Abstract
The highly pathogenic Yersinia enterocolitica strains have a chromosomally encoded type III secretion system (T3SS) that is expressed and functional in vitro only when the bacteria are cultured at 26°C. Mutations that render this system nonfunctional are slightly attenuated in the mouse model of infection only following an oral inoculation and only at early time points postinfection. The discrepancy between the temperature required for the Ysa gene expression and the physiological temperature required for mammalian model systems has made defining the role of this T3SS challenging. Therefore, we explored the use of Drosophila S2 cells as a model system for studying Ysa function. We show here that Y. enterocolitica is capable of infecting S2 cells and replicating intracellularly to high levels, an unusual feature of this pathogen. Importantly, we show that the Ysa T3SS is required for robust intracellular replication. A secretion-deficient mutant lacking the secretin gene, ysaC, is defective in replication within S2 cells, marking the first demonstration of a pronounced Ysa-dependent virulence phenotype. Establishment of S2 cells as a model for Y. enterocolitica infection provides a versatile tool to elucidate the role of the Ysa T3SS in the life cycle of this gastrointestinal pathogen.
INTRODUCTION
Yersinia enterocolitica biovar 1B has two functional type III secretion systems (T3SS). One is the well-characterized Ysc-Yop T3SS, which is essential for virulence in mouse models of infection (reviewed in reference 1). This system is completely contained on an ∼70-kb virulence plasmid and is shared by all three pathogenic yersiniae. Specific functions have been identified for many of the effectors (Yops) of this system and are primarily directed toward inhibiting phagocytosis and blocking immune responses. The Ysa-Ysp T3SS is encoded on the chromosome in a region called the plasticity zone and is present only in the highly virulent Y. enterocolitica biovar 1B strains (2). Of the 15 proteins secreted by the Ysa T3SS under Ysa-inducing growth conditions, 12 are designated Ysps and 3 are Yop effector proteins that are associated with and usually secreted by the plasmid-encoded Ysc T3SS (3). Four Ysps have conserved functional domains, and two of these display their predicted functions in vitro (4). However, unlike the Yops, most of the Ysp effector proteins are unique and have no identified roles (4). Curiously, only one Ysa effector gene, yspA, is located in the same locus as the secretion apparatus genes; the genes encoding the remaining effectors are scattered around the chromosome and are likely to have been acquired by independent horizontal transfer events (4).
In vitro, secretion of Ysps is observed only when the bacteria are cultured at 26°C and in the presence of high concentrations of sodium chloride (∼290 mM) (5, 6). An unusual phosphorelay system, YsrRST, is required for activation of the promoter that controls expression of the ysa apparatus genes (7, 8). RcsB, a response regulator in the Rcs phosphorelay system, is also required to activate this promoter (8). Intriguingly, the majority of the ysa effector genes are coordinately regulated by the same environmental conditions and transcriptional regulators that activate expression of the apparatus genes, despite being scattered around the chromosome (9). This is particularly striking given the assumption that these genes were independently acquired yet apparently have evolved synchronous regulation with the apparatus genes to ensure a complete system that is functional under specific conditions.
Attempts to define the role of the Ysa T3SS in vivo have been challenging, and the function of this system has remained a bit of an enigma. Although the Ysa T3SS is unique to the highly pathogenic Y. enterocolitica 1B strains, mutations that render the Ysa system nonfunctional are only mildly attenuated in the mouse model of oral infection, and the virulence defects are subtle (4, 5, 8). Tissue culture models have provided limited insight, as their use is hampered by the strict low-temperature requirement for expression and function of the Ysa T3SS. While the Ysa-Ysp system does not appear to be a critical virulence factor in the mouse model, the fact that Y. enterocolitica has preserved this intact T3SS and presumably evolved coordinated regulation of independently acquired effector genes would seem to indicate that it is a necessary element during some stage in the life cycle of this gastrointestinal pathogen.
As efforts to study the role of the Ysa T3SS in host systems have been stymied by the conflict between the temperature required to activate the ysa and ysp genes (26°C) and the temperature of host and tissue culture systems (37°C), we turned to a model system compatible with growth at lower temperatures, Drosophila melanogaster S2 cells. S2 cells are a macrophage-like cell line and have been demonstrated as a suitable model for the study of host-pathogen interactions, replicating many phenotypes observed in mouse and human cell lines (10–12). The genetic and cellular tools available for use with these cells are plentiful, and numerous approaches to RNA interference (RNAi) screens have been employed to examine host-pathogen interactions (13–20). In addition, a recent study utilized S2 cells to examine the function of the Yersinia T3SS effector YopJ (21). Therefore, S2 cells seemed to be an ideal host system in which to study the importance of the Ysa T3SS. In this report, we demonstrate for the first time a pronounced role for the Ysa T3SS in an infection model. Elements of this system are required for an intracellular replication process that ultimately kills the S2 cells, while Ysa-deficient strains fail to replicate. In addition to uncovering a potentially important role for this conserved T3SS, this work also provides the basis for numerous studies that will contribute significantly to identifying the function of the Ysa T3SS in the life cycle of Y. enterocolitica.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains and plasmids used in this study are listed in Table 1. Y. enterocolitica strains were grown at 26°C in LB (1% tryptone, 0.5% yeast extract) with 290 mM NaCl (referred to as LB-290). Escherichia coli strains were grown in LB at 37°C. Kanamycin, 50 μg/ml, and nalidixic acid, 20 μg/ml, were added as needed.
Table 1.
Strains and plasmids used in this work
| Strain or plasmid | Relevant genotype or characteristicsa | Reference |
|---|---|---|
| E. coli | Invitrogen | |
| DH5α | F− ϕ80ΔlacZM15 Δ(lacZYA-argF)U169 deoP recA1 endA1 hsdR17(rK− mK−) | |
| S17-1λpir | Tpr Strr recA thi pro hsdR hsdM+ RP4::2-Tc::Mu::Km Tn7; λ pir lysogen | 44 |
| Y. enterocolitica | ||
| JB580v | 8081v (r− m+ Nalr), serotype O:8 | 45 |
| JB580c | JB580v cured of pYVe8081 | Lab strain |
| YVM1178 | JB580v ΔysaC | 31 |
| YVM1184 | JB580c ΔysaC | This work |
| YVM1374 | JB580v ΔyscC | 31 |
| YVM1629 | JB880v ΔysaC ΔyscC | This work |
| YVM1600 | JB580c ΔyspD | This work |
| YVM1370 | JB580v Δinv | This work |
| YVM1390 | JB580c Δinv | This work |
| YVM1520 | JB580c ΔyspA | This work |
| YVM1521 | JB580c ΔyspE | This work |
| YVM1522 | JB580c ΔyspF | This work |
| YVM1523 | JB580c ΔyspI | This work |
| YVM1524 | JB580c ΔyspK | This work |
| YVM1525 | JB580c ΔyspL | This work |
| YVM1528 | JB580c ΔyspM | This work |
| YVM1526 | JB580c ΔyspP | This work |
| YVM1527 | JB580c ΔyspY | This work |
| YVM11511 | JB580c ΔyspAF | This work |
| YVM1550 | JB580c Δysp9 (ΔyspAEFIKLMPY) | This work |
| Plasmids | ||
| pSR47S | Kanr; MobRP4 oriR6K, cloning vector | 24 |
| pGEN-lux | Ampr; source of em7 promoter for pJH026 | 22 |
| pPROBE-gfp[tagless] | Kanr; gfp transcriptional fusion plasmid | 23 |
| pMWO-073 | Specr; low-copy-number expression vector | 25 |
| pJH026 | Kanr; em7 promoter cloned into pPROBE-gfp[tagless] | This work |
| pKW60 | Kanr; yspA in-frame deletion (codons 6–637 deleted) | This work |
| pKW54 | Kanr; yspE in-frame deletion (codons 11–380 deleted) | This work |
| pKW59 | Kanr; yspF in-frame deletion (codons 5–346 deleted) | This work |
| pKW12 | Kanr; yspI in-frame deletion (codons 29–137 deleted) | This work |
| pKW91 | Kanr; yspK in-frame deletion (codons 12–176 deleted) | This work |
| pKW56 | Kanr; yspL in-frame deletion (codons 2–700 deleted) | This work |
| pBF001 | Kanr; yspM in-frame deletion (codons 14–275 deleted) | This work |
| pKW57 | Kanr; yspP in-frame deletion (codons 8–382 deleted) | This work |
| pKW55 | Kanr; yspY in-frame deletion (codons 13–444 deleted) | This work |
| pKW106 | Kanr; yspD in-frame deletion (codons 20–319 deleted) | This work |
| pEll26 | Kanr; inv in-frame deletion (codons 4–833 deleted) | This work |
| pKW132 | Specr; ysaC coding region in pMWO-073 | This work |
| pKW133 | Specr; yspD coding region in pMWO-073 | This work |
Kan, kanamycin; Amp, ampicillin; Spec, spectinomycin.
Plasmid and strain construction.
The plasmids and strains used in this study are listed in Table 1, and the primers are listed in Table 2. Each newly constructed plasmid was confirmed by restriction digest patterns and sequencing. Plasmid pJH026 was constructed as follows. The constitutive em7 promoter was amplified from pGEN-luxCDABE (22) using primers JDH151 and JDH152. The product was cleaved with SalI and BamHI and ligated into those same sites of pPROBE-gfp[tagless] (23).
Table 2.
Primers used in this work
| Name | Sequence (5′–3′)a |
|---|---|
| JDH151 | AAAGTCGACCTCAATCAAGTTATTTTCTTACCAGTC |
| JDH152 | AAAGGATCCACGTATCCTCCAAGCCTGAATTC |
| inv-delA | ACGCGTCGACGCCACTGCAGGTTATTAATCACCAG |
| inv-delB | CGGGATCCTGAATACATTAGTGTACCCCCTTAG |
| inv-delC | CGGGATCCGAGCCTCAATAGTGCTAAATACCAATC |
| inv-delD | ATAAGAATGCGGCCGCCCATGGACACCGGTAAAATTGCCG |
| yspA-delA | GCGTCGAGGAGTATGAGCAGAGCAGCGGC |
| yspA-delB | CGGGATCCCATGATATTAGGCATAACTAATCTTCC |
| yspA-delC | CGGGATCCACAGAGATTGAGTGATGAAACCAG |
| yspA-delD | ATAAGAATGCGGCCGCGGCATATGAGACATAAAGTTCTCC |
| yspE-delA | GCGTCGACGCTCCAGCGCTTTGCCACCATGAC |
| yspE-delB | CGGGATCCCGGGCTACTGCTTTGACTTATCC |
| yspE-delC | CGGGATCCGGCTATACCATTAAGCTCAACGAAG |
| yspE-delD | ATAAGAATGCGGCCGCGGATTACAGAAAGCCATCGATGCC |
| yspF-delA | GCGTCGACGTTTAACACCAAAGCTGAGAACGTG |
| yspF-delB | CGGGATCCTGCTGGCGTCATTAAATGTTGCTC |
| yspF-delC | CGGGATCCCCAACTTCTGCGCCCTCTCTTCC |
| yspF-delD | ATAAGAATGCGGCCGCGTGAATTAGATCTGGCTAAACCTG |
| yspI-delA | ACGCGTCGACCCCAGTGGTTGAGGATAACCACAC |
| yspI-delB | CGGGATCCATTCTTTGAGGTGGATGAGGTATC |
| yspI-delC | CGGGATCCGCTATCTCAGCTAAGCAATCCG |
| yspI-delD | ATAAGAATGCGGCCGCCCTGGCTCAGGGAGCAAATGCAAC |
| yspK-delA | ATAAGAATGCGGCCGCCCGATAAAGGTATTCCCGTCG |
| yspK-delB | CGGGATCCAGGTGTTTGAATAATGGTAGGTG |
| yspK-delC | CGGGATCCGACCGGCGATTACAAATGCGTAC |
| yspK-delD | ATAAGAATGCGGCCGCGCAAGCCCACGCTTAACAGCC |
| yspL-delA | GCGTCGACCTGAGACAAACTTCGGCTTCAGAG |
| yspL-delB | CGGGATCCCATGATTACTCCCTTTAAATATATCC |
| yspL-delC | CGGGATCCTTCAAGTAATAAGTACAGCAGATGCC |
| yspL-delD | ATAAGAATGCGGCCGCAGCTGGTGGATCAAGCCTGGCAAC |
| yspM-delA | ACGCGTCGACATTACCTGTAACAGCGATATACAC |
| yspM-delB | CGGGATCCGGGACAAAAAGATCTGTTGTG |
| yspM-delC | CGGGATCCGCCTATCATATCTGGATATGTTC |
| yspM-delD | ATAAGAATGCGGCCGCGTAAATAACGGCCGATAC |
| yspP-delA | GCGTCGACCTGCGGATCGATATAACCGAATGG |
| yspP-delB | CGGGATCCGGCGATGTTTTTTGTAAGCATTAGTC |
| yspP-delC | CGGGATCCTTATCTGTACCTAAACCTATTTACG |
| yspP-delD | ATAAGAATGCGGCCGCGGTTAGTGATCGATTATTATCGCC |
| yspY-delA | GCGTCGACGGAATCACTGTTCTCTTATCGCG |
| yspY-delB | CGGGATCCGAAATAATCGGCCATAACAATAACCG |
| yspY-delC | CGGGATCCGACCGATAATAACAGTGCTAGTGTG |
| yspY-delD | ATAAGAATGCGGCCGCCCGGCAAAAGTAGCGCAAGCGCC |
| yspD-delA | GCGTCGACCGCTCCAAATGAATCAGGTAGAGG |
| yspD-delB | CGGGATCCCGCGCCCAACATCAGTTCCGGGAC |
| yspD-delC | CGGGATCCCGGCAGGACAATACCAGTTTTGAAACCCTG |
| yspD-delD | ATAAGAATGCGGCCGCCCGTGTTAACGTCTGCTAAGGGGG |
| KW244 | GGGGTACCAAAGTTAAATACAGAGGAGTGATC |
| KW245 | GGGGTACCTCAACAGCGATTATTTCACTACC |
Restriction enzyme sites are underlined.
In-frame gene deletions were constructed as previously described (7). Briefly, fragments of approximately 500 bp upstream and downstream of each gene were independently amplified using primers designated with delA/delB (upstream) or delC/delD (downstream). These fragments were digested with SalI and BamHI (upstream) or BamHI and NotI (downstream), ligated into pSR47S (24), cut with SalI and NotI, and transformed into S17-1 λ pir. The resulting plasmids were verified by restriction digest and sequencing and then introduced into the desired Y. enterocolitica strains by conjugation. Following counterselection on sucrose plates, confirmation of the deleted gene was determined by diagnostic PCR.
Complementation clones for the ysaC and yspD genes were constructed by amplifying the respective gene and cloning into pMWO-073, a low-copy-number vector with the tet operator/promoter inducible with anhydrous-tetracycline (ATc) (25). Primers KW244 and ysaC-delD were used to amplify ysaC, and KW245 and yspD-delD were used to amplify yspD. Each primer adds a restriction site (KpnI on forward and NotI on reverse); the products were cleaved with these enzymes and ligated into pMWO-073 cleaved with these same enzymes. The resulting plasmids, pKW132 (ysaC) and pKW133 (yspD), were verified by restriction digest and sequencing. Expression was induced from these promoters by adding 100 ng/ml ATc.
To cure Y. enterocolitica of the pYV8081 virulence plasmid (pYV), the bacteria were streaked for isolation on LB-MOX agar plates (20 mM sodium oxalate and 20 mM magnesium chloride) and grown at 37°C overnight. Large colonies were selected and passaged on LB-MOX to confirm a pure population of large colonies. Loss of pYV was corroborated by the absence of PCR amplification of genes carried by pYV.
GPA.
For gentamicin protection assay (GPA), Drosophila S2 cells were maintained in Sf-900 II media (Invitrogen) at 26°C without CO2 as described previously (26). The cells were seeded at a concentration of 5 × 105 cells per well in a 24-well tissue culture plate and incubated at 26°C for 16 h before infection. Saturated overnight cultures of Y. enterocolitica carrying pJH026 grown in LB-290 were diluted to a starting optical density at 600 nm (OD600) of 0.2 in 2 ml of LB-290 and grown for 2 h at 26°C on a roller drum to induce the Ysa T3SS. The OD600 of bacterial cultures was measured and used to calculate the volume needed for a multiplicity of infection (MOI) of 10. Bacterial cultures were added to the wells and centrifuged at 215 × g for 5 min to initiate infection. After a 1-hour incubation at 26°C, the medium was replaced with medium containing 100 μg/ml gentamicin (Gent100) to kill extracellular bacteria, and the culture was incubated for 1 h at 26°C. The medium was then replaced again with medium containing 10 μg/ml gentamicin (Gent10), and the culture was incubated until the desired time point. At these time points, the medium was removed and 200 μl 1× phosphate-buffered saline (PBS) was added. Fluorescence (excitation at 485 nm, emission at 528 nm) was measured using a Synergy H1 plate reader (BioTek, Winooski, VT). The relative fluorescence units (RFU) from an uninfected well containing S2 cells was subtracted from the RFU in experimental samples. All strains were tested in quadruplicate on each plate and normalized to the relevant wild-type strain. Each strain was tested at least three times, and data from representative assays are shown. When testing strains with pMWO-073 and derivatives (see Fig. 1B), 100 ng/ml ATc was added to the S2 cell medium to ensure expression during the course of infection.
Fig 1.
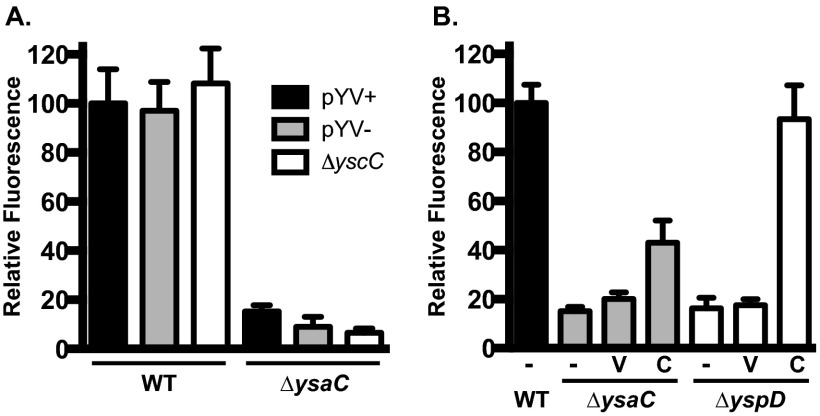
Infection of S2 cells by Y. enterocolitica requires a functional Ysa T3SS. S2 cells were infected with Y. enterocolitica strains at an MOI of 10 and subjected to gentamicin protection assays as described in Materials and Methods. Relative fluorescence units (RFU) measured at 20 h postinfection are averages from 4 wells per strain and normalized to wild type, pYV+ (set to 100%); error bars represent the standard deviations. (A) Strains with mutations that impact T3SS in Y. enterocolitica show that only the Ysa system is required for replication. (B) Both secretion (ΔysaC) and translocation (ΔyspD) components are necessary for replication. Each deletion strain (all pYV−) was tested alone (−, no plasmid) or transformed with pMWO-073 (V, vector) or complementing clone (C, complemented) as indicated below the x axis. Data shown are from a representative assay; each assay was performed a minimum of three times. The complementing clones are pKW132 for ysaC and pKW133 for yspD.
Assay for intracellular bacteria.
Internalized bacteria were measured essentially as described for invasion assays (27). Y. enterocolitica strains were used to infect S2 cells as described for the gentamicin protection assays above. After a 1-h treatment with Gent100, the S2 cells were lysed with 1% Triton X-100 for 5 min, diluted, and plated on LB agar containing nalidixic acid and kanamycin. The percentage of intracellular bacteria was calculated as the number of viable bacteria recovered divided by the inoculum and expressed as percent intracellular bacteria. Because assays were performed on multiple days with some variation in percent intracellular bacteria obtained for the wild-type strain, all comparisons are relative to the average wild-type value obtained from the assay in which the mutants were tested. The relative percent invasion between the wild type and ΔysaC was consistent between assays, validating this approach.
Intracellular bacterial growth curves.
S2 cells were seeded and infected as described for invasion and gentamicin protection assays. At 2, 5, and 8 h postinfection, the S2 growth medium was removed, and the cells were lysed with 1% Triton X-100, diluted, and plated on LB agar plates as described above. Because of the large number of extracellular bacteria at later time points, 100 μl of 10% Triton X-100 was added directly to the growth medium (∼1% final concentration) for the 16- and 20-h samples. Three experiments were performed for 2, 8, and 20 h and two experiments for 5 and 16 h (see Fig. 5B for the data derived from all experiments). Growth rates were calculated from these data using the formula ln(CFU2) − ln(CFU1)/t2 − t1, where t1 is the time in hours and refers to 2 and 5 h and t2 refers to 5 and 8 h (28).
Fig 5.
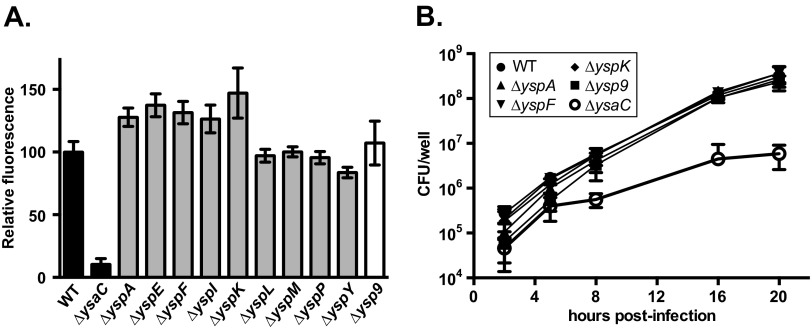
Loss of the known Ysa effector proteins does not impair bacterial replication within S2 cells. A gentamicin protection assay was performed as described for Fig. 1 using pYV− strains carrying in-frame deletions of an individual ysp gene (ΔyspA-P) or all 9 (Δysp9). (A) Endpoint RFU measurement shows that all ysp mutants replicate to wild-type (WT) levels by 20 h postinfection (hpi). Data shown are derived from two independent assays (n ≥ 6). (B) Bacterial growth during infection shows that reduced intracellular bacteria at 2 hpi does not correlate with reduced intracellular growth. CFU were determined at the specified time points as described in Materials and Methods and are shown as averages of 3 assays for 2, 8, and 20 hpi (n ≥ 11) and 2 assays for 5 and 16 hpi (n ≥ 7); error bars represent the standard deviations.
Cytotoxicity assay.
S2 cells were seeded and infected as described above for the invasion and gentamicin protection assays. At 8, 12, or 22 h postinfection, supernatants were transferred to microcentrifuge tubes, centrifuged for 2 min at maximum speed to remove debris, and then transferred to clean microcentrifuge tubes. For samples that were lysed, 100 μl of 10% Triton X-100 was added to the wells and allowed to incubate for 5 min before being transferred and centrifuged as above. Using the d-lactic acid/l-lactic acid kit (R-Biopharm, catalog number 11 112 821 035) with a 300-μl reaction volume, 90 μl of cleared supernatant/lysate was mixed with 133.9 μl buffer, l-lactate solution (8 μg), glutamate-pyruvate transaminase (4.2 U), and water. The reaction was initiated by the addition of NAD (940 μg). Absorbance at 340 nm was measured every 2 min for 1 h using a Tecan infinite M200 plate reader and Magellan version 7.1 software. Lactate dehydrogenase (LDH) activity is represented as the slope of NADH production (ΔOD340/Δtime), where the slope for the uninfected lysed S2 cells was set to 100% and all others normalized to this value.
Fluorescence microscopy.
Prior to seeding S2 cells in 24-well plates, glass coverslips were placed at the bottom of each well, and GPAs were performed as described above. At the desired time point, the medium was removed, and the cells were washed with 1 ml 1× PBS and fixed with 4% paraformaldehyde in 1× PBS. After incubation at 4°C for 15 to 20 min, the coverslips were washed twice with 1× PBS and allowed to dry. Coverslips were mounted on slides using Prolong Gold mounting media with DAPI (4′,6-diamidino-2-phenylindole) (Invitrogen) and cured in the dark at room temperature overnight. At least two fields were selected for each sample, and paired differential interference contrast (DIC), blue fluorescent (DAPI), and green fluorescent protein (GFP) images were obtained for each field to identify whole S2 cells, nuclei, and bacteria, respectively. Exposures were set for wild-type samples and held constant when imaging mutant samples to accurately represent the presence or absence of fluorescent signal. Images were captured using an Olympus BX-60 microscope with a 60× objective equipped with a SPOT-RT slider charge-coupled-device (CCD) camera (Diagnostic Instruments) and processed with iVision imaging software (version 4.0; BioVision Technologies, Exton, PA). Images were prepared for presentation using Adobe Photoshop. Images were obtained and analyzed from at least three independent infections; those shown are from one representative infection.
TEM.
A GPA was performed as described above, except the MOI was increased to 25 to enhance the number of S2 cells containing intracellular Y. enterocolitica. At 4 h postinfection, the cells were fixed in 3% glutaraldehyde–0.15 M sodium phosphate buffer, pH 7.4, for 1 h at room temperature. Following three rinses with 0.15 M sodium phosphate buffer, pH 7.4, the monolayers were postfixed with potassium ferrocyanide-reduced osmium for 1 h at room temperature (1% osmium tetroxide–1.25% potassium ferrocyanide–0.15 M sodium phosphate buffer [29]). After washing with deionized water, the cells were dehydrated using increasing concentrations of ethanol (30%, 50%, 75%, 100%, and 100% for 10 min each) followed by embedment in Polybed 812 epoxy resin (Polysciences, Inc.,Warrington, PA). The cells were sectioned en face to the substrate at 70 nm using a diamond knife. Ultrathin sections were collected on 200 mesh copper grids and stained with 4% aqueous uranyl acetate for 15 min, followed by Reynolds' lead citrate for 7 min (30). Samples were viewed using a LEO EM910 transmission electron microscope (TEM) (Carl Zeiss SMT, Inc., Peabody, MA), operating at an accelerating voltage of 80 kV. Digital images were taken using a Gatan Orius SC 1000 CCD Camera and DigitalMicrograph software (version 3.11.0; Gatan, Inc., Pleasanton, CA).
Statistical analysis.
All experiments were evaluated using an unpaired, two-tailed Student's t test. Analyses were conducted using the statistics feature in Prism version 6.0 (GraphPad Software). The stated P values always refer to comparison with wild-type-infected samples.
RESULTS
The Ysa T3SS is required for infection of S2 cells.
The usefulness of tissue culture cell lines for the study of the Ysa T3SS is abrogated by the low-temperature requirement for expression of genes encoding the system components. Because Drosophila S2 cells grow at 26°C and have proven to be a useful system for the study of other bacterial pathogens, we decided to determine if S2 cells might provide a suitable model for studying the Ysa T3SS. YsaC is the outer membrane secretin protein of the secretion apparatus; deletion of the ysaC gene eliminates Ysa-dependent secretion in vitro and was therefore chosen as a Ysa-deficient strain for these studies (7, 31). To establish whether wild-type or ΔysaC Y. enterocolitica strains could infect S2 cells, we performed gentamicin protection assays (GPA). At 20 to 24 h postinfection, the bacteria were enumerated by plating, and the CFU from wild-type-infected cells averaged >25-fold more bacteria than ΔysaC-infected cells (data not shown; see Fig. 5B). Visual inspection of infected S2 cells by light microscopy prior to lysing for CFU enumeration revealed an unusual result: a large population of extracellular bacteria was observed in wild-type-infected wells, and almost no visible bacteria were observed in the wells infected with ΔysaC. These observations lead to two conclusions: (i) Y. enterocolitica readily infects S2 cells in a manner not observed in mammalian cells, and (ii) the striking difference in growth behavior between the wild-type and ΔysaC strains indicated that the S2 cells could be a suitable model for the study of the Ysa T3SS. To more easily quantify bacterial growth following infection of S2 cells, we transformed the wild-type and ΔysaC strains with a plasmid expressing gfp under the control of a constitutive promoter (pJH026) so that the relative size of the bacterial populations could be inferred by measuring relative fluorescence units (RFU). Consistent with the CFU data, we found that at 20 h postinfection, the wild-type bacteria had proliferated extensively to high numbers (set to 100%), while the number of ΔysaC bacteria remained low (<15%) (Fig. 1A). This indicates that the gfp reporter can be used reliably as an indicator of replication in S2 cell GPAs. Importantly, this is the first demonstration of a pronounced requirement for the Ysa T3SS in an infection model.
All pathogenic Yersinia strains possess a virulence plasmid encoding the Ysc T3SS. This T3SS is essential for virulence of these pathogens in mice and is known to be upregulated in vitro at 37°C under low calcium concentrations (reviewed in reference 1). Although the S2 cell assays are all conducted at 26°C, it has been shown that Yops can be translocated in a Ysc-dependent manner into insect cells at this lower temperature (32). Therefore, we wanted to determine if the Ysc T3SS (encoded by pYV) contributed to the infection of S2 cells. Both the wild-type and ΔysaC strains were cured of the virulence plasmid (pYV) to remove all ysc and yop genes and tested in a GPA. There was no substantial difference between the two Ysa+ strains, as the RFU from the pYV− strain was 97% of the pYV+ strain (Fig. 1A). Similarly, the plasmid-cured ΔysaC strain behaved comparably to its pYV+ counterpart, with 9% of the pYV+ wild-type RFU. It has been shown that three of the proteins secreted through the Ysa T3SS are Ysc effectors (YopE, YopP, and YopN) (3). Curing the strains of pYV also removes these yop genes; in order to assess the potential contribution of these Yops to Ysa-dependent infection of S2 cells, a clean deletion of the yscC gene, encoding the Ysc secretin, was constructed and tested by GPA. This ΔyscC strain was found to behave just as the other Ysa+ strains, yielding RFU at 108% of the wild-type level (Fig. 1A). In addition, a ΔysaC ΔyscC double deletion resulted in a phenotype similar to that of the other Ysa− strains, yielding 7% of the pYV+ wild-type RFU. Thus, neither the Ysc T3SS nor the Yops appear to contribute to the infection of S2 cells.
The cellular impact of type III secreted effectors is typically a result of translocation of these proteins across the host cell membrane. Therefore, we wanted to verify that the observed bacterial replication in S2 cells was a consequence of effector translocation and not just secretion from cytoplasmic bacteria postinternalization. Published data indicate that S2 cells are amenable to translocation (32), and we have observed Ysa-dependent translocation of Ysps into other insect cell lines (data not shown). To assess the importance of Ysp translocation, we performed a GPA with a strain carrying an in-frame deletion of yspD, which encodes one component of the translocon. This strain displayed low RFU levels comparable to those of the ΔysaC strain, with 5% of wild-type RFU (Fig. 1B). A similar result was obtained for another translocon mutant, ΔyspB (data not shown). The low level of fluorescence from both the ΔysaC and ΔyspD strains could be complemented when transformed with plasmids carrying an ATc-inducible copy of the deleted gene (Fig. 1B). These data suggest that translocation, and not just secretion, is required for the bacterial replication phenotype. Taken together, these results demonstrate that infection of S2 cells by Y. enterocolitica (i) requires a fully intact Ysa T3SS (both secretion apparatus and translocon) and (ii) does not require the Ysc T3SS or Yop effectors.
Infection time course evaluation reveals Ysa-dependent intracellular replication.
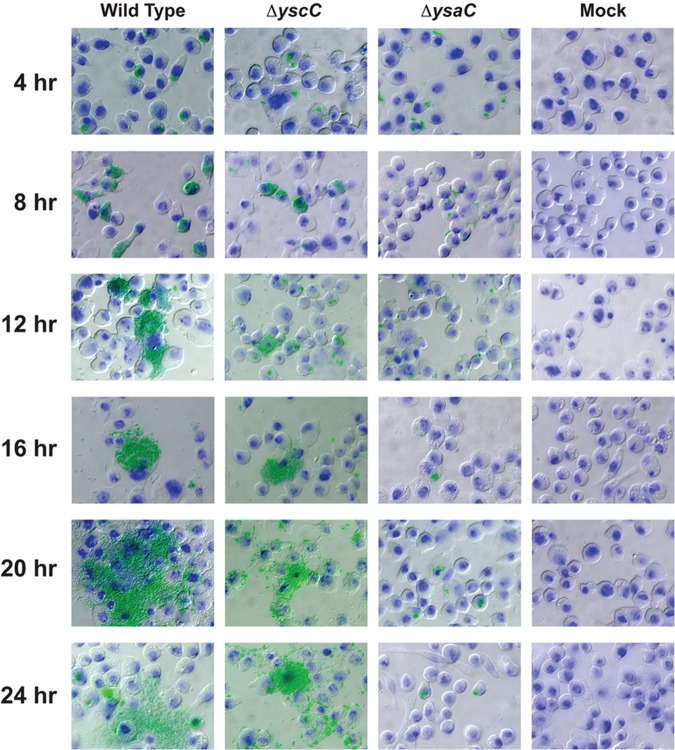
Quantitative fluorescence readings indicated a dramatic difference between wild-type and ΔysaC strains after 20 h of infection but gave no information about the progression of infection. In addition, the presence of extracellular bacteria is a highly unusual attribute of bacteria in a GPA. To gain insight into the various stages of Y. enterocolitica infection in S2 cells, GPAs were performed, and samples were taken every 4 h for fluorescence microscopy. Wild-type, ΔysaC, and ΔyscC strains (all pYV+) were used for this series of experiments to evaluate the contribution of both T3SS (wild type), Ysc T3SS only (ΔysaC), and Ysa only (ΔyscC) to the S2 cell infection progression. At 4 h postinfection, images of S2 cells infected with each strain showed bacteria exclusively in what appears to be an intracellular environment and with comparable bacterial loads (Fig. 2). However, by 8 h postinfection, the cytoplasm of S2 cells infected with Ysa+ strains (wild type and ΔyscC) were filled with bacteria, while those infected with the ΔysaC strain still contained amounts of bacteria similar to that at 4 h. The Ysa+ bacteria clearly demarcated the edges of the S2 cells within which they were confined, suggesting that intracellular bacterial replication had occurred. At 12 and 16 h postinfection, the ΔysaC strain appeared to remain virtually identical to that at 4 h, with modest levels of replication occurring in a few cells. However, replication of wild-type and ΔyscC strains continued to progress, leading to the apparent rupturing of the S2 cells and continued growth as extracellular bacteria (Fig. 2). At the later time points (20 and 24 h), replication of the Ysa+ strains appears to have occurred both intra- and extracellularly, leaving very few healthy or uninfected S2 cells (Fig. 2). The ΔysaC strain showed no noticeable outgrowth and was still predominantly contained within the S2 cells. These time course images provide evidence suggesting that, while both Ysa+ and Ysa− Y. enterocolitica strains survive intracellularly, a functional Ysa T3SS is required for intracellular replication and escape from the S2 cells.
Fig 2.
The Ysa T3SS is necessary for intracellular replication within S2 cells. A gentamicin protection assay was performed using S2 cells seeded on glass coverslips. At the indicated times, samples were washed, fixed, and processed for fluorescence microscopy. Images were taken at a magnification of ×600. Green, gfp-expressing Y. enterocolitica; blue, nuclei stained with DAPI; S2 cells were visualized with differential interference contrast (DIC).
The presence of live extracellular bacteria is a striking phenomenon, especially given that the medium contained 10 μg/ml gentamicin, a dose capable of killing Y. enterocolitica. In a series of control experiments (all performed at 26°C) to test that the extracellular bacteria were not a consequence of differential resistance to gentamicin treatment, we determined that wild-type and ΔysaC strains are equally sensitive to various concentrations of gentamicin and that the 1-h treatment at 100 μg/ml killed >99% of the bacteria (data not shown). However, gentamicin is known to become less active in acidic environments (33), and the S2 culture medium (Sf-900 II) is acidic (pH ∼6.5). To determine whether the low pH reduced the effectiveness of the gentamicin (thus allowing extracellular bacterial growth), we tested the ability of these strains to grow in Sf-900 II as well as in LB at pH 6.3 and unbuffered LB, all with and without various gentamicin concentrations. We found that Y. enterocolitica can grow in the presence of 10 μg/ml gentamicin when the pH of the medium is acidic (data not shown). No growth was detected in the unbuffered LB with gentamicin at 10 μg/ml or higher (lower concentrations were not tested). Higher concentrations of gentamicin were able to prevent bacterial growth even in acidic medium, but these doses were not usable in the GPA because the S2 cells did not tolerate the higher concentrations for the prolonged amount of time required for the assay.
Ysa T3SS promotes internalization of Y. enterocolitica into S2 cells.
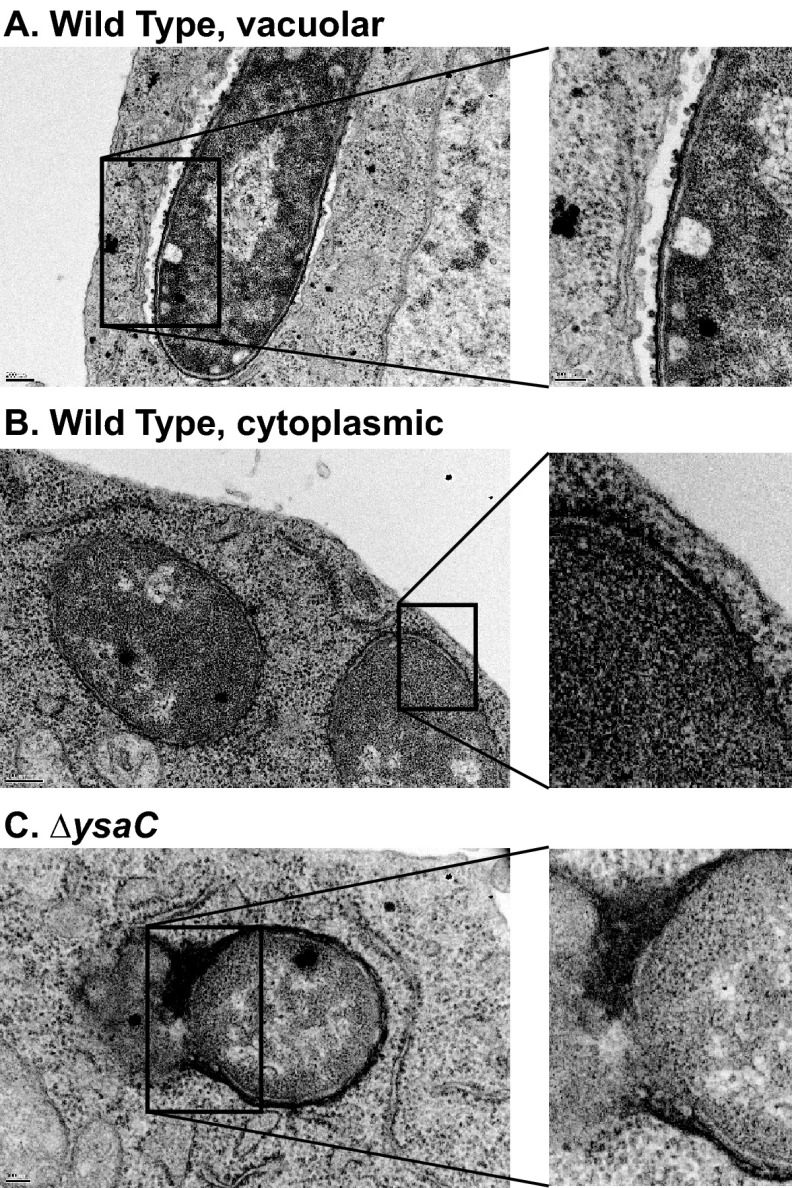
In an effort to more closely examine the intracellular differences between the wild-type and ΔysaC strains and to confirm the intracellular localization of bacteria following infection, transmission electron microscopy was performed on samples harvested at 4 h postinfection. The infection conditions were altered slightly to favor more S2 cells harboring bacteria (see Materials and Methods). In these images, we observed numerous S2 cells with intracellular wild-type bacteria. These bacteria appeared to be either in a single-membrane-bound compartment or cytoplasmic (Fig. 3, top and middle panels). However, very few S2 cells could be found containing ΔysaC bacteria (Fig. 3, bottom panel). Those that were observed were in single-membrane-bound compartments and appeared to be undergoing degradation. These TEM images suggested that the Ysa system plays a role in invasion of S2 cells.
Fig 3.
Transmission electron microscopy reveals Ysa-dependent bacterial internalization. Gentamicin protection assay was performed with pYV− strains at an MOI of 25. At 4 h postinfection, samples were fixed and processed for TEM as described in Materials and Methods. Top panel, wild-type (WT)-infected cell with a bacterium inside a membrane-bound compartment. Middle panel, WT-infected cell with a bacterium that appears to be in the cytoplasm. Bottom panel, ΔysaC-infected cell with a bacterium that appears to be undergoing lysis.
To ascertain if the Ysa T3SS promoted entry of Y. enterocolitica into S2 cells, we measured the number of intracellular bacteria at 2 h postinfection. It was not possible to determine attachment of bacteria to S2 cells independently from internalization into S2 cells; S2 cells are loosely adherent to tissue culture plastics, and attachment assays require extensive washing to remove nonadherent bacteria. Plasmid-cured Y. enterocolitica strains were allowed to infect S2 cells as above, and following the Gent100 treatment, the cells were lysed, diluted, and plated for CFU enumeration. The number of intracellular bacteria was divided by the number of bacteria in the inoculum to determine percent intracellular bacteria. The values obtained for the wild-type strain were typically 2 to 5%, which is quite low compared to that observed for invasion of mammalian epithelial cell lines (typically >10%) (27). However, the percent intracellular bacteria measured for the ΔysaC strain was <1%, consistently measuring about 5-fold less than the wild-type value from experiment to experiment (Fig. 4). Similar decreases were observed for Ysa− strains in the ΔyscC and pYV+ strain backgrounds, indicating that this is a Ysa-specific phenomenon (data not shown).
Fig 4.
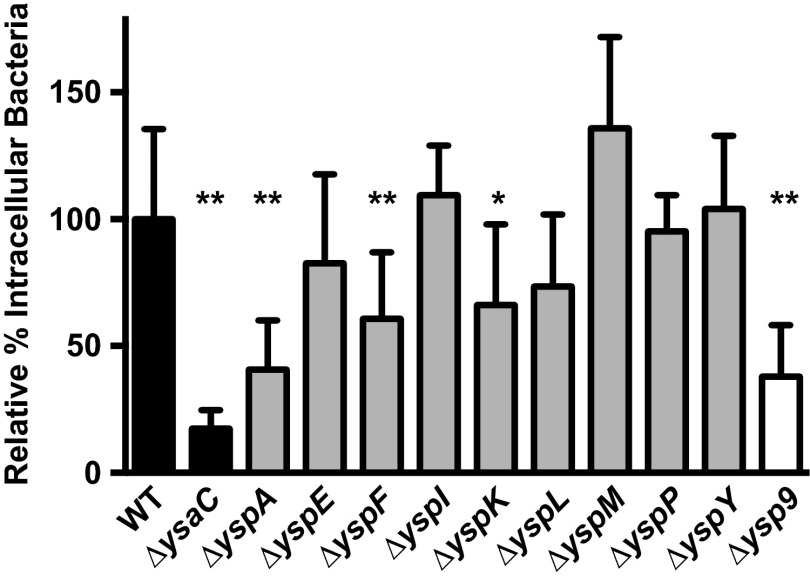
Bacterial internalization is enhanced by the Ysa T3SS. Wild-type and mutant strains were assayed to measure the percentage of internalized bacteria as described in Materials and Methods. The percent intracellular bacteria was determined for each strain and normalized to the average for the wild-type (WT) strain in each assay (WT set to 100%); error bars represent the standard deviations. Strains tested are all pYV− and carry in-frame deletions of a single ysp gene (ΔyspA-P) or all 9 ysp genes (Δysp9). The data are averages of at least 2 assays (n ≥ 8). Student's t test was performed to assess statistical significance. *, P = 0.001; **, P ≤ 0.0001.
The intracellular pathogens Salmonella enterica serovar Typhimurium and Shigella flexneri both use T3SS effectors to promote invasion into their target cells (reviewed in references 34 to 36). Thus, it seemed likely that one or more of the Ysps could similarly promote invasion of Y. enterocolitica into S2 cells. To determine which, if any, of the known effectors played a role in invasion, strains containing in-frame deletions of individual ysp genes were tested for their contribution to the percent intracellular bacteria. Because the experiments testing these mutants were performed on different days, the percent intracellular bacteria determined for each mutant was normalized to that of the wild-type strain to enable comparison. Three effectors appeared to be important for internalization: YspA, YspF, and YspK (Fig. 4). Deletion of each gene showed a statistically significant reduction in the percent intracellular bacteria (P ≤ 0.0001 for ΔyspA and ΔyspF; P = 0.001 for ΔyspK). Loss of any other single ysp gene did not result in a significantly different percent intracellular bacteria compared to the wild type. As effectors present the possible complication of functional redundancy, we also constructed a single strain lacking all nine ysp genes, referred to as Δysp9. This strain showed a reduction in intracellular bacteria relative to the wild type similar to that of the ΔyspA strain (P < 0.0001). Thus, it does not appear that the loss of multiple known effectors had an additive effect on internalization. The more-severe decrease observed with the ΔysaC strain could be a consequence of the inherent variability of this assay, or the structural components themselves may play a role in invasion and/or attachment.
The observed defect in S2 cell invasion by the ΔysaC and certain Δysp strains suggested that active, Ysa-dependent invasion by the bacteria is occurring and that uptake into S2 cells was not simply a consequence of phagocytosis. Y. enterocolitica is an invasive bacterium, and several factors have been shown to contribute to this process, including invasin (27). Knowing that invasin is a crucial factor for the invasion of intestinal M cells (37), we wanted to determine if invasin played a role in the internalization of Y. enterocolitica by S2 cells. A clean deletion of the inv gene was constructed, and Δinv strains with and without the virulence plasmid were transformed with the gfp-expressing plasmid pJH026 and tested in the gentamicin protection and percent intracellular bacteria assays. Both Δinv strains were found to exhibit a percent intracellular bacteria that was the same as, or higher than, that of their wild-type counterpart (data not shown). Unsurprisingly, neither Δinv strain exhibited a marked reduction in RFU compared to its wild-type counterpart, with Δinv pYV+ at 95% and Δinv pYV− at 85% (data not shown). Thus, invasin does not contribute to internalization or intracellular replication of Y. enterocolitica in S2 cells. This is not an altogether surprising result. It is well known that invasin facilitates attachment to M cells by interacting with β1 integrins (38). While the Drosophila genome does contain an ortholog for β1 integrin, we do not know if S2 cells express this gene under these culture conditions or if the region involved in the integrin-invasin interaction is conserved. Thus, invasin would not necessarily be expected to play a role in the invasion of S2 cells.
Known Ysa effector proteins are not important for intracellular replication.
The reduction in intracellular bacteria exhibited by the ΔysaC mutant could contribute to the reduction in the bacterial replication phenotype observed in the GPA. If this is true, then the ΔyspA, ΔyspF, and ΔyspK strains might be expected to be defective for bacterial replication since they are defective for internalization. Because other mechanisms may also be at play, we tested pYV− derivatives of all the ysp deletion strains in the GPA and took RFU measurements at 20 hpi. We observed that none of the individual Ysps were critical for replication within S2 cells, as the RFU from the mutant strains ranged from 84 to 147% of the wild-type strain value (Fig. 5A). Furthermore, the Δysp9 strain exhibited 107% of the wild type RFU (Fig. 5A). At this late time point, there is a significant extracellular population of bacteria that could mask subtle defects in intracellular replication. To circumvent this potential problem, we conducted bacterial growth curves during infections with wild-type, ΔysaC, ΔyspA, ΔyspF, ΔyspK, and Δysp9 strains. A standard GPA was conducted, but at 2, 5, 8, 16, and 20 hpi, samples were lysed and plated for CFU enumeration. At 5 h, all strains show an increase in CFU over the 2-h point (Fig. 5B) and the growth rates were similar, ranging between 0.71 to 0.91 doublings per hour (dph). However, between 5 and 8 hpi, the growth rate of the ΔysaC strain was significantly lower (P = 0.0005) than those of all the other strains, with 0.12 dph compared to a range of 0.38 to 0.55 dph for the other strains. Despite lower percent intracellular bacteria at 2 hpi, the ΔyspA, ΔyspF, ΔyspK, and Δysp9 strains had caught up to the wild-type strain by 8 hpi. Thereafter, all strains except ΔysaC had similar growth rates and CFU values (growth rates were not calculated beyond 8 hpi due to the emergence of extracellular bacteria beyond this time point). These results indicate that, while a functional Ysa T3SS is required for intracellular replication in S2 cells, none of the known Ysps are necessary for replication. This hints at the possibility that Ysa+ strains translocate unidentified Ysa effector proteins that contribute specifically to the ability to replicate within S2 cells. Alternatively, one or more structural components making contact with the host cell could trigger events that are conducive to intracellular replication of this pathogen.
Intracellular replication is correlated with cytotoxicity.
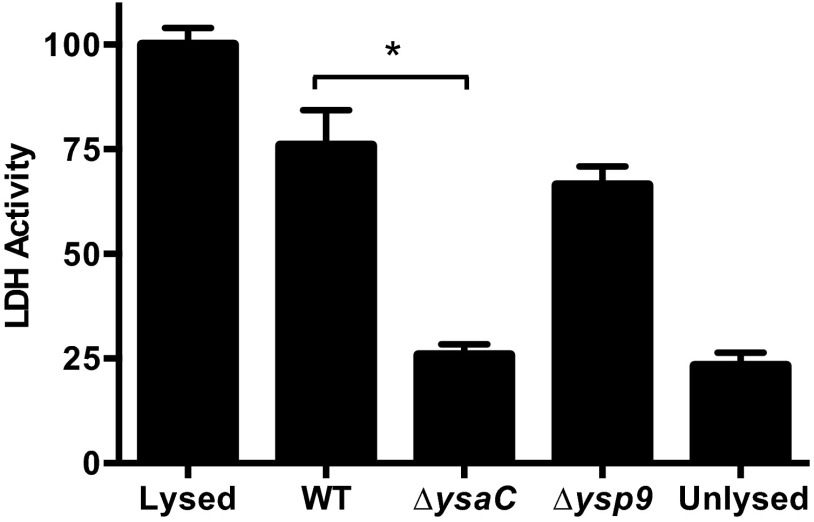
The exact mechanism resulting in extracellular bacteria is not known, but inspection of the microscopy images in Fig. 2 lead us to speculate that it was a consequence of intracellular replication: the bacteria replicate to such high numbers that the cell ruptures when it can hold no more, resulting in cell death. To test whether Ysa+ Y. enterocolitica strains are capable of killing S2 cells, we performed a cytotoxicity assay. Typical colorimetric lactate dehydrogenase (LDH) release assays failed to produce reliable results (data not shown), and this is most likely due to the use of an enzyme (diaphorase) that is sensitive to pH (39) and cannot function properly in the acidic insect cell culture media. We adapted a noncolorimetric kit designed for measuring l-lactic acid such that LDH activity could be determined. NADH, a product of LDH activity, is detected by measuring absorbance at 340 nm; LDH activity is reported as slopes (ΔOD340/Δtime), where the slope for the uninfected lysed S2 cells was set to 100% (maximum) and all others were normalized to this value. At 22 hpi, the LDH activity from wild-type-infected samples was 76% of maximum, whereas the activity from ΔysaC-infected samples was about 25% (Fig. 6). This is just above the LDH levels from unlysed, uninfected cells (23% of maximum). These results indicate that infection with Ysa+ bacteria does indeed result in cell death. Although the Δysp9 strain replicates intracellularly with growth rates similar to those of the wild type, we tested this strain to determine if it had altered cytotoxicity. The LDH activity in samples infected with this strain was about the same as that from wild-type-infected cells at 66% of maximum (Fig. 6). LDH activity from infected cells was not elevated above uninfected, unlysed cells until after 12 hpi (data not shown). These results confirm that infection with Ysa+ bacteria causes cell death and support our model that S2 cell death is a consequence of cell rupture caused by abundant intracellular bacterial replication.
Fig 6.
Infection with Ysa+ bacteria causes S2 cell death. A gentamicin protection assay was performed as described for Fig. 1 using pYV− strains and allowed to proceed for 22 h. LDH activity in supernatants is represented as the slope of NADH production (ΔOD340/Δtime); the slope for the uninfected lysed S2 cells was set to 100%, and all others were normalized to this value. Data shown are averages of at least 2 assays (n ≥ 6). Student's t test was performed to assess statistical significance. *, P ≤ 0.0001.
DISCUSSION
The discovery of the Ysa T3SS was reported in 2000 (5), but its contribution to the Y. enterocolitica life cycle has remained somewhat elusive over the last 12 years. Several studies have shown that it is required for full virulence in mice: ysa mutant strains have a 10-fold increase in the 50% lethal dose (LD50) by an oral route of infection and have a slight defect for colonization of intestinal tissues at early time points postinfection (4, 5, 8). In addition, the results from in vitro assays indicate that the Ysa T3SS induces cytotoxicity against J774.1 cells (8) and Ysa-secreted YopP suppresses tumor necrosis factor alpha (TNF-α) production when the assays are performed at 26°C (3). While these reports certainly indicate that the Ysa T3SS is important for full virulence in mouse model systems, it is not a critical component for pathogenesis in the available mouse models. The subtle measures of attenuation observed with the ysa mutants in mice, combined with the complication of conflicting temperature requirements for Ysa T3SS function and mammalian tissue culture models, have presented challenges to studying the specific role that this system plays in pathogenesis. Drosophila S2 cells have proven to be an excellent model for investigating host-pathogen interactions (40). In addition, S2 cells are cultured at a temperature that is consistent with Ysa gene expression and secretion. Thus, we decided to explore this model as a means to study the Ysa T3SS in hopes that it would shed light on its purpose in the Y. enterocolitica life cycle.
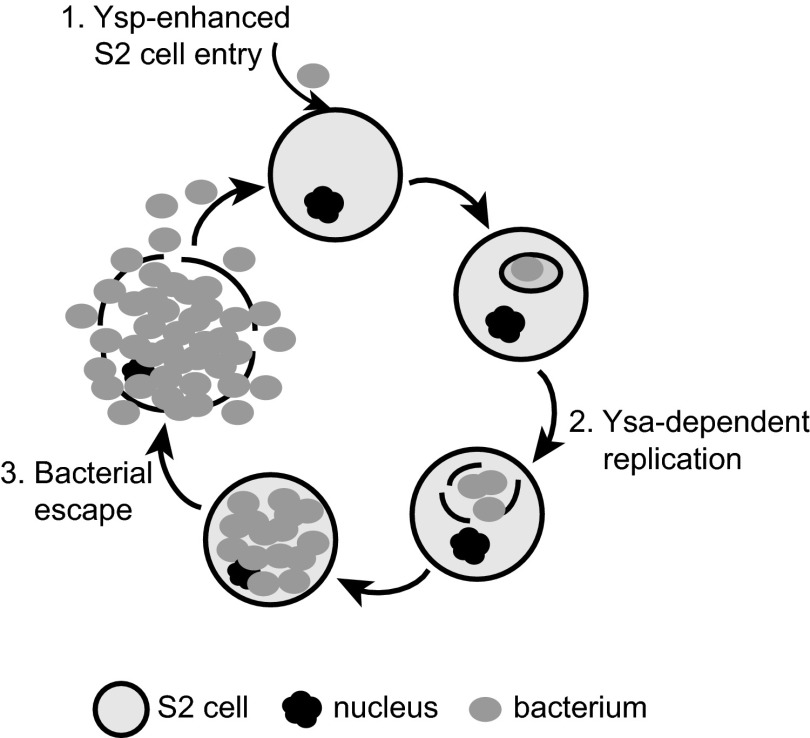
In this report, we show for the first time a pronounced Ysa-dependent phenotype: it is required for infection of Drosophila S2 cells. Infection of S2 cells by Y. enterocolitica is a multistep process that requires (i) bacterial uptake, (ii) intracellular replication, and (iii) escape into the extracellular environment (Fig. 7). The data presented in this report suggest that the Ysa system is important for phases 1 and 2, but the third phase, escape from S2 cells, may not specifically be a Ysa-dependent event, as it could simply occur as the bacterial numbers increase to the point of rupturing the host cell membrane.
Fig 7.
Model of Ysa-dependent Y. enterocolitica infection of S2 cells. The three phases of infection are indicated, showing the influence of a subset of Ysps on internalization (1), replication and presumptive escape from intracellular compartment (2), followed by cell lysis and bacterial escape into the extracellular environment (3), where the cycle can repeat.
The observation that Y. enterocolitica has the capacity to replicate intracellularly is striking, as the pathogenic yersiniae are considered to be primarily extracellular pathogens. Y. enterocolitica and Y. pseudotuberculosis are invasive pathogens and must invade and traverse the M cells that overlay the Peyer's patches (37). It is within this lymph tissue that replication occurs, but primarily in the extracellular environment. There have been several reports showing that Y. enterocolitica can replicate within mouse macrophages (reference 41 and references within). In our lab, we have observed some replication within human and hamster epithelial cell lines (J. D. Hall, K. A. Walker, and V. L. Miller, unpublished results). However, these phenotypes with mammalian cells are likely to be dependent on invasin-mediated invasion and (at least for the epithelial cells) are independent of the Ysa T3SS. The replication phenotype observed in S2 cells is invasin independent and Ysa dependent, suggesting that unique processes are occurring, which are unlike those observed in epithelial cells.
The role of the Ysa T3SS in promoting internalization into S2 cells is likely a secondary role. This notion is derived from the observation that the yspA, yspF, yspK, and ysp9 mutants have a decrease in internalized bacteria, yet these effector mutants have a wild-type replication phenotype. Thus, even at lower initial intracellular numbers, bacteria with deletions in certain ysp genes are still capable of replicating within the S2 cells once internalized (presumably by phagocytosis). Therefore, it appears that the Ysa T3SS is a bifunctional secretion system, acting primarily as an intracellular survival/replication factor, with a secondary role to enhance invasion. Our data indicate that YspA, YspF, and YspK play an active role in invasion, but it is possible that the T3SS structural components also contribute to invasion and/or attachment, as the ΔysaC strain has a more severe defect for internalization. Furthermore, these data indicate that the ability to replicate is not dependent on invasion by the bacterium, as strains with invasion defects had intracellular growth rates similar to those of the wild-type strain. Moreover, the observation that the Δysp9 strain, lacking all known effectors, still has the capacity for normal intracellular replication suggests that there are unidentified ysp genes encoding proteins that contribute to the ability to replicate. It is perhaps a bit naive to assume that all Ysps would be secreted from bacteria grown in laboratory growth medium, and therefore, it is not unreasonable to predict that Y. enterocolitica may express and/or secrete other proteins once in the host intracellular environment. Efforts to determine the Ysa T3SS-dependent proteins that promote intracellular replication within S2 cells are currently being pursued.
Type III secretion effector functions vary from pathogen to pathogen, but many are involved in blocking immune responses and phagocytosis (for extracellular pathogens) or invasion and interfering with phagocyte maturation (for intracellular pathogens) (34–36, 42, 43). Most of the Ysa secreted effectors, the Ysps, are unique proteins with no conserved domains by which to predict their function. YspE, YspK, YspP, and YspM do have conserved domains, but the specific roles in which these proteins perform their functions during infections have not been fully established (4, 31). Here, we present evidence that some of the effectors are important for bacterial entry into S2 cells but that unidentified effector proteins are important for intracellular replication. The observation that there is a strong dependence on the Ysa system for growth within insect cells, paired with the observation that ysa and ysp genes are preferentially expressed at 26°C, also raises the intriguing notion that this T3SS may promote survival in an environmental niche (such as amoebas) or possibly in an insect vector/host. Having established a model system in which a functional Y. enterocolitica Ysa T3SS is necessary for successful infection, we are poised to begin dissecting the contribution of this secretion system to the survival of Y. enterocolitica. Moreover, with the plethora of genetic and cellular techniques available with the S2 cell model, we now have the tools with which to identify the effectors required for intracellular replication and investigate the mechanism(s) used by individual effector proteins in the infection process.
ACKNOWLEDGMENTS
We are indebted to a number of people who helped sustain this project: Brittany Fogarty for construction of the Δysp9 strain, Jordan Walter for construction of pKW106, Stephan Dickgiesser for construction of YVM1600 and initial experiments with this strain, and Patrick Leslie for gentamicin protection assay development. We are grateful for the helpful advice on working with S2 cells given by Stephen Rogers. The contributions by Nikki Wagner and Eric Weening through helpful discussions and thoughtful review of the manuscript are much appreciated. We thank Anthony R. Richardson for advice with the LDH measurements and use of his Tecan platereader and Victoria Madden in the UNC Microscopy Services Laboratory for preparation of samples for transmission electron microscopy and equipment training.
This research was supported in part by National Institutes of Health grant AI063299 awarded to V.L.M. and the UNC SPIRE Postdoctoral Fellowship, NIGMS grant K12GM000678, awarded to J.D.H.
Footnotes
Published ahead of print 29 April 2013
REFERENCES
- 1. Cornelis GR, Boland A, Boyd AP, Geuijen C, Iriarte M, Neyt C, Sory MP, Stainier I. 1998. The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 62:1315–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Thomson NR, Howard S, Wren BW, Holden MTG, Crossman L, Challis GL, Churcher C, Mungall K, Brooks K, Chillingworth T, Feltwell T, Abdellah Z, Hauser H, Jagels K, Maddison M, Moule S, Sanders M, Whitehead S, Quail MA, Dougan G, Parkhill J, Prentice MB. 2006. The complete genome sequence and comparative genome analysis of the high pathogenicity Yersinia enterocolitica strain 8081. PLoS Genet. 2:e206. 10.1371/journal.pgen.0020206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Young BM, Young GM. 2002. Evidence for targeting of Yop effectors by the chromosomally encoded Ysa type III secretion system of Yersinia enterocolitica. J. Bacteriol. 184:5563–5571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Matsumoto H, Young GM. 2006. Proteomic and functional analysis of the suite of Ysp proteins exported by the Ysa type III secretion system of Yersinia enterocolitica Biovar 1B. Mol. Microbiol. 59:689–706 [DOI] [PubMed] [Google Scholar]
- 5. Haller JC, Carlson S, Pederson KJ, Pierson DE. 2000. A chromosomally encoded type III secretion pathway in Yersinia enterocolitica is important in virulence. Mol. Microbiol. 36:1436–1446 [DOI] [PubMed] [Google Scholar]
- 6. Young BM, Young GM. 2002. YplA is exported by the Ysc, Ysa, and flagellar type III secretion systems of Yersinia enterocolitica. J. Bacteriol. 184:1324–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Walker KA, Miller VL. 2004. Regulation of the Ysa type III secretion system of Yersinia enterocolitica by YsaE/SycB and YsrS/YsrR. J. Bacteriol. 186:4056–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Venecia K, Young GM. 2005. Environmental regulation and virulence attributes of the Ysa type III secretion system of Yersinia enterocolitica biovar 1B. Infect. Immun. 73:5961–5977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Walker KA, Miller VL. 2009. Synchronous gene expression of the Yersinia enterocolitica Ysa type III secretion system and its effectors. J. Bacteriol. 191:1816–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Elwell C, Engel JN. 2005. Drosophila melanogaster S2 cells: a model system to study Chlamydia interaction with host cells. Cell. Microbiol. 7:725–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheng LW, Portnoy DA. 2003. Drosophila S2 cells: an alternative infection model for Listeria monocytogenes. Cell. Microbiol. 5:875–885 [DOI] [PubMed] [Google Scholar]
- 12. Santic M, Akimana C, Asare R, Kouokam JC, Atay S, Kwaik YA. 2009. Intracellular fate of Francisella tularensis within arthropod-derived cells. Environ. Microbiol. 11:1473–1481 [DOI] [PubMed] [Google Scholar]
- 13. Cheng LW, Viala JPM, Stuurman N, Wiedemann U, Vale RD, Portnoy DA. 2005. Use of RNA interference in Drosophila S2 cells to identify host pathways controlling compartmentalization of an intracellular pathogen. Proc. Natl. Acad. Sci. U. S. A. 102:13646–13651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Akimana C, Al-Khodor S, Abu Kwaik Y. 2010. Host factors required for modulation of phagosome biogenesis and proliferation of Francisella tularensis within the cytosol. PLoS One 5:e11025. 10.1371/journal.pone.0011025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Agaisse H, Burrack LS, Philips JA, Rubin EJ, Perrimon N, Higgins DE. 2005. Genome-wide RNAi screen for host factors required for intracellular bacterial infection. Science 309:1248–1251 [DOI] [PubMed] [Google Scholar]
- 16. Rämet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RAB. 2002. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature 416:644–648 [DOI] [PubMed] [Google Scholar]
- 17. Philips JA, Rubin EJ, Perrimon N. 2005. Drosophila RNAi screen reveals CD36 family member required for mycobacterial infection. Science 309:1251–1253 [DOI] [PubMed] [Google Scholar]
- 18. Dorer MS, Kirton D, Bader JS, Isberg RR. 2006. RNA interference analysis of Legionella in Drosophila cells: exploitation of early secretory apparatus dynamics. PLoS Pathog. 2:e34. 10.1371/journal.ppat.0020034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Derré I, Pypaert M, Dautry-Varsat A, Agaisse H. 2007. RNAi screen in Drosophila cells reveals the involvement of the Tom complex in Chlamydia infection. PLoS Pathog. 3:1446–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pielage JF, Powell KR, Kalman D, Engel JN. 2008. RNAi screen reveals an Abl kinase-dependent host cell pathway involved in Pseudomonas aeruginosa internalization. PLoS Pathog. 4:e1000031. 10.1371/journal.ppat.1000031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Paquette N, Conlon J, Sweet C, Rus F, Wilson L, Pereira A, Rosadini CV, Goutagny N, Weber ANR, Lane WS, Shaffer SA, Maniatis S, Fitzgerald KA, Stuart L, Silverman N. 2012. Serine/threonine acetylation of TGFβ-activated kinase (TAK1) by Yersinia pestis YopJ inhibits innate immune signaling. Proc. Natl. Acad. Sci. U. S. A. 109:12710–12715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lane MC, Alteri CJ, Smith SN, Mobley HLT. 2007. Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc. Natl. Acad. Sci. U. S. A. 104:16669–16674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Miller WG, Leveau JH, Lindow SE. 2000. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol. Plant Microbe Interact. 13:1243–1250 [DOI] [PubMed] [Google Scholar]
- 24. Merriam JJ, Mathur R, Maxfield-Boumil R, Isberg RR. 1997. Analysis of the Legionella pneumophila fliI gene: intracellular growth of a defined mutant defective for flagellum biosynthesis. Infect. Immun. 65:2497–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Obrist MW, Miller VL. 2012. Low copy expression vectors for use in Yersinia sp. and related organisms. Plasmid 68:33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rogers SL, Rogers GC. 2008. Culture of Drosophila S2 cells and their use for RNAi-mediated loss-of-function studies and immunofluorescence microscopy. Nat. Protoc. 3:606–611 [DOI] [PubMed] [Google Scholar]
- 27. Miller VL, Falkow S. 1988. Evidence for two genetic loci in Yersinia enterocolitica that can promote invasion of epithelial cells. Infect. Immun. 56:1242–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Monod J. 1949. The growth of bacterial cultures. Annu. Rev. Microbiol. 3:371–394 [Google Scholar]
- 29. Russel L, Burguet S. 1977. Ultrastructure of leydig cells as revealed by secondary tissue treatment with a ferrocyanide-osmium mixture. Tissue Cell 9:751–766 [DOI] [PubMed] [Google Scholar]
- 30. Reynolds ES. 1963. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17:208–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Witowski SE, Walker KA, Miller VL. 2008. YspM, a newly identified Ysa type III secreted protein of Yersinia enterocolitica. J. Bacteriol. 190:7315–7325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Boyd AP, Grosdent N, Tötemeyer S, Geuijen C, Bleves S, Iriarte M, Lambermont I, Octave JN, Cornelis GR. 2000. Yersinia enterocolitica can deliver Yop proteins into a wide range of cell types: development of a delivery system for heterologous proteins. Eur. J. Cell Biol. 79:659–671 [DOI] [PubMed] [Google Scholar]
- 33. Baudoux P, Bles N, Lemaire S, Mingeot-Leclercq MP, Tulkens PM, Van Bambeke F. 2007. Combined effect of pH and concentration on the activities of gentamicin and oxacillin against Staphylococcus aureus in pharmacodynamic models of extracellular and intracellular infections. J. Antimicrob. Chemother. 59:246–253 [DOI] [PubMed] [Google Scholar]
- 34. McGhie EJ, Brawn LC, Hume PJ, Humphreys D, Koronakis V. 2009. Salmonella takes control: effector-driven manipulation of the host. Curr. Opin. Microbiol. 12:117–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ibarra JA, Steele-Mortimer O. 2009. Salmonella—the ultimate insider. Salmonella virulence factors that modulate intracellular survival. Cell. Microbiol. 11:1579–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ogawa M, Handa Y, Ashida H, Suzuki M, Sasakawa C. 2008. The versatility of Shigella effectors. Nat. Rev. Microbiol. 6:11–16 [DOI] [PubMed] [Google Scholar]
- 37. Grützkau A, Hanski C, Hahn H, Riecken EO. 1990. Involvement of M cells in the bacterial invasion of Peyer's patches: a common mechanism shared by Yersinia enterocolitica and other enteroinvasive bacteria. Gut 31:1011–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Clark MA, Hirst BH, Jepson MA. 1998. M-cell surface beta1 integrin expression and invasin-mediated targeting of Yersinia pseudotuberculosis to mouse Peyer's patch M cells. Infect. Immun. 66:1237–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kaplan F, Setlow P, Kaplan NO. 1969. Purification and properties of a DPNH-TPNH diaphorase from Clostridium kluyverii. Arch. Biochem. Biophys. 132:91–98 [DOI] [PubMed] [Google Scholar]
- 40. Cherry S. 2008. Genomic RNAi screening in Drosophila S2 cells: what have we learned about host-pathogen interactions? Curr. Opin. Microbiol. 11:262–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pujol C, Bliska JB. 2005. Turning Yersinia pathogenesis outside in: subversion of macrophage function by intracellular yersiniae. Clin. Immunol. 114:216–226 [DOI] [PubMed] [Google Scholar]
- 42. Cornelis GR. 2002. Yersinia type III secretion: send in the effectors. J. Cell Biol. 158:401–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wong ARC, Pearson JS, Bright MD, Munera D, Robinson KS, Lee SF, Frankel G, Hartland EL. 2011. Enteropathogenic and enterohaemorrhagic Escherichia coli: even more subversive elements. Mol. Microbiol. 80:1420–1438 [DOI] [PubMed] [Google Scholar]
- 44. Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kinder SA, Badger JL, Bryant GO, Pepe JC, Miller VL. 1993. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O8 and construction of a transformable R-M+ mutant. Gene 136:271–275 [DOI] [PubMed] [Google Scholar]