Abstract
Histoplasma capsulatum is a respiratory pathogen that infects phagocytic cells. The mechanisms allowing Histoplasma to overcome toxic reactive oxygen molecules produced by the innate immune system are an integral part of Histoplasma's ability to survive during infection. To probe the contribution of Histoplasma catalases in oxidative stress defense, we created and analyzed the virulence defects of mutants lacking CatB and CatP, which are responsible for extracellular and intracellular catalase activities, respectively. Both CatB and CatP protected Histoplasma from peroxide challenge in vitro and from antimicrobial reactive oxygen produced by human neutrophils and activated macrophages. Optimal protection required both catalases, as the survival of a double mutant lacking both CatB and CatP was lower than that of single-catalase-deficient cells. Although CatB contributed to reactive oxygen species defenses in vitro, CatB was dispensable for lung infection and extrapulmonary dissemination in vivo. Loss of CatB from a strain also lacking superoxide dismutase (Sod3) did not further reduce the survival of Histoplasma yeasts. Nevertheless, some catalase function was required for pathogenesis since simultaneous loss of both CatB and CatP attenuated Histoplasma virulence in vivo. These results demonstrate that Histoplasma's dual catalases comprise a system that enables Histoplasma to efficiently overcome the reactive oxygen produced by the innate immune system.
INTRODUCTION
Reactive oxygen species (ROS) are one of the most effective components of the antimicrobial arsenal produced by the innate immune system. Within mammals, a specialized subset of leukocytes that includes monocytes, polymorphonuclear neutrophils (PMNs), dendritic cells, and macrophages produces superoxide anion through assembly of the NADPH oxidase complex (1). Superoxide gives rise to other reactive oxygen molecules, including hydrogen peroxide, which are capable of damaging numerous macromolecules and killing microbes (2). The persistent bacterial and fungal infections that characterize chronic granulomatous disease, which is caused by genetic deficiency in the NADPH oxidase, underscore the critical importance of ROS in combating microbial infections (3, 4).
In order to survive in the mammalian host, successful pathogens must avoid or neutralize host-derived oxidative stresses. Both enzymatic and nonenzymatic strategies are utilized by microbial pathogens of plants and animals to accomplish this task (5–7). Enzymes such as superoxide dismutase and catalase specifically detoxify the ROS molecules superoxide anion and hydrogen peroxide, respectively (7). In the fungal kingdom, nonenzymatic strategies include production of melanin to absorb ROS or reductants such as thioredoxin to mend oxidative protein damage (8–10). While most organisms express a variety of antioxidant factors to cope with metabolically derived intracellular ROS, microbial pathogens must employ additional, often extracellular, factors to defend against ROS produced by host cells and the host environment (11–18). For microbial pathogens that infect ROS-producing phagocytic cells, the ability to defend against phagocyte-derived ROS becomes even more imperative.
Histoplasma capsulatum is one such intracellular fungal pathogen capable of parasitizing phagocytic immune cells. This fungus is found worldwide, with particular prominence in the United States within the Ohio and Mississippi River valleys. Within this area of endemicity, 80% of the population is estimated to have been exposed to Histoplasma (19). Acquisition of Histoplasma occurs upon inhalation of mycelium-produced conidia into the mammalian lung, where the temperature change to 37°C causes conversion of Histoplasma into pathogenic yeast cells (20, 21). Yeast cells encounter both neutrophils and alveolar macrophages. However, the innate immune system alone is unable to control Histoplasma infections. Following uptake of yeasts by phagocytes, the yeasts proliferate within phagosomes until rupture of the host cell and release of yeasts, which are subsequently taken up by neighboring phagocytes. Both immunocompromised and immunocompetent individuals are susceptible to infection, but in most cases, immunocompetent hosts are able to control yeast growth upon activation of the adaptive immune response, which enhances the antifungal response of phagocytes (22).
The substantial interactions between Histoplasma yeasts and host phagocytes make Histoplasma's ability to avoid or detoxify ROS an essential component of its pathogenesis. Although resting macrophages do not generate an oxidative burst in response to Histoplasma yeasts, activation of macrophages or opsonization of yeasts triggers ROS production (17, 23, 24). On the other hand, PMNs readily produce ROS upon encounter with Histoplasma (17, 25–27). Regardless of the cell type and response, Histoplasma yeasts are able to survive the ROS challenge. The molecular mechanisms responsible for this resistance to ROS have largely remained unknown. Recently, we demonstrated that yeasts produce an extracellular superoxide dismutase (Sod3) that protects Histoplasma from macrophage- and PMN-generated superoxide and is required for full virulence within a murine infection model (17).
In addition to Sod3, the Histoplasma genome encodes three catalase proteins that could potentially assist in combating ROS (28). CatA has been reported to be an 80-kDa catalase expressed by mycelia. CatB is an immunodominant 90-kDa extracellular catalase of Histoplasma yeasts also known as M antigen (13, 29–32). The third catalase, CatP, is a 60-kDa catalase (29, 33). CatB has been claimed to be a Histoplasma virulence factor, despite the lack of any genetic evidence for this speculation (28, 34). Likewise, no functional studies on the contribution of intracellular catalases have been performed, but these catalases could also contribute to defense against the membrane-permeant peroxide generated by host phagocytes.
In this study, we examined the roles of the extracellular and intracellular catalases in protecting Histoplasma from host-derived ROS and determined their contribution to Histoplasma virulence. Functional tests were facilitated by generation of Histoplasma strains lacking CatB, CatP, or both proteins. We confirm that CatB and CatP are responsible for the extracellular and intracellular catalase activities of Histoplasma yeast, respectively. Both proteins can protect Histoplasma yeast from hydrogen peroxide in vitro, although CatP appears to be more critical. Each catalase provides a modest degree of protection from phagocyte ROS-dependent killing, yet the loss of either protein singly does not attenuate Histoplasma survival in vivo. Analysis of strains lacking both catalases indicates that CatB and CatP provide redundant functions in defending Histoplasma from host-derived ROS during mammalian infection.
MATERIALS AND METHODS
Fungal strains and culture conditions.
Histoplasma capsulatum strains were derived from the clinical isolate G186AR (ATCC 26027) and are listed in Table 1. Histoplasma cultures were grown and maintained in Histoplasma macrophage medium (HMM) (35). Liquid cultures were grown with constant aeration (200 rpm) until late exponential growth phase. Uracil auxotrophs were grown in medium supplemented with 100 μg/ml uracil. Yeasts were maintained by growth at 37°C with 5% CO2–95% air. For monitoring of yeast growth, liquid culture density was determined by treating yeasts with 1 M NaOH and measuring culture turbidity at 595 nm. Mycelia were grown in static liquid cultures at 25°C in 100% air. For Histoplasma growth on solid medium, HMM was solidified with 0.6% agarose and supplemented with 25 μM FeSO4. Yeasts were transformed with linearized plasmids by electroporation, and Ura+ transformants were selected on HMM (36). For experiments with phagocytes and murine infections, large clumps of yeasts were first removed from yeast suspensions by centrifugation (50 × g), and yeasts were enumerated by hemacytometer.
Table 1.
Histoplasma strains
| Straina | Genotypee | Other designation |
|---|---|---|
| WU8b | ura5-32Δ | Wild type |
| OSU15 | ura5-32Δ sod3-3Δ::hph/pCR468 (URA5, PH2B-gfp:FLAG) | sod3Δ |
| OSU16 | ura5-32Δ catB-2Δ::hph/pCR468 (URA5, PH2B-gfp:FLAG) | catBΔ |
| OSU24 | ura5-32Δ catB-2Δ::hph | catBΔ |
| OSU31c | ura5-32Δ catB-3Δ::apt3 sod3-3Δ::hph | catBΔ sod3Δ |
| OSU45d | ura5-32Δ/pCR468 (URA5, PH2B-gfp:FLAG) | WT |
| OSU46 | ura5-32Δ catB-3Δ::apt3 sod3-3Δ::hph/ pCR468 (URA5, PH2B-gfp:FLAG) | catBΔ sod3Δ |
| OSU51 | ura5-32Δ catB-2Δ::hph/pBY08 (URA5, PH2B-CATB:FLAG) | catBΔ/CATB |
| OSU138 | ura5-32Δ catB-2Δ::hph catP-2Δ::apt3 | catBΔ catPΔ |
| OSU149 | ura5-32Δ catP-1Δ::apt3 | catPΔ |
| OSU157 | ura5-32Δ catP-1Δ::apt3/pCR468 (URA5, PH2B-gfp:FLAG) | catPΔ |
| OSU158 | ura5-32Δ catP-1Δ::apt3/pCR615 (URA5, PH2B-FLAG:CATP) | catPΔ/CATP |
| OSU159 | ura5-32Δ catB-2Δ::hph catP-2Δ::apt3/pCR468 (URA5, PH2B-gfp:FLAG) | catBΔ catPΔ |
Histoplasma RNA isolation and quantitative reverse transcription-PCR (RT-PCR).
Wild-type Histoplasma yeasts were collected by centrifugation (2,000 × g) and resuspended in Ribozol reagent (Amresco, Inc.). Mycelia were collected from static liquid cultures by filtration onto Whatman no. 5 filter paper and scraping of the retained mycelia into Ribozol. RNA was liberated from Histoplasma cells in Ribozol by beating with 0.5-mm-diameter glass beads. RNA was purified by CHCl3 extraction and alcohol precipitation of the aqueous phase. Genomic DNA was removed by two sequential digestions with Turbofree DNase (Ambion), and the absence of DNA was verified by PCR. Five micrograms of total RNA was reverse transcribed using SuperScript III reverse transcriptase (Invitrogen) and random 15-mer primers.
Quantitative PCR (qPCR) was performed on reverse-transcribed RNAs from yeasts and mycelia. Amplification reaction mixtures were assembled using a SYBR-green-based PCR master mix (Bio-Rad) with reverse-transcribed RNA templates (final dilution, 1:200) and 0.5 μM each gene-specific primer (see Table S1 in the supplemental material). Cycling was performed with a realplex thermocycler (Eppendorf) using the following conditions: 95°C for 10 min, followed by 40 cycles of 95°C for 10 s, 55°C for 15 s, and 72°C for 1 min. CATA, CATB, and CATP transcript abundance in yeasts relative to that in mycelia was determined by the ΔΔCT method (where CT is threshold cycle) (37) after normalizing the levels of CAT gene expression to the levels of expression of the RPS15 transcript. To estimate the relative levels of expression of the catalase genes, the normalized CT (ΔCT) numbers were used as an indicator of mRNA abundance. A relative copy number (RCN) value for each gene, determined using the equation 2−ΔCT (38), was used to estimate abundance, and was then compared to the RCN of ACT1 as a common reference point, which was set at 100%. Statistical analyses were performed on the normalized CT values using Student's t test on the results for RNA from three biological replicate cultures of yeasts and mycelia.
Generation and validation of Histoplasma strains with catalase gene deletions.
To generate the catBΔ or catPΔ strains, positive and negative selections were used to enrich for yeasts with a deletion of the CATB- or CATP-coding region by allelic replacement (39). WU8 uracil-auxotrophic yeasts were transformed with the URA5 plasmid pBY19 or pEH71. pBY19 and pEH71contain a hygromycin resistance gene (hph) or a G418 resistance gene (apt3) flanked by 1.8 kb of sequence upstream and downstream of the CATB- or CATP-coding sequence, respectively. Multiple Ura+ transformants were passaged three times in liquid HMM containing uracil and either 150 μg/ml hygromycin or 100 μg/ml G418. Selection for deletion alleles and loss of the URA5 plasmids was done by plating yeast cultures on HMM containing uracil, 150 μg/ml hygromycin or 100 μg/ml G418, and 1 mg/ml 5-flurourotic acid (5-FOA). Hygromycin- or G418-resistant, 5-FOA-resistant colonies were picked and screened for the loss of CATB or CATP by PCR. Using similar procedures, construction of the catBΔ catPΔ and the catBΔ sod3Δ double mutant strains was done by disrupting the CATP gene in the catBΔ background and by disrupting the CATB gene in the sod3Δ background using plasmids pEH71 and pBY23, respectively. To restore uracil prototrophy, mutant strains were transformed with pCR468, a URA5 plasmid with a green fluorescent protein (GFP) expression cassette. The catB and catP deletions were complemented by transforming the mutant yeasts with plasmids pBY08 and pCR615, which express the CATB and CATP genes from the constitutive H2B promoter, respectively.
To confirm the construction of catalase-deficient and catalase-complemented lines, genotypes were initially verified by PCR using genomic DNA isolated from each strain. PCR amplification of the CATB gene, CATP gene, hph cassette, or apt3 cassette utilized 0.5 μM primers specific for each gene (see Table S1 in the supplemental material). The competence of the Histoplasma genomic DNA template for PCR was verified by amplification of the ACT1 gene. Loss of CatB protein was validated by separation of culture filtrate proteins (5 μg total protein) by SDS-PAGE, followed by silver staining to visualize proteins. The presence or absence of CatP in mutant strains was determined by immunoblotting. Custom antibodies to CatP were generated by immunization of rabbits (Open Biosystems) with Escherichia coli-produced full-length Histoplasma CatP protein. For production of the immunogen, the Histoplasma CATP sequence was cloned into a C-terminal 6× His vector (pET28a; EMD Biosciences) and transformed into E. coli. The 6× His-tagged CatP protein was purified from E. coli lysates by immobilized cobalt affinity chromatography. For immunoblotting, 10 μg of Histoplasma total cellular lysate proteins was separated by SDS-PAGE and transferred to nitrocellulose. Immunoblotting was performed with the anti-CatP immune serum. No Histoplasma cellular proteins were detected by preimmune serum (data not shown).
Assay of extracellular and intracellular catalase activity.
Histoplasma yeast cultures were grown in liquid HMM at 37°C with aeration until they reached late exponential/early stationary growth phase. Three biological replicate yeast cultures were used, and the relative number of yeast cells was normalized by comparison of optical densities. For extracellular catalase activity, cell-free supernatant and yeast cell fractions were separated by centrifugation (16,000 × g). For yeast cell lysates, cultures were pelleted, washed twice with phosphate-buffered saline (PBS), and suspended in PBS. Cytoplasmic protein was released by bead beating yeasts with 0.5-mm-diameter glass beads. Protein concentrations were determined using the Bradford method with bovine serum albumin protein as the standard.
Catalase activity assays for extracellular and intracellular samples were performed by spectrophotometrically measuring their ability to reduce the H2O2 concentration over time. For the soluble extracellular fractions, normalized amounts of clarified culture supernatants were brought to 200 μl with 50 mM Na-phosphate buffer (pH 7.2), before the addition of 800 μl of 10 mM H2O2 in 50 mM Na-phosphate buffer (pH 7.2). Hydrogen peroxide destruction was measured at 240 nm with correction at 595 nm every 30 s for 5 min and compared to that in a control containing buffer and HMM only. For cell-associated catalase activity, H2O2 destruction was determined after incubation of yeasts with a buffered H2O2 solution and removal of yeast cells by passage through a 0.22-μm-pore-size filter Spin-X column (Corning) before spectrophotometric measurement at 240 nm. Because of the additional manipulations before reading, all steps and incubations were carried out at 4°C to slow enzymatic destruction of H2O2. For intracellular catalase activity determination, cellular lysates (12 μg of total cytosolic protein) were added to the Na-phosphate-buffered H2O2 solution, and the H2O2 destruction was measured by taking spectrophotometric readings every 30 s for 5 min. For relative comparison of soluble extracellular to cell-associated extracellular activities, the decrease in H2O2 concentration was determined for supernatant and yeast samples incubated at room temperature for 5 min, and the results were normalized to the H2O2 decrease in the absence of CatB using samples from the catBΔ mutant. Statistical analyses were performed on three biological replicates for each strain using Student's t test and data points at 5 min.
Determination of in vitro sensitivity to hydrogen peroxide.
Sensitivity to H2O2 in vitro was determined by application of H2O2-saturated filter disks to growing Histoplasma yeasts. Histoplasma yeasts (1 × 107) were spread on 10-cm-diameter HMM plates. After inoculation of plates, sterile filter disks treated with 20 μl of 0, 75, 150, or 300 mM H2O2 in PBS were applied and the plates were incubated at 37°C with 5% CO2–95% air to allow Histoplasma growth. After 6 days, the plates were photographed and the areas lacking yeast growth were measured using ImageJ software (http://rsbweb.nih.gov/ij/) after binary thresholding of the images to define the areas of clearing around the filter disks. Statistical analyses on the area of the clearing zones were performed for three biological replicates for each strain using Student's t test.
Phagocyte isolation and infection with Histoplasma yeasts.
PMNs were isolated from the blood of healthy human donors through venipuncture as previously described (40, 41). Briefly, 10 ml of blood was collected into syringes containing 250 U heparin and diluted into 10 ml 0.9% saline solution. PMNs were harvested by density sedimentation (40 min at 400 × g) after overlaying onto 4.5 ml Ficoll-Paque PLUS (GE Healthcare). Erythrocytes were removed by resuspension of cells in 6 ml 0.9% saline, followed by addition of 6 ml of 3% Dextran (500 kDa) and incubation at 4°C for 20 min to allow the heavier red blood cells to settle. The upper PMN-containing layer was removed, and cells were collected by centrifugation (15 min at 800 × g). Residual erythrocytes were lysed by the addition of 10 ml cold H2O for 20 s before the addition of 10 ml of 1.8% saline solution to restore osmotic balance.
For infection of PMNs, PMNs were collected by centrifugation (10 min at 800 × g) and washed in Hanks balanced salt solution (lacking Ca2+ and Mg2+) before enumeration by hemacytometer. Viability of cells was verified to be greater than 98% by trypan blue staining. Autologous serum was prepared from separate blood samples utilizing coagulation and centrifugation (460 × g). PMNs were seeded at 2 × 105 cells per well of a 96-well plate in Dulbecco modified Eagle medium plus 10% autologous serum, 2 × 104 Histoplasma yeast cells were added to each PMN-containing well, and the cells were coincubated for 4 h at 37°C with 5% CO2–95% air. To determine yeast viability, culture medium was removed and the PMNs were lysed by addition of cold H2O and scraping of the wells with a pipette tip. Lysates were serially diluted and plated on solid HMM to determine viable CFU numbers. Relative survival of yeasts was calculated by comparing infected wells to wells containing yeast only. Statistical analyses were performed using Student's t test on three biological replicate tests.
For infection of human monocyte-derived macrophages, peripheral blood was collected from healthy human donors by venipuncture and macrophages were differentiated from monocytes (42). One hundred milliliters of blood was collected, and four 20-ml fractions were each diluted into 15 ml of 0.9% saline before they were added to 14 ml of Ficoll-Paque PLUS (GE Healthcare). Buffy coats were collected into conical tubes following density sedimentation (40 min at 400 × g), and volumes were brought to 50 ml with cold PBS. Cells were pelleted by centrifugation (15 min at 460 × g and 4°C), the four fractions were resuspended in RPMI 1640 and pooled, and cells were enumerated by hemacytometer. Peripheral blood cells in RPMI 1640 with 20% autologous serum were added to Teflon dishes (Savilex) at 2 × 106 cells/ml and incubated at 37°C with 5% CO2–95% air for 5 days to allow differentiation of monocytes into macrophages.
For infection of macrophages, cells were collected after 5 days by washing of the Teflon dishes and recovery of the cells by centrifugation (10 min at 130 × g and 4°C). Cell suspensions were adjusted to 4 × 106 cells/ml in RPMI 1640 with 10% autologous serum after enumeration, and 200 μl (8 × 104 macrophages) was added to the wells of a 96-well plate and incubated at 37°C with 5% CO2–95% air. After 2 h, nonadherent cells were removed by washing the wells three times with PBS. For cytokine activation, macrophage monolayers were first treated with 1,000 U/ml gamma interferon (IFN-γ) in RPMI 1640 with 10% autologous serum for 2 days. Infection of macrophages with yeasts was done by addition of 2 × 103 yeasts to each monolayer and incubation for 4 h at 37°C with 5% CO2–95% air. Macrophages were lysed by adding cold H2O and scraping the wells with a pipette tip. Lysates were diluted and plated on solid HMM to enumerate the number of viable Histoplasma CFU. Survival was determined by comparing the number of CFU from infected wells to the number from wells containing yeast in the absence of macrophages. Statistical analyses were performed using Student's t test on three biological replicate tests.
Determination of fungal burdens in murine tissue following respiratory infection.
C57BL/6 male mice (NCI) were infected intranasally with 2 × 104 yeast cells suspended in HMM. Histoplasma yeast cells were enumerated by hemacytometer following centrifugal removal of yeast aggregates (50 × g). The actual number of CFU in the inoculum delivered was computed by plating serial dilutions of the inoculum suspensions on solid medium. At 4-day incremental time points following infection, lung tissue and spleen tissue were collected from infected mice and homogenized in 5 ml or 3 ml HMM, respectively. Serial dilutions of the homogenates were plated on solid HMM to determine the number of viable fungal CFU in each organ. Statistical analyses were performed on four replicate infections at each time point using Student's t test.
All animal experiments were performed in compliance with the National Research Council's Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee at Ohio State University (protocol 2007A0241). Human cells were obtained from healthy volunteers after obtaining written informed consent and HIPAA authorization. Human subjects research was approved by the Biomedical Sciences Institutional Review Board at Ohio State University (protocol 2008H0242).
RESULTS
Pathogenic-phase Histoplasma cells express CATB and CATP.
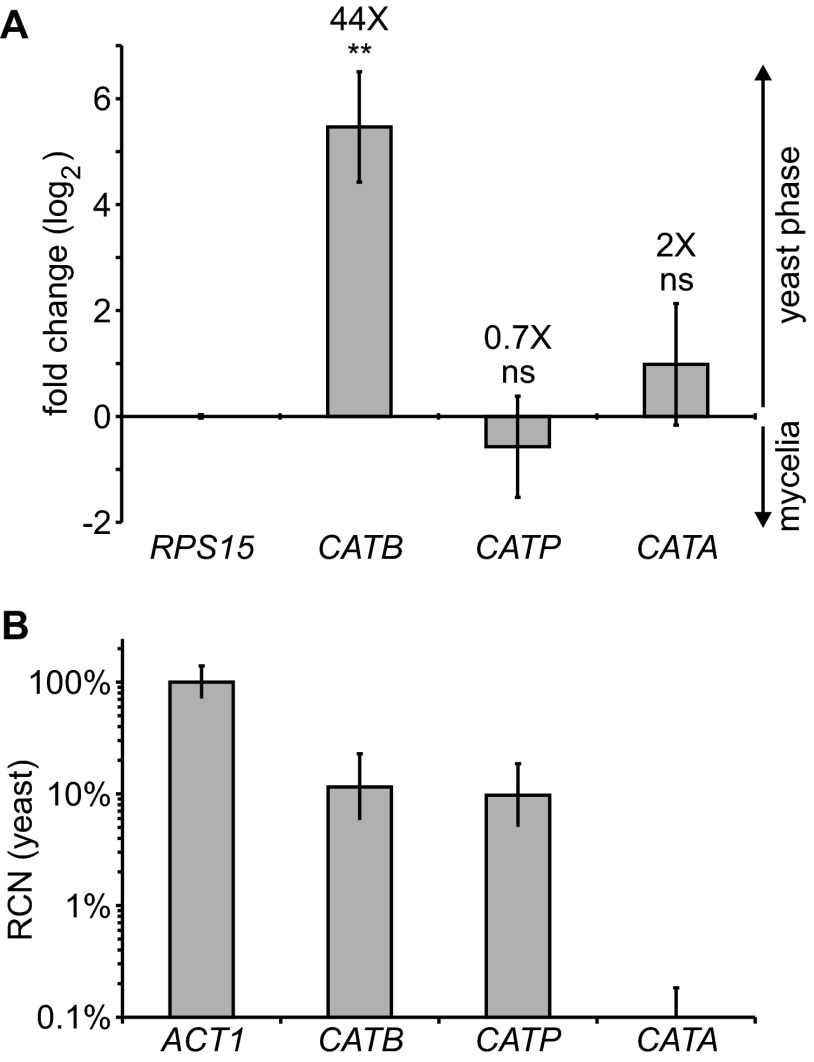
To identify catalases with the potential to protect Histoplasma from oxidative stress related to infection, we determined which catalase genes were expressed by pathogenic-phase cells. The Histoplasma genome encodes three catalase proteins: CatB, CatP, and CatA (28). We previously showed that CATB expression is enriched in pathogenic-phase cells. This pathogenic-phase-restricted expression suggests that its role is linked to pathogenesis (43). We used quantitative RT-PCR to determine the CATB, CATP, and CATA expression profile in the pathogenic yeast cells and in the avirulent mycelia. In contrast to CATB, CATP and CATA did not show phase-dependent regulation (Fig. 1A). CATA expression was barely detectable by RT-PCR. Since the preceding analysis indicated only the relative enrichment in the yeast or mycelial phase but did not show to what degree the genes were expressed in yeasts, we compared the catalase mRNA levels to the level of the actin (ACT1) housekeeping gene. Both the CATB gene and the CATP gene were expressed at reasonably detectable levels in yeast cells (Fig. 1B). However, yeasts expressed comparatively little CATA, implying that CatA does not contribute to ROS defenses of pathogenic yeast cells.
Fig 1.
CATB and CATP are the major Histoplasma catalase genes expressed, but only CATB shows pathogenic-phase regulation. (A) Yeast- and mycelium-phase expression profiles for the extracellular catalase CATB gene and the CATP and CATA catalase genes. Relative expression of each catalase gene was determined by quantitative PCR of reverse-transcribed RNA from yeasts and mycelia after normalization to the level of expression of the RPS15 gene. Data represent the log (base 2) value of the fold change in expression of the CATB, CATP, and CATA genes in yeasts compared to mycelia. The fold change (yeast-phase compared to mycelium-phase expression) is indicated by the values above the respective columns. Error bars represent standard deviations of three biological replicate RNA samples. Asterisks represent significant differences between yeast and mycelial phases determined by Student's t test (ns, not significant; **, P < 0.01). (B) Estimation of the relative level of CATB, CATP, and CATA expression in yeasts. Expression levels were approximated by use of the RCN value, which was calculated from the RPS15-normalized cycle thresholds for each gene and compared to the value for RPS15-normalized actin (ACT1), set at 100%.
CatB and CatP represent the predominant extracellular and intracellular catalase activities of Histoplasma yeasts.
To facilitate functional characterization of Histoplasma catalases, we created Histoplasma strains which lacked CatB and CatP. Deletion alleles of CATB and CATP were created by replacing the coding region with a hygromycin (hph) or a G418 (apt3) resistance gene, respectively (Table 1). For each deletion strain, a gene-complemented line was created by transforming the mutants with a linear plasmid that expressed either CATB or CATP from a constitutive promoter. Gene deletions and complementation lines were confirmed by PCR using gene-specific or deletion-allele-specific primers (see Fig. S1 in the supplemental material). To address potential functional redundancy between CatB and CatP, a double mutant strain with deletions of both the CATB and CATP genes was also generated. The catalase-deletion strains grew at rates similar to the rate for wild-type Histoplasma in vitro in rich growth medium (data not shown).
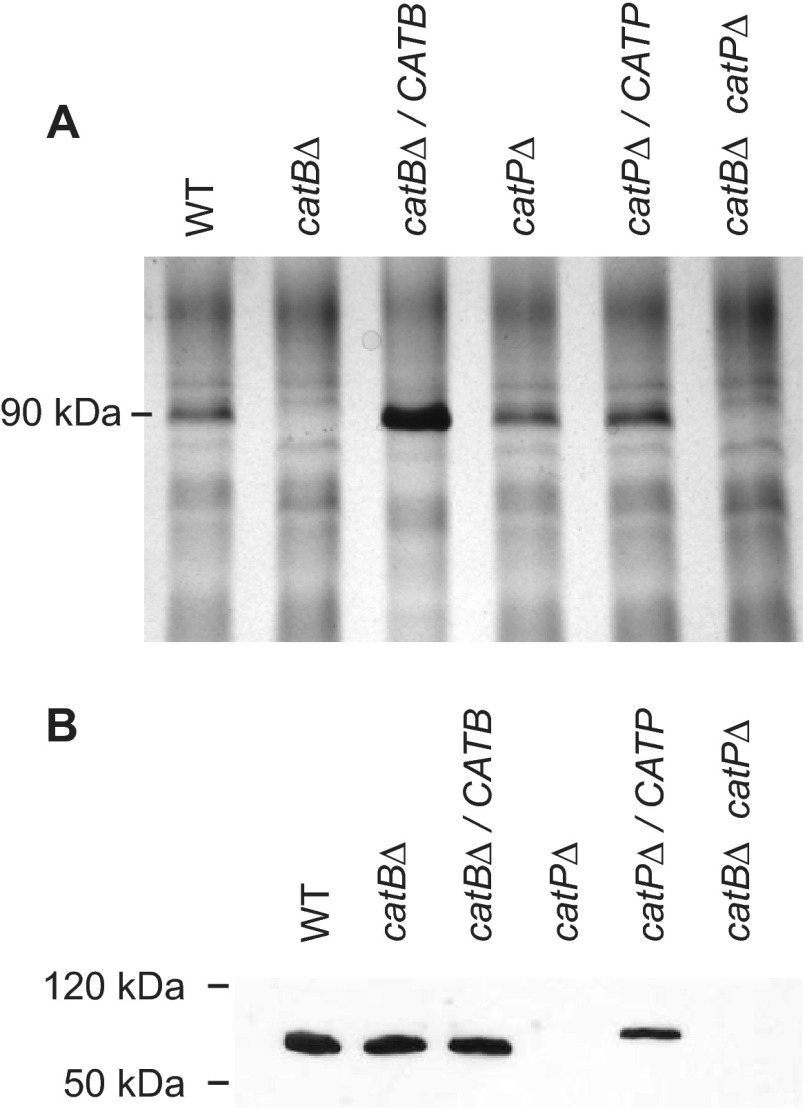
The loss of catalase proteins was verified by visualizing CatB and CatP proteins in the mutant and complemented strains. Analysis of extracellular proteins by silver staining following electrophoretic separation of yeast culture filtrates through polyacrylamide showed loss of a 90-kDa protein specifically in single and double mutant strains with the catB deletion allele (Fig. 2A). The identity of this 90-kDa band as CatB was further confirmed by the increased abundance of the protein band by overexpression of the CATB gene from a strong, constitutive promoter in the CATB-complemented line (catBΔ/CATB). Due to the complexity of the intracellular protein population, yeast cellular lysates were probed for the presence of the CatP protein by immunoblotting with CatP-specific antisera (Fig. 2B). The 60-kDa CatP protein was absent from lysates derived from the catPΔ strain and the catBΔ catPΔ double mutant, whereas CatP was easily detected in lysates from the wild type and the catBΔ mutant. Complementation of the CatP deficiency by expression of the CATP gene (catPΔ/CATP) restored the presence of the CatP protein in yeast cells. The slightly slower migration of CatP in the complemented line was due to the addition of a FLAG epitope to CatP. Thus, mutant strains were depleted of their respective catalase. Furthermore, compensatory production of catalase enzymes was not apparent, as the levels of CatP in the catBΔ mutant and of CatB in the catPΔ mutant were not noticeably changed from those in the wild-type strain. CATA was also not upregulated in the CatB- and CatP-deficient yeasts (data not shown).
Fig 2.
Deletion of the CATB and CATP genes specifically results in loss of extracellular CatB and intracellular CatP, respectively. (A) Analysis of extracellular CatB production. Total culture filtrate proteins from wild type, the catalase mutants, and their corresponding complemented strains were separated by SDS-PAGE, and proteins were visualized by silver staining to monitor production of the 90-kDa CatB protein. (B) Analysis of intracellular CatP production. Cellular lysates were prepared from wild type, the catalase mutants, and their corresponding complemented strains. Evidence of CatP protein in lysates was shown by immunoblotting of lysate proteins with a custom antibody to Histoplasma CatP. WT, wild type (OSU45); catBΔ, catB mutant (OSU16); catBΔ/CATB, complemented catB mutant (OSU51); catPΔ, catP mutant (OSU157); catPΔ/CATP, complemented catP mutant (OSU158); catBΔ catPΔ, catB catP double mutant (OSU159).
To determine if CatB was the source of extracellular catalase activity produced by Histoplasma yeast, we examined the H2O2-degrading capacity of extracellular fractions of the CatB-deficient mutants. Cell-free culture supernatants from wild-type Histoplasma yeast readily destroyed hydrogen peroxide, as determined by the decrease in the absorbance at 240 nm of an H2O2-containing solution (Fig. 3A). However, yeast culture supernatants collected from the catBΔ and catBΔ catPΔ mutants showed virtually no ability to degrade H2O2. Catalase activity was restored by complementing the catB deletion strain with a copy of the CATB gene. Importantly, loss of CatP did not impact the extracellular catalase activity, as the catalase activity in culture supernatants from the catPΔ mutant was not statistically different from that in supernatants from the wild type.
Fig 3.
CatB and CatP are specifically responsible for extracellular and intracellular catalase activity, respectively. (A) Extracellular cell-free (soluble) catalase activity. Culture filtrates harvested from wild-type and catalase-deficient yeasts were tested for catalase activity by monitoring the ability of the culture filtrate proteins to destroy H2O2. Relative H2O2 destruction was measured as the decrease in absorbance (Abs) at 240 nm over time at 25°C. (B) Extracellular catalase activity associated with yeast cells. The ability of yeasts to eliminate H2O2 was monitored by incubation of washed yeast cells with H2O2 and quantitation of the H2O2 remaining by determination of the absorbance at 240 nm. At 1-min intervals, yeasts were removed by centrifugation through 0.45-μm-pore-size filters and the absorbance of the clarified solution was determined. The assay was performed at 4°C to slow the enzyme kinetics to allow experimental manipulations. (C) Intracellular catalase activity of yeast cells. Cellular lysates were tested for catalase activity by addition of total yeast lysates to 10 mM H2O2 and monitoring the decrease in H2O2 by determination of the absorbance at 240 nm over time at 25°C. Error bars represent standard deviations from three biological replicate samples. Asterisks represent significant differences from the wild type strain determined by Student's t test (ns, not significant; **, P < 0.01; ***, P < 0.001). WT, wild type (OSU45); catBΔ, catB mutant (OSU16); catBΔ/CATB, complemented catB mutant (OSU51); catPΔ, catP mutant (OSU157); catPΔ/CATP, complemented catP mutant (OSU158); catBΔ catPΔ, catB catP double mutant (OSU159).
We used the catBΔ mutant to test if CatB associated with the yeast cell surface also contributes to Histoplasma yeasts' ability to break down exogenous H2O2. Previous studies showed that in addition to soluble CatB protein, wild-type yeasts also have CatB associated with the yeast cells (32). Consistent with the cell association of CatB, washed, intact Histoplasma yeasts could destroy H2O2 (Fig. 3B). This activity was dependent on production of the CatB protein, as catBΔ and catBΔ catPΔ mutant yeast cells did not degrade exogenous H2O2. Additionally, the extracellular catalase activity associated with cells was not affected by loss of CatP. Complementation of CatB deficiency with CATB restored peroxide destruction, confirming that CatB is the source of the extracellular catalase activity. The cell-associated catalase kinetic assays were performed at 4°C due to the manipulations required to remove yeast cells before spectrophotometric readings. To determine the relative amount of catalase activity associated with cells compared to that present in the culture filtrate, endpoint assays (5 min) were performed at room temperature with equivalent amounts of yeasts from which supernatant samples were derived. Comparison of the H2O2 degradation rates between wild type and the catBΔ mutant yeast and culture filtrates showed that (i) all of the extracellular catalase activity is produced by CatB and (ii) roughly half of the total extracellular catalase activity is found in the cell-free supernatant and half is cell associated (data not shown).
In contrast to the CatB-dependent extracellular catalase activity, CatP was responsible for all the intracellular catalase activity of Histoplasma yeast. Previously, Howard showed that cellular lysates of Histoplasma yeasts are capable of degrading hydrogen peroxide (44). To determine if CatP produces the observed intracellular catalase activity, CatP-deficient yeast cells were lysed and the clarified lysates were examined for the ability to destroy hydrogen peroxide (Fig. 3C). Lysates from wild-type yeast and from catBΔ yeast readily degraded hydrogen peroxide in vitro. However, cell lysates from the catPΔ mutant strain were completely unable to degrade peroxide, unless a plasmid copy of CATP was expressed. The CatP activity in the complemented line was only about one-third of that in wild type (Fig. 3C). Lysates from the catBΔ catPΔ double mutant were indistinguishable from those from the catPΔ single mutant and from buffer alone. These data indicate that CatP is the source of intracellular hydrogen peroxide-degrading activity and that CatB does not contribute to the intracellular activity. Furthermore, the lack of extracellular catalase activity in CatB-deficient strains and the lack of intracellular catalase activity in CatP-deficient strains further prove that CatA does not contribute to the catalase activity of Histoplasma yeast cells, consistent with its lack of expression (Fig. 1).
CatB and CatP protect Histoplasma from H2O2 in vitro.
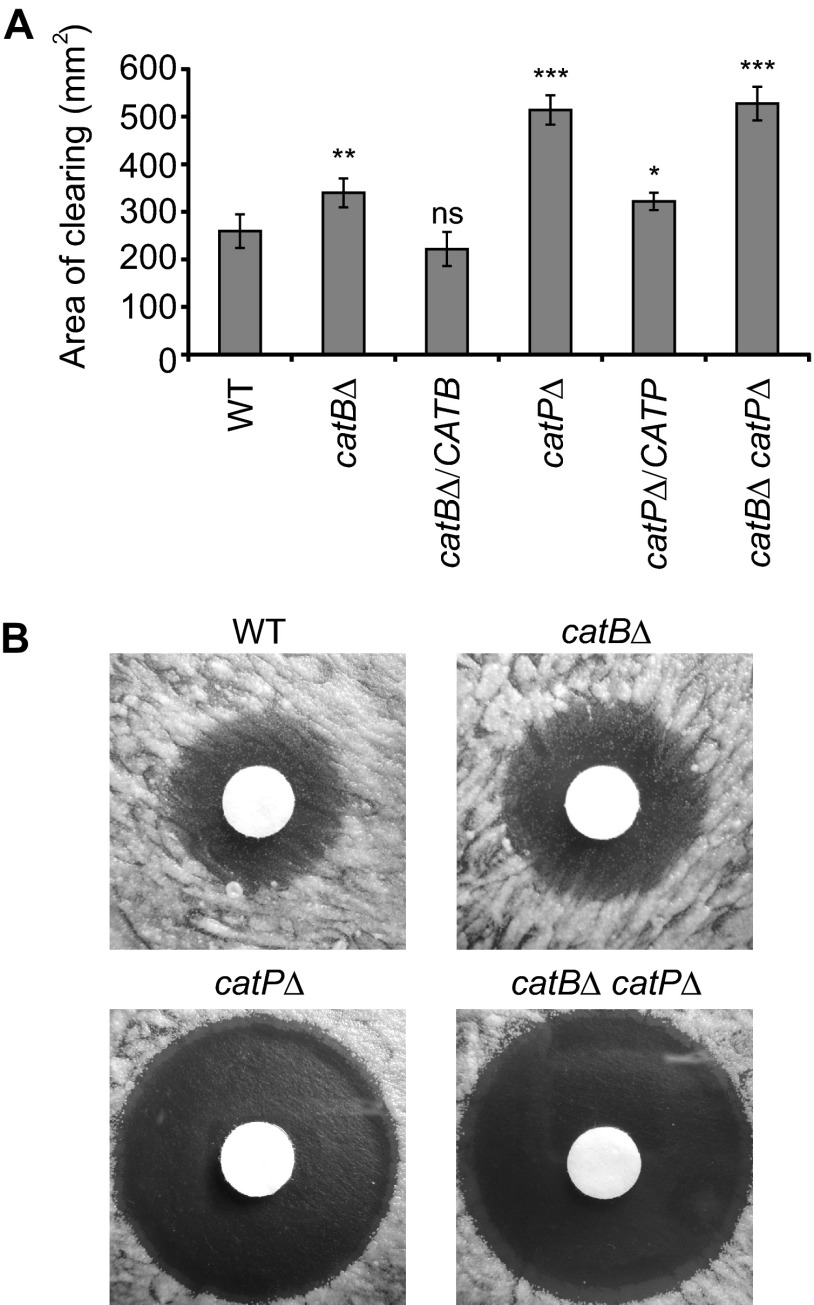
To determine if CatB and/or CatP can defend Histoplasma yeasts against peroxide in vitro, CatB- and CatP-deficient yeasts were tested for their resistance to hydrogen peroxide using a disk diffusion assay. Increasing sensitivity of Histoplasma yeasts was scored as the increased area around filter disks saturated with hydrogen peroxide in which there was negligible yeast growth (Fig. 4A). Wild-type Histoplasma yeasts expressing both CatB and CatP catalases showed the lowest sensitivity to hydrogen peroxide in vitro. Loss of CatB slightly increased the sensitivity of catBΔ mutant yeast, and H2O2 resistance was restored upon complementation of the catBΔ mutant with CATB. Loss of CatP was the most detrimental to yeast, as the catPΔ mutant had the largest area of clearing. The loss of both CatB and CatP catalases resulted in peroxide sensitivity nearly identical to that of the single catPΔ mutant, indicating that, although CatB can provide some protection, the major source of in vitro peroxide defense is the CatP catalase. Complementation of the CatP deficiency largely restored yeast resistance to hydrogen peroxide. Interestingly, zones of clearing for strains lacking CatP were completely clear of yeast growth, but regardless of the size of the clearing zone, strains with CatP activity had microcolonies of surviving yeast (Fig. 4B). Gradation in the concentration of hydrogen peroxide applied to the disks led to a dose response in the area cleared of yeast growth around the disk (data not shown). The ratio of the area cleared around the disk between mutant strains and the wild type was relatively constant across the range of hydrogen peroxide concentrations tested (data not shown).
Fig 4.
CatB and CatP provide protection to yeasts from H2O2 in vitro. Resistance of wild-type and catalase-deficient yeasts to killing by H2O2. H2O2 killing of yeast was determined by a filter disk diffusion assay on solid medium using filters with 300 mM H2O2. (A) Quantitation of the sensitivity to H2O2. The zone of clearing around the disks was determined as the total area lacking yeast cell growth. Data presented are the average areas of clearing for three replicate tests, with error bars representing standard deviations. Asterisks represent significant differences from the wild-type strain determined by Student's t test (ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001). (B) Images of Histoplasma yeast growth around H2O2-saturated disks showing the presence of microcolonies of surviving yeasts in zones of clearing for wild type and for CatB-deficient strains but not for strains lacking CatP (catPΔ or catBΔ catPΔ strains). WT, wild type (OSU45); catBΔ, catB mutant (OSU16); catBΔ/CATB, complemented catB mutant (OSU51); catPΔ, catP mutant (OSU157); catPΔ/CATP, complemented catP mutant (OSU158); catBΔ catPΔ, catB catP double mutant (OSU159).
CatB and CatP are both required to protect Histoplasma from phagocytes.
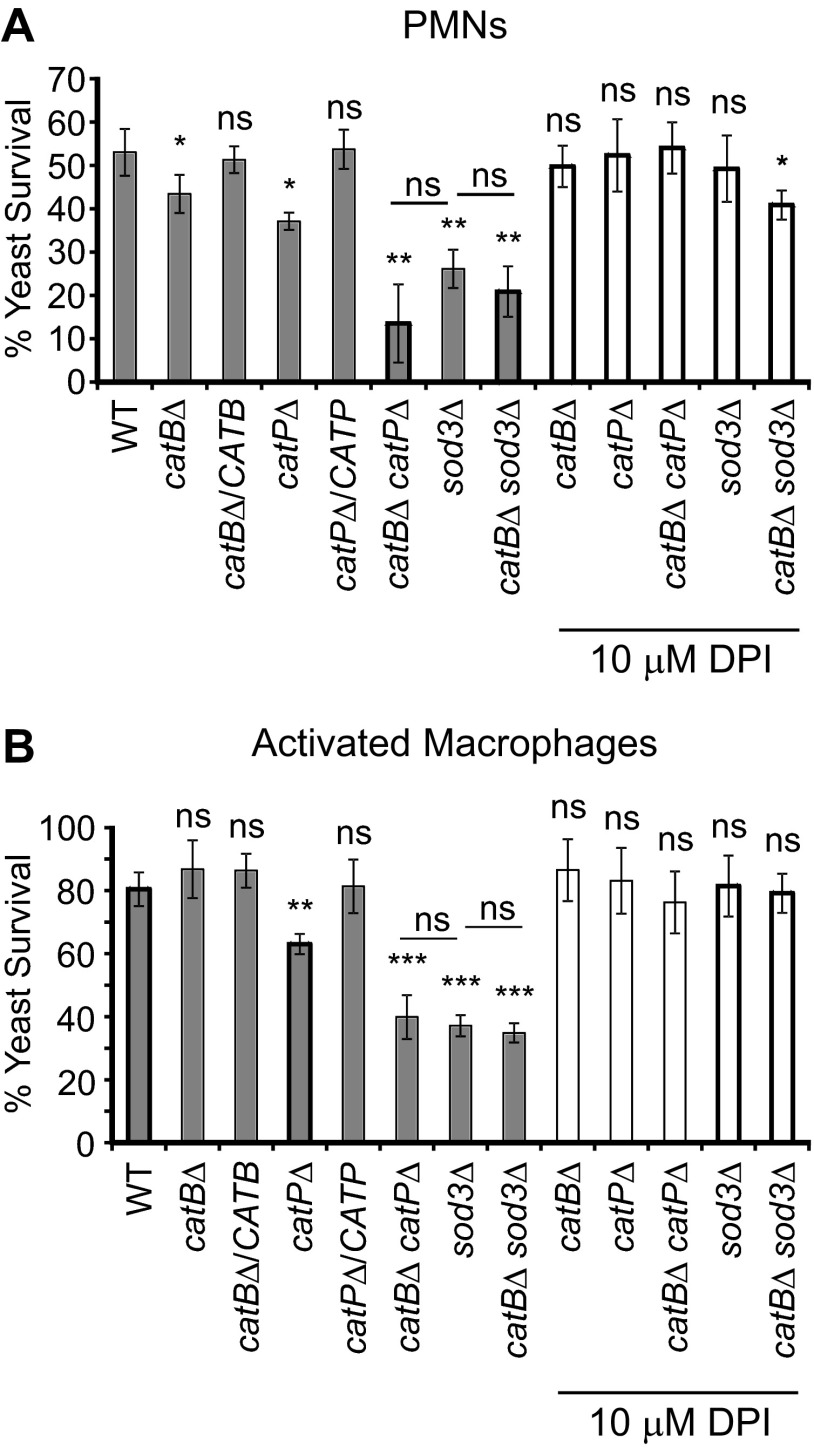
To functionally test the role of CatB and CatP catalases in defending Histoplasma from oxidative killing by PMNs, CatB- and CatP-deficient yeast cells were coincubated with human PMNs and yeast viability was determined. Previous reports indicated that Histoplasma yeasts are able to resist oxidative killing by phagocytic cells, including PMNs (17, 25, 26). When added to human PMNs in culture, 54% of the wild-type yeasts were viable after 4 h (Fig. 5A). Loss of CatB modestly impaired Histoplasma's ability to survive these fungicidal cells; survival was decreased to 80% of that of wild type (43% versus 54%). Likewise, the loss of CatP diminished Histoplasma's ability to survive PMN challenge, although to a greater extent than loss of CatB; loss of CatP decreased survival to 69% of that of wild type (37% versus 54%). Complementation of either catalase deficiency restored yeast survival to that of wild type. Loss of both CatB and CatP activities caused a considerable survival defect to 26% of wild-type yeast survival (14% versus 54%). This suggests that CatB and CatP act redundantly to optimally defend Histoplasma against peroxide produced by human PMNs. The decreased survival of the catBΔ, the catPΔ, and the catBΔ catPΔ mutants compared to the survival of wild-type yeast was reversed by addition of the phagocyte oxidase inhibitor diphenylene iodinium (DPI), indicating that the enhanced sensitivity of the mutant strains is due to an inability to resist PMN-produced reactive oxygen (Fig. 5A).
Fig 5.
CatP and CatB can provide protection to Histoplasma yeasts against phagocyte-produced reactive oxygen. (A) Survival of Histoplasma yeasts after coincubation with human PMNs. Wild-type and catalase- and/or superoxide dismutase-deficient mutant yeasts were added to PMNs isolated from peripheral human blood. Relative yeast survival (mean ± standard deviation) was determined by enumeration of the number of viable CFU after 4 h. In some experiments, DPI was added 24 h prior to yeast addition to inactivate the phagocyte NADPH oxidase. (B) Survival of Histoplasma yeast after infection of activated human macrophages. Wild-type, catalase, and superoxide dismutase (Sod3) mutants were added to IFN-γ-activated macrophages derived from human peripheral blood monocytes. Relative yeast survival (mean ± standard deviation) was determined by enumeration of viable CFU after 4 h. In some experiments, DPI was added 24 h prior to yeast addition to inactivate the phagocyte NADPH oxidase. Asterisks represent significant differences from survival of the wild-type strain determined by Student's t test of three replicates (ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001). Additionally, statistical significance was determined for the Sod3-deficient mutant compared to the double mutant lacking both the extracellular superoxide dismutase and the extracellular catalase. WT, wild type (OSU45); catBΔ, catB mutant (OSU16); catBΔ/CATB, complemented catB mutant (OSU51); catPΔ, catP mutant (OSU157); catPΔ/CATP, complemented catP mutant (OSU158); catBΔ catPΔ, catB catP double mutant (OSU159); sod3Δ, sod3 mutant (OSU15); catBΔ sod3Δ, catB sod3 double mutant (OSU46).
Since the primary host cell for Histoplasma yeast during mammalian infection is the macrophage, we determined if CatB and/or CatP was required for Histoplasma's ability to survive and grow within these phagocytes. For these experiments, human monocyte-derived macrophages were stimulated with IFN-γ to enhance the phagocyte oxidative burst response to Histoplasma yeast, since resting human macrophages are unable to induce oxidative killing of yeast (17, 45–47). The CatB-deficient catBΔ strain had no defect in survival compared to wild type after infection of macrophages (Fig. 5B). Loss of CatP in the catPΔ mutant yeasts modestly reduced survival to 79% of wild-type survival (63% versus 80%). Resistance to macrophage killing was restored by expression of the CATP gene in the complemented mutant. However, loss of both CatB and CatP substantially decreased yeast survival following infection of macrophages; survival of catBΔ catPΔ mutant yeast was 50% of wild-type yeast survival (40% versus 80%). As with PMNs, killing of catalase-deficient yeasts by macrophages was prevented by treatment of macrophages with DPI, indicating that killing of catalase-deficient yeasts was predominantly ROS mediated. These results indicate that CatB and CatP serve redundant functions with respect to defense against peroxide stress and that either is largely sufficient to protect Histoplasma yeast from killing by macrophages.
As Histoplasma's extracellular superoxide dismutase (Sod3) also contributes to the resistance of Histoplasma to killing by host ROS (17), we tested whether loss of the extracellular catalase, CatB, further increased the sensitivity of yeast to PMNs and activated macrophages in the absence of extracellular Sod3. Both CatB and Sod3, as extracellular enzymes, could contribute to protection of yeast from extracellular ROS, and a double catBΔ sod3Δ mutant was constructed to test this. Compared to the survival of the sod3Δ mutant strain, the catBΔ sod3Δ mutant survival after coincubation with PMNs (Fig. 5A) or with activated human macrophages (Fig. 5B) was not further reduced. Thus, there was no additive defect if CatB was depleted from a strain already lacking Sod3. DPI treatment of PMNs and macrophages alleviated the sensitivity of the sod3Δ and catBΔ sod3Δ mutants to phagocyte killing, confirming that the effects of catalase and superoxide dismutase deficiency on yeast viability result from the lack of defenses against PMN- and macrophage-produced ROS.
CatB is not required in vivo for Histoplasma virulence.
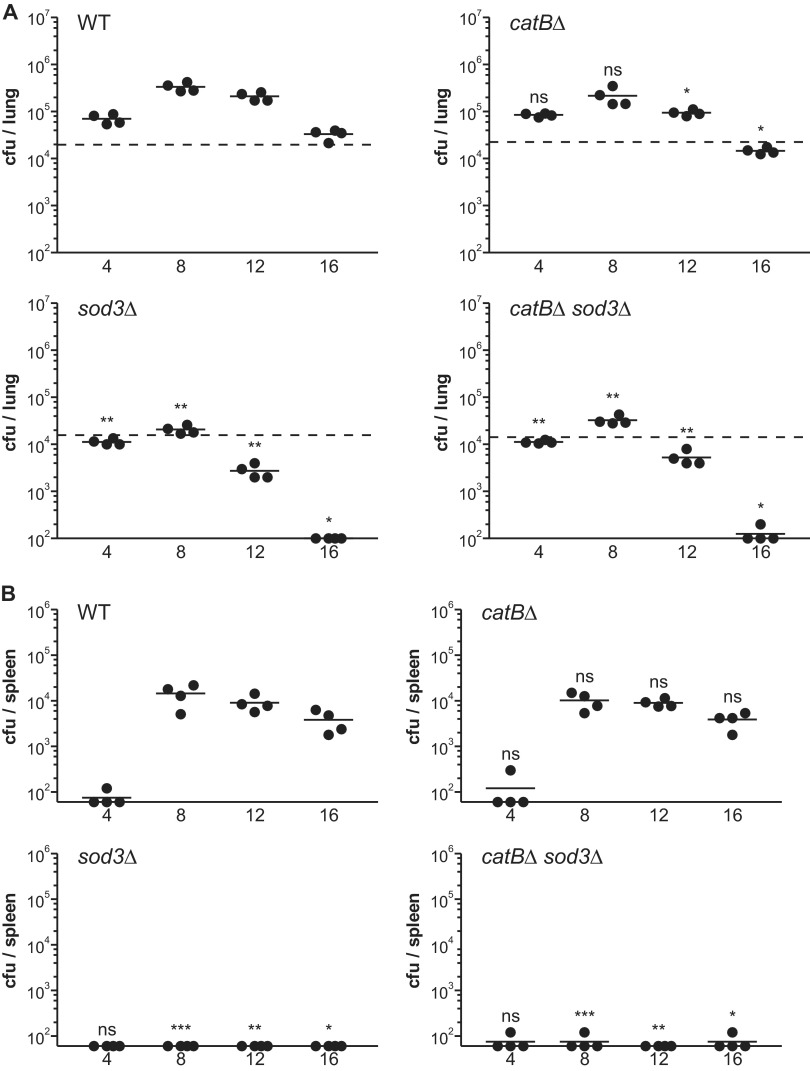
To determine if the pathogenic-phase-regulated, extracellular CatB catalase is necessary for full Histoplasma virulence in vivo, CatB-deficient yeasts were tested for their ability to establish respiratory infection and disseminate to the spleen. Following inhalation of a sublethal inoculum, wild-type Histoplasma yeasts survived and replicated within lung tissue, reaching a maximum fungal burden (33-fold above the inoculum) at between 8 and 12 days postinfection (Fig. 6A). Extrapulmonary dissemination of the infection to the spleen occurred beginning at day 4 postinfection and reached a peak fungal burden on day 8 (Fig. 6B). Subsequent activation of cell-mediated immunity caused a decline of the fungal burden in the lungs and spleen after day 8 postinfection. Despite speculations that CatB is a virulence factor, the loss of CatB, surprisingly, did not impair Histoplasma yeasts' ability to infect the lung (Fig. 6A) or to disseminate (Fig. 6B). The rate of clearing of CatB-deficient yeasts from the lung was slightly increased from the rate of clearing of wild-type yeasts, although this was only barely at statistical significance and did not manifest in a repeat experiment (Fig. 7A). Thus, CatB appears to be dispensable for murine lung infection.
Fig 6.
Sod3 deficiency, but not CatB deficiency, attenuates Histoplasma virulence in mice. (A) Pulmonary fungal burden kinetics after sublethal lung infection by Histoplasma yeast. Wild-type C57BL/6 mice were infected intranasally with 2 × 104 wild-type, CatB-deficient, Sod3-deficient, or doubly CatB- and Sod3-deficient yeasts. At 4-day intervals postinfection, fungal burdens were determined by quantitative plating of lung homogenates for determination of the number of Histoplasma CFU. The lower limit of detection was 102 CFU. Dashed horizontal lines indicate the actual number of CFU delivered in the inoculum. (B) Dissemination kinetics of Histoplasma yeasts after sublethal lung infection. Wild-type C57BL/6 mice were infected intranasally with 2 × 104 wild-type, CatB-deficient, Sod3-deficient, or CatB- and Sod3-deficient yeasts. At 4-day intervals postinfection, splenic fungal burdens were determined by quantitative plating of spleen homogenates for determination of the number of Histoplasma CFU. The lower limit of detection was 60 CFU. No yeasts had disseminated from the lung to the spleen at day 0. Each data point represents the CFU counts per organ from an individual animal (n = 4). Horizontal bars represent the mean fungal burden. Asterisks indicate statistically significant differences from wild type determined by Student's t test (ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001). WT, wild type (OSU45); catBΔ, catB mutant (OSU16); sod3Δ, sod3 mutant (OSU15); catBΔΔ sod3Δ, catB sod3 double mutant (OSU46).
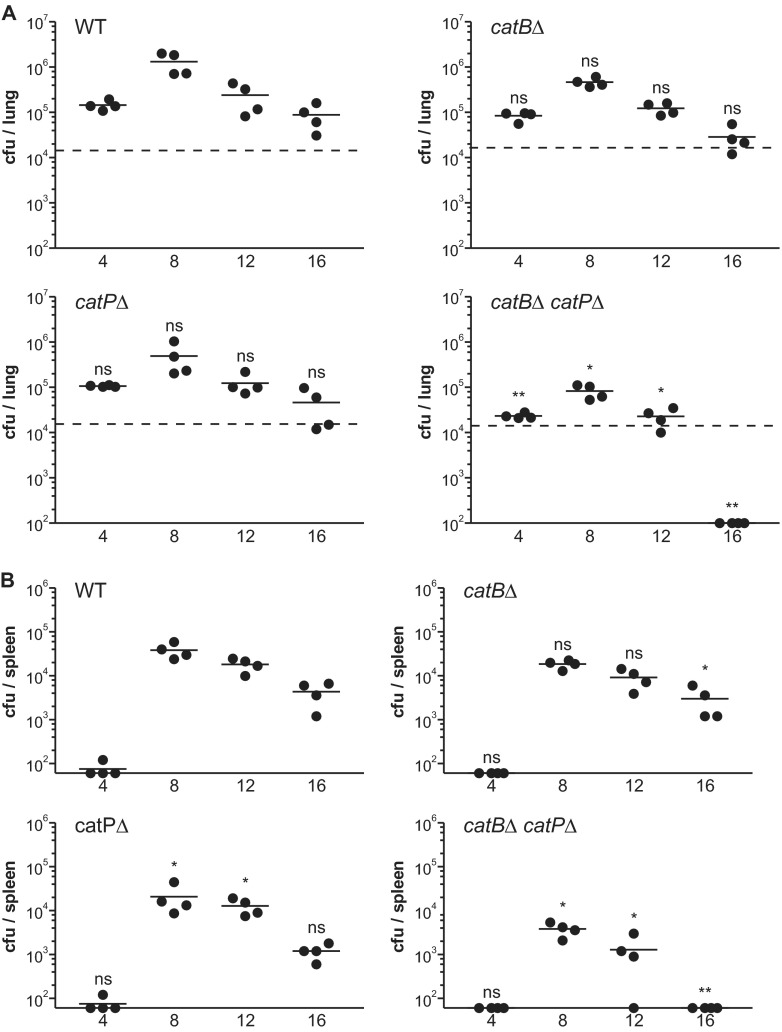
Fig 7.
CatB and CatP provide redundant catalase functions necessary for Histoplasma virulence in vivo. (A) Pulmonary fungal burden kinetics after sublethal lung infection by Histoplasma yeast. Wild-type C57BL/6 mice were infected intranasally with 2 × 104 wild-type, CatB-deficient, CatP-deficient, or double mutant yeasts lacking both catalases. At 4-day intervals postinfection, fungal burdens were determined by quantitative plating of lung homogenates for determination of the number of Histoplasma CFU. The lower limit of detection was 102 CFU. Dashed horizontal lines indicate the actual number of CFU delivered in the inoculum. (B) Dissemination kinetics of Histoplasma yeast after sublethal lung infection. Wild-type C57BL/6 mice were infected intranasally with 2 × 104 wild-type, CatB-deficient, CatP-deficient, or double catalase mutant yeasts. At 4-day intervals postinfection, splenic fungal burdens were determined by quantitative plating of spleen homogenates for determination of the number of Histoplasma CFU. The lower limit of detection was 60 CFU. No yeasts had disseminated from the lung to the spleen at day 0. Each data point represents CFU counts per organ from an individual animal (n = 4). Horizontal bars represent the mean fungal burden. Asterisks indicate statistically significant differences from wild type determined by Student's t test (ns, not significant; *, P < 0.05; **, P < 0.01). WT, wild type (OSU45); catBΔ, catB mutant (OSU16); catPΔ, catP mutant (OSU157); catBΔ catPΔ, catB catP double mutant (OSU159).
As Sod3 provides substantial in vivo defense against phagocyte-derived ROS (17), we tested whether loss of CatB function in the ROS-sensitive sod3Δ strain would reveal an important CatB function in vivo. Similar to results with cultured phagocyte infections, sod3Δ mutant yeasts were attenuated in their ability to survive in the lung, showing a decline in fungal burden from the inoculum at day 4 and only a net 1.7-fold overall increase by day 8 (Fig. 6A). Dissemination of the sod3Δ mutant was practically absent (Fig. 6B). The catBΔ sod3Δ double mutant lacking both extracellular superoxide dismutase and catalase functions was similarly attenuated (Fig. 6A and B). In fact, the defect in lung infection and dissemination was nearly identical between the sod3Δ single mutant and the catBΔ sod3Δ double mutant at all time points, indicating that the additional loss of CatB in the Sod3-deficient strain does not further impair Histoplasma virulence in vivo.
Redundant CatB and CatP functions are required for full Histoplasma virulence.
The unanticipated finding that CatB alone is not required for Histoplasma virulence could be explained by redundant ROS-protective functions provided by the intracellular CatP catalase. To determine if CatP and CatB act redundantly, we compared the infection kinetics of catBΔ or catPΔ single mutant strains with that of the double mutant lacking both catalases. Neither the loss of CatP alone nor the loss of CatB alone negatively affected the virulence of Histoplasma, as the mutant strains established similar fungal burdens as wild type in the lung (Fig. 7A). These results of an independent test of the catBΔ mutant were highly comparable to those obtained previously (Fig. 6A). Rates of clearance from the lung for singly CatP- and CatB-deficient strains were not significantly different from those for wild type. Thus, CatP and CatB individually are not critical to Histoplasma survival and growth within the lung. A similar lack of attenuation was apparent from fungal burdens in spleen (Fig. 7B).
In contrast to the results for the single catBΔ and catPΔ mutants, the loss of both CatB and CatP catalases significantly reduced lung infection and dissemination of Histoplasma yeast (Fig. 7A and B). Net survival and replication of the catBΔ catPΔ double mutant strain in lungs were 6.2- to 16-fold lower than those of wild type before the yeasts were cleared by day 16 postinfection. This defect of Histoplasma yeasts lacking both catalases was similar to the infection kinetics of Sod3-deficient yeasts, although the Sod3-deficient strain was more attenuated (Fig. 6A). A similar trend in the fungal burdens was observed in spleens when the catBΔ catPΔ double mutant was compared to the catBΔ and catPΔ single mutants (Fig. 7B). Thus, CatB and CatP can functionally compensate for each other during infection and together provide for greater Histoplasma yeast survival in vivo.
DISCUSSION
The ability to avoid or detoxify antimicrobial ROS produced by phagocytes in response to infection is a prerequisite for microbial pathogens, particularly those pathogens that successfully survive within ROS-generating phagocytes. The optimal strategy is to prevent the production of reactive oxygen compounds. To accomplish this, Histoplasma yeasts minimize exposure of immunostimulatory cell wall β-glucans by concealing them beneath an α-linked glucan polysaccharide (48). Other fungal pathogens use different mechanisms to achieve the same end with various degrees of success. For example, Cryptococcus yeasts produce a polysaccharide capsule that hides β-glucan and mannan pathogen-associated molecular patterns (49, 50), and Aspergillus conidial β-glucans are not revealed until germination (51, 52). However, avoidance of detection is not perfect, and entry into host cells, particularly by phagocytosis, can be accompanied by ROS production. Thus, before uptake into phagocytes, intracellular fungal pathogens must have preformed antioxidant systems to destroy phagocyte-derived ROS to ensure fungal survival. In this study, we demonstrate that CatB and CatP comprise an effective, redundant approach that enables Histoplasma yeasts to optimally contend with peroxide stress as a component of host-derived ROS.
Although the Histoplasma genome has three catalase genes, only CatB and CatP are expressed by Histoplasma yeasts. Lack of CATA expression in yeasts by quantitative RT-PCR is consistent with a previous analysis of the CAT genes by Northern blotting (28). However, Johnson et al. reported CATA expression by mycelia, which we did not observe (28). This difference is due to the different genetic backgrounds used: CATA expression is 10- to 20-fold higher in G217B (the strain used by Johnson et al. [28]) than in G186A (the strain characterized in this study) by quantitative RT-PCR (data not shown). Regardless of the strain, CATA is not significantly expressed by pathogenic Histoplasma yeasts. By phylogenetic analysis, CatP belongs to the peroxisomal class of catalase enzymes (28). Our results show that CatP accounts specifically for the intracellular catalase activity of Histoplasma yeasts, consistent with this prediction. CatB belongs to the B clade of catalases, which are found only in Ascomycetes. Opposite from the findings for CatP, CatB is responsible for all the extracellular catalase activity of Histoplasma yeasts, consistent with its secretion (43) and cell wall association (32). In vitro, roughly half of the CatB activity is secreted into the surrounding medium and half remains associated with the yeast cell. The mechanism retaining CatB on the cell surface is unknown, and no predicted attachment motifs (e.g., glycosylphosphatidylinositol anchors) are evident from the primary amino acid sequence. Virtually all extracellular and intracellular catalase activities are eliminated by deletion of the CATB and CATP genes, respectively, indicating that CatB and CatP, but not CatA, account for the catalase activities of Histoplasma yeasts.
The extracellular location of CatB and its restricted expression by pathogenic-phase yeasts imply that its purpose is to combat exogenous peroxide stress produced by the host. Both Cryptococcus and Candida albicans lack extracellular catalase activity (53, 54). Aspergillus fumigatus, like Histoplasma, produces a glycosylated catalase (Cat1) with a signal sequence sufficient to direct its secretion (55). Loss of CatB function increases the sensitivity of Histoplasma yeasts to H2O2 in vitro, showing that H2O2 can be antimicrobial to Histoplasma yeasts and that CatB can detoxify this ROS molecule. However, loss of CatB has no deleterious consequences for Histoplasma virulence in vivo and only minor effects on yeast survival against cultured phagocytes. This discrepancy between in vitro and in vivo phenotypes for catalase mutants is not without precedent; the loss of catalase proteins does not attenuate virulence for bacterial pathogens such as Neisseria gonorrhoeae (56), Salmonella enterica serovar Typhimurium (57), or Haemophilus influenzae (58). A similar situation exists for fungal pathogens; elimination of the Aspergillus fumigatus conidial catalase (CatA) increases conidial sensitivity to H2O2 in vitro but does not impact conidial killing in vivo (59). Similarly, Cryptococcus cells lacking multiple catalases are completely virulent in a mouse inhalation model (54). Normal virulence is also manifest for Candida glabrata yeasts lacking the Cta1 catalase (60) and Magnaporthe oryzae cells lacking the CpxB catalase (61).
One possibility for this discrepancy is that the H2O2 concentration produced by phagocytes in vivo may differ substantially from fungicidal levels in vitro. Alternatively, reactive oxygen compounds other than peroxide may be more relevant to fungicidal mechanisms in vivo. In Histoplasma, loss of the Sod3 superoxide dismutase increases the sensitivity of yeasts to ROS-mediated phagocyte killing and attenuates virulence in vivo (17). In this study, we show that additional loss of CatB from Sod3-deficient yeasts does not further increase yeast sensitivity to phagocyte ROS in vitro or virulence in vivo (Fig. 6). As peroxide is derived from superoxide by either enzymatically catalyzed or spontaneous dismutation, the attenuation of Sod3-deficient yeasts indicates that the CatB (and CatP) catalase(s) is not sufficient to protect Histoplasma from host ROS. This confirms that although superoxide leads to production of peroxide, superoxide is a major fungicidal immune defense molecule that Histoplasma must combat and does so specifically through Sod3. Nevertheless, Histoplasma virulence requires protection from peroxide stress as well, since yeasts lacking both CatB and CatP (i.e., the catBΔ catPΔ mutant) are attenuated, even though they express Sod3. The similarity of the catBΔ sod3Δ mutant's survival to the catBΔ single mutant's survival indicates that Histoplasma can still eliminate peroxide stress in the absence of CatB, despite elevated ROS. This suggests that Histoplasma has additional peroxide detoxification mechanisms (e.g., CatP).
A second factor contributing to the contrasting requirement for catalases in vitro and in vivo is the existence of redundant or compensatory peroxide-detoxifying mechanisms. Similar to Aspergillus and Cryptococcus, Histoplasma's multiple catalases create possible redundancy. Our results indicate that intracellular CatP can supply sufficient peroxide-destroying defense functions in the absence of extracellular CatB. The loss of CatP results in a more severe defect in response to peroxide in vitro and more severe survival defects when Histoplasma is cocultured with phagocytic cells than loss of CatB. This suggests that CatP provides more catalase-based ROS defense. However, like CatB-deficient yeasts, CatP-deficient yeasts are fully virulent in vivo. Only loss of both catalases attenuates virulence, consistent with CatB and CatP serving redundant defense functions during infection. It is unlikely that Histoplasma CatA contributes to yeast ROS defenses, as CatA is not expressed by yeasts. Furthermore, the lack of single mutant phenotypes is not explained by compensatory upregulation of other catalases; CatB and CatP production is not altered in the catPΔ and catBΔ single mutants, respectively (Fig. 2). In Aspergillus, Cat1 and Cat2 mycelial catalases are redundant, with the double mutant showing a 2-fold increase in sensitivity to H2O2 compared to that of single catalase mutants (59). In contrast to the catalase redundancy of Aspergillus, Cryptococcus, and Histoplasma, Candida albicans has only a single intracellular catalase (Cat1), and loss of its function eliminates all catalase activity, impairs survival of Candida against PMNs, and attenuates virulence in a candidemia model (53, 62). The lack of attenuation of most fungal catalase mutants is often ascribed to noncatalase enzymes that reduce intracellular ROS (e.g., peroxiredoxins [63], glutaredoxins [64], cytochrome c peroxidase [65], alternative oxidase [66, 67]) and/or ROS scavengers (e.g., melanin [7], mannitol [68], capsule [69], and trehalose [70, 71]). The current findings revealed by Histoplasma yeasts lacking both CatB and CatP and the data for Candida albicans mutants lacking Cat1 highlight the critical role of catalases in pathogenesis for these fungi, whereas other fungal pathogens likely rely on other non-catalase-based mechanisms to combat host ROS.
In some fungal systems, oxidative stress increases the expression of catalases and other antioxidant systems (62, 70, 72–74). This response is often mediated by AP1-class transcription factors, and loss of AP1 leads to enhanced sensitivity of fungi to peroxide and can attenuate virulence (60, 74–77). However, the rapid lethal action of host-derived ROS leaves little time for adaptive responses. Since Histoplasma is predominantly associated with phagocytes, the synthesis of catalases by yeasts before interaction with ROS-producing host cells before the host-pathogen interaction makes intuitive sense. Consistent with this, exogenous peroxide does not upregulate CatB and CatP expression (28). The only evidence for Histoplasma CatB regulation is increased CATB expression when iron concentrations increase, presumably to prevent the formation of hydroxyl radicals by destroying the predecessor molecule, hydrogen peroxide (78).
The redundant functions of Histoplasma CatB and CatP provide an optimal system for protection against host-derived ROS. However, the roles of CatB and CatP go beyond simple redundancy in detoxifying peroxide. Extracellular phagocyte-produced ROS would first encounter and be detoxified by extracellular Sod3 and CatB defense factors. Unlike short-lived superoxide anion, stable hydrogen peroxide that escapes destruction by CatB is membrane permeant and can diffuse into the yeast cell and affect intracellular targets. However, the Histoplasma intracellular CatP catalase comprises a second line of defense, degrading this now intracellular peroxide challenge and thereby protecting Histoplasma yeast from cellular damage. The appearance of microcolonies of yeasts that escape H2O2-mediated destruction in the disk diffusion assay lends support to this model. Only microcolonies of wild-type and CatB-deficient yeasts appear in the zones of clearing because intracellular CatP enables some yeasts to survive by continual detoxification of peroxide that diffuses into cells. On the other hand, microcolonies never appeared with either of the CatP-deficient strains; although CatB largely detoxifies the extracellular peroxide stress, peroxide that diffuses into cells is not eliminated without CatP. Consequently, no yeasts that lack CatP escape killing in the clearing zone around the peroxide-saturated disk. Thus, this spatial separation of individual catalases, while singly not required to protect Histoplasma in vivo, nonetheless (i) enables Histoplasma to efficiently detoxify the majority of ROS before it can cause cellular damage and (ii) provides a backup system to degrade any ROS that eludes the primary extracellular defense system. As a consequence, Histoplasma's dual catalases enable yeasts to readily survive infection in the presence of ROS-producing phagocytes, thereby facilitating Histoplasma pathogenesis.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by research grant AI083335 from the National Institutes of Health and a predoctoral research fellowship to E. D. Holbrook from the Public Health Preparedness for Infectious Diseases Program at Ohio State University.
Footnotes
Published ahead of print 15 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00173-13.
REFERENCES
- 1. DeLeo FR, Allen LA, Apicella M, Nauseef WM. 1999. NADPH oxidase activation and assembly during phagocytosis. J. Immunol. 163:6732–6740 [PubMed] [Google Scholar]
- 2. Fang FC. 2011. Antimicrobial actions of reactive oxygen species. mBio 2(5):00141–11. 10.1128/mBio.00141-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cohen MS, Isturiz RE, Malech HL, Root RK, Wilfert CM, Gutman L, Buckley RH. 1981. Fungal infection in chronic granulomatous disease. The importance of the phagocyte in defense against fungi. Am. J. Med. 71:59–66 [DOI] [PubMed] [Google Scholar]
- 4. Soler-Palacin P, Margareto C, Llobet P, Asensio O, Hernandez M, Caragol I, Espanol T. 2007. Chronic granulomatous disease in pediatric patients: 25 years of experience. Allergol. Immunopathol. (Madr.) 35:83–89 [DOI] [PubMed] [Google Scholar]
- 5. Fairfield AS, Abosch A, Ranz A, Eaton JW, Meshnick SR. 1988. Oxidant defense enzymes of Plasmodium falciparum. Mol. Biochem. Parasitol. 30:77–82 [DOI] [PubMed] [Google Scholar]
- 6. Cabiscol E, Tamarit J, Ros J. 2000. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int. Microbiol. 3:3–8 [PubMed] [Google Scholar]
- 7. Hamilton AJ, Holdom MD. 1999. Antioxidant systems in the pathogenic fungi of man and their role in virulence. Med. Mycol. 37:375–389 [DOI] [PubMed] [Google Scholar]
- 8. Liu L, Tewari RP, Williamson PR. 1999. Laccase protects Cryptococcus neoformans from antifungal activity of alveolar macrophages. Infect. Immun. 67:6034–6039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ruijter GJ, Bax M, Patel H, Flitter SJ, van de Vondervoort PJ, de Vries RP, vanKuyk PA, Visser J. 2003. Mannitol is required for stress tolerance in Aspergillus niger conidiospores. Eukaryot. Cell 2:690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alp S. 2010. Melanin and its role on the virulence of Cryptococcus neoformans. Mikrobiyol. Bul. 44:519–526 (In Turkish.) [PubMed] [Google Scholar]
- 11. Garre V, Tenberge KB, Eising R. 1998. Secretion of a fungal extracellular catalase by Claviceps purpurea during infection of rye: putative role in pathogenicity and suppression of host defense. Phytopathology 88:744–753 [DOI] [PubMed] [Google Scholar]
- 12. Fang FC, DeGroote MA, Foster JW, Baumler AJ, Ochsner U, Testerman T, Bearson S, Giard JC, Xu Y, Campbell G, Laessig T. 1999. Virulent Salmonella typhimurium has two periplasmic Cu, Zn-superoxide dismutases. Proc. Natl. Acad. Sci. U. S. A. 96:7502–7507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zancope-Oliveira RM, Reiss E, Lott TJ, Mayer LW, Deepe GS., Jr 1999. Molecular cloning, characterization, and expression of the M antigen of Histoplasma capsulatum. Infect. Immun. 67:1947–1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhu X, Gibbons J, Garcia-Rivera J, Casadevall A, Williamson PR. 2001. Laccase of Cryptococcus neoformans is a cell wall-associated virulence factor. Infect. Immun. 69:5589–5596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Frohner IE, Bourgeois C, Yatsyk K, Majer O, Kuchler K. 2009. Candida albicans cell surface superoxide dismutases degrade host-derived reactive oxygen species to escape innate immune surveillance. Mol. Microbiol. 71:240–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang N, Zhang S, Borchert S, Richardson K, Schmid J. 2011. High levels of a fungal superoxide dismutase and increased concentration of a PR-10 plant protein in associations between the endophytic fungus Neotyphodium lolii and ryegrass. Mol. Plant Microbe Interact. 24:984–992 [DOI] [PubMed] [Google Scholar]
- 17. Youseff BH, Holbrook ED, Smolnycki KA, Rappleye CA. 2012. Extracellular superoxide dismutase protects Histoplasma yeast cells from host-derived oxidative stress. PLoS Pathog. 8:e1002713. 10.1371/journal.ppat.1002713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zamocky M, Droghetti E, Bellei M, Gasselhuber B, Pabst M, Furtmuller PG, Battistuzzi G, Smulevich G, Obinger C. 2012. Eukaryotic extracellular catalase-peroxidase from Magnaporthe grisea—biophysical/chemical characterization of the first representative from a novel phytopathogenic KatG group. Biochimie 94:673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Edwards LB, Acquaviva FA, Livesay VT, Cross FW, Palmer CE. 1969. An atlas of sensitivity to tuberculin, PPD-B, and histoplasmin in the United States. Am. Rev. Respir. Dis. 99(Suppl):1–132 [PubMed] [Google Scholar]
- 20. Ajello L. 1971. The medical mycological iceberg. HSMHA Health Rep. 86:437–448 [PMC free article] [PubMed] [Google Scholar]
- 21. Mukhopadhyay S, Katzenstein AL. 2010. Biopsy findings in acute pulmonary histoplasmosis: unusual histologic features in 4 cases mimicking lymphomatoid granulomatosis. Am. J. Surg. Pathol. 34:541–546 [DOI] [PubMed] [Google Scholar]
- 22. Kroetz DN, Deepe GS. 2012. The role of cytokines and chemokines in Histoplasma capsulatum infection. Cytokine 58:112–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eissenberg LG, Goldman WE. 1987. Histoplasma capsulatum fails to trigger release of superoxide from macrophages. Infect. Immun. 55:29–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wolf JE, Kerchberger V, Kobayashi GS, Little JR. 1987. Modulation of the macrophage oxidative burst by Histoplasma capsulatum. J. Immunol. 138:582–586 [PubMed] [Google Scholar]
- 25. Schnur RA, Newman SL. 1990. The respiratory burst response to Histoplasma capsulatum by human neutrophils. Evidence for intracellular trapping of superoxide anion. J. Immunol. 144:4765–4772 [PubMed] [Google Scholar]
- 26. Kurita N, Brummer E, Yoshida S, Nishimura K, Miyaji M. 1991. Antifungal activity of murine polymorphonuclear neutrophils against Histoplasma capsulatum. J. Med. Vet. Mycol. 29:133–143 [PubMed] [Google Scholar]
- 27. Kurita N, Terao K, Brummer E, Ito E, Nishimura K, Miyaji M. 1991. Resistance of Histoplasma capsulatum to killing by human neutrophils. Evasion of oxidative burst and lysosomal-fusion products. Mycopathologia 115:207–213 [DOI] [PubMed] [Google Scholar]
- 28. Johnson CH, Klotz MG, York JL, Kruft V, McEwen JE. 2002. Redundancy, phylogeny and differential expression of Histoplasma capsulatum catalases. Microbiology 148:1129–1142 [DOI] [PubMed] [Google Scholar]
- 29. Pine L, Malcolm GB, Gross H, Gray SB. 1978. Evaluation of purified H and M antigens of histoplasmin as reagents in the complement fixation test. Sabouraudia 16:257–269 [DOI] [PubMed] [Google Scholar]
- 30. Hamilton AJ, Bartholomew MA, Figueroa J, Fenelon LE, Hay RJ. 1990. Evidence that the M antigen of Histoplasma capsulatum var. capsulatum is a catalase which exhibits cross-reactivity with other dimorphic fungi. J. Med. Vet. Mycol. 28:479–485 [PubMed] [Google Scholar]
- 31. Kauffman CA. 2007. Histoplasmosis: a clinical and laboratory update. Clin. Microbiol. Rev. 20:115–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guimaraes AJ, Hamilton AJ, de M Guedes HL, Nosanchuk JD, Zancope-Oliveira RM. 2008. Biological function and molecular mapping of M antigen in yeast phase of Histoplasma capsulatum. PLoS One 3:e3449. 10.1371/journal.pone.0003449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reiss E, Hutchinson H, Pine L, Ziegler DW, Kaufman L. 1977. Solid-phase competitive-binding radioimmunoassay for detecting antibody to the M antigen of histoplasmin. J. Clin. Microbiol. 6:598–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Albuquerque PC, Nakayasu ES, Rodrigues ML, Frases S, Casadevall A, Zancope-Oliveira RM, Almeida IC, Nosanchuk JD. 2008. Vesicular transport in Histoplasma capsulatum: an effective mechanism for trans-cell wall transfer of proteins and lipids in ascomycetes. Cell. Microbiol. 10:1695–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Worsham PL, Goldman WE. 1988. Quantitative plating of Histoplasma capsulatum without addition of conditioned medium or siderophores. J. Med. Vet. Mycol. 26:137–143 [PubMed] [Google Scholar]
- 36. Woods JP, Heinecke EL, Goldman WE. 1998. Electrotransformation and expression of bacterial genes encoding hygromycin phosphotransferase and beta-galactosidase in the pathogenic fungus Histoplasma capsulatum. Infect. Immun. 66:1697–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3:1101–1108 [DOI] [PubMed] [Google Scholar]
- 38. Gavrilin MA, Bouakl IJ, Knatz NL, Duncan MD, Hall MW, Gunn JS, Wewers MD. 2006. Internalization and phagosome escape required for Francisella to induce human monocyte IL-1beta processing and release. Proc. Natl. Acad. Sci. U. S. A. 103:141–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sebghati TS, Engle JT, Goldman WE. 2000. Intracellular parasitism by Histoplasma capsulatum: fungal virulence and calcium dependence. Science 290:1368–1372 [DOI] [PubMed] [Google Scholar]
- 40. Kouoh F, Gressier B, Luyckx M, Brunet C, Dine T, Ballester L, Cazin M, Cazin JC. 2000. A simple method for isolating human and rabbit polymorphonuclear neutrophils (PMNs). Biol. Pharm. Bull. 23:1382–1383 [DOI] [PubMed] [Google Scholar]
- 41. Nauseef WM. 2007. Isolation of human neutrophils from venous blood. Methods Mol. Biol. 412:15–20 [DOI] [PubMed] [Google Scholar]
- 42. Schlesinger LS. 1993. Macrophage phagocytosis of virulent but not attenuated strains of Mycobacterium tuberculosis is mediated by mannose receptors in addition to complement receptors. J. Immunol. 150:2920–2930 [PubMed] [Google Scholar]
- 43. Holbrook ED, Edwards JA, Youseff BH, Rappleye CA. 2011. Definition of the extracellular proteome of pathogenic-phase Histoplasma capsulatum. J. Proteome Res. 10:1929–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Howard DH. 1983. Studies on the catalase of Histoplasma capsulatum. Infect. Immun. 39:1161–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Newman SL, Bullock WE. 1994. Interaction of Histoplasma capsulatum yeasts and conidia with human and animal macrophages. Immunol. Ser. 60:517–532 [PubMed] [Google Scholar]
- 46. Brummer E, Kurita N, Yoshida S, Nishimura K, Miyaji M. 1991. Killing of Histoplasma capsulatum by gamma-interferon-activated human monocyte-derived macrophages: evidence for a superoxide anion-dependent mechanism. J. Med. Microbiol. 35:29–34 [DOI] [PubMed] [Google Scholar]
- 47. Fleischmann J, Wu-Hsieh B, Howard DH. 1990. The intracellular fate of Histoplasma capsulatum in human macrophages is unaffected by recombinant human interferon-gamma. J. Infect. Dis. 161:143–145 [DOI] [PubMed] [Google Scholar]
- 48. Rappleye CA, Eissenberg LG, Goldman WE. 2007. Histoplasma capsulatum alpha-(1,3)-glucan blocks innate immune recognition by the beta-glucan receptor. Proc. Natl. Acad. Sci. U. S. A. 104:1366–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kozel TR, Gotschlich EC. 1982. The capsule of Cryptococcus neoformans passively inhibits phagocytosis of the yeast by macrophages. J. Immunol. 129:1675–1680 [PubMed] [Google Scholar]
- 50. Vecchiarelli A, Retini C, Pietrella D, Monari C, Tascini C, Beccari T, Kozel TR. 1995. Downregulation by cryptococcal polysaccharide of tumor necrosis factor alpha and interleukin-1 beta secretion from human monocytes. Infect. Immun. 63:2919–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hohl TM, Feldmesser M, Perlin DS, Pamer EG. 2008. Caspofungin modulates inflammatory responses to Aspergillus fumigatus through stage-specific effects on fungal beta-glucan exposure. J. Infect. Dis. 198:176–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Leal SM, Jr, Cowden S, Hsia YC, Ghannoum MA, Momany M, Pearlman E. 2010. Distinct roles for Dectin-1 and TLR4 in the pathogenesis of Aspergillus fumigatus keratitis. PLoS Pathog. 6:e1000976. 10.1371/journal.ppat.1000976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wysong DR, Christin L, Sugar AM, Robbins PW, Diamond RD. 1998. Cloning and sequencing of a Candida albicans catalase gene and effects of disruption of this gene. Infect. Immun. 66:1953–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Giles SS, Stajich JE, Nichols C, Gerrald QD, Alspaugh JA, Dietrich F, Perfect JR. 2006. The Cryptococcus neoformans catalase gene family and its role in antioxidant defense. Eukaryot. Cell 5:1447–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Calera JA, Paris S, Monod M, Hamilton AJ, Debeaupuis JP, Diaquin M, Lopez-Medrano R, Leal F, Latge JP. 1997. Cloning and disruption of the antigenic catalase gene of Aspergillus fumigatus. Infect. Immun. 65:4718–4724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Soler-Garcia AA, Jerse AE. 2007. Neisseria gonorrhoeae catalase is not required for experimental genital tract infection despite the induction of a localized neutrophil response. Infect. Immun. 75:2225–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Buchmeier NA, Libby SJ, Xu Y, Loewen PC, Switala J, Guiney DG, Fang FC. 1995. DNA repair is more important than catalase for Salmonella virulence in mice. J. Clin. Invest. 95:1047–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vergauwen B, Herbert M, Van Beeumen JJ. 2006. Hydrogen peroxide scavenging is not a virulence determinant in the pathogenesis of Haemophilus influenzae type b strain Eagan. BMC Microbiol. 6:3. 10.1186/1471-2180-6-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Paris S, Wysong D, Debeaupuis JP, Shibuya K, Philippe B, Diamond RD, Latge JP. 2003. Catalases of Aspergillus fumigatus. Infect. Immun. 71:3551–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cuellar-Cruz M, Briones-Martin-del-Campo M, Canas-Villamar I, Montalvo-Arredondo J, Riego-Ruiz L, Castano I, De Las Penas A. 2008. High resistance to oxidative stress in the fungal pathogen Candida glabrata is mediated by a single catalase, Cta1p, and is controlled by the transcription factors Yap1p, Skn7p, Msn2p, and Msn4p. Eukaryot. Cell 7:814–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tanabe S, Ishii-Minami N, Saitoh K, Otake Y, Kaku H, Shibuya N, Nishizawa Y, Minami E. 2011. The role of catalase-peroxidase secreted by Magnaporthe oryzae during early infection of rice cells. Mol. Plant Microbe Interact. 24:163–171 [DOI] [PubMed] [Google Scholar]
- 62. Nakagawa Y. 2008. Catalase gene disruptant of the human pathogenic yeast Candida albicans is defective in hyphal growth, and a catalase-specific inhibitor can suppress hyphal growth of wild-type cells. Microbiol. Immunol. 52:16–24 [DOI] [PubMed] [Google Scholar]
- 63. Missall TA, Pusateri ME, Lodge JK. 2004. Thiol peroxidase is critical for virulence and resistance to nitric oxide and peroxide in the fungal pathogen, Cryptococcus neoformans. Mol. Microbiol. 51:1447–1458 [DOI] [PubMed] [Google Scholar]
- 64. Chaves GM, Bates S, Maccallum DM, Odds FC. 2007. Candida albicans GRX2, encoding a putative glutaredoxin, is required for virulence in a murine model. Genet. Mol. Res. 6:1051–1063 [PubMed] [Google Scholar]
- 65. Giles SS, Perfect JR, Cox GM. 2005. Cytochrome c peroxidase contributes to the antioxidant defense of Cryptococcus neoformans. Fungal Genet. Biol. 42:20–29 [DOI] [PubMed] [Google Scholar]
- 66. Akhter S, McDade HC, Gorlach JM, Heinrich G, Cox GM, Perfect JR. 2003. Role of alternative oxidase gene in pathogenesis of Cryptococcus neoformans. Infect. Immun. 71:5794–5802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ruiz OH, Gonzalez A, Almeida AJ, Tamayo D, Garcia AM, Restrepo A, McEwen JG. 2011. Alternative oxidase mediates pathogen resistance in Paracoccidioides brasiliensis infection. PLoS Negl. Trop. Dis. 5:e1353. 10.1371/journal.pntd.0001353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chaturvedi V, Wong B, Newman SL. 1996. Oxidative killing of Cryptococcus neoformans by human neutrophils. Evidence that fungal mannitol protects by scavenging reactive oxygen intermediates. J. Immunol. 156:3836–3840 [PubMed] [Google Scholar]
- 69. Zaragoza O, Chrisman CJ, Castelli MV, Frases S, Cuenca-Estrella M, Rodriguez-Tudela JL, Casadevall A. 2008. Capsule enlargement in Cryptococcus neoformans confers resistance to oxidative stress suggesting a mechanism for intracellular survival. Cell. Microbiol. 10:2043–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Alvarez-Peral FJ, Zaragoza O, Pedreno Y, Arguelles JC. 2002. Protective role of trehalose during severe oxidative stress caused by hydrogen peroxide and the adaptive oxidative stress response in Candida albicans. Microbiology 148:2599–2606 [DOI] [PubMed] [Google Scholar]
- 71. Martinez-Esparza M, Aguinaga A, Gonzalez-Parraga P, Garcia-Penarrubia P, Jouault T, Arguelles JC. 2007. Role of trehalose in resistance to macrophage killing: study with a tps1/tps1 trehalose-deficient mutant of Candida albicans. Clin. Microbiol. Infect. 13:384–394 [DOI] [PubMed] [Google Scholar]
- 72. Gonzalez-Parraga P, Hernandez JA, Arguelles JC. 2003. Role of antioxidant enzymatic defences against oxidative stress H(2)O(2) and the acquisition of oxidative tolerance in Candida albicans. Yeast 20:1161–1169 [DOI] [PubMed] [Google Scholar]
- 73. Enjalbert B, MacCallum DM, Odds FC, Brown AJ. 2007. Niche-specific activation of the oxidative stress response by the pathogenic fungus Candida albicans. Infect. Immun. 75:2143–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kusch H, Engelmann S, Albrecht D, Morschhauser J, Hecker M. 2007. Proteomic analysis of the oxidative stress response in Candida albicans. Proteomics 7:686–697 [DOI] [PubMed] [Google Scholar]
- 75. Alarco AM, Raymond M. 1999. The bZip transcription factor Cap1p is involved in multidrug resistance and oxidative stress response in Candida albicans. J. Bacteriol. 181:700–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lessing F, Kniemeyer O, Wozniok I, Loeffler J, Kurzai O, Haertl A, Brakhage AA. 2007. The Aspergillus fumigatus transcriptional regulator AfYap1 represents the major regulator for defense against reactive oxygen intermediates but is dispensable for pathogenicity in an intranasal mouse infection model. Eukaryot. Cell 6:2290–2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lin CH, Yang SL, Chung KR. 2009. The YAP1 homolog-mediated oxidative stress tolerance is crucial for pathogenicity of the necrotrophic fungus Alternaria alternata in citrus. Mol. Plant Microbe Interact. 22:942–952 [DOI] [PubMed] [Google Scholar]
- 78. Hwang LH, Seth E, Gilmore SA, Sil A. 2012. SRE1 regulates iron-dependent and -independent pathways in the fungal pathogen Histoplasma capsulatum. Eukaryot. Cell 11:16–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Marion CL, Rappleye CA, Engle JT, Goldman WE. 2006. An alpha-(1,4)-amylase is essential for alpha-(1,3)-glucan production and virulence in Histoplasma capsulatum. Mol. Microbiol. 62:970–983 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.