Microarray data accession number.
The transcriptomic data have been deposited in the gene expression omnibus database (http://www.ncbi.nlm.nih.gov/geo/) and can be accessed using the accession number GSE40681.
RESULTS AND DISCUSSION
Isolation of hypermotile variant strains.
Contrary to their original designation as nonmotile, a number of A. baumannii strains have been shown to participate in various forms of motility when grown under appropriate conditions (3). Motility of strain A. baumannii ATCC 17978 is evident on semisolid Luria-Bertani (LB) medium containing <0.5% agar (3, 10, 12), with concentrations of agar above this level inhibiting the phenotype. However, we observed two distinct morphologies in a cryopreserved ATCC 17978 stock cultured on LB medium containing 1% agar. The colony morphology of one variant appeared nonmotile as per wild-type (WT) ATCC 17978, whereas the other displayed a motile appearance, which was designated a hypermotile phenotype. To examine the motility of these variants, five individual hypermotile colonies were inoculated into the center of both LB and Mueller-Hinton (MH) media containing different concentrations of agar. The phenotype was most distinct using MH medium containing 0.25% agar, conditions nonpermissive for motility of WT cells. Whereas four out of five hypermotile variants were equally motile and covered the entire surface of the plate within 8 h (migration, ∼45 mm), one hypermotile variant was found to be delayed by approximately 2 h compared to the other hypermotile variants. This indicated that at least two distinct variants of the parental strain were isolated. To examine potential genotypic differences that may explain the altered phenotypic characteristics, one of the hypermotile strains displaying motility on semisolid MH medium, designated 17978hm, and the parent WT ATCC 17978 strain were selected for further analyses.
Sequence analysis of the hypermotile A. baumannii ATCC 17978 derivatives.
The genetic differences between the A. baumannii strains 17978hm and ATCC 17978 were assessed by sequencing both their genomes using Illumina BeadArray technology. Sequence reads were assembled using the software program Velvet 1.1 (43), generating 245 contigs for strain 17978hm and 292 contigs for strain ATCC 17978. Whole-genome alignments were generated using the program Mauve, and the ATCC 17978 genome sequence reported by Smith et al. (CP000521) (30) was included for comparative purposes.
The most striking finding from comparative analyses of the parental and 17978hm genome sequences was an insertion sequence (IS) element found in the hns (A1S_0268) locus of strain 17978hm (Fig. 1A). In other bacteria, H-NS has been shown to act as a global repressor that preferentially binds AT-rich DNA sequences (36, 37). Spontaneous insertional inactivation of hns, as observed in strain 17978hm, has also been found in Mycobacterium smegmatis (45). Interestingly, the M. smegmatis H-NS mutant strain was also found to display a hypermotile phenotype (39). The H-NS protein is well conserved between Acinetobacter strains; the ATCC 17978 H-NS protein sequence is 100% identical to that from strains AYE and SDF and 92% identical to H-NS from Acinetobacter baylyi ADP1. The ATCC 17978 genome encodes only a single copy of hns, and to date, strain AB058 is the only A. baumannii strain found to harbor two hns-like genes (data not shown).
Fig 1.
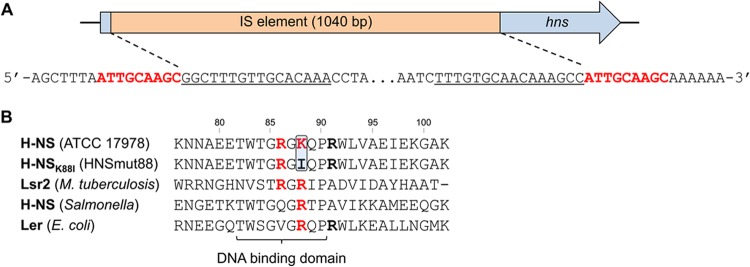
Genetic characteristics of hypermotile A. baumannii strains. (A) Using whole-genome sequencing of the ATCC 17978 and 17978hm strains, an insertion sequence (IS) element was identified in a position unique to A. baumannii strain 17978hm. The sequence as found in strain 17978hm has been provided; the target sites of the IS element are shown in red, and the inverted repeat section of the IS element is underlined. (B) Amino acid sequences of the DNA binding region of the H-NS-like proteins from strain ATCC 17978, HNSmut88, Mycobacterium tuberculosis, Salmonella, and E. coli are displayed. Positively charged residues that may be involved in DNA interactions are shown in red. The arginine (R) residues positioned outside the DNA binding domain are in bold. The lysine (K) in position 88 in strain ATCC 17978 mutated to an isoleucine (I) in strain HNSmut88 is boxed in blue.
Analysis of the DNA sequence revealed that transposition of an IS element into hns in strain 17978hm had occurred with nine nucleotides of the hns target sequence being duplicated and now flanking the termini of the IS element (Fig. 1A). The sequence of this IS element exhibited >99% identity to an IS element harboring A1S_0628, which encodes a putative transposase in strain ATCC 17978, raising the possibility that the IS element identified within the hns locus of strain 17978hm was generated by a transposition event. Two similar transposases found in strain 17978hm and ATCC 17978, designated A1S_2554 and A1S_1172, showed 97% and 84% sequence identity to A1S_0628, respectively, and as such are unlikely to be the originator of the insertion.
As mentioned above, a total of five hypermotile variants were isolated in this study. The hns gene of the four other hypermotile strains was PCR amplified and sequenced. Three of these were found to possess an insertion disruption identical to that seen in strain 17978hm and also displayed a hypermotile phenotype similar to that of 17978hm (data not shown). The hns gene of the variant displaying a less-pronounced level of hypermotility (motility delayed by 2 h on semisolid MH medium compared to that of strain 17978hm) contained a single nucleotide polymorphism, resulting in a lysine-to-isoleucine substitution at position 88 of the H-NS protein (H-NSK88I). Interestingly, lysine 88 is part of the DNA-binding domain of H-NS-like proteins (46) (Fig. 1B). Lacking this positively charged residue may have resulted in an altered affinity for regulatory binding sites and consequently reduced repression of the genes located downstream. As mentioned, the A. baumannii strain expressing the H-NSK88I protein, designated HNSmut88, displayed an intermediate hypermotile phenotype compared to that of strain 17978hm, which was investigated by comparing the positively charged residues in the DNA binding domain to those in other H-NS-like proteins, such as Lsr2 from Mycobacterium tuberculosis, H-NS from Salmonella, and Ler from Escherichia coli (46, 47). Whereas the Salmonella H-NS and E. coli Ler proteins contain a single positively charged residue in the DNA binding domain, M. tuberculosis Lsr2 and A. baumannii H-NS contain two; in A. baumannii H-NS, these are lysine and arginine at positions 88 and 86, respectively (Fig. 1B). Previous mutagenesis studies in M. tuberculosis have shown a significant level of Lsr2 DNA binding in mutants where only a single positively charged residue in the DNA binding domain was removed (47). However, double Lsr2 mutations or mutation of the single charged residue present in the DNA binding domain of Salmonella H-NS resulted in a complete lack of binding (47). Therefore, arginine 86 may be sufficient to maintain significant binding affinity of H-NSK88I to certain regulatory targets, explaining the intermediate motility phenotype observed in this strain. Although the A. baumannii H-NS and E. coli Ler proteins possess an arginine adjacent to the DNA-binding domain (Fig. 1B), when it is present as the sole positively charged residue in this region of the protein, it does appear to be insufficient for maintaining the ability to bind its targets (46).
To confirm that the altered phenotypes observed in the hypermotile A. baumannii variants were a result of mutations in hns, the WT hns gene was cloned into the E. coli-Acinetobacter shuttle vector pWH1266, producing pWH0268, and used for complementation in the mutant strains. Motility assays revealed that introduction of pWH0268 into A. baumannii strains 17978hm and HNSmut88 resulted in loss of the hypermotile phenotype on solid LB medium and semisolid MH medium, returning the phenotype to that observed for WT cells (data not shown). Due to the reduced severity of phenotypic changes observed in HNSmut88 compared to those in 17978hm, only the ATCC 17978 and 17978hm strains were assessed in subsequent phenotypic and transcriptional analyses.
Adherence characteristics.
Biofilm formation is a complex process initiated by attachment to a surface, followed by the formation of a multilayered biomass containing secondary structures. It was hypothesized that the adherence characteristics of strain 17978hm may differ from that of WT cells since changes in motility could affect biofilm formation, as previously reported for P. aeruginosa (18). However, adherence to the surface of a polystyrene microtiter tray showed no major differences between the WT and hypermotile strains (data not shown). In contrast, pellicle formation, i.e., the biofilm at the air-liquid interface, appeared significantly higher for 17978hm than for the WT strain or its complemented derivative 17978hm (pWH0268) (Fig. 2A). The level of pellicle biomass in strain 17978hm (pWH0268) was higher than that observed for the WT strain, which could be a result of loss of the complementation plasmid over the 72-h incubation period. Pellicle formation was observed in LB medium only at 25°C and not at 37°C (data not shown), which corroborates data from similar studies of Acinetobacter spp. (6). Attempts to demonstrate pellicle formation by strains grown in MH medium were unsuccessful. During static culturing for pellicle formation, it was apparent that the planktonic growth of the pellicle forming 17978hm cells was lower than that of the non-pellicle-forming WT cells (OD600 = ∼0.15 and ∼0.50, respectively). The differences in planktonic growth may be due to a reduction of oxygen levels in the growth medium of strain 17978hm as a result of the high oxygen dependency of the pellicle, a phenomenon described previously (48).
Fig 2.
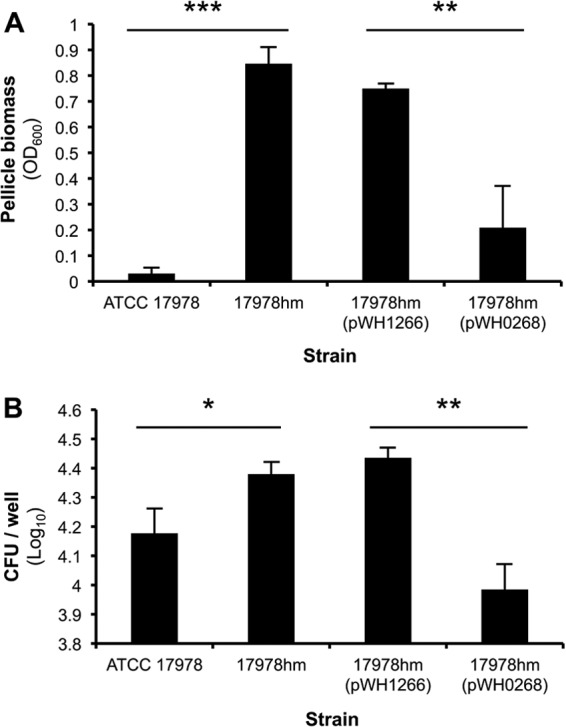
Adherence characteristics of strains ATCC 17978 and 17978hm. (A) Pellicles of A. baumannii strains ATCC 17978 and 17978hm were examined after incubation for 72 h at 25°C, and statistically significant (P < 0.001; two-tailed Student's t test) differences are indicated by asterisks (**, P < 0.05; ***, P < 0.001). Experiments were performed at least three times, and error bars represent the standard deviations. (B) The CFU in log10 values of A. baumannii ATCC 17978, 17978hm, 17978hm (pWH1266), and 17978hm (pWH0268), recovered from a washed A549 cell culture after incubation for 4 h, were enumerated. Significant differences between ATCC 17978 and 17978hm (P < 0.05), and 17978hm(pWH1266) and 17978hm(pWH0268) (P < 0.005) were observed using a two-tailed Student t test and are indicated by asterisks (*, P < 0.005; **, P < 0.05). Error bars show the standard errors of the means.
Adherence to abiotic and biotic surfaces appears to be mediated by different molecular mechanisms in most A. baumannii strains and as such does not always correlate (3, 5, 49). Therefore, despite similar abiotic adherence levels observed in microtiter trays, adherence to biotic surfaces by strains ATCC 17978, 17978hm, 17978hm (pWH1266), and 17978hm (pWH0268) was investigated. A549 pneumocytes were selected for these experiments to mimic adherence to the epithelial layer of the human lung (50). After the bacteria were incubated in conjunction with pneumocytes for 4 h, the 17978hm cells adhered to and potentially intracellularly located in the washed A549 eukaryotic cells were enumerated, and the number was found to be significantly higher than the number of ATCC 17978 cells (∼1.6-fold; P < 0.05) (Fig. 2B). Interestingly, adherence levels of 17978hm (pWH0268) cells were lower than those for WT cells, indicating a possible dose-dependent effect due to a difference in the plasmid copy number (35), where H-NS may be more abundant in the complemented cells. The increased adherence potential of strain 17978hm to human pneumocytes may indicate that 17978hm cells have higher persistence levels during infection and more specifically pneumonia. Further work using a mouse pneumonia model may reveal whether the differences seen in adherence to pneumocytes play a biological significant role in the disease process.
The hypermotile variant possesses an increased virulence potential.
Since 17978hm showed a significantly increased ability to adhere to eukaryotic cells, its disease potential was investigated further by examining strains ATCC 17978 and 17978hm for their ability to kill C. elegans nematodes. Measurable death of the C. elegans nematodes was observed between 72 and 144 h of incubation on either WT or 17978hm cells (Fig. 3). A significant difference between the percentage of live nematodes incubated with either WT or 17978hm cells was observed at 120 h and 144 h (Fig. 3). At both time points, death rates for nematodes incubated with 17978hm cells that were approximately 20% higher than rates for those incubated with WT cells were observed (Fig. 3). The experiment was terminated after 144 h, since the remaining live nematodes were consuming the disintegrated dead nematodes at time points beyond 144 h, making it difficult to accurately determine the percentage of live versus dead nematodes during the latter part of the experiment (data not shown).
Fig 3.
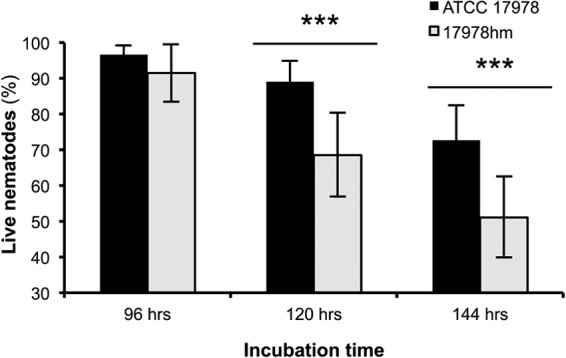
Examination of the virulence potentials of ATCC 17978 and 17978hm. The virulence potentials of A. baumannii strains ATCC 17978 and 17978hm were examined by counting live versus dead C. elegans nematodes grown on a lawn of the respective A. baumannii test strains. Significant differences in killing were observed at 120 h and 144 h as determined by Student's t test and are indicated by asterisks (***, P < 0.001). The error bars show the standard deviations (n = 4).
Cell surface hydrophobicity and fatty acid composition.
The differences between the phenotypes of the WT and hypermotile A. baumannii cells described above may be related to changes at the cell surface. Therefore, the hydrophobicity of WT and 17978hm cells was investigated using the microbial adhesion to hydrocarbons (MATH) test (39); an increase in the hydrophobicity index (HI) of 17978hm cells (HI = 65%) compared to that for WT cells (HI = 44%) was observed (Table 1). When a WT copy of hns (carried on pWH0268) was introduced into 17978hm cells, the hydrophobicity decreased significantly, returning to levels lower than those observed for ATCC 17978 cells.
Table 1.
Cell surface hydrophobicity and fatty acid composition
| Strain | Hydrophobicity index (SD) | Fatty acid (%) |
||
|---|---|---|---|---|
| 15:0 | 16:0 | 17:0 | ||
| ATCC 17978 | 44 (13.1) | 1.9 | 29.1 | 3.4 |
| 17978hm | 65 (6.4) | 2.8 | 25.4 | 6.9 |
| 17978hm(pWH1266) | 64 (3.3) | 2.4 | 24.4 | 5.2 |
| 17978hm(pWH0268) | 14 (0.1) | 0.7 | 31.4 | 2.6 |
To investigate a potential cause for the change in cell surface hydrophobicity, the total fatty acid compositions of WT and 17978hm cells and 17978hm(pWH1266) and 17978hm(pWH0268) cells were determined. In a study of Listeria innocua, decreases in the ratio between C15 and C17 saturated fatty acids were linked to increases in cell surface hydrophobicity (51). Similarly, in this study, the percentage of C17 fatty acid was significantly higher in the hypermotile strains, which may explain the differences observed in cell surface hydrophobicity (Table 1). Major changes in the concentrations of other fatty acids were not observed.
Transcriptomic analysis of the motile versus nonmotile populations.
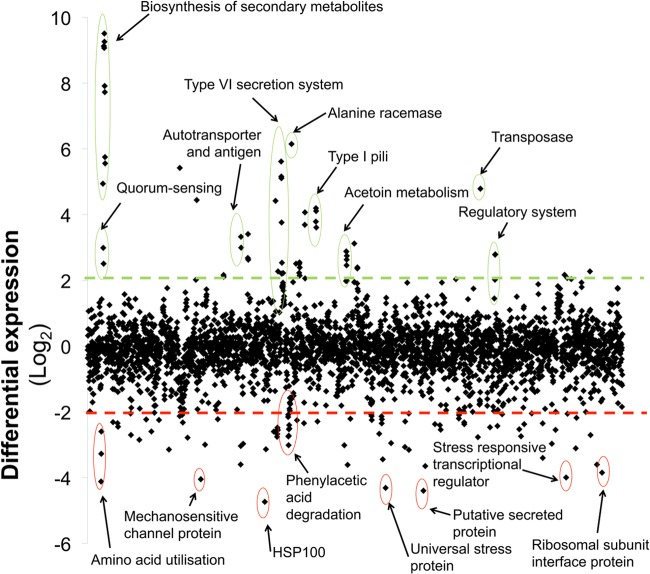
Comparative analysis of the transcriptomes of cells in distinct lifestyles may provide information about the molecular mechanisms and regulatory pathways responsible for driving a population into a certain mode of living. Therefore, we employed a whole-genome microarray to examine the effect of the hns inactivation on the transcriptome of A. baumannii. Major differences between the transcriptomes of the hypermotile 17978hm and nonmotile WT cells were observed (Fig. 4). More than 4-fold-differential expression was seen for 152 genes, of which 91 were down- and 61 upregulated (see Table S2 in the supplemental material) (GEO:GSE40681). The most striking differences were observed in the quorum-sensing-regulated genes encoding homoserine lactone synthase (A1S_0109) and the homoserine lactone responsive regulator (A1S_0111), which were both heavily upregulated. Furthermore, an adjacent cluster involved in the production of secondary metabolites, putatively lipopeptides or polyketides, was upregulated by more than 100-fold. Both quorum sensing and the adjacent cluster have been shown to play an important role in motility of A. baumannii (12).
Fig 4.
Overview of the transcriptional differences between motile and nonmotile A. baumannii cells. Comparative transcriptomics are displayed as the differential expression (in log2 values) of strains 17978hm (motile) and ATCC 17978 (nonmotile) harvested from semisolid MH medium (0.25% agar). Diamond markers indicate the differential expression levels of all predicted open reading frames of the ATCC 17978 genome and are sorted on the x axis according to the locus tag. The dashed lines indicate 4-fold (log2 = 2) differential expression; upregulated genes in motile populations are located above the green line and downregulated genes below the red line. Examples of differentially expressed genes, such as the genes involved in biosynthesis of quorum-sensing signals and those encoding proteins of the phenylacetic acid degradation pathway, are indicated.
Various genes encoding surface-presented structures were also heavily upregulated in A. baumannii strain 17978hm. The autotransporter Ata (A1S_1032) was overexpressed by approximately 10-fold. In a recent report, Ata was found to play a major role in adherence and virulence of A. baumannii strain ATCC 17978 (52), potentially explaining the increased virulence levels of strain 17978hm observed in the C. elegans killing assays described above. This gene appears to be transcriptionally linked to A1S_1033 (8-fold upregulation), the product of which shares homology with putative OmpA-like proteins (20–24). Even though there is no evidence of a role for type I pili in promotion of surface translocation in A. baumannii, four genes encoding a novel type I pilus cluster (A1S_1507-A1S_1510) were upregulated by more than 10-fold. Other phenotypes, such as increased adherence to human epithelial cells or increased pellicle formation, may be associated with overexpression of type I pili in the hypermotile strain. A gene cluster predicted to encode a type VI secretion system (A1S_1292-1311) was found to be upregulated in the motile population. Type VI secretion systems can contribute to bacterial pathogenicity, as observed for P. aeruginosa (53); however, the role of these systems in A. baumannii is yet to be elucidated (54). It would appear not to be essential for full virulence in a number of isolates, since comparative genomic analyses revealed that four open reading frames encoding proteins of unknown function have replaced the type VI secretion system gene cluster in A. baumannii strain D1279779 (accession no. AERZ00000000). The type VI secretion system may also be nonfunctional in strain 1656-2 (CP001921) (55), since it contains an insertion element in the gene homologous to A1S_1302. Furthermore, the gene cluster coding for the type VI secretion system can also be found in the nonpathogenic A. baumannii strain SDF (CU468230). Therefore, the function of the type VI secretion system in A. baumannii requires further investigation.
Interestingly, genes functioning in fatty acid biosynthesis, including fabG (A1S_0524) and fabF (A1S_0525), were found to be overexpressed by more than 2-fold in strain 17978hm. FabF is responsible for fatty acid elongation, which occurs in steps of two carbon additions, i.e., C13 to C15 to C17 (56). Thus, these increases in transcription levels may be related to the increase of C17 observed in 17978hm cells compared to findings for WT cells (Table 1).
Numerous genes functioning in metabolism, such as those encoding members of the phenylacetic acid degradation pathway, were found to be downregulated in the motile population (Fig. 4). Downregulation of metabolic pathways was also observed in a transcriptional investigation of swarming P. aeruginosa cells (57). A Phenotype MicroArray (Biolog, Inc.) analysis using MicroPlates PM01 and PM02A was performed to obtain a comprehensive depiction of carbon source utilization by the Acinetobacter strains 17978hm and ATCC 17978. The most pronounced dissimilarities between these strains were observed in the presence of l-threonine or d-malic acid, in which WT ATCC 17978 cells showed more active respiration than 17978hm cells (see Table S3 in the supplemental material). The upregulation of a putative threonine efflux transporter (A1S_3397) in 17978hm may be associated with a reduction in available threonine and consequently lower respiratory levels seen in this strain. Changes in transcription levels of genes encoding proteins involved in d-malic acid metabolism were not observed (data not shown).
Identification of potential H-NS targets in the ATCC 17978 genome.
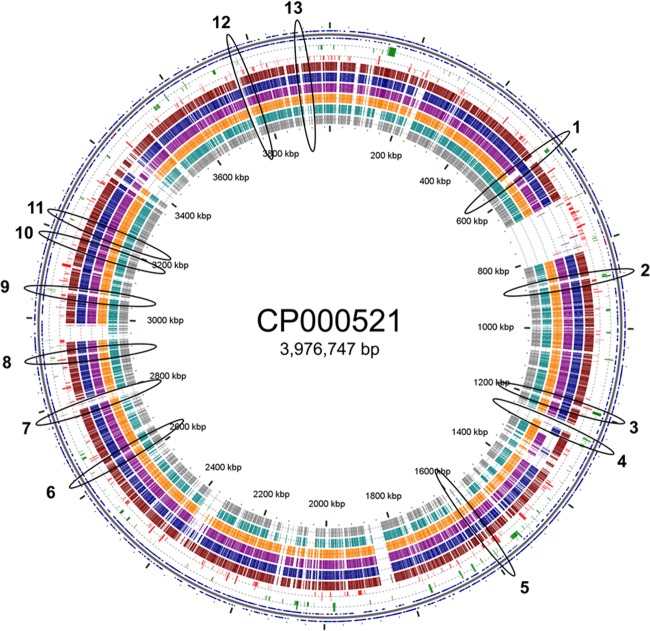
Although not previously examined for A. baumannii, H-NS is a well-studied protein for many bacterial genera (58, 59). Sequence homology between H-NS proteins from different bacteria is often low (<30%); however, the targets appear to be similar. Horizontally acquired genetic material is often AT rich and is therefore a likely target for H-NS proteins (59). Hence, the H-NS proteins are also known as xenogeneic silencers. A common example of a horizontally acquired genomic region targeted by H-NS encodes the type VI secretion system (60). Indeed, this cluster (A1S_1292-A1S_1312) was found to be heavily upregulated in strain 17978hm (Fig. 4). Based on the potential for H-NS to act as a xenogeneic silencer, other potential regulatory targets were identified encoded within novel genomic regions. Transcriptomic data were overlaid with the results of BLASTP comparisons of CP000521 (ATCC 17978) with six other Acinetobacter genomes, those of A. baumannii AB0057, ACICU, AYE, AB307-0294, and SDF and A. baylyi ADP1, on a circular representation of the ATCC 17978 genome (Fig. 5). A correlation was seen between upregulation and lack of conservation of the respective genes/gene clusters. These nonconserved upregulated loci were subsequently analyzed using a whole-genome alignment generated in MAUVE (61) containing the genomes described above. Thirteen potential horizontally acquired H-NS targets (Fig. 5), which included a surface protein (A1S_0745) and the autotransporter adhesin (A1S_1032) described above, were identified. Interestingly, in E. coli, transcription of a gene encoding the autotransporter protein UpaC was recently found to be repressed by H-NS (62), consistent with our findings. These surface-presented proteins, potentially regulated by H-NS, may play a role in the phenotypic alterations of strain 17978hm. The S-adenosyl-l-methionine-dependent methyltransferase (A1S_2744), also potentially regulated by H-NS, is involved in methylation of proteins, lipids, DNA, and RNA and therefore in controlling a wide range of cellular processes (63). The putative threonine efflux transporter protein described above (A1S_3397), also appeared to be under regulatory control of H-NS (Fig. 5). Overall, these analyses suggested that various horizontally acquired genome regions are targets for transcriptional repression by H-NS in A. baumannii and that inactivation of H-NS results in upregulation of the genes within these regions.
Fig 5.
Bioinformatic analysis of the putative H-NS targets in the ATCC 17978 genome. Transcriptome results were mapped onto a circular representation of A. baumannii ATCC 17978 (CP000521) using the software program CGview. Upregulated genes are represented in green and downregulated genes in red; half-sized bars equal 2- to 4-fold differential expression, and full-sized bars indicate expression of >4-fold. To identify potentially horizontally acquired genomic regions, comparative BLASTP analyses between CP000521 (outer ring) and A. baumannii AB0057 (brown), ACICU (blue), AYE (purple), AB307-0294 (orange), SDF (turquoise), and A. baylyi ADP1 (gray) are included. Various upregulated genes or gene clusters not fully conserved between CP000521 and other genomes were identified; these were subsequently examined using Mauve. Based on CGview and Mauve analyses, the following 13 upregulated genes or gene clusters were found to be horizontally acquired and therefore putative H-NS target sites: 1, A1S_0519-A1S_0525, fatty acid biosynthesis; 2, A1S_0745, bacterial surface protein; 3, A1S_1032-A1S_1033, Ata and a putative antigen; 4, A1S_1078-A1S_1079, hypothetical protein and dichlorophenol hydroxylase; 5, A1S_1357, alanine racemase; 6, A1S_2271, RNA splicing ligase; 7, A1S_2396, transcriptional regulator; 8, A1S_2509, putative chaperone; 9, A1S_2648-A1S_2649, putative regulatory proteins; 10, A1S_2744, S-adenosyl-l-methionine-dependent methyltransferase; 11, A1S_2789, metallopeptidase; 12, A1S_3273, putative secreted protein; 13, A1S_3397, lysine (LysE) or the homoserine/threonine resistance (RhtB) protein.
Although H-NS is known to bind high-AT-percentage regions, the global GC-skew analysis shown in the circular genome figure did not reveal such an association with transcriptional upregulation (Fig. 5). This may be due to the generally low GC percentage of the A. baumannii ATCC 17978 genome (∼39%). However, when examining the upstream regions of heavily overexpressed genes, a correlation between high AT content and transcriptional upregulation in strain 17978hm was observed. The GC percentages of various potential H-NS targets were as low as 20% (Fig. 6), which is significantly lower than the average GC percentage of the ATCC 17978 genome-wide intergenic regions (∼35%).
Fig 6.
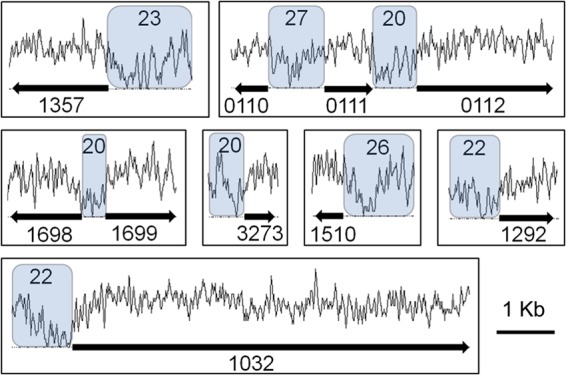
GC percentage of the putative H-NS targets. The GC-content plots are shown above schematic representations of genes differentially expressed in the hns mutant strain, 17978hm, with their respective locus tag number displayed underneath. The average GC-percentage has been calculated over the regions located upstream of these highly upregulated genes and is highlighted in blue. The GC-content plots were derived from the A. baumannii ATCC 17978 (CP000521) genome displayed in UGENE v1.10.1 (UniPro). These regions have a GC percentage significantly lower than the average genome-wide intergenic regions (∼35%) and may form suitable H-NS binding sites.
The genomic region harboring the gene encoding homoserine lactone synthase (A1S_0109) and the cluster involved in biosynthesis of polyketides/lipopeptides (A1S_0112-A1S_0119) possessed two regulatory sites with a low GC percentage (Fig. 6). In particular, the upstream region of the biosynthesis cluster (GC content of 20%) may be targeted by H-NS. Previously, Clemmer et al. showed that the A1S_0112-A1S_0119 cluster is under the control of quorum-sensing signals in the form of acyl-homoserine lactones via AbaR (12).
Other overexpressed genes with a putative H-NS binding region upstream included the above-described autotransporter Ata and putative ompA-like gene (A1S_1032-A1S_1033), the type VI secretion system (A1S_1292-A1S_1312), a novel type I pilus cluster (A1S_1510-A1S_1507), a cluster involved in lipoate synthesis and acetoin metabolism (A1S_1698-A1S_1704), and a small putatively secreted protein (A1S_3273) (Fig. 6).
Alanine racemases are ubiquitous in prokaryotes and are responsible for racemization of l- and d-alanine. Like most other prokaryotes, A. baumannii strains possess two alanine racemases, DadX (A1S_0096), mediating conversion of d- to l-alanine, and Alr (A1S_2176), which facilitates l- to d-alanine racemization. However, A. baumannii ATCC 17978 also contains a second Alr-like alanine racemase encoded by A1S_1357. This heavily upregulated gene in strain 17978hm (>70-fold) appeared to have been horizontally acquired and also possessed an AT-rich upstream region, making it a likely target for transcriptional repression by H-NS (Fig. 5 and 6).
Transcriptional analysis of putative H-NS-regulated genes.
We investigated the transcription levels of nine differentially expressed genes in the complemented strain pair, 17978hm (pWH1266) and 17978hm (pWH0268). These genes are potential targets of H-NS based on previously described findings or the bioinformatic analysis performed in this study. Seven of these genes, A1S_0111, A1S_0112, A1S_1032, A1S_1292, A1S_1510, A1S_1699, and A1S_3273, were highly upregulated (>4-fold), and two, A1S_0095 and A1S_1336, were highly downregulated (>4-fold), as determined by our microarray data. Comparative qRT-PCR analysis examining the control 17978hm (pWH1266) and complemented 17978hm (pWH0268) strains, using the oligonucleotides listed in Table S1 in the supplemental material, showed significant up- or downregulation of the respective genes that were found to be significantly differentially expressed in strain 17978hm compared to expression in the WT strain by microarray analysis (Table 2). It is noteworthy that three of the investigated genes are known to be coregulated by other proteins, A1S_0111 and A1S_0112 by AbaR and A1S_0095 by Lrp, and therefore it is difficult to assess the magnitude of the effect that H-NS exerts on these genes. Nevertheless, successful complementation of the hypermotile variants at phenotypic and transcriptional levels was shown, confirming that inactivation of H-NS resulted in various alterations, including an increased virulence potential and transcriptional changes observed in the hypermotile mutants.
Table 2.
Transcriptional profiling of putative H-NS-regulated genes
| Locus tag | Gene product | Differential expression level, fold (SD) |
|
|---|---|---|---|
| 17978hm(pWH1266) vs 17978hm(pWH0268)a | 17978hm vs ATCC 17978b | ||
| A1S_0111 | AbaR | 5.5 (1.9) | 5.7 |
| A1S_0112 | Lipopeptide/polyketide | 100.4 (23.4) | 558.6 |
| A1S_1032 | Ata | 5.3 (1.8) | 10.0 |
| A1S_1292 | Type VI secretion system | 359.3 (56.1) | 49.0 |
| A1S_1510 | Type I pili | 86.6 (6.4) | 12.2 |
| A1S_1699 | Acetoin metabolism | 5.0 (0.1) | 7.4 |
| A1S_3273 | Putative peptide signal | 82.0 (39.4) | 4.8 |
| A1S_0095 | d-Amino acid dehydrogenase | −4.9 (0.6) | −17.4 |
| A1S_1336 | Phenylacetic acid degradation | −20.3 (5.2) | −4.2 |
Expression levels were determined using qRT-PCR, and differential expression was measured using the ΔΔCT method (62).
Expression levels were determined using microarray analysis (see Table S2 in the supplemental material).
Summary.
This article describes for the first time the role of H-NS in A. baumannii. We employed a broad range of phenotypic and genotypic characterization methods to gain insight into A. baumannii virulence mechanisms and the role that H-NS plays in their regulation. Phenotypic characterization showed that hydrophobicity, adherence, and motility are likely to be coregulated in strain ATCC 17978. Furthermore, these features may be associated with virulence based on data from a nematode killing assay and increased binding capacity for the A549 eukaryotic lung cell line. Our analyses also provided evidence that the cellular fatty acid composition is linked to changes in cell surface hydrophobicity, which subsequently may alter the adherence, motility, and virulence characteristics. Although these phenotypes appear to be coregulated in strain ATCC 17978, it is known that A. baumannii strains show major variation in their motility and adherence phenotypes (3, 7). Therefore, the role of H-NS is likely to be distinct in other A. baumannii strains, which correlates with the function of H-NS as a regulator of nonconserved genomic regions.
Transcriptomic analysis of 17978hm and its parent strain on semisolid media identified molecular mechanisms that may be responsible for the phenotypic changes described above. The autotransporter encoded by ata, which has been proven to play a role in adherence and virulence (52), was heavily upregulated. Furthermore, the type VI secretion system and a type I pilus were found to be upregulated, and these surface-presented protein structures are known to affect adherence and virulence in other Gram-negative pathogens, such as P. aeruginosa and E. coli. Significantly, upregulation of genes encoding the fatty acid biosynthetic proteins FabG and FabF may explain the increase of C17 fatty acids in the hypermotile mutant compared to findings for the WT strain. Although FabG has previously been defined as a nucleoid-associated protein for other bacterial genera (64), the regulatory control of fabG and fabF in the H-NS-deficient strain requires further examination.
Bioinformatic analyses using a number of fully sequenced A. baumannii genomes assisted in identification of putative H-NS targets. Various genomic regions that were characterized as horizontally acquired were transcriptionally repressed by H-NS in the WT strain. Furthermore, examination of the upstream regions of highly expressed genes showed that H-NS is likely to bind AT-rich DNA regions, as observed for H-NS in other bacterial genera. The bioinformatics approach applied here could also be of use when examining the role of H-NS in A. baumannii strains other than ATCC 17978.
In summary, the phenotypic, genomic, and transcriptomic analyses carried out using strain ATCC 17978 and its hypermotile derivatives 17978hm and HNSmut88 revealed a significant role for H-NS in the regulation of A. baumannii persistence- and virulence-associated genes.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by project grant 535053 to M.H.B. and I.T.P. from the National Health and Medical Research Council, Australia. B.A.E. is the recipient of a School of Biological Sciences Endeavor International Postgraduate Research Scholarship, and K.A.H. is supported by an APD fellowship from the Australian Research Council (DP110102680).
We thank David Apps and Melissa Gregory for assistance with fatty acid analyses.
Footnotes
Published ahead of print 6 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00065-13.
REFERENCES
- 1. Bergogne-Berezin E, Towner KJ. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 9:148–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gordon NC, Wareham DW. 2010. Multidrug-resistant Acinetobacter baumannii: mechanisms of virulence and resistance. Int. J. Antimicrob. Agents 35:219–226 [DOI] [PubMed] [Google Scholar]
- 3. Eijkelkamp BA, Stroeher UH, Hassan KA, Papadimitrious MS, Paulsen IT, Brown MH. 2011. Adherence and motility characteristics of clinical Acinetobacter baumannii isolates. FEMS Microbiol. Lett. 323:44–51 [DOI] [PubMed] [Google Scholar]
- 4. Lee JC, Koerten H, van den Broek P, Beekhuizen H, Wolterbeek R, van den Barselaar M, van der Reijden T, van der Meer J, van de Gevel J, Dijkshoorn L. 2006. Adherence of Acinetobacter baumannii strains to human bronchial epithelial cells. Res. Microbiol. 157:360–366 [DOI] [PubMed] [Google Scholar]
- 5. de Breij A, Dijkshoorn L, Lagendijk E, van der Meer J, Koster A, Bloemberg G, Wolterbeek R, van den Broek P, Nibbering P. 2010. Do biofilm formation and interactions with human cells explain the clinical success of Acinetobacter baumannii? PLoS One 5:e10732. 10.1371/journal.pone.0010732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Marti S, Rodriguez-Bano J, Catel-Ferreira M, Jouenne T, Vila J, Seifert H, De E. 2011. Biofilm formation at the solid-liquid and air-liquid interfaces by Acinetobacter species. BMC Res. Notes 4:5. 10.1186/1756-0500-4-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McQueary CN, Actis LA. 2011. Acinetobacter baumannii biofilms: variations among strains and correlations with other cell properties. J. Microbiol. 49:243–250 [DOI] [PubMed] [Google Scholar]
- 8. Pour NK, Dusane DH, Dhakephalkar PK, Zamin FR, Zinjarde SS, Chopade BA. 2011. Biofilm formation by Acinetobacter baumannii strains isolated from urinary tract infection and urinary catheters. FEMS Immunol. Med. Microbiol. 62:328–338 [DOI] [PubMed] [Google Scholar]
- 9. Rodriguez-Bano J, Marti S, Soto S, Fernandez-Cuenca F, Cisneros JM, Pachon J, Pascual A, Martinez-Martinez L, McQueary C, Actis LA, Vila J. 2008. Biofilm formation in Acinetobacter baumannii: associated features and clinical implications. Clin. Microbiol. Infect. 14:276–278 [DOI] [PubMed] [Google Scholar]
- 10. Eijkelkamp BA, Hassan KA, Paulsen IT, Brown MH. 2011. Investigation of the human pathogen Acinetobacter baumannii under iron limiting conditions. BMC Genomics 12:126. 10.1186/1471-2164-12-126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mussi MA, Gaddy JA, Cabruja M, Arivett BA, Viale AM, Rasia R, Actis LA. 2010. The opportunistic human pathogen Acinetobacter baumannii senses and responds to light. J. Bacteriol. 192:6336–6345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Clemmer KM, Bonomo RA, Rather PN. 2011. Genetic analysis of surface motility in Acinetobacter baumannii. Microbiology 157:2534–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Han X, Kennan RM, Davies JK, Reddacliff LA, Dhungyel OP, Whittington RJ, Turnbull L, Whitchurch CB, Rood JI. 2008. Twitching motility is essential for virulence in Dichelobacter nodosus. J. Bacteriol. 190:3323–3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Alarcon I, Evans DJ, Fleiszig SM. 2009. The role of twitching motility in Pseudomonas aeruginosa exit from and translocation of corneal epithelial cells. Invest. Ophthalmol. Vis. Sci. 50:2237–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stewart RM, Wiehlmann L, Ashelford KE, Preston SJ, Frimmersdorf E, Campbell BJ, Neal TJ, Hall N, Tuft S, Kaye SB, Winstanley C. 2011. Genetic characterization indicates that a specific subpopulation of Pseudomonas aeruginosa is associated with keratitis infections. J. Clin. Microbiol. 49:993–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Winstanley C, Kaye SB, Neal TJ, Chilton HJ, Miksch S, Hart CA. 2005. Genotypic and phenotypic characteristics of Pseudomonas aeruginosa isolates associated with ulcerative keratitis. J. Med. Microbiol. 54:519–526 [DOI] [PubMed] [Google Scholar]
- 17. Zolfaghar I, Evans DJ, Fleiszig SM. 2003. Twitching motility contributes to the role of pili in corneal infection caused by Pseudomonas aeruginosa. Infect. Immun. 71:5389–5393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Klausen M, Aaes-Jorgensen A, Molin S, Tolker-Nielsen T. 2003. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol. Microbiol. 50:61–68 [DOI] [PubMed] [Google Scholar]
- 19. Cerqueira GM, Peleg AY. 2011. Insights into Acinetobacter baumannii pathogenicity. IUBMB Life 63:1055–1060 [DOI] [PubMed] [Google Scholar]
- 20. Choi CH, Lee JS, Lee YC, Park TI, Lee JC. 2008. Acinetobacter baumannii invades epithelial cells and outer membrane protein A mediates interactions with epithelial cells. BMC Microbiol. 8:216. 10.1186/1471-2180-8-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gaddy JA, Tomaras AP, Actis LA. 2009. The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect. Immun. 77:3150–3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Choi CH, Lee EY, Lee YC, Park TI, Kim HJ, Hyun SH, Kim SA, Lee SK, Lee JC. 2005. Outer membrane protein 38 of Acinetobacter baumannii localizes to the mitochondria and induces apoptosis of epithelial cells. Cell Microbiol. 7:1127–1138 [DOI] [PubMed] [Google Scholar]
- 23. McConnell MJ, Pachon J. 2011. Expression, purification, and refolding of biologically active Acinetobacter baumannii OmpA from Escherichia coli inclusion bodies. Protein Expr. Purif. 77:98–103 [DOI] [PubMed] [Google Scholar]
- 24. Ofori-Darko E, Zavros Y, Rieder G, Tarle SA, Van Antwerp M, Merchant JL. 2000. An OmpA-like protein from Acinetobacter spp. stimulates gastrin and interleukin-8 promoters. Infect. Immun. 68:3657–3666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Luke NR, Sauberan SL, Russo TA, Beanan JM, Olson R, Loehfelm TW, Cox AD, St Michael F, Vinogradov EV, Campagnari AA. 2010. Identification and characterization of a glycosyltransferase involved in Acinetobacter baumannii lipopolysaccharide core biosynthesis. Infect. Immun. 78:2017–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jacobs AC, Hood I, Boyd KL, Olson PD, Morrison JM, Carson S, Sayood K, Iwen PC, Skaar EP, Dunman PM. 2010. Inactivation of phospholipase D diminishes Acinetobacter baumannii pathogenesis. Infect. Immun. 78:1952–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Russo TA, MacDonald U, Beanan JM, Olson R, MacDonald IJ, Sauberan SL, Luke NR, Schultz LW, Umland TC. 2009. Penicillin-binding protein 7/8 contributes to the survival of Acinetobacter baumannii in vitro and in vivo. J. Infect. Dis. 199:513–521 [DOI] [PubMed] [Google Scholar]
- 28. Russo TA, Luke NR, Beanan JM, Olson R, Sauberan SL, MacDonald U, Schultz LW, Umland TC, Campagnari AA. 2010. The K1 capsular polysaccharide of Acinetobacter baumannii strain 307-0294 is a major virulence factor. Infect. Immun. 78:3993–4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gaddy JA, Arivett BA, McConnell MJ, Lopez-Rojas R, Pachon J, Actis LA. 2012. Role of acinetobactin-mediated iron acquisition functions in the interaction of Acinetobacter baumannii strain ATCC 19606T with human lung epithelial cells, Galleria mellonella caterpillars, and mice. Infect. Immun. 80:1015–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smith MG, Gianoulis TA, Pukatzki S, Mekalanos JJ, Ornston LN, Gerstein M, Snyder M. 2007. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 21:601–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Henry R, Vithanage N, Harrison P, Seemann T, Coutts S, Moffatt JH, Nation RL, Li J, Harper M, Adler B, Boyce JD. 2012. Colistin-resistant, lipopolysaccharide-deficient Acinetobacter baumannii responds to lipopolysaccharide loss through increased expression of genes involved in the synthesis and transport of lipoproteins, phospholipids, and poly-beta-1,6-N-acetylglucosamine. Antimicrob. Agents Chemother. 56:59–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Daniel C, Haentjens S, Bissinger MC, Courcol RJ. 1999. Characterization of the Acinetobacter baumannii Fur regulator: cloning and sequencing of the fur homolog gene. FEMS Microbiol. Lett. 170:199–209 [DOI] [PubMed] [Google Scholar]
- 33. Bhargava N, Sharma P, Capalash N. 2010. Quorum sensing in Acinetobacter: an emerging pathogen. Crit. Rev. Microbiol. 36:349–360 [DOI] [PubMed] [Google Scholar]
- 34. Gaddy JA, Actis LA. 2009. Regulation of Acinetobacter baumannii biofilm formation. Future Microbiol. 4:273–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tomaras AP, Flagler MJ, Dorsey CW, Gaddy JA, Actis LA. 2008. Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology 154:3398–3409 [DOI] [PubMed] [Google Scholar]
- 36. Rimsky S. 2004. Structure of the histone-like protein H-NS and its role in regulation and genome superstructure. Curr. Opin. Microbiol. 7:109–114 [DOI] [PubMed] [Google Scholar]
- 37. Lang B, Blot N, Bouffartigues E, Buckle M, Geertz M, Gualerzi CO, Mavathur R, Muskhelishvili G, Pon CL, Rimsky S, Stella S, Babu MM, Travers A. 2007. High-affinity DNA binding sites for H-NS provide a molecular basis for selective silencing within proteobacterial genomes. Nucleic Acids Res. 35:6330–6337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics 77:71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rosenberg M, Gutnick D, Rosenberg E. 1980. Adherence of bacteria to hydrocarbons: a simple method for measuring cell-surface hydrophobicity. FEMS Microbiol. Lett. 9:29–33 [Google Scholar]
- 40. Gregory MK, See VH, Gibson RA, Schuller KA. 2010. Cloning and functional characterisation of a fatty acyl elongase from southern bluefin tuna (Thunnus maccoyii). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 155:178–185 [DOI] [PubMed] [Google Scholar]
- 41. Rozen S, Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365–386 [DOI] [PubMed] [Google Scholar]
- 42. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 43. Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hunger M, Schmucker R, Kishan V, Hillen W. 1990. Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene 87:45–51 [DOI] [PubMed] [Google Scholar]
- 45. Arora K, Whiteford DC, Lau-Bonilla D, Davitt CM, Dahl JL. 2008. Inactivation of lsr2 results in a hypermotile phenotype in Mycobacterium smegmatis. J. Bacteriol. 190:4291–4300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cordeiro TN, Schmidt H, Madrid C, Juarez A, Bernado P, Griesinger C, Garcia J, Pons M. 2011. Indirect DNA readout by an H-NS related protein: structure of the DNA complex of the C-terminal domain of Ler. PLoS Pathog. 7:e1002380. 10.1371/journal.ppat.1002380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gordon BR, Li Y, Cote A, Weirauch MT, Ding P, Hughes TR, Navarre WW, Xia B, Liu J. 2011. Structural basis for recognition of AT-rich DNA by unrelated xenogeneic silencing proteins. Proc. Natl. Acad. Sci. U. S. A. 108:10690–10695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liang Y, Gao H, Chen J, Dong Y, Wu L, He Z, Liu X, Qiu G, Zhou J. 2010. Pellicle formation in Shewanella oneidensis. BMC Microbiol. 10:291. 10.1186/1471-2180-10-291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. de Breij A, Gaddy J, van der Meer J, Koning R, Koster A, van den Broek P, Actis L, Nibbering P, Dijkshoorn L. 2009. CsuA/BABCDE-dependent pili are not involved in the adherence of Acinetobacter baumannii ATCC19606(T) to human airway epithelial cells and their inflammatory response. Res. Microbiol. 160:213–218 [DOI] [PubMed] [Google Scholar]
- 50. Talbot UM, Paton AW, Paton JC. 1996. Uptake of Streptococcus pneumoniae by respiratory epithelial cells. Infect. Immun. 64:3772–3777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Moorman MA, Thelemann CA, Zhou S, Pestka JJ, Linz JE, Ryser ET. 2008. Altered hydrophobicity and membrane composition in stress-adapted Listeria innocua. J. Food Prot. 71:182–185 [DOI] [PubMed] [Google Scholar]
- 52. Bentancor LV, Camacho-Peiro A, Bozkurt-Guzel C, Pier GB, Maira-Litran T. 2012. Identification of Ata, a multifunctional trimeric autotransporter of Acinetobacter baumannii. J. Bacteriol. 194:3950–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordonez CL, Lory S, Walz T, Joachimiak A, Mekalanos JJ. 2006. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312:1526–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Weber BS, Miyata ST, Iwashkiw JA, Mortensen BL, Skaar EP, Pukatzki S, Feldman MF. 2013. Genomic and functional analysis of the type VI secretion system in Acinetobacter. PLoS One 8:e55142. 10.1371/journal.pone.0055142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shin JH, Lee HW, Kim SM, Kim J. 2009. Proteomic analysis of Acinetobacter baumannii in biofilm and planktonic growth mode. J. Microbiol. 47:728–735 [DOI] [PubMed] [Google Scholar]
- 56. Zhang YM, Rock CO. 2008. Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol. 6:222–233 [DOI] [PubMed] [Google Scholar]
- 57. Tremblay J, Deziel E. 2010. Gene expression in Pseudomonas aeruginosa swarming motility. BMC Genomics 11:587. 10.1186/1471-2164-11-587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kahramanoglou C, Seshasayee AS, Prieto AI, Ibberson D, Schmidt S, Zimmermann J, Benes V, Fraser GM, Luscombe NM. 2011. Direct and indirect effects of H-NS and Fis on global gene expression control in Escherichia coli. Nucleic Acids Res. 39:2073–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dorman CJ, Kane KA. 2009. DNA bridging and antibridging: a role for bacterial nucleoid-associated proteins in regulating the expression of laterally acquired genes. FEMS Microbiol. Rev. 33:587–592 [DOI] [PubMed] [Google Scholar]
- 60. Bernard CS, Brunet YR, Gueguen E, Cascales E. 2010. Nooks and crannies in type VI secretion regulation. J. Bacteriol. 192:3850–3860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14:1394–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Allsopp LP, Beloin C, Ulett GC, Valle J, Totsika M, Sherlock O, Ghigo JM, Schembri MA. 2012. Molecular characterization of UpaB and UpaC, two new autotransporter proteins of uropathogenic Escherichia coli CFT073. Infect. Immun. 80:321–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sun W, Xu X, Pavlova M, Edwards AM, Joachimiak A, Savchenko A, Christendat D. 2005. The crystal structure of a novel SAM-dependent methyltransferase PH1915 from Pyrococcus horikoshii. Protein Sci. 14:3121–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ohniwa RL, Ushijima Y, Saito S, Morikawa K. 2011. Proteomic analyses of nucleoid-associated proteins in Escherichia coli, Pseudomonas aeruginosa, Bacillus subtilis, and Staphylococcus aureus. PLoS One 6:e19172. 10.1371/journal.pone.0019172 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.