Abstract
This study examined the in vitro interaction between Mycobacterium leprae, the causative agent of leprosy, and human alveolar and nasal epithelial cells, demonstrating that M. leprae can enter both cell types and that both are capable of sustaining bacterial survival. Moreover, delivery of M. leprae to the nasal septum of mice resulted in macrophage and epithelial cell infection in the lung tissue, sustaining the idea that the airways constitute an important M. leprae entry route into the human body. Since critical aspects in understanding the mechanisms of infection are the identification and characterization of the adhesins involved in pathogen-host cell interaction, the nude mouse-derived M. leprae cell surface-exposed proteome was studied to uncover potentially relevant adhesin candidates. A total of 279 cell surface-exposed proteins were identified based on selective biotinylation, streptavidin-affinity purification, and shotgun mass spectrometry; 11 of those proteins have been previously described as potential adhesins. In vitro assays with the recombinant forms of the histone-like protein (Hlp) and the heparin-binding hemagglutinin (HBHA), considered to be major mycobacterial adhesins, confirmed their capacity to promote bacterial attachment to epithelial cells. Taking our data together, they suggest that the airway epithelium may act as a reservoir and/or portal of entry for M. leprae in humans. Moreover, our report sheds light on the potentially critical adhesins involved in M. leprae-epithelial cell interaction that may be useful in designing more effective tools for leprosy control.
INTRODUCTION
Leprosy is a chronic infectious disease caused by Mycobacterium leprae, an acid-fast, rod-shaped obligate intracellular bacillus. The disease remains a major health problem in several developing countries, with new cases being documented every year (1). M. leprae mainly affects the coldest parts of the human body such as the skin, nasal mucosa, and peripheral nerves. The disease manifests as a spectrum of clinical forms, with the tuberculoid and lepromatous leprosy forms occupying the opposite poles. Patients with tuberculoid leprosy, also known as paucibacillary leprosy, develop a strong, specific cell-mediated immunity to M. leprae antigens which is able to confine the disease to a few well-defined lesions with well-developed granuloma and rare acid-fast bacilli. At the other end of the spectrum, however, the lepromatous/multibacillary patients show no apparent immunological resistance to M. leprae, resulting in numerous nodular lesions. Biopsy specimens of these lesions reveal abundant foamy dermal macrophages containing a large amount of bacilli (2).
The nose and skin are considered the main routes of M. leprae infection and transmission (3–5). Nasal mucosa involvement is observed in early leprosy even before lesions become apparent in the skin, nerves, and other parts of the body (6), suggesting that the airways are the primary infection sites. In support of this idea, M. leprae DNA has been detected by PCR in nasal swabs and nasal mucosa biopsy specimens of healthy individuals living in areas of endemicity (7–9). Moreover, the presence of acid-fast bacilli in the lungs of mice after M. leprae airborne infection implies that the lungs may also be a portal of entry (10). In lepromatous patients with a high bacteriological index (BI), the nasal mucosa is typically so heavily colonized that these patients are known to shed large numbers (an average of 107 per day) of viable bacteria through their nasal passages (11).
Thus, since it is likely that M. leprae interaction with airway epithelial cells plays a relevant role during the natural course of leprosy, studies on this subject deserve far more attention. Epithelial cells are important contributors to the mucosal innate response by producing an array of signal molecules responsible for modulating the adaptive immune response to microbial pathogens (for a review, see reference 12). A greater understanding of M. leprae interaction with these cells could lead to the design of more effective preventive tools to control the spread of the disease.
In the present study, the in vitro M. leprae infection of human alveolar cells, nasal epithelial cell lines, and human primary nasal epithelial cells was performed. It was seen that M. leprae actually invades these cells and that epithelial cells are capable of sustaining bacterial survival. The surface proteome of nude mouse-derived M. leprae was determined, allowing the identification of potentially relevant molecules for M. leprae-epithelial cell interaction. Among these proteins, it was confirmed that the heparin-binding hemagglutinin (HBHA) and the histone-like protein (Hlp), previously described as two major mycobacterial adhesins (13, 14), are exposed on the surface of M. leprae and mediate bacterial adhesion to epithelial cells.
MATERIALS AND METHODS
Cell lines and culture conditions.
The A549 human alveolar pneumocyte type II cell line (ATCC CCL185; ATCC, Manassas, VA) was cultured in Dulbecco's modified Eagle's medium (DMEM)-F12 (ATCC) supplemented with 10% (vol/vol) serum-inactivated fetal bovine serum (FBS; Gibco, Grand Island, NY). The RPMI 2650 human epithelial nasal septal cell line (ATCC CCL30) was grown in Eagle's minimum essential medium (EMEM; Gibco) with 10% FBS. Cells were maintained in culture and, for the assays, were detached from the plastic via the use of Tryple Express (1×) with phenol red (Gibco) at 37°C. The cells were then centrifuged at 2,000 × g for 10 min at 4°C, counted in a Neubauer chamber, and plated onto tissue culture wells or flasks at 37°C in a 5% CO2 atmosphere.
Isolation of human primary nasal epithelial cells.
The procedures described in this study were approved by the Pedro Ernesto University Hospital, the State University of Rio de Janeiro, and the Oswaldo Cruz Foundation (FIOCRUZ) Ethical Committee located in Rio de Janeiro, RJ, Brazil. All participants provided their written consent. Primary nasal epithelial cells were isolated from the nasal polyps of patients undergoing polypectomy for nasal clearing. These cells were then cultured as previously described (15). Briefly, the explants were washed with RPMI 1640 medium (Gibco) plus an antibiotic combination of 100 μg/ml penicillin, 100 μg/ml streptomycin, and 25 μg/ml amphotericin B. Next, the explants were cut into 2-to-3-mm-thick specimens and seeded onto type I collagen-coated circular glass coverslips in 24-well culture plates containing a defined RPMI 1640 medium (supplemented with 1 μg/ml insulin, 1 μg/ml transferrin, 10 ng/ml epithelial growth factor, 0.5 μg/ml hydrocortisone, and 10 ng/ml retinoic acid).
Cultures were incubated at 37°C in a humidified 5% CO2-air mixture. Under these conditions, epithelial cells migrated from the explants, forming an outgrowth area around them. In 7 days, the cultures were ready for use. The medium was replaced every 2 days of culture. To determine culture purity, the cells were labeled with cytokeratin-19 (CK-19) (Santa Cruz, Santa Cruz, CA) since this protein is present in the vast majority of the epithelial cells in the nasal cavity (16). Primary cells were fixed with 4% (wt/vol) paraformaldehyde (PFA) for 20 min at 4°C and permeabilized with 0.25% (vol/vol) Triton X-100 in phosphate-buffered saline (PBS) for 40 min at room temperature (RT). The cells were subsequently blocked with 5% (vol/vol) normal goat serum–PBS (blocking solution) overnight at 4°C. Afterwards, the cells were incubated with IgG1 mouse monoclonal primary antibody against human CK-19 (1:100) in blocking solution for 3 h at RT, washed 5 times with PBS, and then incubated for 2 h at RT with Alexa Fluor 555 goat anti-mouse IgG (Molecular Probes, Eugene, OR) at a dilution rate of 1:100 in blocking solution. A control isotype (IgG1) was used as the control. Cell nuclei were labeled with 4′,6′diamidino-2-phenylindole (DAPI) (Molecular Probes) (1:10,000) for 1 min. The purity of epithelial cells was assessed on an Observer.Z1 inverted fluorescence microscope (Zeiss, Oberkochen, Germany) in a set of 300 cells.
Bacterial strains and growth conditions.
Live M. leprae (Thai-53 strain) isolated from infected nude mouse food pads was kindly provided by James Krahenbuhl (National Hansen's Disease Program, Laboratory Research Branch, Louisiana State University, Baton Rouge, LA) through the National Institute of Allergy and Infectious Diseases (NIAID) (Bethesda, MD) (contract no. 155262). For proteomic analysis, the bacteria were treated or not with 0.1 N sodium hydroxide (NaOH) before shipment, as previously described (17). In the other assays, only NaOH-treated bacteria were used. The Mycobacterium smegmatis mc2155 strain was grown in Middlebrook 7H9 broth (Difco, Detroit) with 0.05% (vol/vol) Tween 80 and supplemented with 10% (vol/vol) OADC (oleic acid, albumin, dextrose, and catalase; Becton, Dickinson and Company, Franklin Lakes, NJ). The mycobacteria were grown at 37°C under conditions of agitation until they reached an optical density at 600 nm (OD600) of 0.8. To determine CFU, the bacteria were cultivated in Middlebrook 7H11 (Difco) supplemented with 10% OADC and 0.5% (vol/vol) glycerol at 37°C for 4 days.
Mycobacterium staining.
M. leprae was stained with fluorescent dyes using a PKH26/67 fluorescent cell linker minikit for general cell membrane labeling according to the recommendation of the manufacturer (Sigma, St. Louis, MO). Briefly, M. leprae bacilli (1 × 109) were suspended in 1.0 ml of the provided “diluent C” and then stained for 2 min at RT with a 1:250 dilution of either PKH26 (red) or PKH67 (green) dye. The staining was halted after 2 min by adding an equal volume of PBS. The suspension was washed (10,000 × g for 5 min) twice in PBS, and the numbers of bacteria were recounted following staining by the Shepard's direct count method (18).
Determination of M. leprae viability.
The membrane integrity of individual M. leprae bacteria in suspension was evaluated via the use of a Live/Dead BacLight bacterial viability kit (Molecular Probes). M. leprae bacteria (2 × 107) were washed twice (10,000 × g for 5 min) in PBS and incubated for 15 min at RT in 6 μM Syto9 (green dye) and 30 μM propidium iodide (PI) (red dye). The bacteria were washed twice with PBS, and the pellet was suspended in 20 μl of 10% (vol/vol) glycerol in normal saline solution followed by placement of 5 μl of the suspension on a glass slide with an 18-mm2 glass coverslip. The dead and live bacteria were enumerated by direct counting of the fluorescent green and red bacilli under a fluorescence microscope using the appropriate single-bandpass filter sets. The excitation/emission maxima are 480 nm/500 nm for Syto9 and 490 nm/635 nm for PI. A total of 200 bacteria were counted in duplicate.
In vitro binding and invasion assays.
RPMI 2650 and A549 cells were infected with live M. leprae at a multiplicity of infection (MOI) of 10 at different time intervals: 2 h, 4 h, 6 h, and 24 h. Unless otherwise mentioned, all experiments performed with live M. leprae were conducted at 33°C to preserve bacterial viability (19). The cells were then washed 6 times with PBS and fixed with 4% PFA, after which they were permeabilized with 0.1% Triton X-100 for 5 min and blocked with 1% bovine serum albumin (BSA)–PBS for 30 min. Cells were labeled for 20 min with fluorescein isothiocyanate (FITC)-phalloidin (Sigma) (1:40 dilution)–1% BSA–PBS and washed 3 times with PBS to remove the excess dye. Cell nuclei were stained with DAPI (1:10,000). The ability of M. leprae to invade and attach epithelial cells was investigated by confocal microscopy. At a determined time point, the sum of the numbers of adherent and internalized bacteria represented the total number of bacteria that interacted with the cells. A set of 300 cells was examined for the presence of M. leprae. In addition, the percentage of infected cells was calculated as follows: number of infected cells/300 cells × 100. Live and dead bacteria were compared with respect to their ability to bind and invade epithelial cells.
Prior to these experiments, an aliquot of freshly harvested M. leprae was heat killed at 60°C for 30 min (19). The cells were infected with live and dead bacteria at 33°C for 24 h at an MOI of 10. M. leprae binding and invasion in epithelial cells were determined by confocal microscopy after FITC-phalloidin and DAPI staining. To evaluate the effect of the cytoskeleton inhibitors on bacterial internalization, monolayers of A549 and RPMI 2650 cells were pretreated with 3 μM cytochalasin B (Sigma) or 10 μM colchicine (Enzo Life Sciences, Farmingdale, NY) 1 h prior to infection. Next, the cells were infected with M. leprae for 24 h at 33°C. The inhibitors were present during the entire assay. Control cultures were treated with dimethyl sulfoxide (DMSO), the vehicle used for cytochalasin B and colchicine solutions. Subsequently, the cells were washed 6 times, fixed with 4% PFA, and labeled with FITC-phalloidin and DAPI as described above.
Finally, to determine whether M. leprae is able to bind and invade human primary nasal epithelial cells, they were incubated with gamma-irradiated bacteria for 2 h at 37°C at an MOI of 10. For analysis by confocal microscopy, the cells were both immune labeled with anti-CK19 and labeled with DAPI as described above.
Determination of cellular phagocytic activity.
To verify the uptake activity of RPMI 2650 and A549 cells, 1-μm-diameter red fluorescent beads were used (Molecular Probes). Briefly, the beads were incubated with cells for 2 to 6 h at two different temperatures: 4°C and 37°C. The cells were then washed 6 times with PBS and detached with a cell scraper. The percentages of labeled cells were determined by flow cytometry (BD Accuri C6 flow cytometer; BD Biosciences, San Jose, CA). Labeled cells at 4°C were considered to have attached beads only, while labeled cells at 37°C were considered to have both attached and internalized beads. To determine the percentage of cells with internalized beads, the following formula was used: % cells with internalized beads = % labeled cells at 37°C − % labeled cells at 4°C. Moreover, the median fluorescence intensity (MFI) of cells with internalized beads was determined as follows: MFIinternalizedbeads = MFI37°C − MFI4°C.
M. leprae survival assays within epithelial cells.
RPMI 2650 and A549 cells (2 × 106 per well of a 6-well plate) were infected with live M. leprae at 33°C at an MOI of 10 at different time points: 1, 4, 8, or 10 days. By the same token, A549 cells were infected with M. leprae at 37°C at the same time points. Prior to infection, M. leprae viability (also referred to as “initial viability”) was determined. M. leprae bacilli were simultaneously incubated in cell-free media as a control. Cells were washed 6 times with PBS to remove any noninternalized bacteria and then lysed with 10% glycerol–1% Triton X-100–1% Tween 20 (lysis buffer) for 5 min to release intracellular bacteria. In addition, unattached bacteria were recovered during the washing steps (extracellular M. leprae). A broken-cell extract was centrifuged for 1 min at 300 × g to remove cell debris. The supernatant was harvested and centrifuged again at 13,500 × g for 10 min and the pellet washed 3 times to remove the lysis buffer. Finally, M. leprae cells were stained using a Live/Dead BacLight bacterial viability kit. The percentage of M. leprae intracellular viability was compared with the viability of extracellular bacteria and M. leprae cell-free media. A total of 200 bacteria were counted in duplicate to calculate viability.
Assays with recombinant M. leprae proteins.
The recombinant M. leprae proteins heparin-binding hemagglutinin (HBHA) and histone-like protein (Hlp) were obtained as described previously (20, 21). Red polystyrene fluorescent beads (1.0 μm diameter) were coated with recombinant M. leprae Hlp (50 μg/ml), M. leprae HBHA (50 μg/ml), or BSA (50 μg/ml) for 3 h at RT in 0.2 M carbonate-bicarbonate buffer (pH 9.6). The beads were washed 3 times to remove unbound protein and then blocked with 2% BSA for 2 h. A549 and RPMI 2650 cells (7 × 104 cells per well) were cultured in 24-well plates containing glass slips. The beads were then added to the cells (100:1 ratio) for 1 h at 37°C. Cells were washed 6 times with PBS and fixed with 4% PFA for 20 min at 4°C. The cells were observed using phase-contrast microscopy and white light to visualize the cells and a fluorescence filter with appropriate single-bandpass filter sets to see the beads. The excitation/emission maxima were 480 nm/500 nm. In this assay, 300 cells were counted in duplicate. Alternatively, M. smegmatis bacteria were pretreated with these proteins. The bacteria were first centrifuged at 16,000 × g for 10 min and then washed with PBS to remove any remaining media. This procedure was performed 2 more times. The bacteria were subsequently treated with 50 μg/ml of Hlp or HBHA for 2 h at 37°C and then centrifuged at 16,000 × g for 10 min and washed with PBS to remove unbound proteins. The washing step was repeated 2 more times. Confluent cells (105 per well in 24-well plates) were infected with 106 CFU bacteria at 37°C. The bacteria were released after incubation with 500 μl of 0.1% Triton X-100–PBS for 5 min. The viable bacteria were serially diluted in PBS and then plated onto 7H11 agar plates at 37°C for 4 days for CFU quantification. To check whether Hlp and HBHA of M. leprae were able to bind to the cell surface of M. smegmatis, an enzyme-linked immunosorbent assay (ELISA) was performed. The bacteria were suspended with carbonate-bicarbonate buffer (pH 9.6), and 100 μl was added to 96-well plates at 4°C overnight. Untreated bacteria were used as controls. The wells were washed 4 times with PBS and blocked with 2% BSA–PBS for 2 h at 37°C. After blocking, the wells were washed four times and then incubated with primary mouse monoclonal antibody VEF4 against HBHA (1:1,000) in a blocking solution for 2 h at 37°C. To detect Hlp, the wells were incubated with 5G9 primary mouse monoclonal antibody under the same conditions described for antibody VEF4. Subsequently, secondary anti-mouse peroxidase (1:1,000) was used to detect the immunocomplex for 1 h at 37°C. Secondary antibody was diluted in the blocking solution. Tetramethyl benzidine (TMB; 100 μl) was added to the wells, and the reaction was stopped with 50 μl of 2.5 N sulfuric acid. The plates were read at a wavelength of 450 nm by using an automatic microplate-scanning spectrophotometer (Spectramax 190; Molecular Devices).
Transmission electron microscopy.
For electron microscopy, RPMI 2650 and A549 monolayers infected with M. leprae at 24 h were washed 6 times with PBS and then detached with a cell scraper. Subsequently, the cells were washed 3 times with cold 0.2 M potassium phosphate buffer (pH 7.4) and fixed with 2.5% (vol/vol) glutaraldehyde and 1.5% PFA (diluted in the potassium phosphate buffer) overnight at 4°C. After the fixation process, the cells were washed again in the same buffer. The samples were postfixed in cold 1% osmium tetroxide–phosphate buffer for 90 min and dehydrated in an ascending ethanol series and embedded in epoxy resin. Ultrathin sections (80 mm) were cut out of blocks, mounted on grids, and stained with uranyl acetate and lead citrate before examination via the use of a FEI Tecnai G2 transmission electron microscope operating at 80 kV. The images were captured at the Oregon University High Tech Extension Service (Corvallis, OR).
Confocal microscopy.
Preparations were examined using a Zeiss LSM 510 Meta confocal microscope equipped with a Plan Apochromat 40× or 100× objective (Carl Zeiss, Thornwood, NY) and a CoolSNAP-Pro CF digital camera in conjunction with Axion Vision Version 4.7.2 software (Carl Zeiss). The images were edited using AxioVision software. Images were acquired, colored, and merged via the use of LSM 510 Zeiss software. Argon and neon-helium lasers (Mellets Griot) emitting at 488, 543, and 633 nm were used. Pictures of 20 to 40 confocal planes through the cell (z-stack) with a step size of 0.2 to 0.3 μm were taken with a 1,000× objective every 30 s for 5 to 10 min via the use of LSM image software. For some experiments, z-stacks of images were captured, processed, and rendered by the use of an LSM imaging system (Carl Zeiss).
Mouse infection.
C57BL/6 strain mice were briefly anesthetized with isoflurane, and 10 μl of bacterial suspension (107 cells) was delivered to each nostril with a protein gel-loading tip. The course of infection was followed at different time points: 4 h, 4 days, 10 days, and 20 days. Euthanasia was performed using CO2 exposure. A total of 20 mice were used in this study: 6 mice were used for each of the first 2 time points and 4 mice were used for each of the latter time points. The heads and lungs were recovered and fixed in 1% formaldehyde. Following fixation, the heads were decalcified and processed as described previously (22). Then, the head and lung tissues were embedded in paraffin and stained using Ziehl-Neelsen stain and hematoxylin and eosin (H&E), as previously reported (23). This study was approved by Oregon State University Animal Care and Use Proposal (ACUP/4309).
Biotinylation and purification of M. leprae cell surface-exposed proteins.
Live, freshly harvested M. leprae bacilli were treated or not with O.1 N NaOH and then incubated with 1 mg/ml of NHS-LC-biotin (Pierce) for 20 min at 23°C. The reaction was stopped by the use of 5 mM glycine–PBS, and bacterial cells were washed 3 times with PBS. Bacteria were lysed, and biotinylated proteins were purified with streptavidin-magnetic microspheres, as previously described (24). Samples were resuspended in 50 μl ammonium bicarbonate and digested with Trypsin Gold and ProteaseMAX (Promega, Madison WI), according to the manufacturer's instructions. Nonbiotinylated bacteria were included as negative controls to eliminate unspecific background and endogenously biotinylated proteins. Data from negative controls were pooled to create a master list of false-positive identifications. These proteins were then subtracted from the experimental data sets. The negative-control master list is included as Table S4 in the supplemental material.
Peptide purification and LC-tandem mass spectrometry (LC-MS/MS) analysis.
Digested peptides were purified and desalted on Vivapure C18 microspin columns according to the instructions of the manufacturer (Sartorius, Goettingen, Germany). The samples were dehydrated by the use of a speed vacuum and suspended in 500 ng/μl of 5% acetonitrile (ACN)–0.1% formic acid (FA). Peptides were separated using liquid chromatography (LC) with a NanoAcquity UltraPerformance LC (UPLC) system (Waters) and then delivered to an LTQ Velos dual-pressure linear ion-trap mass spectrometer (Thermo Fisher) by electrospray ionization with a captive spray source (Microm Biosciences). A binary gradient system consisting of solvent A (0.1% aqueous FA) and solvent B (ACN containing 0.1% FA) was used as described previously (24). A normalized collision energy value of 30% was adopted.
Database search.
An M. leprae database was created by combining protein sequences from M. leprae strain Thai-53 (taxon no. 1769; 2,327 sequences) and M. leprae strain Br4923 (taxon no. 561304; 1,599 sequences) obtained from UniProt. The duplicate entries were removed, resulting in a total of 2,430 proteins. Reversed databases were used to estimate error thresholds (25). The database sequences along with their reversed sequences were appended to 179 common contaminant sequences and their reversed forms to create a final database of 5,218 sequences. Database processing was performed via the use of the python scripts available at http://www.ProteomicAnalysisWorkbench.com. The average parent ion-mass tolerance value was 2.5 Da. The monoisotopic fragment ion-mass tolerance value was 1.0 Da. The ion series used in scoring were b and y. A static modification of +57 Da was added for all cysteine residues. Variable modifications of +16 Da on methionine residues and 339.2 Da (biotin) on lysine residues were also allowed, with a maximum of 3 modifications per peptide. RAW data from the mass spectrometer were converted into DTA files representing individual MS2 spectra using extract_msn.exe (version 5.0; Thermo Fisher). The group scan minimum count was 1, and a minimum of 25 ions were required while the mass tolerance for combining DTAs was set at a very small value (0.0001 Da). A linear discriminant transformation was used to improve the identification sensitivity of the SEQUEST analysis (25, 26); SEQUEST scores were combined with the linear discriminant function scores; and discriminant score histograms were created separately for each peptide charge state (1+, 2+, and 3+), number of tryptic termini (0, 1, or 2), and modification state (unmodified or M +16 modified). The score histograms for reversed matches were used to estimate the peptide false-discovery rates (FDR) and set score thresholds for each peptide class that achieved the desired peptide FDR (typically 1% unless noted otherwise). Peptide-to-protein mapping and protein filtering were performed using PAW_results_6.py (version 6.1). The in-house Python scripts have been described previously (27). The overall probability of protein identification was calculated by dividing the total number of MS/MS spectra matching the protein by the total number of matching MS/MS spectra unique to the protein. The surface-exposed M. leprae proteins were functionally classified according to the Leproma database annotations. The Tuberculist database was also used to determine M. tuberculosis homologs.
Statistical analysis.
To compare three or more groups under different conditions, one-way analysis of variance (ANOVA) was used followed by the Bonferroni posttest. For conducting comparisons between two sets of conditions, a Student t test was used. Data analysis was performed using the GraphPad InStat program (GraphPad Software, San Diego, CA), and P values < 0.05 were considered statistically significant.
RESULTS
M. leprae attaches to and enters nasal and alveolar epithelial cells.
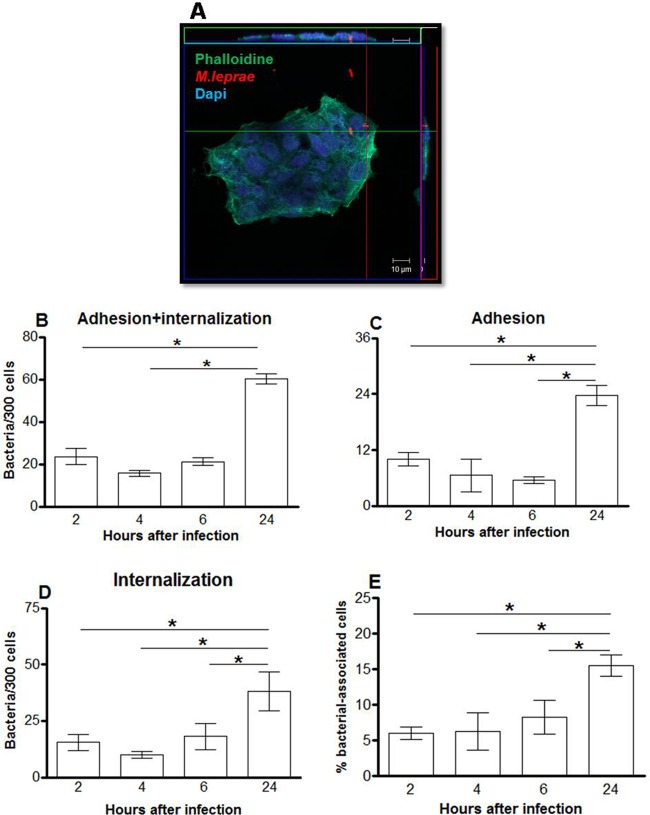
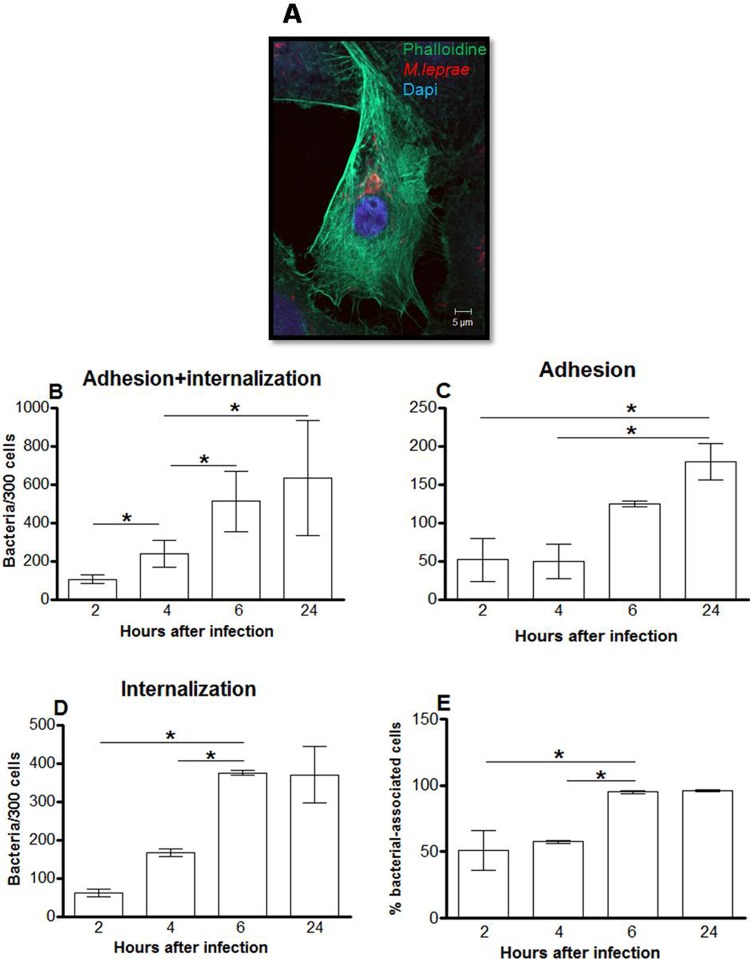
Confocal microscopy was used to investigate whether M. leprae is able to interact with nasal epithelial cells (RPMI 2650) and alveolar epithelial cells (A549). The results are expressed as the numbers of associated (adherent plus internalized) bacteria in a set of 300 cells (Fig. 1 and 2). Live M. leprae bacilli were able to attach to and enter both cell types. However, while only very few bacilli were able to interact with RPMI 2650 cells even after 24 h of incubation (61.6 bacteria ± 2.3/300 cells), the amount of associated bacteria in A549 cells was much greater at the same time point (633.2 bacteria ± 134.3/300 cells) (Fig. 1B and 2B). In RPMI 2650 cells, no increase in binding or invasion was observed between 2 h and 6 h of incubation, even though the number of internalized as well as adherent bacteria roughly doubled at 24 h (Fig. 1). On the other hand, the number of bacteria that adhered to and entered A549 cells significantly increased over the same time period, reaching saturation as early as 6 h postinfection (Fig. 2D). Similar results were obtained when the percentages of cells with associated bacteria were determined. At 2 h postinfection, 6% ± 0.9% and 51.3% ± 8.6% of bacterium-associated RPMI 2650 and A549 cells, respectively, were observed. An increase in the level of RPMI 2650 cells with associated bacteria was seen only 24 h postinfection (15.5% ± 1.5%), whereas most A549 cells showed associated bacteria at 6 h after infection (95% ± 0.5%) (Fig. 1E and 2E).
Fig 1.
M. leprae enters the nasal epithelial cell lineage RPMI 2650. Nasal epithelial cells were infected with live M. leprae at an MOI of 10 at 33°C at different time points. The bacteria were labeled with PKH26 (red), and the monolayers were stained with DAPI (blue) and FITC-phalloidin (green). (A) Representative confocal image of 4 independent experiments showing live M. leprae bacilli interacting with RPMI 2650 cells at 24 h of infection. (B, C, and D) Numbers of bacteria that interact with (B), adhere to (C), and enter (D) nasal epithelial cells. (E) Percentages of bacterium-associated cells. Data represent the means ± standard deviations (SD) of the results of 4 experiments performed in duplicate. *, P < 0.05 was considered statistically significant.
Fig 2.
M. leprae enters the alveolar epithelial cell lineage A549. Alveolar epithelial cells were infected with live M. leprae at an MOI of 10 at 33°C at increasing incubation times. The bacteria were labeled with PKH26 (red), and the cells were stained with DAPI (blue) and FITC-phalloidin (green). (A) Representative fluorescent image of 4 independent experiments showing live M. leprae bacilli interacting with A549 cells at 24 h of infection. (B, C, and D) Numbers of bacteria that interact with (B), adhere to (C), and enter (D) alveolar epithelial cells. (E) Percentages of bacterium-associated cells. Data represent the means ± standard deviations of the results of 4 experiments performed in duplicate. *, P < 0.05 was considered statistically significant.
It was then decided to compare the interactions of inert particles of latex beads with RPMI 2650 and A549 cells. Both the nasal and alveolar cells were incubated with fluorescent beads at 37°C and 4°C followed by the determination of the percentages of cells with adherent (4°C) as well as internalized beads (% labeled cells at 37°C − % labeled cells at 4°C) by flow cytometry. A significant difference was observed only after 2 h of incubation, and the difference dissipated at 6 h (see Fig. S1A and C in the supplemental material). Similar results were obtained when the median fluorescence intensity (MFI) was determined (Fig. S1D). A549 and RPMI 2650 cells displayed similar levels of MFI after 2 and 6 h of incubation, indicating that the two types of cells engage in similar endocytic activities. Thus, the higher association of M. leprae to alveolar epithelial cells at 6 h and later time points cannot be explained by the higher endocytic activity of these cells, suggesting higher intrinsic affinity on the part of this pathogen with these cells.
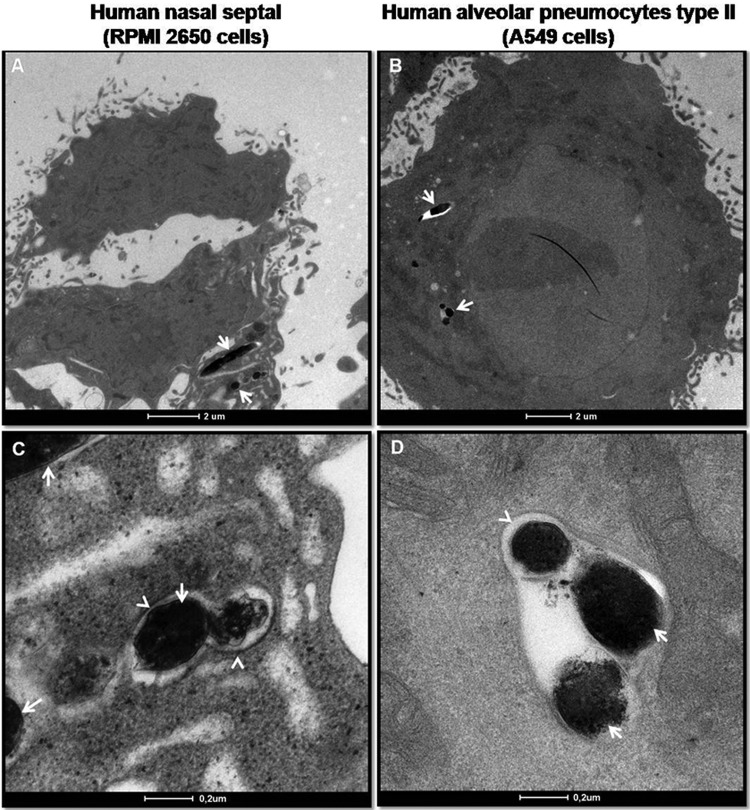
Finally, transmission electron microscopy was performed to visualize intracellular M. leprae. A549 and RPMI 2650 cells were infected with live M. leprae for 24 h and then processed for electron microscopy. As shown in Fig. 3, M. leprae (arrows) can be seen inside membrane-bound compartments (arrowheads) in both cell types but was present at higher numbers in the A549 cells. In some images, bacteria were seen to apparently float freely within the cytoplasm. Therefore, these data confirm the results obtained by confocal microscopy showing that M. leprae is indeed able to effectively enter airway epithelial cells.
Fig 3.
Transmission electron micrographs of M. leprae-infected epithelial cells. Epithelial cells were infected with live M. leprae for 24 h at 33°C. RPMI 2650 (A and C) and A549 (B and D) cells were fixed, processed, and visualized by transmission electron microscopy. The arrows point to bacteria; the arrowheads point to the membrane-bound compartments.
Ex vivo and in vivo M. leprae infection of airway epithelial cells.
As a next step, human primary nasal epithelial cells were successfully isolated from nasal polyps (Fig. 4A and B), and the ability of M. leprae (arrow) to interact with these cells was investigated by confocal microscopy (Fig. 4C). M. leprae was able to enter primary nasal epithelial cells after 2 h of interaction (21.2 ± 7 bacteria/300 cells) (Fig. 4D). Longer incubation times (6 h and 24 h) did not increase the number of associated bacteria (data not shown).
Fig 4.
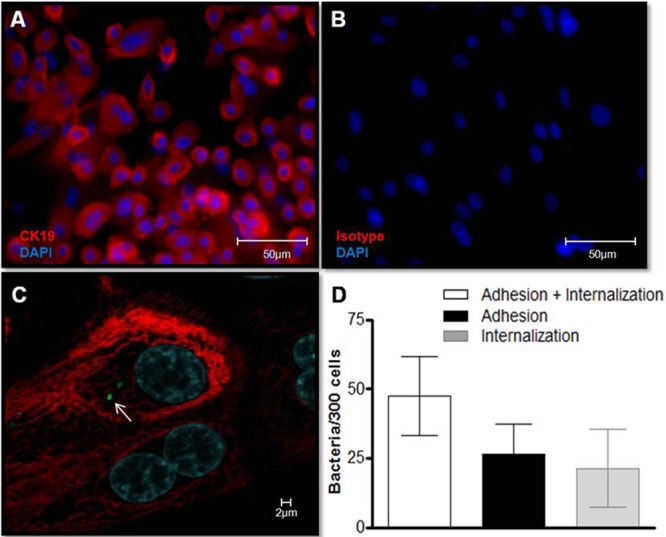
M. leprae enters human primary nasal epithelial cells. (A) Primary nasal epithelial cells were isolated from nasal polyps, and culture purity was determined by immunostaining with anti-cytokeratin-19 (CK-19) (red). Cell nuclei were labeled with DAPI (blue). (B) Isotype control. (C and D) Cells were infected with prelabeled PKH67 gamma-irradiated M. leprae at an MOI of 10 at 37°C for 2 h. (C) Representative confocal image of 4 independent experiments showing nasal primary cells infected with M. leprae (green; arrow). (D) The numbers of bacteria that interact with, adhere to, and enter primary nasal epithelial cells were determined. Data represent the means ± standard deviations of the results of 4 experiments performed in duplicate.
To further validate these in vitro findings, C57BL/6 mice were challenged intranasally with M. leprae to evaluate whether the bacteria interact in vivo with airway epithelial cells. As summarized in Table 1, only a few bacteria were observed in only a couple of animals at early postinfection time points. M. leprae was found in 2 of 6 mice in parabronchiolar and bronchiole lumens and also in terminal bronchioles at 4 h of infection and in 1 of 6 mice on day 4 of infection. Interestingly, some acid-fast bacteria (arrows) were observed inside macrophages and, apparently, in pulmonary epithelial cells (Fig. 5). At later time points, no acid-fast bacilli were found in the lungs. However, no mycobacteria were seen in nasal mucosa even at earlier time points and, in fact, no visible lesions were found in the nose or lungs at any time point. These results suggest that airway epithelial cells, especially lung cells, are susceptible to infection during in vivo airborne exposure to M. leprae.
Table 1.
Detection of M. leprae in the respiratory tract of C57BL/6 mice after intranasal challenge with 107 bacilli
| Time of infection | Total no. of mice | No. of mice infected with visible acid-fast bacilli | Infected tissues and cells (Ziehl-Nielssen staining) | H&E staining result |
|---|---|---|---|---|
| 4 h | 6 | 2 | Acid-fast bacilli in terminal bronchioles and circumjacent lung tissue; bacilli were observed inside epithelial cells and macrophages | No visible lesions |
| 4 days | 6 | 1 | Infected macrophages in lung tissue | No visible lesions |
| 10 days | 4 | 0 | No visible lesions | |
| 20 days | 4 | 0 | No visible lesions |
Fig 5.
M. leprae infects respiratory tract cells in vivo. C57BL/6 mice were intranasally challenged with M. leprae. Airway and lungs were monitored by histological examination for detection of bacteria and lesions. The images show terminal bronchiole and circumjacent lung tissue after 4 h of infection. Acid-fast bacilli (arrows) in luminal macrophages (A) and in shed epithelial cells (B) are shown. Bars, 10 μm (A) and 5 μm (B).
M. leprae internalization is a passive process dependent on host-cell cytoskeleton machinery.
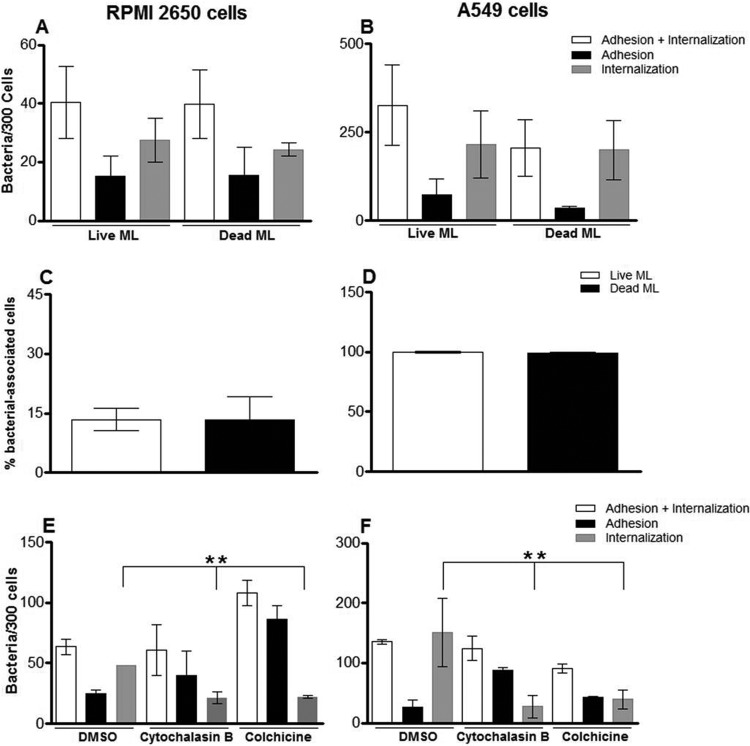
Since M. leprae is able to enter epithelial cells, which seems to occur during the early stages of airborne exposure to the pathogen, the mechanisms involved in the M. leprae entrance process were investigated. To determine whether M. leprae internalization in epithelial cells is an active or passive process, M. leprae was heat killed at 60°C for 30 min and then used to interact with nasal and alveolar cells. After 24 h of infection, the samples were analyzed by confocal microscopy and the numbers of binding and internalized bacteria in 300 cells were determined (Fig. 6). It was shown that M. leprae viability did not affect bacterial attachment and uptake, since the differences between live and dead bacteria were not statistically significant in these sets of cells (P = 0.3068 for RPMI 2650 cells [Fig. 6A] and P = 0.6267 for A549 cells [Fig. 6B]). In addition, the percentage of cells infected with live or dead M. leprae was calculated (Fig. 6C and D). However, no difference between the percentages of cells associated with live and with dead bacteria was found. In the context of RPMI 2650 cells, 12.5% ± 2.8% were infected with live M. leprae whereas 13.3% ± 5.8% showed internalized dead bacteria (P = 0.846) (Fig. 6C). In the context of A549 cells, practically all cells showed (either live or dead) internalized M. leprae (P = 0.5185) (Fig. 6D). These results indicate that M. leprae enters epithelial cells through a passive process that depends on its ability to adhere to these cells via the use of preexisting surface bacterial components.
Fig 6.
M. leprae enters airway epithelial cells in a passive cytoskeleton-dependent manner. (A to D) Live and heat-killed M. leprae interaction with RPMI 2650 (A and C) and A549 (B and D) cells for 24 h at 33°C. The bacteria were labeled with PKH26, and the numbers of bacteria interacting with the cells and the percentages of bacterium-associated cells were determined by fluorescence microscopy. (E and F) Effect of cytoskeleton inhibitors on M. leprae uptake by RPMI 2650 cells (E) and A549 cells (F). Cells were pretreated with DMSO (drug vehicle), colchicine, or cytochalasin B for 1 h and during the 24 h of the assay at 33°C. Data represent the means ± standard deviations of the results of 3 experiments performed in duplicate. **, P < 0.01.
The host-cell cytoskeletal involvement in the bacterial uptake process was then examined by treating these cells with the drugs cytochalasin B and colchicine. Cytochalasin B is a fungal metabolite that binds to actin, causing inhibition of polymerization and depolymerization (28). Likewise, colchicine is an alkaloid that completely inhibits microtubule self-assembly (29). These drugs have been extensively administered in an attempt to more fully understand the roles played by actin and tubulin in bacterial invasion (30–32). The effect of these drugs was observed at 24 h of infection since at that time point most of the bacteria would be located inside the cells. Our data showed that the number of M. leprae inside the cells decreased by half when the RPMI 2650 lineage was treated with cytochalasin B (Fig. 6E; RPMI 2650 + DMSO = 48 bacteria ± 2.7/300 cells and RPMI 2650 + cytochalasin B = 21.25 bacteria ± 2.5/300 cells [P = 0.002]). The same effect was also observed when the cells were treated with colchicine (Fig. 6E; RPMI 2650 + DMSO = 48 bacteria ± 2.7/300 cells and RPMI 2650 + colchicine = 22 bacteria ± 0.5/300 cells [P = 0.001]). In A549 cells, treatment with these inhibitors also decreased M. leprae entry (Fig. 6F; A549 + DMSO = 150.8 bacteria ± 28.8/300 cells and A549 ± cytochalasin B = 27.5 bacteria ± 9.2/300 cells [P = 0.001]; A549 + colchicine = 39.2 bacteria ± 7.7/300 cells [P = 0.001]). In summary, M. leprae adhesion was not affected by the inhibitors, showing that their use affected only bacterial internalization. Moreover, as monitored by trypan blue staining, no toxic effect of the inhibitors was observed (data not shown). These results are strong indicators that M. leprae enters nasal and alveolar epithelial cells in a microtubule- and microfilament-dependent manner.
The intracellular environment of nasal and alveolar epithelial cells supports M. leprae survival.
Another important issue to investigate in the context of M. leprae interaction with epithelial cells was the capacity of these cells to support bacterial survival. Cultures were infected with live M. leprae, and bacterial viability was monitored for up to 10 days via the use of a Live/Dead BacLight bacterial viability kit. The viability of intracellular bacteria was compared with that of bacteria recovered from the washing steps (extracellular M. leprae) and with that of bacteria kept in cell-free medium (axenic medium). These assays were performed at 33°C since this temperature has been shown to be optimal for in vitro M. leprae survival (19).
Intracellular M. leprae in RPMI 2650 cells showed significantly higher viability on day 10 of infection than the M. leprae kept in a cell-free medium (66.4% ± 2.5% viability versus 33.76% ± 3.9%; P = 0.0004) (Fig. 7A). The viability of extracellular M. leprae was the same as that of M. leprae incubated in a cell-free medium. In A549 cells, on days 8 and 10, the viability of intracellular M. leprae was significantly higher than the viability of extracellular bacteria (77.29% ± 4.2% viability versus 52% ± 4.8% [P = 0.017] and 77.48% ± 4.87% versus 47.2% ± 4.6% [P = 0.0014], respectively) (Fig. 7B). Again, similar viability results were observed when extracellular M. leprae and M. leprae kept in axenic medium were compared. No differences were observed at earlier time points of incubation. Moreover, when the viability of intracellular bacteria on day 10 was compared with the viability on day 1 of infection, no significant difference (P = 0.1) was found, suggesting that the epithelial cells were able to maintain M. leprae viability for at least 10 days.
Fig 7.
M. leprae survives inside epithelial cells. Intra- and extracellular M. leprae viability in RPMI 2650 and A549 cells was determined by the use of a Live/Dead kit. (A) RPMI 2650 cells at 33°C. (B) A549 cells at 33°C. (C) A549 cells at 37°C. Intracellular M. leprae bacilli (black bars) were recovered by lysing buffer, while extracellular bacteria (gray bars) were recovered after washing steps. In parallel, M. leprae bacilli were incubated in cell-free medium (white bar). Bacteria were stained by the use of a Live/Dead BacLight Bacterial Viability kit and enumerated by direct counting via fluorescence microscopy at a magnification of ×1,000. The data represent the mean values ± SD of the results of five experiments with duplicate samples from each one. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
These data suggest that the airway epithelial cells may act as a reservoir and/or portal of entry for M. leprae. However, the normal temperature in human lungs is 37°C, an unfavorable condition for M. leprae survival (19). So, to validate the pulmonary epithelial cells as a colonizable tissue during M. leprae infection, identical viability assays with A549 cells were conducted at 37°C instead of 33°C (Fig. 7C). Interestingly, the viability of both intracellular and extracellular M. leprae bacilli was close to zero on day 10 of infection at 37°C. However, about 47.7% ± 9.5% of intracellular bacteria remained viable after 4 days of incubation, in contrast to the viability of 7.7% ± 4.9% (P = 0.02) displayed by extracellular bacteria under the same conditions. Bacteria in a cell-free medium also presented a faster decrease in viability than intracellular M. leprae.
Altogether, our data suggest that nasal epithelial cells may constitute an appropriate intracellular niche for M. leprae survival whereas pulmonary epithelial cells may act as a portal of entry but also as a bacterial reservoir.
The M. leprae cell-surface proteome reveals an array of potential adhesin candidates.
Bacteria express adhesive molecules on their surfaces that promote interaction with host-cell receptors or with soluble macromolecules and their subsequent attachment to and successful colonization of the host. Due to their critical role in infection, the molecular characterization of adhesins has been a successful strategy in designing new therapeutic tools and/or vaccine candidates for the control of several infectious diseases (33).
So, to identify the potential M. leprae adhesins involved in the interaction with nasal and alveolar epithelial cells, a cell-surface proteomic analysis of M. leprae isolated from nude mice footpads was performed. Both crude and NaOH-treated M. leprae bacilli were submitted to analysis. A total of 279 surface proteins were identified. Among these proteins, 85 were found solely in crude M. leprae, 79 were detected only in the NaOH-treated bacteria, and 115 were identified in both samples (Fig. 8). The identified proteins are listed in Tables S1, S2, and S3 in the supplemental material.
Fig 8.
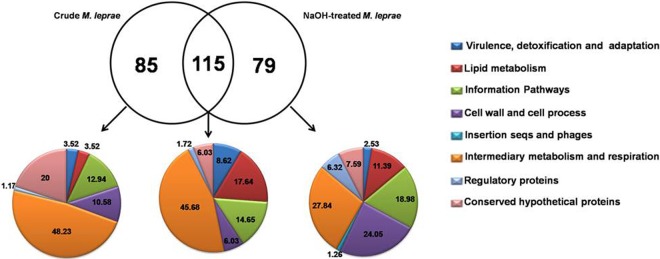
Functional grouping and intercross of surface-exposed proteins identified in crude and NaOH-treated M. leprae. A Venn diagram illustrating overlap between surface proteins identified in NaOH-treated and crude M. leprae is shown. Proteins identified exclusively in the NaOH-treated bacilli, only in the crude bacilli, or simultaneously found in both samples were functionally classified according to the Leproma database (http://genolist.pasteur.fr/Leproma/). Functional categories are expressed in percentages. seqs, sequences.
Seven of the identified proteins, ML0458c, ML1553, ML1795, ML2088c, ML2346, ML2347 and ML2498, do not have homologs in M. tuberculosis (34). Two proteins not previously described in the M. leprae proteomic analysis were identified: a possible oxidoreductase (ML2276c; ml2276c) and a conserved hypothetical protein (ML2309; ml2309) (see Table S3 in the supplemental material). Figure 8 also details the functional categories of these proteins. Note the greater enrichment of proteins categorized as participants in the cell wall and cell processes in addition to lipid metabolism as opposed to those classified as being involved in the intermediary metabolism and respiration observed in NaOH-treated versus crude bacteria.
Table 2 lists 11 M. leprae surface-exposed proteins previously identified as potential mycobacterial adhesins. Most (8 proteins) were identified in both crude and NaOH-treated bacteria. Of note was the confirmed presence of 2 well-known mycobacterial adhesins, Hlp (ML1683c; hupB) and HBHA (ML2454c; hbhA), on the M. leprae surface. Another interesting finding was the presence of mammalian cell entry 1A (mce1A) (ML2589; mce1A) on the surface of M. leprae. In contrast to the wild-type phenotype, recombinant Escherichia coli expressing the M. tuberculosis mce1A protein has been shown to invade epithelial cells (35). Furthermore, previous studies with recombinant mce1A of M. leprae showed that this protein increased polystyrene-bead interaction with nasal and bronchial epithelial cells but not with dermal keratinocytes, reinforcing the idea that the mce1A gene product has the capacity to mediate M. leprae entry into respiratory epithelial cells (36). Superoxide dismutase (ML0072c; sodA) has also been suggested to be potentially able to interact with human respiratory epithelial cells in the context of M. avium (37).
Table 2.
Putative surface-exposed adhesins identified in crude and NaOH-treated M. leprae bacillia
| Leproma ID | Gene | Protein description | FC |
|---|---|---|---|
| ML0072c | sodA | Probable superoxide dismutase, SodA | 0 |
| ML0317 | groEL2 | Heat shock protein Hsp65 family | 0 |
| ML0381 | groEL1 | 60-kDa chaperonin 1 GroEL1 | 0 |
| ML2589 | mce1A | MCE family protein | 0 |
| ML2496c | dnaK | Probable chaperone protein DnaK | 0 |
| ML2028 | fbpB | Secreted antigen 85B | 1 |
| ML1683c | hupB | Probable DNA-binding protein | 2 |
| ML1877c | tuf | Iron-regulated elongation factor | 2 |
| ML2454c | hbhA | Possible hemagglutinin | 3 |
| ML2069 | glcB | Malate synthase | 7 |
| ML0255 | eno | Probable enolase | 7 |
The leproma identification (ID) number and the functional category (FC) data were obtained from the Leproma World-Wide Web Server (http://genolist.pasteur.fr/Leproma/).
Malate synthase (ML2069), an enzyme involved in the glyoxylate pathway, was also found on the M. leprae surface. This protein has been shown to localize on the M. tuberculosis cell surface and to bind laminin acting as an adhesin (38–41). Antigen 85B (ML2028), also detected on the M. leprae surface in confirmation of previous findings (42, 43), has been described as a fibronectin-binding protein (FBP) (44, 45). A study using the nasal epithelial cell RPMI 2650 lineage showed that treatment with fibronectin increased M. leprae adherence to this cell. In addition, the β1 chain of integrins is also implicated in the binding of M. leprae to nasal epithelial cells (46).
Several chaperones with potential adhesin properties were also found on the M. leprae surface (Table 2). Besides binding to human monocytes, the GroEL2 homologue was found on both the cell wall and surface of M. avium (47). DnaK and GroEL1 have been described as being involved in mediating M. bovis BCG adhesion to dendritic and macrophages cells via interaction with a C-type lectin known as DC-SIGN (48). DnaK and GroEL2 have also been shown to be present within the M. tuberculosis capsule (49), but only GroEL2 was found to bind to macrophages. In another study, it was shown that M. tuberculosis DnaK, Ag85B, and the iron-regulated elongation factor Tu (Ef-Tu) were able to bind and activate plasminogen (50). Plasminogen can be converted into plasmin, which, in turn, is capable of degrading the extracellular matrix and basal membrane components. Many bacteria are able to bind and convert plasminogen into plasmin, thereby facilitating pathogen dissemination (51). Finally, ML0255, a probable enolase, was found on the M. leprae surface and was included as a potential adhesion, based on recent findings indicating that Plasmodium spp. are able to bind plasminogen (52).
M. leprae Hlp and HBHA can mediate mycobacterial entry into epithelial cells.
Two mycobacterial proteins, HBHA and Hlp, have been intensely studied and shown to mediate bacterial adhesion to epithelial cells (13, 20, 53). Since we confirmed by proteomic data that HBHA and Hlp are surface-exposed proteins in M. leprae, we investigated whether recombinant M. leprae proteins would be able to bind to A549 and RPMI 2650 cells.
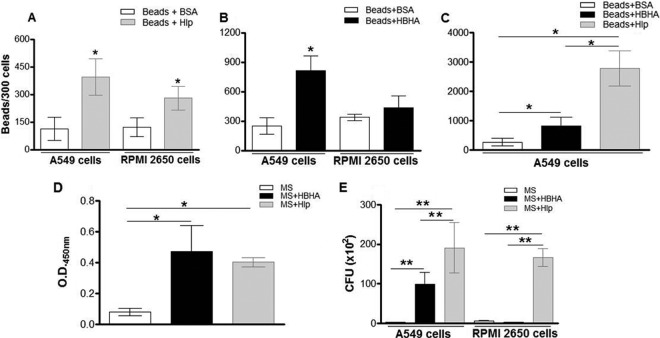
As a first step, assays with fluorescent beads coated with the recombinant forms of Hlp (Hlp-Beads) and HBHA (HBHA-Beads) were done. As negative controls, the beads were treated with BSA (BSA-Beads). As shown in Fig. 9A, the capacity of Hlp-Beads to bind to nasal epithelial RPMI 2650 cells was significantly higher than the binding capacity of BSA-Beads (280.0 ± 26.27 Hlp-beads versus 120.9 ± 17.76 BSA-Beads). Similarly, while only 113.3 ± 31.18 BSA-Beads were found to be attached to A549 cells, 394.8 ± 49.15 Hlp-Beads were found in association with these cells in identical assays. The differences in the numbers of Hlp-Beads attached to RPMI 2650 and A549 cells were not statistically significant (P = 0.89). Independent assays were then performed with HBHA-coated beads. Interestingly, HBHA was able to enhance bead interaction only with A549 cells (813 ± 76.05 HBHA-Beads versus 248.3 ± 48.7 BSA-Beads) and not with RPMI 2650 cells (432.5 ± 60 HBHA-Beads versus 336 ± 16.6 BSA-Beads) (Fig. 9B). Moreover, when assays were run to compare the adhesive properties of HBHA- and Hlp-coated beads in relation to A549 cells simultaneously, Hlp was significantly more adhesive than HBHA (Fig. 9C).
Fig 9.
M. leprae recombinant Hlp and HBHA mediate the mycobacterial attachment to airway epithelial cells. (A and B) Fluorescent beads coated with Hlp (A), with HBHA (B), or with BSA (negative control) were incubated with RPMI 2650 and A549 cells for 1 h at 37°C. Cells were then washed and fixed. The numbers of beads associated with these cells were counted in a fluorescence microscope in a set of 300 cells using a magnification of ×400. (C) Fluorescent beads coated with Hlp, HBHA, or BSA were incubated simultaneously with A549 cells for 1 h at 37°C. (D) M. smegmatis (MS) was pretreated or not with Hlp (MS+Hlp) or HBHA (MS+HBHA), and an ELISA was performed with anti-Hlp or anti-HBHA, respectively, in order to monitor the binding of these proteins to the bacterial surface. (E) MS and Hlp or MS and HBHA were used to infect epithelial cells for 2 h at 37°C. Internalized bacteria were released with lysis buffer for CFU counting. The data represent the mean values ± SD of the results of at least three experiments performed in duplicate. *, P < 0.05; **, P < 0.01.
To confirm these results, similar assays were then conducted with Mycobacterium smegmatis precoated with Hlp (MS + Hlp) or with HBHA (MS + HBHA). The binding of Hlp and HBHA to the M. smegmatis surface was confirmed by an ELISA performed with specific antibodies against these proteins (Fig. 9D). Subsequently, the levels of binding of precoated and control bacteria to RPMI 2650 and A549 cells at 2 h were determined (Fig. 9E). As expected, MS + Hlp bacteria showed an enhanced ability to bind to nasal epithelial cells in contrast to the untreated bacteria (MS = 5.8 ± 0.6 × 102 bacteria; MS + Hlp = 156.7 ± 21.3 × 102 bacteria [P = 0.0008]) (Fig. 9E). The same was observed in the context of A549 cells (MS = 2.6 ± 0.4 × 102 bacteria; MS + Hlp = 190.3 ± 8 × 102 bacteria [P = 0.0018]) (Fig. 9E). Regarding HBHA, precoated bacteria also showed a higher adhesive capacity with respect to A549 (MS = 2.6 ± 0.4 × 102 bacteria; MS + HBHA = 99.0 ± 10 × 102 bacteria [P = 0.0014]). However, in similarity to the results obtained with coated beads, HBHA-coated M. smegmatis did not show any enhanced adherence capacity compared to control bacteria in the context of RPMI 2650 cells (MS = 5.8 ± 0.6 × 102 bacteria; MS + HBHA = 2.2 ± 0.1 × 102 bacteria [P = 0.389]) (Fig. 9E). In addition, as previously shown with beads, Hlp displayed a higher binding capacity to A549 cells than HBHA, confirming that Hlp seems to interact more avidly with A549 cells than HBHA.
Collectively, these results show that Hlp and HBHA may act as adhesins during M. leprae-mucosal airway interaction. Additionally, while Hlp may mediate interaction with both nasal and alveolar epithelia, HBHA seems to be restricted to mediating the bacterial attachment to alveolar epithelial cells.
DISCUSSION
Studies regarding the interaction of M. leprae with airway epithelial cells, although rare to date, are of great interest since they could shed more light on the critical events occurring during the early stages of M. leprae-human host interaction.
In the present study, in vitro assays were performed to investigate the capacity of M. leprae to both infect and survive within nasal and alveolar epithelial cells. Moreover, the capacity of M. leprae to colonize the respiratory tract after intranasal infection of mice was analyzed. For the first time, a proteomic analysis of nude mouse-derived M. leprae was carried out using a biotinylation approach to selectively identify surface-exposed molecules for the purpose of mapping potential adhesin candidates.
In our view, the most significant findings of our study are the following: (i) M. leprae was able to invade both nasal and alveolar epithelial cell lineages, but alveolar cells were infected more efficiently; (ii) ex vivo and in vivo studies, respectively, performed with primary human nasal epithelial cells and intranasal-infected immunocompetent mice confirmed the capacity of M. leprae to interact with respiratory tract cells during the early stages of airborne exposure; (iii) epithelial cells were able to sustain bacterial viability for at least 10 days postinfection; and (iv) the proteome of the M. leprae surface was defined, revealing a complex profile of proteins, including a group of potential adhesin candidates such as HBHA and Hlp, which may play a major role in the interaction of the pathogen with the respiratory tract.
Although M. leprae was able to interact with the nasal RPMI 2650 cell lineage as well as primary nasal cells, the number of bacteria capable of binding to and invading these cells was very small compared to the number of those binding to A549 cells. While this discrepancy is not due to differences in the intrinsic endocytic activities of these cells, which were shown to be similar, there is a suggestion that M. leprae itself may display a higher capacity to enter the alveolar epithelial cells than to enter their nasal counterparts. Likewise, the kinetics involved in the invasion of nasal cells appears to be slower than that of the A549 cells, especially in view of the fact that an increased bacterial association during the first 6 h of interaction did not materialize and a decidedly clear increase in A549 at early time points did occur. Nonetheless, M. leprae numbers inside RPMI 2650 cells doubled at 24 h of infection, suggesting a possible modulation of cell receptor expression. The capacity of M. leprae to interact with RPMI 2650 cells has been previously described (46). Interestingly, in a previous study, the percentage of M. leprae-infected A549 cells was similar to the percentage of cells infected with M. tuberculosis (54), suggesting that M. leprae is as efficient as M. tuberculosis at binding to and entering these cells. In addition, a previous study suggests that the M. tuberculosis binding and invasion capacities in alveolar epithelial cells might reflect mycobacterial virulence (55). As seen with M. tuberculosis, intracellular M. leprae bacteria were observed both as isolated individual cells and in clusters in membrane compartment structures, suggesting that, in A549 cells, the bacteria are located inside vesicles.
The present study also showed that heat-killed M. leprae, similarly to live bacteria, was able to bind and invade nasal and alveolar epithelial cells. This suggests that M. leprae entry into epithelial cells does not require newly synthesized bacterial products but that the molecules already present on the surface are sufficient. Interestingly, previous studies have shown that M. avium invades epithelial cells through mechanisms that depend on bacterial viability (56).
Furthermore, similarly to what has been observed in the context of M. tuberculosis (54), it was shown that M. leprae enters epithelial cells in microfilament- and microtubule-dependent manners. This differs from some invasive bacterial entry mechanisms in the epithelium, in which the actin cytoskeleton, but not the microtubule network, has been shown to play a major role (30, 57, 58), raising the speculation that M. leprae displays at least two different mechanisms of epithelium invasion.
Our data also show that the bacilli are able to survive within nasal epithelial cells, suggesting that these cells may constitute an important niche and source of M. leprae infection. Using a mouse model, the capacity of M. leprae to interact with respiratory tract cells during the early stages of airborne exposure was confirmed in our study. However, this activity was observed only during early time points of infection when acid-fast bacilli were found in the lungs but not in the nose.
This observation is also in correlation with the higher affinity of M. leprae for alveolar epithelial cells than for nasal epithelial cells observed in our in vitro assays. Similar results were detected after the airborne infection of the thymectomized-irradiated CBA mouse strain with M. leprae (10). In this study, bacilli were undetectable in the nasal homogenates, although bacteria were found in the lungs of 100% of the mice immediately after exposure to M. leprae aerosols. After 24 months of infection, one-third of the mice developed disseminated infection, implying that the lungs are the primary portal of entry of M. leprae after airborne exposure (10). Indeed, based on our survival assays, it is speculated that, due to the 37°C temperature of human lungs, M. leprae can survive there for a relatively short period of time and that the alveolar epithelial cells and macrophages may primarily serve as a portal of entry and not as a residing niche for M. leprae multiplication. This hypothesis takes into account the absence of lung involvement in clinically established lepromatous leprosy.
In contrast, histopathological studies of the septum and turbinates of lepromatous leprosy patients have amply confirmed the density of bacilli in these tissues, the nose being the major escape route for the bacilli (59). As demonstrated in our survival experiments, the nasal mucosa temperature of 33°C favors M. leprae survival. Even so, the evidence has so far sustained the idea that nasal infection arises from the systemic spread of M. leprae instead of from the small numbers of bacilli retained during airborne infection (10). Nonetheless, the nose cannot be excluded as a possible portal of entry, and it is noteworthy that several reports using the mouse model favor this possibility (60, 61).
The present study also made an important contribution in defining the surface proteomics of M. leprae. An extensive M. leprae study in proteomics has included the mapping of cell-wall-associated proteins (62), but all were performed with preparations of M. leprae isolated from frozen tissues of infected armadillos, which typically display low viability. Therefore, a new proteomic study focusing on surface-exposed proteins became imperative in order to identify molecules with potentially relevant roles in bacterial pathogenesis located at the interface of M. leprae and the human being.
Our idea was to identify the specific adhesins and molecules involved in the entry of M. leprae in airway epithelial cells. Investigations were carried out with high-quality, freshly isolated M. leprae bacteria from athymic nude-mouse footpads having over 90% viability in conjunction with a biotinylation approach that had been successfully applied to selective labeling and purification of surface-exposed proteins of M. avium (24). For comparison, NaOH-treated and crude M. leprae preparations were analyzed. The former was used to remove contaminant proteins derived from mouse tissues and the cytosol of lysed bacteria (17). Moreover, NaOH treatment might favor the biotinylation of less abundant cell-surface proteins, which are sometimes masked by contaminant proteins.
Indeed, in NaOH-treated versus crude bacteria, our results showed an enrichment of the proteins involved in cell wall and cell processes and lipid metabolism over those involved in intermediary metabolism and respiration. Moreover, most proteins on the surface of NaOH-treated bacteria have been previously described in the cell wall fraction of M. leprae, while most proteins in crude bacteria have been previously described in the cytosolic fraction (see Tables S1 and S3 in the supplemental material) (62, 63). In addition, a small set of proteins related to cell wall processes and lipid metabolism was identified only in crude bacteria, suggesting that these proteins are somehow affected and lost during treatment with NaOH.
The proteomic analysis presented here revealed that the M. leprae surface is composed of a highly complex mixture of proteins. Many of these are cytosolic proteins with no predictive signal peptide or cell-wall anchoring motif (see Tables S1, S2, and S3 in the supplemental material). These results are in agreement with a previous characterization of the M. avium surface proteome that also revealed an abundance of cytosolic proteins represented on the bacterial surface (24). In addition, the cell surface proteome of M. smegmatis likewise revealed a solid set of cytosolic proteins such as ribosomal proteins, the Ef-Tu, and glyceraldehyde-3-phosphate dehydrogenase (64).
Glycolytic and other cytosolic enzymes have also been shown to localize on the surface of several other pathogens, and postulations regarding possible alternative functions of these proteins at this location have been made. As such, as listed in Table 2, several enzymes found on the M. leprae surface have been shown to mediate the pathogen colonization of host cells and facilitate the establishment of infection (65–70).
Among the many identified adhesin candidates on the surface of M. leprae, the potential roles of two of them, i.e., Hlp and HBHA, in bacterium-epithelial cell interactions were further explored. Hlp and HBHA are well known as extracellular matrix-binding proteins. While HBHA binds heparan sulfate (13, 20), Hlp has been described as a multifunctional adhesin able to interact with laminin, hyaluronic acid, heparan sulfate, and collagen (14, 53, 71–73). Previous studies have shown the in vivo expression of both proteins by M. leprae (62, 71). A previous study also showed the presence of Hlp in armadillo-derived M. leprae by immunoelectron microscopy (14). But, to our knowledge, this is the first time they have been shown to be surface exposed in freshly isolated M. leprae from athymic nude-mice as originally speculated.
The present study also confirmed that Hlp and HBHA M. leprae recombinant homologs are able to mediate the interaction of beads and mycobacteria with airway epithelial cells, as had been previously shown in the context of M. bovis BCG (53, 74). The higher capacity of Hlp but not HBHA to bind both A549 and RPMI 2650 cells may be explained by the capacity of Hlp to interact with a larger number of host-cell proteins than HBHA. It is also possible to infer that RPMI 2650 cells express a small amount of heparan sulfate on the cell surface, justifying the inability of the HBHA protein to interact with these cells.
In conclusion, based on the considerations mentioned above, we hypothesize that during the subclinical infection and early stages of leprosy, respiratory epithelial cells may constitute a niche for bacterial retention and survival and a potential portal of entry for M. leprae. Although our data indicate that Hlp and HBHA can potentially mediate the M. leprae attachment to airway epithelial cells, the complex proteome of the bacterial surface demonstrated here suggests that M. leprae displays an armamentarium of adhesins involved in its interaction with epithelial cells. In reality, such cell-surface glycolipids as PGL-I have been seen to play a role as adhesins (75) but may also contribute to M. leprae-epithelial cell interaction. Altogether, our data suggest that the airway epithelium plays an important role as a primary site of M. leprae infection in humans. A clearer understanding of M. leprae interaction with these cells could contribute to the design of more effective preventive tools for leprosy control.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by CNPq/Brazil (individual grants to M.C.V.P.; C.A.M.S. was a recipient of fellowships from CNPq and Capes/Fulbright). This work was also supported by the Foundation for Microbiology and Infectious Diseases (L.E.B.).
We are grateful to James Krahenbuhl for providing viable M. leprae and Anne-Marie Girard of the Confocal Microscopy Facility, Center for Genome Research and Biocomputing, Oregon State University, Corvallis, OR, for furnishing the assisted images. Mass spectrometric analysis was performed by the Oregon Health and Science University, Proteomics Shared Resources, with partial support from the NIAID (core grants no. P30EY01572 and P30CA069533). We also thank Thais Almeida and Rodrigo Avellar for providing the nasal polyps, Ashok Reddy of Proteomics Shared Resources for his technical support, and Judy Grevan for editing the text.
Footnotes
Published ahead of print 13 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00147-13.
REFERENCES
- 1. WHO 2012. Global leprosy situation, August. Wkly. Epidemiol. Rec. 87:317–328 [PubMed] [Google Scholar]
- 2. Scollard DM, Adams LB, Gillis TP, Krahenbuhl JL, Truman RW, Williams DL. 2006. The continuing challenges of leprosy. Clin. Microbiol. Rev. 19:338–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McDougall AC, Rees RJ, Weddell AG, Kanan MW. 1975. The histopathology of lepromatous leprosy in the nose. J. Pathol. 115:215–226 [DOI] [PubMed] [Google Scholar]
- 4. Job CK. 1990. Nasal mucosa and abraded skin are the two routes of entry of M. leprae. Star 49:1 [Google Scholar]
- 5. Job CK, Jayakumar J, Kearney M, Gillis TP. 2008. Transmission of leprosy: a study of skin and nasal secretions of household contacts of leprosy patients using PCR. Am. J. Trop. Med. Hyg. 78:518–521 [PubMed] [Google Scholar]
- 6. Suneetha S, Arunthathi S, Job A, Date A, Kurian N, Chacko CJ. 1998. Histological studies in primary neuritic leprosy: changes in nasal mucosa. Lepr. Rev. 69:358–366 [DOI] [PubMed] [Google Scholar]
- 7. Klatser PR, van Beers S, Madjid B, Day B, de Wit MY. 1993. Detection of Mycobacterium leprae nasal carriers in populations for which leprosy is endemic. J. Clin. Microbiol. 31:2947–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chan PC, Reyes LM, Demerre-Lopez B, Gonzaga EM, De lós Santos MFA, Gillis TP. 1997. Leprosy infection and disease in the national capital region. Philippine J. Microbiol. Infect. Dis. 26:159–162 [Google Scholar]
- 9. Patrocínio LG, Goulart IM, Goulart LR, Patrocínio JA, Ferreira FR, Fleury RN. 2005. Detection of Mycobacterium leprae in nasal mucosa biopsies by the polymerase chain reaction. FEMS Immunol. Med. Microbiol. 44:311–316 [DOI] [PubMed] [Google Scholar]
- 10. Rees RJW, McDougall AC. 1977. Airborne infection with M. leprae in mice. J. Med. Microbiol. 10:63–68 [DOI] [PubMed] [Google Scholar]
- 11. Davey TF, Rees RJ. 1974. The nasal discharge in leprosy: clinical and bacteriological aspects. Lepr. Rev. 45:121–134 [DOI] [PubMed] [Google Scholar]
- 12. Mayer AK, Dalpke AH. 2007. Regulation of local immunity by airway epithelial cells. Arch. Immunol. Ther. Exp. (Warsz) 55:353–362 [DOI] [PubMed] [Google Scholar]
- 13. Menozzi FD, Rouse JH, Alavi M, Sharp ML, Muller J, Bischofffl R, Brennan MJ, Locht C. 1996. Identification of a heparin-binding hemagglutinin present in mycobacteria. J. Exp. Med. 184:993–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shimoji Y, Ng V, Matsumura K, Fischetti VA, Rambukkana A. 1999. A 21-kDa surface protein of Mycobacterium leprae binds peripheral nerve laminin-2 and mediates Schwann cell invasion. Proc. Natl. Acad. Sci. U. S. A. 96:9857–9862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chevillard M, Hinnrasky J, Zahm JM, Plotkowski MC, Puchelle E. 1991. Proliferation, differentiation and ciliary beating of human respiratory ciliated cells in primary culture. Cell Tissue Res. 264:49–55 [DOI] [PubMed] [Google Scholar]
- 16. Hicks W, Jr, Ward R, Edelstein D, Hall L, III, Albino A, Hard R, Asch B. 1995. Cytokeratin expression in human respiratory epithelium of nasal polyps and turbinates. Cell Biol. Int. 19:301–306 [DOI] [PubMed] [Google Scholar]
- 17. Lahiri R, Randwana B, Krahenbuhl J. 2005. Effects of purification and fluorescent staining on viability of Mycobacterium leprae. Int. J. Lepr. Other Mycobact Dis. 73:194–202 [PubMed] [Google Scholar]
- 18. Shepard CC, McRae DHA. 1968. Method for counting acid-fast bacteria. Int. J. Lepr. Other Mycobact. Dis. 36:78–82 [PubMed] [Google Scholar]
- 19. Lahiri R, Randwana B, Krahenbuhl J. 2005. Application of a viability-staining method for Mycobacterium leprae derived from the athymic (nu/nu) mouse foot pad. J. Med. Microbiol. 54:235–242 [DOI] [PubMed] [Google Scholar]
- 20. de Lima CS, Marques MA, Debrie AS, Almeida EC, Silva CA, Brennan PJ, Sarno EN, Menozzi FD, Pessolani MC. 2009. Heparin-binding hemagglutinin (HBHA) of Mycobacterium leprae is expressed during infection and enhances bacterial adherence to epithelial cells. FEMS Microbiol. Lett. 292:162–169 [DOI] [PubMed] [Google Scholar]
- 21. de Melo Marques MA, Mahapatra S, Nandan D, Dick T, Sarno EN, Brennan PJ, Vidal Pessolani MC. 2000. Bacterial and host derived cationic proteins bind alpha2-laminins and enhance Mycobacterium leprae attachment to human Schwann cells. Microbes Infect. 2:1407–1417 [DOI] [PubMed] [Google Scholar]
- 22. Preece A. 1972. A manual for histologic technicians, 3rd ed Little, Brown & Co., Boston, MA [Google Scholar]
- 23. Kim SY, Goodman JR, Petrofsky M, Bermudez LE. 1998. Mycobacterium avium infection of gut mucosa in mice associated with late inflammatory response and intestinal cell necrosis. J. Med. Microbiol. 47:725–731 [DOI] [PubMed] [Google Scholar]
- 24. McNamara M, Tzeng SC, Maier C, Zhang L, Bermudez LE. 2012. Surface proteome of “Mycobacterium avium subsp. hominissuis” during the early stages of macrophage infection. Infect. Immun. 80:1868–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Elias JE, Gygi SP. 2007. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4:207–214 [DOI] [PubMed] [Google Scholar]
- 26. Keller A, Nesvizhskii AI, Kolker E, Aebersold R. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74:5383–5392 [DOI] [PubMed] [Google Scholar]
- 27. Wilmarth PA, Riviere MA, David LL. 2009. Techniques for accurate protein identification in shotgun proteomic studies of human, mouse, bovine, and chicken lenses. J. Ocul. Biol. Dis. Infor. 2:223–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cooper JA. 1987. Effects of cytochalasin and phalloidin on actin. J. Cell Biol. 105:1473–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Borisy GG, Taylor EW. 1967. The mechanism of action of colchicine. Colchicine binding to sea urchin eggs and the mitotic apparatus. J. Cell Biol. 34:535–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Finlay BB, Falkow S. 1988. Comparison of the invasion strategies used by S. choleraesuis, Shigella flexneri, and Yersinia enterocolitica to enter cultured animal cells: endosome acidification is not required for bacterial invasion or intracellular replication. Biochimie 70:1089–1099 [DOI] [PubMed] [Google Scholar]
- 31. Moulder JW. 1985. Comparative biology of intracellular parasitism. Microbiol. Rev. 49:298–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fleiszig SMJ, Zaidi TS, Pier GB. 1995. Pseudomonas aeruginosa invasion of and multiplication within corneal epithelial cells in vitro. Infect. Immun. 63:4072–4077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kline KA, Fälker S, Dahlberg S, Normark S, Henriques-Normark B. 2009. Bacterial adhesins in host-microbe interactions. Cell Host Microbe 5:580–592 [DOI] [PubMed] [Google Scholar]
- 34. Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, III, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544 [DOI] [PubMed] [Google Scholar]
- 35. Arruda S, Bomfim G, Knights R, Huima-Byron T, Riley LW. 1993. Cloning of an M. tuberculosis DNA fragment associated with entry and survival inside cells. Science 261:1454–1457 [DOI] [PubMed] [Google Scholar]
- 36. Sato N, Fujimura T, Masuzawua M, Yogi Y, Kanoh M, Riley LW, Katsuoka K. 2007. Recombinant Mycobacterium leprae protein associated with entry into mammalian cells of respiratory and skin components. J. Dermatol. Sci. 46:101–110 [DOI] [PubMed] [Google Scholar]
- 37. Reddy VM, Kumar B. 2000. Interaction of Mycobacterium avium complex with human respiratory epithelial cells. J. Infect. Dis. 181:1189–1193 [DOI] [PubMed] [Google Scholar]
- 38. Laal S, Samanich KM, Sonnenberg MG, Zolla-Pazner S, Phadtare JM, Belisle JT. 1997. Human humoral responses to antigens of Mycobacterium tuberculosis: immunodominance of high-molecular-mass antigens. Clin. Diagn. Lab. Immunol. 4:49–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sonnenberg MG, Belisle JT. 1997. Definition of Mycobacterium tuberculosis culture filtrate proteins by two-dimensional polyacrylamide gel electrophoresis, N-terminal amino acid sequencing and electrospray mass spectrometry. Infect. Immun. 65:4515–4524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Samanich K, Belisle JT, Laal S. 2001. Homogeneity of antibody responses in tuberculosis patients. Infect. Immun. 69:4600–4609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kinhikar AG, Vargas D, Li H, Mahaffey SB, Hinds L, Belisle JT, Laal S. 2006. Mycobacterium tuberculosis malate synthase is a laminin-binding adhesin. Mol. Microbiol. 60:999–1013 [DOI] [PubMed] [Google Scholar]
- 42. Rambukkana A, Das PK, Burggraaf JD, Faber WR, Teeling P, Krieg S, Thole JE, Harboe M. 1992. Identification and characterization of epitopes shared between the mycobacterial 65-kilodalton heat shock protein and the actively secreted antigen 85 complex: their in situ expression on the cell wall surface of Mycobacterium leprae. Infect. Immun. 60:4517–4527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pessolani MC, Brennan PJ. 1992. Mycobacterium leprae produces extracellular homologs of the antigen 85 complex. Infect. Immun. 60:4452–4459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Abou-Zeid C, Ratliff TL, Wiker HG, Harboe M, Bennedsen J, Rook GA. 1988. Characterization of fibronectin-binding antigens released by Mycobacterium tuberculosis and Mycobacterium bovis BCG. Infect. Immun. 56:3046–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kitaura H, Ohara N, Naito M, Kobayashi K, Yamada T. 2000. Fibronectin-binding proteins secreted by Mycobacterium avium. APMIS 108:558–564 [DOI] [PubMed] [Google Scholar]
- 46. Byrd SR, Gelber R, Bermudez LE. 1993. Roles of soluble fibronectin and β-1 integrin receptors in the binding of Mycobacterium leprae to nasal epithelial cells. Clinic Immunol. Immunopathol. 69:266–271 [DOI] [PubMed] [Google Scholar]
- 47. Rao SP, Ogata K, Morris SL, Catanzaro A. 1994. Identification of a 68 kd surface antigen of Mycobacterium avium that binds to human macrophages. J. Lab. Clin. Med. 123:526–535 [PubMed] [Google Scholar]
- 48. Carroll MV, Sim RB, Bigi F, Jäkel A, Antrobus R, Mitchell DA. 2010. Identification of four novel DC-SIGN ligands on Mycobacterium bovis BCG. Protein Cell 1:859–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hickey TBM, Ziltener HJ, Speert DP, Stokes RW. 2010. Mycobacterium tuberculosis employs Cpn60.2 as an adhesin that binds CD43 on the macrophage surface. Cell. Microbiol. 12:1634–1647 [DOI] [PubMed] [Google Scholar]
- 50. Xolalpa W, Vallecillo AJ, Lara M, Mendoza-Hernandez G, Comini M, Spallek R, Singh M, Espitia C. 2007. Identification of novel bacterial plasminogen-binding proteins in the human pathogen Mycobacterium tuberculosis. Proteomics 7:3332–3341 [DOI] [PubMed] [Google Scholar]
- 51. Lähteenmäki K, Edelman S, Korhonen TK. 2005. Bacterial metastasis: the host plasminogen system in bacterial invasion. Trends Microbiol. 13:79–85 [DOI] [PubMed] [Google Scholar]
- 52. Ghosh AK, Jacobs-Lorena M. 2011. Surface-expressed enolases of Plasmodium and other pathogens. Mem. Inst. Oswaldo Cruz 106(Suppl 1):85–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Aoki K, Matsumoto S, Hirayama Y, Wada T, Ozeki Y, Niki M, Domenech P, Umemori K, Yamamoto S, Mineda A, Matsumoto M, Kobayashi K. 2004. Extracellular mycobacterial-binding protein 1 participates in mycobacterium-lung epithelial cell interaction through hyaluronic acid. J. Biol. Chem. 279:39798–39806 [DOI] [PubMed] [Google Scholar]
- 54. Bermudez LE, Goodman J. 1996. Mycobacterium tuberculosis invades and replicates within type II alveolar cells. Infect. Immun. 64:1400–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ashiru OT, Pillay M, Sturm AW. 2010. Adhesion to and invasion of pulmonary epithelial cells by the F15/LAM4/KZN and Beijing strains of Mycobacterium tuberculosis. J. Med. Microbiol. 59(Pt 5):528–533 [DOI] [PubMed] [Google Scholar]
- 56. Mapother ME, Songer JG. 1984. In vitro interaction of Mycobacterium avium with intestinal epithelial cells. Infect. Immun. 45:67–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Finlay BB, Ruschkowski S, Dedhar S. 1991. Cytoskeletal rearrangement accompanying Salmonella entry into epithelial cells. J. Cell Sci. 99:283–296 [DOI] [PubMed] [Google Scholar]
- 58. Jones BD, Paterson HF, Hall A, Falkow S. 1993. Salmonella typhimurium induces membrane ruffling by a growth factor receptor independent mechanism. Proc. Natl. Acad. Sci. U. S. A. 90:10390–10394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Job CK, Karat AB, Karat S. 1966. The histopathological appearance of leprous rhinitis and pathogenesis of septal perforation in leprosy. J. Laryngol. Otol. 80:718–732 [DOI] [PubMed] [Google Scholar]
- 60. McDermott-Lancaster RD, McDougall AC. 1990. Mode of transmission and histology of M. leprae infection in nude mice. Int. J. Exp. Pathol. 71:689–700 [PMC free article] [PubMed] [Google Scholar]
- 61. Sethna KB, Birdi TJ, Antia NH. 1997. Adherence of Mycobacterium leprae to the nasal mucosa is influenced by surface integrity and viability. J. Biosci. 22:575–583 [Google Scholar]
- 62. Marques MA, Neves-Ferreira AG, da Silveira EK, Valente RH, Chapeaurouge A, Perales J, da Silva Bernardes R, Dobos KM, Spencer JS, Brennan PJ, Pessolani MC. 2008. Deciphering the proteomic profile of Mycobacterium leprae cell envelope. Proteomics 8:2477–2491 [DOI] [PubMed] [Google Scholar]
- 63. Marques MA, Espinosa BJ, Xavier da Silveira EK, Pessolani MC, Chapeaurouge A, Perales J, Dobos KM, Belisle JT, Spencer JS, Brennan PJ. 2004. Continued proteomic analysis of Mycobacterium leprae subcellular fractions. Proteomics 4:2942–2953 [DOI] [PubMed] [Google Scholar]
- 64. He Z, De Buck J. 2010. Cell wall proteome analysis of Mycobacterium smegmatis strain MC2 155. BMC Microbiol. 10:121. 10.1186/1471-2180-10-121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pancholi V, Chhatwal GS. 2003. Housekeeping enzymes as virulence factors for pathogens. Int. J. Med. Microbiol. 293:391–401 [DOI] [PubMed] [Google Scholar]
- 66. Pancholi V, Fischetti VA. 1992. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J. Exp. Med. 176:415–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pancholi V, Fischetti VA. 1997. Regulation of the phosphorylation of human pharyngeal cell proteins by group A streptococcal surface dehydrogenase: signal transduction between streptococci and pharyngeal cells. J. Exp. Med. 186:1633–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gozalbo D, Gil-Navarro I, Azorin I, Renau-Piqueras J, Martinez JP, Gil ML. 1998. The cell wall-associated glyceraldehyde-3-phosphate dehydrogenase of Candida albicans is also a fibronectin and laminin binding protein. Infect. Immun. 66:2052–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chhatwal GS. 2002. Anchorless adhesins and invasins of Gram-positive bacteria: a new class of virulence factors. Trends Microbiol. 10:205–208 [DOI] [PubMed] [Google Scholar]
- 70. Bergmann S, Rohde M, Chhatwal GS, Hammerschmidt S. 2001. Alpha-enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol. Microbiol. 40:1273–1287 [DOI] [PubMed] [Google Scholar]
- 71. Soares de Lima C, Zulianello L, Marques MA, Kim H, Portugal MI, Antunes SL, Menozzi FD, Ottenhoff TH, Brennan PJ, Pessolani MC. 2005. Mapping the laminin-binding and adhesive domain of the cell surface-associated Hlp/LBP protein from Mycobacterium leprae. Microbes Infect. 7:1097–1109 [DOI] [PubMed] [Google Scholar]
- 72. Portugal ML, Todeschini AR, de Lima CS, Silva CA, Mohana-Borges R, Ottenhoff TH, Mendonça-Previato L, Previato JO, Pessolani MC. 2008. Characterization of two heparan sulphate-binding sites in the mycobacterial adhesin Hlp. BMC Microbiol. 8:75. 10.1186/1471-2180-8-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Dias AA, Raze D, de Lima CS, Marques MA, Drobecq H, Debrie AS, Ribeiro-Guimarães ML, Biet F, Pessolani MC. 2012. Mycobacterial laminin-binding histone-like protein mediates collagen-dependent cytoadherence. Mem. Inst. Oswaldo Cruz 107(Suppl 1):174–182 [DOI] [PubMed] [Google Scholar]
- 74. Pethe K, Alonso S, Biet F, Delogu G, Brennan MJ, Locht C, Menozzi FD. 2001. The heparin-binding haemagglutinin of M. tuberculosis is required for extrapulmonary dissemination. Nature 412:190–194 [DOI] [PubMed] [Google Scholar]
- 75. Ng V, Zanazzi G, Timpl R, Talts JF, Salzer JL, Brennan PJ, Rambukkana A. 2000. Role of the cell wall phenolic glycolipid-1 in the peripheral nerve predilection of Mycobacterium leprae. Cell 103:511–524 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.