Abstract
Porphyromonas gingivalis is a major pathogen in periodontal disease and is associated with immune dysbiosis. In this study, we found that P. gingivalis did not induce the expression of the T-cell chemokine IP-10 (CXCL10) from neutrophils, peripheral blood mononuclear cells (PBMCs), or gingival epithelial cells. Furthermore, P. gingivalis suppressed gamma interferon (IFN-γ)-stimulated release of IP-10, ITAC (CXCL11), and Mig (CXCL9) from epithelial cells and inhibited IP-10 secretion in a mixed infection with the otherwise stimulatory Fusobacterium nucleatum. Inhibition of chemokine expression occurred at the level of gene transcription and was associated with downregulation of interferon regulatory factor 1 (IRF-1) and decreased levels of Stat1. Ectopic expression of IRF-1 in epithelial cells relieved P. gingivalis-induced inhibition of IP-10 release. Direct contact between P. gingivalis and epithelial cells was not required for IP-10 inhibition. These results highlight the immune-disruptive potential of P. gingivalis. Suppression of IP-10 and other Th1-biasing chemokines by P. gingivalis may perturb the balance of protective and destructive immunity in the periodontal tissues and facilitate the pathogenicity of oral microbial communities.
INTRODUCTION
Periodontal diseases are a group of microbially driven inflammatory infections that, in severe cases, result in the destruction of the periodontal tissues with eventual exfoliation of the tooth. Periodontal diseases are among the most common infections of humans, and in the United States over one-half of the adult population experience symptoms of the disease (1). Communities of organisms working in concert are responsible for the initiation and progression of disease. Successful periodontal pathogens can disrupt host innate immunity and interact synergistically with other constituents of the periodontal microbial community (2–4).
Porphyromonas gingivalis, a Gram-negative anaerobe, is strongly associated with severe and chronic cases of periodontal disease (5, 6). The organism possesses a number of attributes that contribute to its success as a periodontal pathogen. These include adhesion and colonization of the available substrata in the oral cavity, production of proteolytic enzymes that target the structural integrity of the periodontium, and avoidance of immune responses (2, 6–8). In addition, the virulence of P. gingivalis is elevated in the context of a community with accessory pathogens such as the oral streptococci (4, 9), and the organism functions in the role of a keystone pathogen in that it can elevate the virulence of the entire community (10). P. gingivalis can invade and survive within gingival epithelial cells, and an intracellular location physically sequesters the organism from recognition by the immune system (11). Intracellularly, P. gingivalis impacts host cell gene expression, resulting in the modulation of expression of immune effectors (12, 13). In particular, through the action of a serine phosphatase (SerB) that interferes with host cell signaling, P. gingivalis can suppress production and secretion of the neutrophil chemokine interleukin 8 (IL-8), a phenomenon known as localized chemokine paralysis (14, 15). Moreover, P. gingivalis can antagonize IL-8 secretion from epithelial cells induced by other important periodontal bacteria such as Fusobacterium nucleatum (14). Indeed, P. gingivalis has a number of stealth-like properties and is capable of evading or suppressing many aspects of innate and acquired immunity. The organism can produce an atypical lipopolysaccharide with a 4-acyl monophosphorylated lipid A moiety that can potently antagonize Toll-like receptor 4 (16, 17). Survival of P. gingivalis in the gingival compartment is enhanced by the complement C5 convertase-like activity of its gingipains and the induction of a subversive cross talk between C5a receptor (C5aR) and Toll-like receptor 2 (18), which can impede the killing capacity of leukocytes (19, 20).
The periodontal host response is highly complex and involves both protective and destructive elements (21, 22). T cells are present in diseased tissues (22), although the role of the individual effector Th subsets is debatable (21). Studies in humans led to one widely held view that Th1 cells and their cytokines (such as IL-12 and gamma interferon [IFN-γ]) predominate in early and stable periodontal lesions, while Th2 cells are associated with disease progression, consistent with the B-cell nature of the progressive lesion (23). More recently recognized Th17 cells have been implicated in bone loss through activation of osteoclasts (24). Further development of this model accommodates both protective and destructive roles for T cells. Cytokines and other effectors produced by Th1 and Th17 cells can increase levels of receptor activator of NF-κB ligand (RANKL) and matrix metalloproteinases (MMPs), thus contributing to tissue destruction (25). Conversely, these effectors also recruit and activate neutrophils and monocytes, which serve to control the bacterial challenge. The production of Th2-dependent antibodies and cytokines may then dampen the Th1-dependent destructive events.
T cells are recruited to sites of inflammation by distinct sets of chemokines. For example, IFN-γ-inducible protein-10 (IP-10, or CXCL10), interferon-inducible T-cell alpha chemoattractant (ITAC, or CXCL11), and monokine induced by IFN-γ (Mig, or CXCL9) all induce T-cell migration through binding to the CXCR3 receptor (26). As CXCR3 is found predominantly on Th1 cells, IP-10, ITAC, and Mig are Th1-biasing chemokines (26). P. gingivalis can impact T-cell lineage responses to evade or suppress adaptive T-cell responses (23, 27, 28). In addition, P. gingivalis can inhibit the expression and accumulation of IL-2, which promotes T-cell proliferation (29). Indeed, it is well established that P. gingivalis can modulate the synthesis of cytokines by host cells (6, 30) and, moreover, that the manifold proteases of P. gingivalis can degrade various cytokines (31, 32). However, the effect of P. gingivalis on production and accumulation of T-cell chemokines is not well understood. In this study, we examined the production and secretion of IP-10, ITAC, and Mig by epithelial cells in response to P. gingivalis infection.
MATERIALS AND METHODS
Bacterial and eukaryotic cell culture.
P. gingivalis ATCC 33277 was cultured in Trypticase soy broth supplemented with yeast extract (1 mg/ml), hemin (5 μg/ml), and menadione (1 μg/ml). F. nucleatum ATCC 25586 was cultured in brain heart infusion broth supplemented with hemin (5 μg/ml) and menadione (1 μg/ml). Both species were grown anaerobically at 37°C. Human telomerase immortalized gingival keratinocytes (TIGKs) (33), derived from the gingival epithelium, were routinely cultured at 37°C and 5% CO2 in keratinocyte serum-free medium (Invitrogen) supplemented with 0.4 mM calcium chloride, 25 μg/ml bovine pituitary extract (BPE), and 0.2 ng/ml epidermal growth factor (EGF) (TIGKM). Human neutrophils and peripheral blood mononuclear cells (PBMCs) were isolated from the blood of healthy donors using plasma-Percoll gradients as previously described (34) and approved by the Institutional Review Board of the University of Louisville. Neutrophil and PBMC fractions were collected separately, and Wright-stained cytospins of each isolated fraction showed that over 95% of cells were neutrophils or PBMCs, respectively. A trypan blue exclusion assay indicated that greater than 97% cells were viable. Neutrophils or PBMCs were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, 10 U/ml of penicillin, and 10 μg/ml of streptomycin (Invitrogen) in a 37°C and 5% CO2 environment.
ELISAs for IP-10, ITAC, and Mig.
TIGKs at 70% confluence, neutrophils (4 × 106 cells/ml), or PBMCs (4 × 106cells/ml) were stimulated with bacteria (multiplicity of infection [MOI], 10 for neutrophils and PBMCs and 100 for TIGKs, unless otherwise stated) or Escherichia coli lipopolysaccharide (LPS) (100 ng/ml; Sigma-Aldrich). Supernatants were collected and centrifuged at 4,000 × g to remove bacteria. For IFN-γ stimulation, after 2 h of incubation with bacteria, TIGKs were washed with TIGKM and extracellular bacteria killed with 200 μg/ml metronidazole and 300 μg/ml gentamicin. Cells were subjected to 5 ng/ml IFN-γ for 18 h, and supernatants were collected and centrifuged at 4,000 × g for 10 min. Levels of IP-10, ITAC, Mig, and IL-1β in TIGK supernatants (diluted 1:30) were assayed using Quantikine sandwich enzyme-linked immunosorbent assay (ELISA) kits (R & D Systems) and in neutrophil and PBMC supernatants (diluted 1:30) by Milliplex MAP Multiplex kits (EMD Millipore) according to the manufacturers' instructions. A transwell system (Costar) was used to prevent direct contact between P. gingivalis (upper compartment) and epithelial cells (lower compartment).
Quantitative reverse transcription-PCR (qRT-PCR).
Total RNA from TIGKs was isolated with TRIzol (Invitrogen) and purified with the Qiagen miRNeasy kit, according to the manufacturer's instructions. cDNA was synthesized (2 μg RNA/reaction volume) using a High Capacity RNA-cDNA kit (Applied Biosystems). Real-time RT-PCRs used TaqMan FAST universal PCR mastermix and gene expression assays for IP-10, ITAC, Mig, IRF-1, and Stat1 (Applied Biosystems). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a housekeeping control, and negative RT reactions were included in each assay. An Applied Biosystems StepOne plus cycler with StepOne software V2.2.2 was employed, with the autocalculated threshold cycle selected. The cycle threshold (CT) values were determined, and mRNA expression levels were normalized to GAPDH and expressed relative to controls following the 2−ΔΔCT method.
Western immunoblotting.
TIGKs were lysed with 10% trichloroacetic acid in buffer consisting of 2 M thiourea, 7 M urea, 3% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), and 1% Triton X-100. After centrifugation at 17,400 × g for 30 min at 4°C, the supernatant was separated by SDS-PAGE and electroblotted onto nitrocellulose. Membranes were blocked with TBST (Tris-buffered saline and 0.1% Tween 20) containing 5% nonfat dried milk and incubated with primary antibodies (rabbit monoclonal anti-Stat1 and rabbit monoclonal anti-phospho-Stat1 Y701 from Cell Signaling Technology; mouse monoclonal anti-β-actin from Sigma-Aldrich). Antigen-antibody binding was detected using horseradish peroxidase-conjugated secondary antibodies (goat anti-mouse IgG and goat anti-rabbit IgG; Cell Signaling Technology) followed by ECL Western Blotting Substrate (Thermo Scientific). Images were acquired with Image Lab Software version 3.0 (Bio-Rad).
Transfection of TIGK cells.
TIGK cells were transfected with pCMV-IRF-1 (Panomics) or empty vector control at 2 μg/105 cells using siPORT NeoFX transfection agent (Applied Biosystems). Following 24 h in transfection media, cells were returned to TIGKM for a further 24 h prior to assay.
Confocal laser scanning microscopy.
TIGKs on glass coverslips were washed twice in phosphate-buffered saline (PBS) and fixed for 10 min in 4% paraformaldehyde. Permeabilization was with 0.2% Triton X-100 for 10 min at room temperature, prior to blocking in 10% goat serum for 20 min. IRF-1 was detected by reacting with primary antibody (Cell Signaling) at a 1:100 dilution for 1 h, followed by Alexa-647-conjugated anti-rabbit secondary antibody (1:200) for 1 h in the dark. Following a 20-min block in 0.1% goat serum, actin was labeled using 1:100 fluorescein isothiocyanate (FITC)-phalloidin (Sigma-Aldrich) for 40 min at room temperature. After 4 washes in PBS, coverslips were mounted using ProLong Gold with DAPI (4′,6-diamidino-2-phenylindole) mounting medium (Invitrogen). Images were acquired on an Olympus Fluoview inverted fluorescence microscope, and analyzed using Fluoview FV10-ASW 1.7 software (Olympus), using a 60× oil immersion objective. z-stacks were obtained (10 layers/stack, 0.2-μm intervals) through the z axis of cells (3 z-stacks/coverslip).
RESULTS
P. gingivalis infection inhibits the secretion of T-cell chemokines.
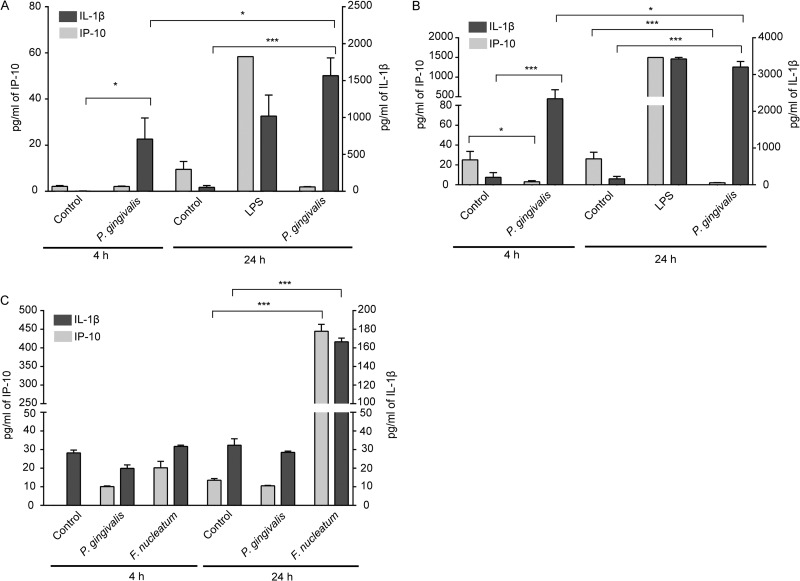
We sought to establish whether infection of host cells with P. gingivalis would affect the release of cytokines associated with T-cell recruitment. As shown in Fig. 1A and B, P. gingivalis failed to induce release of IP-10 from neutrophils or PBMCs up to 24 h after infection. In contrast, secretion of IP-10 was induced by E. coli LPS, and P. gingivalis was capable of stimulating the release of IL-1β, indicating that the low levels of IP-10 were not the result of overall metabolic suppression by P. gingivalis. To examine IP-10 secretion from gingival epithelial cells (TIGKs), F. nucleatum at an MOI of 100 was used as the positive control, as epithelial cells do not respond well to LPS in serum-free medium. P. gingivalis did not induce IP-10 secretion from TIGKs, whereas F. nucleatum stimulated the release of IP-10 and IL-1β (Fig. 1C).
Fig 1.
P. gingivalis does not stimulate the release of IP-10. Human neutrophils (A), PBMCs (B), or TIGKs (C) were stimulated with P. gingivalis or F. nucleatum at an MOI of 10 (A and B) or 100 (C) or with E. coli LPS (100 ng/ml) or were left unstimulated (Control). Supernatants were collected at 4 and 24 h poststimulation, and IP-10 or IL-1β levels were measured. Data in panels A and B are expressed as means with standard errors of results from six donors. Data in panel C are means ± standard deviations (SD) (n = 3) and are representative of three independent experiments. ***, P < 0.0001; *, P < 0.05, by the t test.
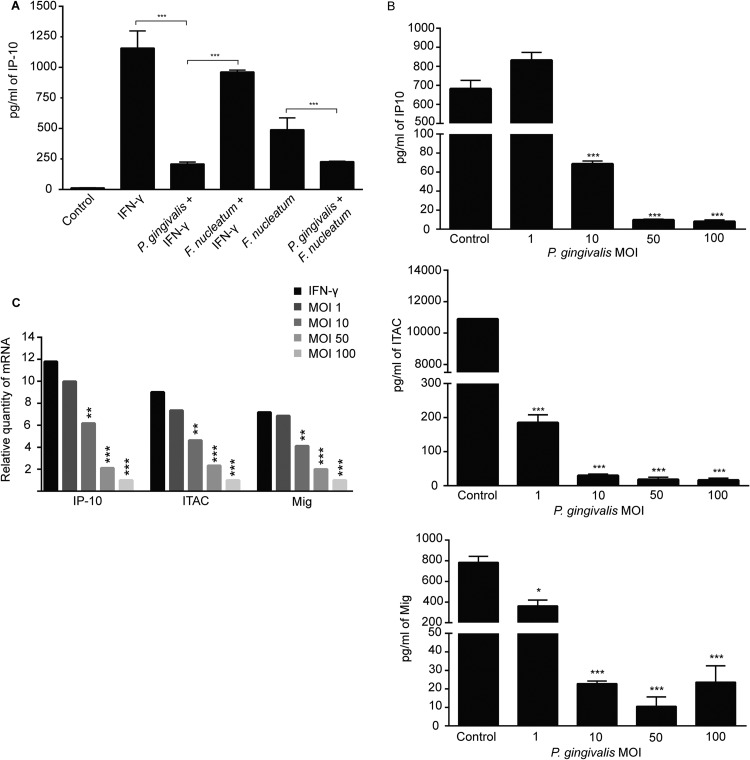
We elected to study the mechanism of IP-10 suppression by P. gingivalis in the stable gingival epithelial cell line TIGK, and to mimic more closely the in vivo situation, cells infected with P. gingivalis or F. nucleatum were stimulated with IFN-γ to induce production and secretion of IP-10. P. gingivalis suppressed production of IP-10 (Fig. 2A), whereas F. nucleatum had no effect on IFN-γ-stimulated IP-10 levels. When IP-10 was induced by infection with F. nucleatum, P. gingivalis also suppressed chemokine secretion. Hence, P. gingivalis is capable of specifically antagonizing the production of IP-10 induced either by cytokines or by other bacteria.
Fig 2.
P. gingivalis antagonizes production and release of IP-10, ITAC, and Mig. (A) TIGK cells were infected with P. gingivalis and/or F. nucleatum (MOI, 100) and stimulated with IFN-γ (5 ng/ml) or left unstimulated. Controls were uninfected and unstimulated. IP-10 levels in culture supernatants were analyzed by ELISA. Data are means with standard deviations (n = 3) and are representative of three independent experiments. ***, P < 0.001 by the Tukey-Kramer multiple-comparison test. (B) Secretion of IP-10, ITAC, and Mig from TIGKs infected with P. gingivalis at MOIs of 1 to 100. Supernatants from IFN-γ-stimulated TIGKs were analyzed by ELISA. Data are means with standard deviations (n = 3) and are representative of three independent experiments. ***, P < 0.001; *, P < 0.05 by the pairwise Bonferroni test. (C) IP-10, ITAC, and Mig transcriptional activity was measured by qRT-PCR of mRNA from TIGKs infected with P. gingivalis (MOI, 100) or left uninfected and stimulated with IFN-γ. Data were normalized to GAPDH mRNA and are expressed relative to uninfected, unstimulated controls. Results are representative of 3 independent assays. **, P < 0.01; ***, P < 0.001 by the Tukey-Kramer multiple-comparison test.
Next, the ability of P. gingivalis to inhibit secretion of the related T-cell chemokines ITAC and Mig, along with the dose-response dependency, was investigated in TIGK cells. P. gingivalis prevented accumulation of ITAC and Mig as well as IP-10 in IFN-γ stimulated cells (Fig. 2B). P. gingivalis significantly reduced levels of IP-10 at MOIs of 10, 50, and 100, while an MOI of 1 was insufficient to prevent secretion. Levels of ITAC and Mig were reduced to background levels by P. gingivalis at MOIs of 10, 50, and 100, while an MOI of 1 significantly reduced the amounts of these chemokines but to a lesser degree.
Inhibition of T-cell chemokines occurs at the transcriptional level.
To determine whether P. gingivalis affects T-cell chemokine gene regulation and to ensure that reduced chemokine levels are not the result of proteolytic degradation, we examined the mRNA levels of IP-10, ITAC, and Mig following P. gingivalis infection using quantitative RT-PCR. Figure 2C shows that transcription of genes encoding all three chemokines was ablated by P. gingivalis.
Collectively, these results indicate that P. gingivalis has a specific ability to antagonize production of the major Th1 T-cell chemokines IP-10, ITAC, and Mig by inhibiting gene transcription.
P. gingivalis infection reduces expression of IRF-1.
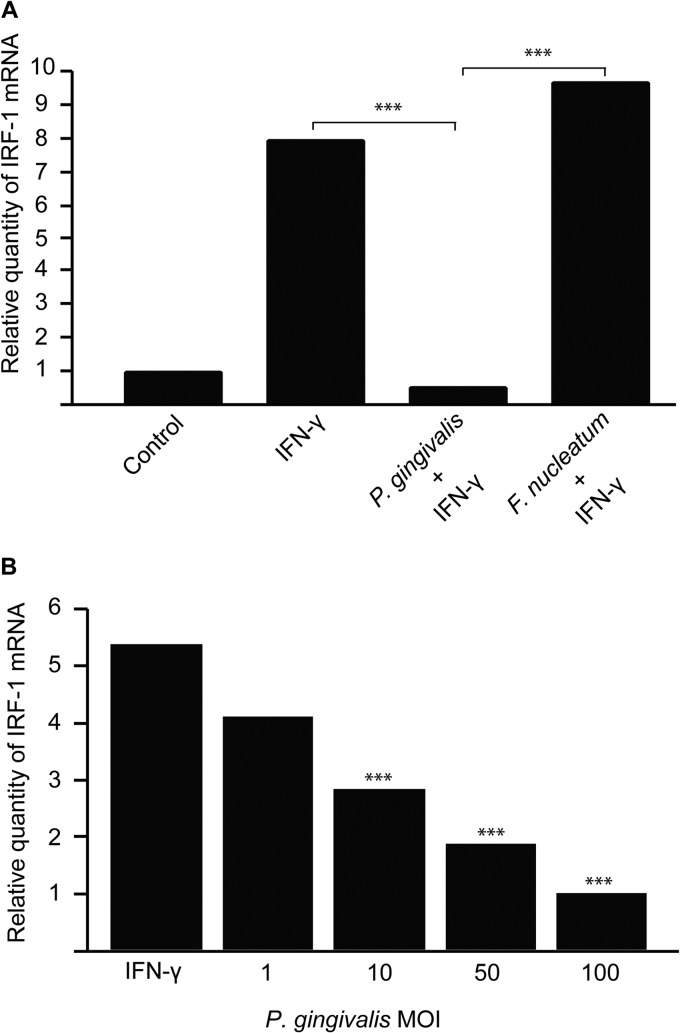
Transcription of IP-10, ITAC, and Mig is controlled by a number of regulatory factors, including NF-κB and IRF-1. P. gingivalis can suppress NF-κB activation in gingival epithelial cells through the action of a secreted serine phosphatase, SerB (15). However, infection of IFN-γ-stimulated TIGKs with a SerB mutant of P. gingivalis did not relieve inhibition of chemokine expression (not shown), indicating that regulatory factors other than NF-κB are involved in P. gingivalis-induced chemokine suppression. Hence, we turned our attention to IRF-1, a transcription factor that can itself be controlled at the transcriptional level (35). To determine whether IRF-1 expression differed in P. gingivalis-infected cells, quantitative RT-PCR for IRF-1 mRNA was performed. IRF-1 mRNA levels were reduced over 8-fold in P. gingivalis-infected, IFN-γ-stimulated cells but were unaltered in F. nucleatum-infected cells (Fig. 3A). This inhibition was dose responsive, with IRF-1 expression levels negatively correlated to the P. gingivalis MOI, and inhibition was observed over an MOI range that also inhibits expression of IP-10, ITAC, and Mig (Fig. 3B).
Fig 3.
IRF-1 mRNA levels are decreased in TIGKs following P. gingivalis infection. (A) IRF-1 expression was measured by qRT-PCR of mRNA from TIGKs infected with P. gingivalis or F. nucleatum at an MOI of 100 or left uninfected and stimulated with IFN-γ (5 ng/ml). Controls were uninfected and unstimulated. Data were normalized to GAPDH mRNA and are expressed relative to controls. Results are representative of 3 independent assays. (B) IRF-1 mRNA levels in TIGKs infected with P. gingivalis at MOIs of 1 to 100 or left uninfected and stimulated with IFN-γ. ***, P < 0.001 by the pairwise t test.
Stat1 expression is reduced in P. gingivalis-infected TIGKs.
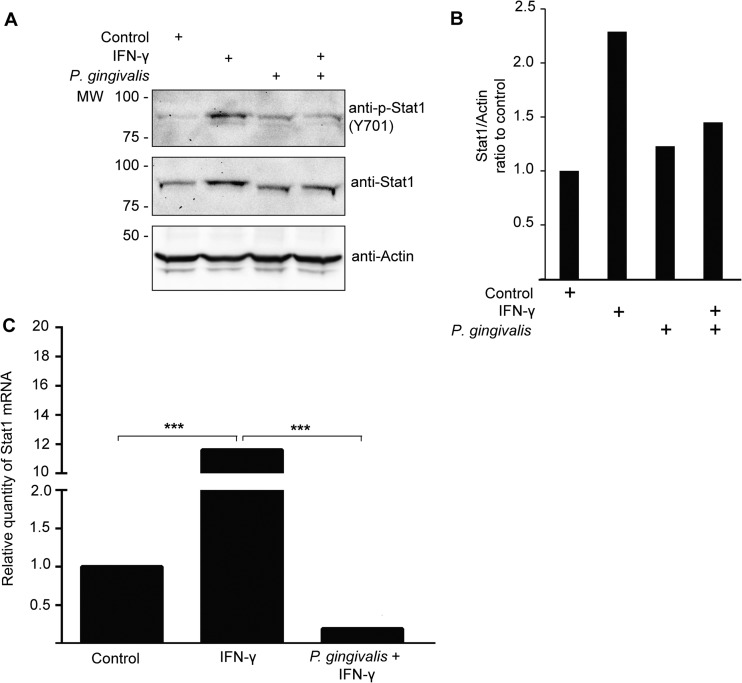
Stat1 is known to be an activator of IRF-1 transcription (35). We therefore investigated whether expression of Stat1 was modulated in TIGKs following P. gingivalis infection. Immunoblot analysis (Fig. 4A and B) demonstrated that IFN-γ did not induce an increase in Stat1 levels in P. gingivalis-infected cells; however, the Stat1 that was present was phosphorylated. This finding suggested that P. gingivalis may repress the production of Stat1, and to investigate this possibility, qRT-PCR was performed. As shown in Fig. 4C, infection with P. gingivalis reduced the amount of Stat1 mRNA present in TIGKs. These results reveal that Stat1 may be involved in regulating IRF-1 expression, ultimately leading to the inhibition of T-cell-activating chemokines in TIGKs infected with P. gingivalis.
Fig 4.
P. gingivalis reduces Stat1 levels in TIGKs. (A) Western blot analysis of Stat1 and phospho-Stat1 (p-Stat1) levels in P. gingivalis-infected TIGK cells. Whole-cell lysates (20 μg protein) of P. gingivalis-infected or uninfected TIGK cells stimulated with IFN-γ or left unstimulated were examined by Western blotting with antibodies to Stat1 or p-Stat1. Actin was used as a loading control. Data are representative of 4 independent experiments. MW, molecular weight (in thousands). (B) Scanning densitometry showing relative amounts of Stat1 and p-Stat1 in infected TIGKs expressed relative to uninfected controls in panel A. (C) qRT-PCR of mRNA for Stat1 in TIGKs infected with P. gingivalis (MOI, 100) and stimulated with IFN-γ or left unstimulated. Data were normalized to GAPDH mRNA and are expressed relative to control uninfected, unstimulated cells. Data are representative of 3 independent experiments. ***, P < 0.001 by the Tukey-Kramer multiple-comparison test.
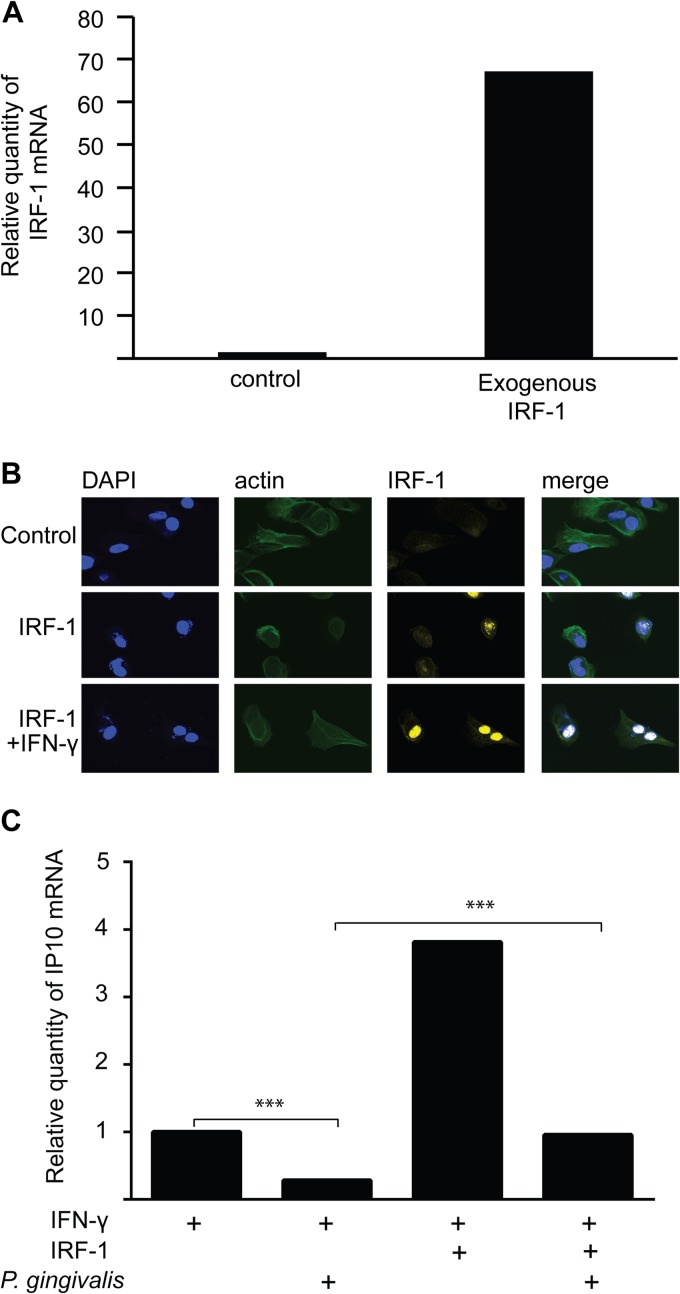
Exogenous IRF-1 alleviates IP-10 inhibition in P. gingivalis-infected TIGK cells.
To corroborate the role of IRF-1 in P. gingivalis-mediated suppression of IP-10, ITAC, and Mig, IRF-1 was ectopically expressed in TIGK cells. Increased expression of IRF-1 in transfected cells was confirmed by qRT-PCR (Fig. 5A) and confocal laser scanning microscopy (CLSM) (Fig. 5B). Cells were subsequently infected with P. gingivalis and stimulated with IFN-γ prior to RNA isolation. Quantitative RT-PCR for IP-10 expression (Fig. 5C) showed that exogenous IRF-1 ablated the reduction in IP-10 mRNA produced by P. gingivalis in IFN-γ-stimulated cells. These results provide further evidence that inhibition of IP-10 by P. gingivalis is mediated through the action of IRF-1.
Fig 5.
Exogenous IRF-1 alleviates the inhibition of IP-10 expression in P. gingivalis-infected cells. TIGK cells were transiently transfected with IRF-1 or vector only (control). (A) IRF-1 expression was measured by qRT-PCR of mRNA from TIGKs. Data were normalized to GAPDH mRNA and are expressed relative to control. Results are representative of 3 independent assays. (B) IRF-1-transfected TIGK cells were labeled with IRF-1 antibodies (yellow) and analyzed by CSLM. Actin was labeled with phalloidin (green), and nuclei were stained with DAPI (blue). Merged images are shown in the right panels. Magnification, ×63. Data shown are maximum projections through z-stacks (15 slices/stack). (C) IP-10 mRNA was measured by qRT-PCR in TIGKs transfected with exogenous IRF-1 or empty vector, infected with P. gingivalis (MOI, 100) or left uninfected and stimulated with IFN-γ (5 ng/ml). Data were normalized to GAPDH mRNA and are expressed relative to the IFN-γ-stimulated control transfected cells. Results are representative of 3 independent assays. ***, P < 0.001 by the Tukey-Kramer multiple-comparison test.
Direct cell-to-cell contact is not required for IP-10 suppression.
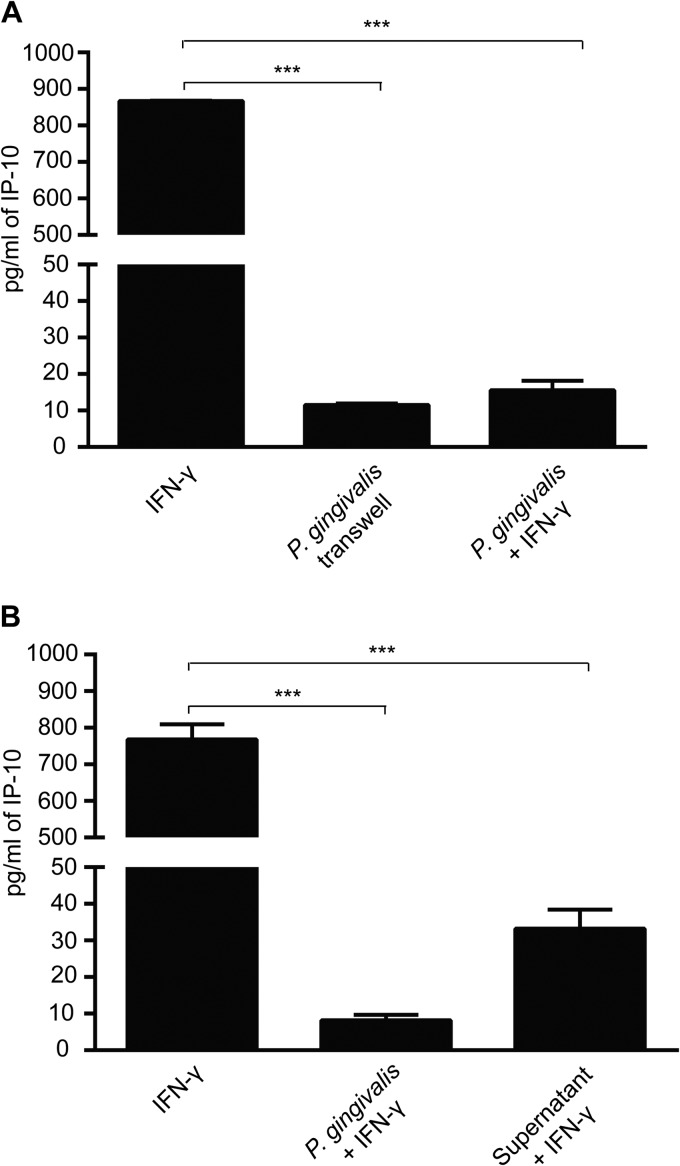
To begin to examine the basis for P. gingivalis inhibition of IP-10, we investigated the requirement for direct cell-to-cell contact. When P. gingivalis was inoculated into upper transwell chambers and epithelial cells in the lower chamber were stimulated with IFN-γ, inhibition of IP-10 production was unaffected (Fig. 6A). Hence, direct contact between P. gingivalis and epithelial cells is not necessary for IP-10 suppression, and the effect may be mediated by a P. gingivalis-secreted molecule. To verify the existence of a secreted factor, culture supernatant from P. gingivalis-infected cells was collected, P. gingivalis cells were removed, and the bacterium-free medium was added to uninfected epithelial cells. This conditioned medium was also capable of abrogating IFN-γ-induced IP-10 secretion (Fig. 6B).
Fig 6.
Direct contact between P. gingivalis and TIGKs is not required for IP-10 inhibition. (A) TIGKs were cultured in the lower chamber of a transwell plate, and P. gingivalis (MOI, 100) or medium only was added to the upper chamber. Cells were stimulated with IFN-γ (5 ng/ml) or left unstimulated. IP-10 levels in culture supernatants from the lower chamber were measured by ELISA. Data are means with standard deviations (n = 3) and are representative of three independent experiments. ***, P < 0.001 by the Tukey-Kramer multiple-comparison test. (B) TIGKs were treated with filtered supernatants from P. gingivalis-infected (MOI, 100) or uninfected cells and stimulated with IFN-γ (5 ng/ml). Infection with P. gingivalis (MOI, 100) and IFN-γ stimulation is the positive control. Data are means with standard deviations (n = 3) and are representative of three independent experiments. ***, P < 0.001 by the Tukey-Kramer multiple-comparison test.
DISCUSSION
P. gingivalis interactions with the host immune system, while complex and multithreaded, ultimately result in immune dysbiosis, which contributes to periodontal tissue destruction (2, 3, 10). In this study, we show that P. gingivalis does not stimulate release of IP-10 from neutrophils, PBMCs, or gingival epithelial cells. Moreover, P. gingivalis can inhibit the production of IFN-γ-stimulated IP-10, ITAC, and Mig by preventing transcription of the corresponding genes. Regulation of mRNA was associated with a reduction in the amount of the Stat1-IRF-1 transcriptional regulators, and ectopically expressed IRF-1 relieved inhibition of IP-10 by P. gingivalis. Furthermore, P. gingivalis was able to inhibit IP-10 production in the context of a mixed infection with F. nucleatum, a common oral organism that does not suppress chemokine production.
IP-10, ITAC, and Mig are ligands for the CXCR3 inflammatory chemokine receptor, which is highly expressed on Th1 CD4+ cells. Additionally, CXCR3 is expressed on effector CD8+ T cells and on innate lymphocytes including monocytes, NK cells, and NKT cells and on dendritic and B cells (26, 36). Ligand-CXCR3 engagement results in activation and trafficking of the receptor-expressing cells. The wide range of cells expressing CXCR3 suggests that IP-10, ITAC, and Mig play important roles in T-cell lymphopoiesis and the development of innate immunity and inflammation. Indeed, IP-10-deficient mice are defective in the generation and trafficking of effector T cells, demonstrating the importance of IP-10 for T-cell responses in vivo (37). Alterations in CXCR3 ligand levels are associated with chronic inflammatory conditions, autoimmune and neoplastic disease, and the pathogenesis of infectious diseases such as Helicobacter pylori infections (36). The role of IP-10, ITAC, and Mig in periodontal disease may vary according to disease stage and severity. Kabashima et al. (38) reported expression of IP-10-producing and CXCR3-positive cells in inflamed gingival tissues but not in healthy controls, and Garlet et al. (39) found that the expression of IP-10 and CXCR3 was significantly more prevalent in aggressive periodontitis than in chronic periodontitis or in healthy subjects. In contrast, Gemmell et al. (40) showed that the levels of IP-10 were inversely correlated with periodontal inflammation. Hence, P. gingivalis-mediated disruption of the gradient of CXCR3 ligands in gingival tissues and the consequent reduction in the proportion of Th1 cells may be an early event in the immune dysbiosis that characterizes periodontal destruction.
In addition to their role in regulating immune responses, direct antimicrobial activity against a variety of Gram-positive and Gram-negative bacteria has been established for IP-10, ITAC, and Mig (41–43). The chemokines may exert antimicrobial activity in a manner similar to that of defensins or interact with specific bacterial components such as ABC transporters to mediate direct microbial killing. Another consequence of P. gingivalis suppression of chemokine production may therefore be to reduce the overall antimicrobial activity in the local environment and facilitate bacterial overgrowth. IP-10 can also induce apoptosis (44), and suppression of IP-10 by P. gingivalis is consistent with the antiapoptotic properties of this organism (45, 46).
IRF-1 is a transcriptional regulator that displays functional diversity in the regulation of cellular responses by targeting genes possessing an interferon-stimulated response element (ISRE) in the promoter region, including IP-10, ITAC, and Mig (26). IP-10, ITAC, and Mig are thus all controlled by IRF-1, although the affinities for each gene can differ. IRF-1 itself can be transcriptionally regulated, and consistent with this, our results demonstrated that P. gingivalis inhibits the production of IRF-1 mRNA. Stat1 phosphorylation and activation constitute one pathway that drives IRF-1 synthesis (35), and P. gingivalis reduced the amounts of Stat1 in epithelial cells. Thus, one means by which P. gingivalis suppresses production of IP-10, ITAC, and Mig is through reducing the levels of components of the Stat1-IRF-1 transcriptional activation pathway. Our earlier work has shown that P. gingivalis can activate Stat3 in gingival epithelial cells (45), and hence the organism displays an exquisite selectivity in the manipulation of host cell signal transduction pathways in the establishment of immune dysbiosis. Ectopic expression of exogenous IRF-1 relieved inhibition of IP-10 by P. gingivalis, corroborating the involvement of IRF-1 in IP-10 regulation. Stat1 may also regulate chemokine expression directly, as in addition to regulating IRF1, Stat1 activation induces expression of IP-10 (47).
The ability of P. gingivalis to suppress IP-10 production is consistent with previous studies that have shown poor or no induction of this chemokine from host cells. Neither biofilm nor planktonic P. gingivalis was capable of inducing IP-10 from oral keratinocytes (48), and the organism does not stimulate IP-10 production from human monocyte-derived dendritic cells (49). In addition, P. gingivalis stimulation of splenic T cells and CD11b(+) cells induces IL-10, which suppress IFN-γ production by T cells (28). Thus, P. gingivalis can suppress both IFN-γ and IFN-γ-dependent chemokines.
The identity of the P. gingivalis effector molecule remains to be determined. Mutants lacking the major or minor fimbriae of P. gingivalis did not show a loss of IP-10 inhibition (our unpublished data). Other studies have reported that the arginine-specific gingipain of P. gingivalis can suppress IP-10 in gingival fibroblasts stimulated with T cells (50). As IP-10 mRNA levels were reduced by the protease, the role of the enzyme extends beyond proteolytic degradation of IP-10. Further, peptidylarginine deiminase (PPAD) produced by P. gingivalis can inhibit IRF-1 mRNA expression in fibroblasts (51). In the current study, we found that physical separation of P. gingivalis from epithelial cells did not affect the inhibition of IP-10 production. Furthermore, P. gingivalis conditioned medium contained the active factor, consistent with a role for either gingipains or PPAD. Secretion of an effector by P. gingivalis would be a means by which P. gingivalis could provide blanket inhibition of T-cell recruitment for the entire microbial community. Interrogation of the P. gingivalis secretome is under way to test the involvement of gingipains or PPAD and to identify the active component.
In the gingival compartment, a major lifestyle of P. gingivalis is as a constituent of a mixed-species community. Emerging concepts of P. gingivalis virulence hold that the organism is a keystone pathogen that can elevate the virulence of the entire community through induction of immune dysbiosis (52). The ability of the organism to cause localized chemokine paralysis with respect to IL-8 synthesis is an important component of dysregulation of innate immunity. Our finding that P. gingivalis suppresses T-cell chemokine secretion from myeloid and epithelial cells by a transcriptional mechanism extends the target range of chemokine paralysis. Inhibition of chemokine synthesis in the context of a mixed infection shows that the influence of P. gingivalis supersedes that of other common periodontal bacteria.
ACKNOWLEDGMENTS
We thank the NIH for support through DE11111, DE17921 (R.J.L.), and HL087924 (S.M.U.).
Footnotes
Published ahead of print 15 April 2013
REFERENCES
- 1. Eke PI, Dye BA, Wei L, Thornton-Evans GO, Genco RJ. 2012. Prevalence of periodontitis in adults in the United States: 2009 and 2010. J. Dent. Res. 91:914–920 [DOI] [PubMed] [Google Scholar]
- 2. Darveau RP, Hajishengallis G, Curtis MA. 2012. Porphyromonas gingivalis as a potential community activist for disease. J. Dent. Res. 91:816–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hajishengallis G, Lamont RJ. 2012. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol. Oral Microbiol. 27:409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Whitmore SE, Lamont RJ. 2011. The pathogenic persona of community-associated oral streptococci. Mol. Microbiol. 81:305–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Byrne SJ, Dashper SG, Darby IB, Adams GG, Hoffmann B, Reynolds EC. 2009. Progression of chronic periodontitis can be predicted by the levels of Porphyromonas gingivalis and Treponema denticola in subgingival plaque. Oral Microbiol. Immunol. 24:469–477 [DOI] [PubMed] [Google Scholar]
- 6. Lamont RJ, Jenkinson HF. 1998. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62:1244–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kuboniwa M, Lamont RJ. 2010. Subgingival biofilm formation. Periodontol. 2000 52:38–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wright CJ, Burns LH, Jack AA, Back CR, Dutton LC, Nobbs AH, Lamont RJ, Jenkinson HF. 2013. Microbial interactions in building of communities. Mol. Oral Microbiol. 28:83–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Daep CA, Novak EA, Lamont RJ, Demuth DR. 2011. Structural dissection and in vivo effectiveness of a peptide inhibitor of Porphyromonas gingivalis adherence to Streptococcus gordonii. Infect. Immun. 79:67–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hajishengallis G, Darveau RP, Curtis MA. 2012. The keystone-pathogen hypothesis. Nat. Rev. Microbiol. 10:717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tribble GD, Lamont RJ. 2010. Bacterial invasion of epithelial cells and spreading in periodontal tissue. Periodontol. 2000 52:68–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Handfield M, Mans JJ, Zheng G, Lopez MC, Mao S, Progulske-Fox A, Narasimhan G, Baker HV, Lamont RJ. 2005. Distinct transcriptional profiles characterize oral epithelium-microbiota interactions. Cell. Microbiol. 7:811–823 [DOI] [PubMed] [Google Scholar]
- 13. Handfield M, Baker HV, Lamont RJ. 2008. Beyond good and evil in the oral cavity: insights into host-microbe relationships derived from transcriptional profiling of gingival cells. J. Dent. Res. 87:203–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Darveau RP, Belton CM, Reife RA, Lamont RJ. 1998. Local chemokine paralysis, a novel pathogenic mechanism for Porphyromonas gingivalis. Infect. Immun. 66:1660–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hasegawa Y, Tribble GD, Baker HV, Mans JJ, Handfield M, Lamont RJ. 2008. Role of Porphyromonas gingivalis SerB in gingival epithelial cell cytoskeletal remodeling and cytokine production. Infect. Immun. 76:2420–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Coats SR, Berezow AB, To TT, Jain S, Bainbridge BW, Banani KP, Darveau RP. 2011. The lipid A phosphate position determines differential host Toll-like receptor 4 responses to phylogenetically related symbiotic and pathogenic bacteria. Infect. Immun. 79:203–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jain S, Darveau RP. 2010. Contribution of Porphyromonas gingivalis lipopolysaccharide to periodontitis. Periodontol. 2000 54:53–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hajishengallis G, Lambris JD. 2011. Microbial manipulation of receptor crosstalk in innate immunity. Nat. Rev. Immunol. 11:187–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang M, Krauss JL, Domon H, Hosur KB, Liang S, Magotti P, Triantafilou M, Triantafilou K, Lambris JD, Hajishengallis G. 2010. Microbial hijacking of complement-toll-like receptor crosstalk. Sci. Signal. 3:ra11. 10.1126/scisignal.2000697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liang S, Krauss JL, Domon H, McIntosh ML, Hosur KB, Qu H, Li F, Tzekou A, Lambris JD, Hajishengallis G. 2011. The C5a receptor impairs IL-12-dependent clearance of Porphyromonas gingivalis and is required for induction of periodontal bone loss. J. Immunol. 186:869–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gaffen SL, Hajishengallis G. 2008. A new inflammatory cytokine on the block: re-thinking periodontal disease and the Th1/Th2 paradigm in the context of Th17 cells and IL-17. J. Dent. Res. 87:817–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Teng YT. 2006. Protective and destructive immunity in the periodonticum. Part 2. T-cell-mediated immunity in the periodontium. J. Dent. Res. 85:209–219 [DOI] [PubMed] [Google Scholar]
- 23. Gemmell E, Yamazaki K, Seymour GJ. 2007. The role of T cells in periodontal disease: homeostasis and autoimmunity. Periodontol. 2000 43:14–40 [DOI] [PubMed] [Google Scholar]
- 24. Sato K, Suematsu A, Okamoto K, Yamaguchi A, Morishita Y, Kadono Y, Tanaka S, Kodama T, Akira S, Iwakura Y, Cua DJ, Takayanagi H. 2006. Th17 functions as an osteoclastogenic helper T cell subset that links T cell activation and bone destruction. J. Exp. Med. 203:2673–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Garlet GP. 2010. Destructive and protective roles of cytokines in periodontitis: a re-appraisal from host defense and tissue destruction viewpoints. J. Dent. Res. 89:1349–1363 [DOI] [PubMed] [Google Scholar]
- 26. Groom JR, Luster AD. 2011. CXCR3 ligands: redundant, collaborative and antagonistic functions. Immunol. Cell Biol. 89:207–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moutsopoulos NM, Kling HM, Angelov N, Jin W, Palmer RJ, Nares S, Osorio M, Wahl SM. 2012. Porphyromonas gingivalis promotes Th17 inducing pathways in chronic periodontitis. J. Autoimmun. 39:294–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gaddis DE, Maynard CL, Weaver CT, Michalek SM, Katz J. 2013. Role of TLR2-dependent IL-10 production in the inhibition of the initial IFN-gamma T cell response to Porphyromonas gingivalis. J. Leukoc. Biol. 93:21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Khalaf H, Bengtsson T. 2012. Altered T-cell responses by the periodontal pathogen Porphyromonas gingivalis. PLoS One 7:e45192. 10.1371/journal.pone.0045192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sandros J, Karlsson C, Lappin DF, Madianos PN, Kinane DF, Papapanou PN. 2000. Cytokine responses of oral epithelial cells to Porphyromonas gingivalis infection. J. Dent. Res. 79:1808–1814 [DOI] [PubMed] [Google Scholar]
- 31. Fitzpatrick RE, Wijeyewickrema LC, Pike RN. 2009. The gingipains: scissors and glue of the periodontal pathogen, Porphyromonas gingivalis. Future Microbiol. 4:471–487 [DOI] [PubMed] [Google Scholar]
- 32. Guo Y, Nguyen KA, Potempa J. 2010. Dichotomy of gingipains action as virulence factors: from cleaving substrates with the precision of a surgeon's knife to a meat chopper-like brutal degradation of proteins. Periodontol. 2000 54:15–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Moffatt-Jauregui CE, Robinson B, de Moya AV, Brockman RD, Roman AV, Cash MN, Culp DJ, Lamont RJ. Establishment and characterization of a telomerase immortalized human gingival epithelial cell line. J. Periodontal Res., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Uriarte SM, Rane MJ, Luerman GC, Barati MT, Ward RA, Nauseef WM, McLeish KR. 2011. Granule exocytosis contributes to priming and activation of the human neutrophil respiratory burst. J. Immunol. 187:391–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shultz DB, Rani MR, Fuller JD, Ransohoff RM, Stark GR. 2009. Roles of IKK-beta, IRF1, and p65 in the activation of chemokine genes by interferon-gamma. J. Interferon Cytokine Res. 29:817–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu M, Guo S, Hibbert JM, Jain V, Singh N, Wilson NO, Stiles JK. 2011. CXCL10/IP-10 in infectious diseases pathogenesis and potential therapeutic implications. Cytokine Growth Factor Rev. 22:121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dufour JH, Dziejman M, Liu MT, Leung JH, Lane TE, Luster AD. 2002. IFN-gamma-inducible protein 10 (IP-10; CXCL10)-deficient mice reveal a role for IP-10 in effector T cell generation and trafficking. J. Immunol. 168:3195–3204 [DOI] [PubMed] [Google Scholar]
- 38. Kabashima H, Yoneda M, Nagata K, Hirofuji T, Maeda K. 2002. The presence of chemokine (MCP-1, MIP-1alpha, MIP-1beta, IP-10, RANTES)-positive cells and chemokine receptor (CCR5, CXCR3)-positive cells in inflamed human gingival tissues. Cytokine 20:70–77 [DOI] [PubMed] [Google Scholar]
- 39. Garlet GP, Martins W, Jr, Ferreira BR, Milanezi CM, Silva JS. 2003. Patterns of chemokines and chemokine receptors expression in different forms of human periodontal disease. J. Periodontal Res. 38:210–217 [DOI] [PubMed] [Google Scholar]
- 40. Gemmell E, Carter CL, Seymour GJ. 2001. Chemokines in human periodontal disease tissues. Clin. Exp. Immunol. 125:134–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Crawford MA, Lowe DE, Fisher DJ, Stibitz S, Plaut RD, Beaber JW, Zemansky J, Mehrad B, Glomski IJ, Strieter RM, Hughes MA. 2011. Identification of the bacterial protein FtsX as a unique target of chemokine-mediated antimicrobial activity against Bacillus anthracis. Proc. Natl. Acad. Sci. U. S. A. 108:17159–17164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang D, Chen Q, Hoover DM, Staley P, Tucker KD, Lubkowski J, Oppenheim JJ. 2003. Many chemokines including CCL20/MIP-3alpha display antimicrobial activity. J. Leukoc. Biol. 74:448–455 [DOI] [PubMed] [Google Scholar]
- 43. Cole AM, Ganz T, Liese AM, Burdick MD, Liu L, Strieter RM. 2001. Cutting edge: IFN-inducible ELR-CXC chemokines display defensin-like antimicrobial activity. J. Immunol. 167:623–627 [DOI] [PubMed] [Google Scholar]
- 44. Zhang HM, Yuan J, Cheung P, Chau D, Wong BW, McManus BM, Yang D. 2005. Gamma interferon-inducible protein 10 induces HeLa cell apoptosis through a p53-dependent pathway initiated by suppression of human papillomavirus type 18 E6 and E7 expression. Mol. Cell. Biol. 25:6247–6258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mao S, Park Y, Hasegawa Y, Tribble GD, James CE, Handfield M, Stavropoulos MF, Yilmaz O, Lamont RJ. 2007. Intrinsic apoptotic pathways of gingival epithelial cells modulated by Porphyromonas gingivalis. Cell. Microbiol. 9:1997–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nakhjiri SF, Park Y, Yilmaz O, Chung WO, Watanabe K, El-Sabaeny A, Park K, Lamont RJ. 2001. Inhibition of epithelial cell apoptosis by Porphyromonas gingivalis. FEMS Microbiol. Lett. 200:145–149 [DOI] [PubMed] [Google Scholar]
- 47. Qian C, An H, Yu Y, Liu S, Cao X. 2007. TLR agonists induce regulatory dendritic cells to recruit Th1 cells via preferential IP-10 secretion and inhibit Th1 proliferation. Blood 109:3308–3315 [DOI] [PubMed] [Google Scholar]
- 48. Peyyala R, Kirakodu SS, Novak KF, Ebersole JL. 2012. Oral microbial biofilm stimulation of epithelial cell responses. Cytokine 58:65–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jotwani R, Pulendran B, Agrawal S, Cutler CW. 2003. Human dendritic cells respond to Porphyromonas gingivalis LPS by promoting a Th2 effector response in vitro. Eur. J. Immunol. 33:2980–2986 [DOI] [PubMed] [Google Scholar]
- 50. Oido-Mori M, Rezzonico R, Wang PL, Kowashi Y, Dayer JM, Baehni PC, Chizzolini C. 2001. Porphyromonas gingivalis gingipain-R enhances interleukin-8 but decreases gamma interferon-inducible protein 10 production by human gingival fibroblasts in response to T-cell contact. Infect. Immun. 69:4493–4501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pyrc K, Milewska A, Kantyka T, Sroka A, Maresz K, Koziel J, Nguyen KA, Enghild JJ, Knudsen AD, Potempa J. 2013. Inactivation of epidermal growth factor by Porphyromonas gingivalis as a potential mechanism for periodontal tissue damage. Infect. Immun. 81:55–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, McIntosh ML, Alsam A, Kirkwood KL, Lambris JD, Darveau RP, Curtis MA. 2011. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe 10:497–506 [DOI] [PMC free article] [PubMed] [Google Scholar]