Abstract
Enterococcus faecalis is a commensal bacterium found in the gastrointestinal tract of most mammals, including humans, and is one of the leading causes of nosocomial infections. One of the hallmarks of E. faecalis pathogenesis is its unusual ability to tolerate high concentrations of lysozyme, which is an important innate immune component of the host. Previous studies have shown that the presence of lysozyme leads to the activation of SigV, an extracytoplasmic function (ECF) sigma factor in E. faecalis, and that the deletion of sigV increases the susceptibility of the bacterium toward lysozyme. Here, we describe the contribution of Eep, a membrane-bound zinc metalloprotease, to the activation of SigV under lysozyme stress by its effects on the stability of the anti-sigma factor RsiV. We demonstrate that the Δeep mutant phenocopies the ΔsigV mutant in lysozyme, heat, ethanol, and acid stress susceptibility. We also show, using an immunoblot analysis, that in an eep deletion mutant, the anti-sigma factor RsiV is only partially degraded after lysozyme exposure, suggesting that RsiV is processed by unknown protease(s) prior to the action of Eep. An additional observation is that the deletion of rsiV, which results in constitutive SigV expression, leads to chaining of cells, suggesting that SigV might be involved in regulating cell wall-modifying enzymes important in cell wall turnover. We also demonstrate that, in the absence of eep or sigV, enterococci bind significantly more lysozyme, providing a plausible explanation for the increased sensitivity of these mutants toward lysozyme.
INTRODUCTION
Enterococcus faecalis is a commensal organism present in the mammalian gastrointestinal system (1). Over the past few decades, E. faecalis has arisen as one of the leading causes of nosocomial infection (2). Its role as an opportunistic pathogen is strengthened by the mobile genetic elements it harbors, which are often responsible for conferring resistance to a broad range of antibiotics, including vancomycin (3). In addition, E. faecalis is known to demonstrate a heightened ability to survive in the presence of environmental stress factors, such as increased temperature, acidic pH, and oxidative stress (4). In addition to persistence in the presence of the aforementioned stress factors, previous studies have shown that E. faecalis is also highly resistant to lysozyme (5). This high-level resistance to lysozyme (>62 mg/ml) is predominantly attributed to the extracytoplasmic function (ECF) sigma factor SigV (5). ECF sigma factors are sequestered by membrane-bound anti-sigma factors and rendered inactive in the absence of a given external stress. Under stress-inducing conditions, the anti-sigma factors are degraded by membrane and cytosolic proteases, leading to the activation of ECF sigma factors in a process referred to as regulated intramembrane proteolysis (RIP) (6).
RIP has been shown to play an important role in multiple transmembrane signaling processes associated with increased virulence and environmental fitness (7). In Escherichia coli, DegS, a site 1 protease, and RseP, a site 2 protease, have been shown to degrade the anti-sigma factor RseA in response to environmental stress, thus mediating the release of SigE (8). A similar mechanism is displayed by Bacillus subtilis, in which PrsW and RasP are the site 1 and site 2 protease, respectively (9). In E. faecalis, neither a site 1 nor a site 2 protease involved in the processing of ECF sigma factors has been identified to date. The release of SigV from RsiV is thought to require proteolytic cleavage of RsiV. A candidate site 2 protease was predicted to be Eep, as it possesses many of the characteristics of membrane-localized site 2 proteases (10). We hypothesized that Eep would play an important role in the regulated intramembrane proteolysis of RsiV leading to the activation of SigV. Here, we show that Eep is essential for the complete degradation of RsiV, which in turn is essential for the activation of SigV. Furthermore, the deletion of eep retards the ability of the SigV regulon to respond to lysozyme and several other stresses, which likely explains the significant contribution that Eep makes during infection (11).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The pertinent bacterial strains and plasmids used in the current study are listed in Tables 1 and 2. Strains were cultured in Todd-Hewitt broth (THB) and grown at 37°C unless otherwise indicated. Escherichia coli ElectroTen-Blue (Stratagene, La Jolla, CA) and E. faecalis V583 were used for the maintenance and propagation of plasmid constructs. ElectroTen-Blue clones were cultured aerobically in Luria-Bertani (LB) broth at 37°C, and E. faecalis V583 and FA2-2-derived strains were cultured in THB at 37°C. The antibiotics used for selection included chloramphenicol at 10 μg ml−1 and spectinomycin at 150 μg ml−1 for E. coli and chloramphenicol at 15 μg ml−1 and spectinomycin at 500 μg ml−1 for E. faecalis. Transformation of plasmids into E. faecalis was done as described previously (12).
Table 1.
Bacterial strains used in this study
| Strain | Genotype or description | Reference or source |
|---|---|---|
| V583 | Parental strain | 48 |
| FA2-2 | Parental strain | 14 |
| SV03 | V583 Δeep | This study |
| SV05 | FA2-2 Δeep | This study |
| SV07 | V583 ΔsigV | This study |
| SV08 | SV03(pSV24), complemented eep mutant, Specr | This study |
| SV17 | SV03(pSV04), empty vector control, Specr | This study |
| SV09 | V583(pSV23), GFP-RsiV fusion, Specr | This study |
| SV10 | SV03(pSV23) GFP-RsiV fusion, Specr | This study |
| SV11 | V583(pSV14), sigV promoter fusion to lacZ, Specr | This study |
| SV12 | SV03(pSV14), sigV promoter fusion to lacZ, Specr | This study |
| SV13 | SV07(pSV14), sigV promoter fusion to lacZ, Specr | This study |
| SV14 | V583 ΔrsiV | This study |
| SV15 | SV14(pSV14), sigV promoter fusion to lacZ, Specr | This study |
| WM01 | FA2-2 ΔsigV | This study |
| WM02 | V583 Δeep ΔrsiV | This study |
| VI50 | V583 ΔpgdA | This study |
| VI60 | V583(pVI16), pgdA promoter fusion to lacZ, Specr | This study |
| VI61 | SV03(pVI16), pgdA promoter fusion to lacZ, Specr | This study |
| VI62 | SV07(pVI16), pgdA promoter fusion to lacZ, Specr | This study |
| SV16 | SV14(pVI16), pgdA promoter fusion to lacZ, Specr | This study |
Table 2.
Plasmid constructs used in this study
| Plasmid | Description | Reference |
|---|---|---|
| pLT06 | Deletion vector, chloramphenicol resistance | 13 |
| pSV03 | pLT06 containing engineered eep deletion (∼2-kb EcoRI/PstI fragment) | This study |
| pSV15 | pLT06 containing engineered sigV deletion (∼2-kb EcoRI/PstI fragment) | This study |
| pML28 | pAT28 derivative containing the aph promoter | 18 |
| pSV17 | pML28 derivative containing rsiV with an N-terminal Flag tag | This study |
| pSV23 | pSV17 derivative containing a gfp-rsiV fusion under the constitutive aph promoter | This study |
| pTCVLac-Spec | Shuttle vector for promoter fusion studies, Ermr Specr | 16 |
| pKS12A | A derivative of pTCVLac-Spec in which the erythromycin methylase gene was removed by AflII digestion, Specr | This study |
| pMV158GFP | gfp-containing plasmid, Tetr | 19 |
| pSV04 | A derivative of pTCVLac-Spec in which the lacZ and erythromycin methylase genes were removed by SalI digestion, Specr | This study |
| pSV24 | pSV04 containing full-length eep under the native eep promoter | This study |
| pVI15 | pLT06 containing engineered pgdA deletion (∼2-kb EcoRI/PstI fragment) | This study |
| pWM07 | pLT06 containing engineered rsiV deletion (∼2-kb BamHI/PstI fragment) | This study |
| pSV14 | pKS12A containing sigV promoter region (1,234-bp EcoRI/BamHI fragment) | This study |
| pVI16 | pKS12A containing pgdA promoter region (1,005-bp EcoRI/BamHI fragment) | This study |
Construction of E. faecalis in-frame deletion mutants.
In-frame deletions of sigV, rsiV, pgdA, and eep individually and the double deletion mutant of rsiV and eep in E. faecalis were done using plasmids derived from pLT06 (13), an E. coli enterococcal temperature-sensitive cloning vector that possesses selectable and counterselectable markers that aid in the selection of mutants containing the targeted deletions. The primers used for all the deletions are listed in Table 3. Flanking regions (∼1 kb) from both the 5′ and 3′ ends of sigV, rsiV, pgdA, and eep were PCR amplified by using the primers listed in Table 3. For the construction of the pSV03 plasmid (eep deletion), primers EepP1 and EepP2 were used to amplify the ∼1-kb region flanking the 5′ end of eep on the V583 genome. Primers EepP3 and EepP4 were used to amplify the ∼1-kb region flanking the 3′ end of the eep region. The EepP1 and EepP2 primers contained EcoRI and XbaI sites, respectively, and the EepP3 and EepP4 primers contained XbaI and SphI sites, respectively. Each product was cut with the respective restriction enzymes and ligated to pLT06 cut with EcoRI and SphI prior to electroporation into E. coli ElectroTen-Blue cells. Confirmation of the appropriate clones was performed by restriction digest and sequence analysis. An analogous approach was used for the construction of pSV15 (sigV deletion), pWM07 (rsiV deletion), and pVI15 (pgdA deletion). Purified plasmids from E. coli cells were electroporated into electrocompetent V583 cells. Strains SV03 (V583 Δeep), SV07 (V583 ΔsigV), SV14 (V583 ΔrsiV), and VI50 (V583 ΔpgdA) were generated by following the protocol as previously described (13). The eep deletion allele was designed such that the first 2 and the last 7 codons remained (98% deleted). For sigV, the deletion allele consisted of the initial 6 and last 7 codons (92% deleted). The rsiV deletion allele included the first 6 and last 7 codons (96% deleted), while the pgdA deletion allele possessed the first 3 and the last codon (99% deleted). To create the double deletion of rsiV and eep, electrocompetent SV03 cells were transformed with pWM07, and the deletion mutant designated WM02 (V583 Δeep ΔrsiV) was created similarly (13). To rule out strain differences associated with V583, we also created deletion mutants for eep and sigV in the FA2-2 (14) strain background. FA2-2 is a plasmid-free strain derived from the nonhemolytic, nonproteolytic clinical isolate JH2 (15). FA2-2 was transformed with pSV03 (Δeep) and pSV15 (ΔsigV), and following plasmid integration and excision events, the deletion strains SV05 (FA2-2 Δeep) and WM01 (FA2-2 ΔsigV) were created.
Table 3.
Oligonucleotides used in this study
| Primer | Sequence (5′–3′)a |
|---|---|
| EepP1 | GAGAGGATCCGACATCAATGACACGTTGCC |
| EepP2 | CTCTTCTAGAGTTTTCATAGATGTCCTTCTTC |
| EepP3 | GAGATCTAGAACTTGGAACGATATTCAACGC |
| EepP4 | CTCTGCATGCCCACGAACCAAGACCATTAC |
| EepUp | TCTTTGGTGTACGGAGGACA |
| EepDown | GCATCTTCTTCTGTTGCTTC |
| Eep 5′ | GAGAGAATTCACTTTCATGGTAGCACATGTC |
| Eep 3′ | CTCTGGATCCCTGTTTCATTAAAACTCTCCTC |
| SigVP1 | GAGAGAATTCGAGCAGATTCGGAACTTTGAG |
| SigVP2 | CTCTGGATCCAACCATCGATTTCTGGAACCT |
| SigVP3 | GAGAGGATCCCTCCGAAGTCTATTGAATTAGT |
| SigVP4 | CTCTCTGCAGCAACTGACTTGGTTAGGTCAG |
| SigVUp | GTCACACATTGGCTTATAAGG |
| SigVDown | GCCACTTCTTCTTCGTTTCC |
| PgdAP1 | GAGAGAATTCGCTTGATTTGCTTGCAGTGC |
| PgdAP2 | CTCTGGATCCATGTCGCATACTTTCACTCCT |
| PgdAP3 | GAGAGGATCCTAGAGCAACTCGGAGCAC |
| PgdAP4 | CTCTCTGCAGGCCACCTTATGATCCAAGAG |
| PgdAUp | TCGCTTGGCTACTGTTGTGC |
| PgdADown | TTGCGAATACTCCTGAAGTAC |
| RsiVP1 | GAGAGGATCCCGGTATCTGTTGTTAATGGTG |
| RsiVP2 | CTTCTTCTAGACCGTTGGCAACGGTTGTTG |
| RsiVP3 | GAGATCTAGATGATTCCTGATCAAGTCATTG |
| RsiVP4 | CTCTCTGCAGCAATGACTTGGTCGTTGCTG |
| RsiVUp | CCGAGGAAGTCCTGCAAGG |
| RsiVDown | TCACTAATGGTAATGGTTGATC |
| RsiV5′ | GACTACAAAGACGATGACGACAAGCATATGGAAGATTTTGTAAAAAGTGTG |
| RsiV3′ | CTCTGCATGCGTCGCGTGTTTTTTACTGAGT |
| GFP5′ | GAGAGGATCCAAGGAGGAAAAACATATGAGTA |
| GFP3′ | CTCTATTAATTTTGTATAGTTCATCCATGCC |
| FLAGTAG | GAGAGGATCCAAGGAGGATTTATAGATGGATTATAAGGATCATGATTATAAGGATCATGATATCGACTACAAAGACGATGACGACAAG |
The underlined sequences denote restriction sites that were added to the template-directed sequences to facilitate cloning.
Complementation of eep deletion mutant.
An in-frame eep deletion in E. faecalis V583 was complemented with full-length eep under the control of the native eep promoter region in a pSV04 vector background and was designated pSV24. The eep complement was amplified from the V583 genome with primers Eep5′ and Eep3′ (Table 3) and subsequently inserted as an EcoRI/BamHI fragment into pSV04 cut with EcoRI/BamHI. To construct pSV04, plasmid pTCVLac-Spec (16) was digested with SalI and self-ligated to release the SalI fragment containing the lacZ gene. Plasmid pSV24 was transformed into SV03 (V583 Δeep), and phenotypic complementation was confirmed by a lysozyme MIC assay.
MIC assay for determining lysozyme sensitivity.
The MICs of lysozyme against strains V583, SV03 (V583 Δeep), SV07 (V583 ΔsigV), SV14 (V583 ΔrsiV), VI50 (V583 ΔpgdA), WM02 (V583 Δeep ΔrsiV), and SV08 (SV03 with Eep complementation vector), along with FA2-2 and its isogenic derivatives SV05 (FA2-2 Δeep) and WM01 (FA2-2 ΔsigV), were determined by 2-fold serial dilution of a 250-mg/ml lysozyme stock in LB broth to achieve a series of lysozyme concentrations ranging from 0 mg/ml to 62.5 mg/ml in a 96-well microtiter plate. Briefly, the strains were grown as standing cultures at 37°C in LB broth overnight to reach stationary phase. These overnight cultures (∼108 CFU/ml) were diluted 1:100 in fresh LB, and then 100 μl was added to 100 μl of the LB containing serially diluted lysozyme such that each well contained an initial inoculum of ∼105 CFU. The plate was then incubated at 37°C for 24 h before the results were documented. SV08 (SV03 with eep complementation vector, pSV24) and SV17 (SV03 with empty vector pSV04) were grown overnight in the presence of 500 μg ml−1 spectinomycin for plasmid maintenance.
Settling and chaining assays.
V583, SV14 (V583 ΔrsiV), SV03 (V583 Δeep), SV07 (V583 ΔsigV), and WM02 (Δeep ΔrsiV) were grown in THB overnight at 37°C with and without lysozyme at 1 mg/ml and photographed to observe the settling phenotype, which is indicated by growth of the bacterium at the bottom of the test tube. The upper layer of the growth medium becomes transparent as cells settle and grow on the bottom of the tube. Liquid cultures from the respective strains were also Gram stained and photographed to observe chaining (Nikon Eclipse 80i with a 100× oil immersion objective).
Miller assay using strains containing PsigV-lacZ and PpgdA-lacZ reporter fusion plasmids.
To investigate the transcriptional activity of known SigV-dependent promoters, we created sigV and pgdA promoter fusions to a lacZ reporter in plasmid pKS12A, a derivative of pTCV-Lac Spec (16), in which a small AflII fragment containing the erythromycin methylase gene was deleted. pSV14 (PsigV-lacZ) and pVI16 (PpgdA-lacZ) were created by amplifying the promoter regions of sigV with primers SigVP1 and SigV2 and the pgdA promoter with primers PgdAP1 and PgdAP2. The promoter regions for both these plasmids were defined based on the known transcriptional start sites for both sigV and pgdA (4). These promoters contain the consensus SigV promoter recognition sequence (5′ TGAAAC-N17-CGTC 3′), and we included an additional ∼1-kb region (1,188 bp for sigV and 914 bp for pgdA) upstream from the transcriptional start site to provide additional genetic context for the promoter fusion studies. Primers were engineered with EcoRI and BamHI restriction sites to facilitate cloning into pKS12A. The resulting vectors, pSV14 and pVI16, were transformed into strains V583, SV03 (V583 Δeep), SV07 (V583 ΔsigV), and SV14 (V583 ΔrsiV). The new strains, SV11 [V583(pSV14)], SV12 [SV03(pSV14)], SV13 [SV07(pSV14)], SV15 [SV14(pSV14)], VI60 [V583(pVI16)], VI61 [SV03(pVI16)], VI62 [SV07(pVI16)], and SV16 [SV14(pVI16)], were grown overnight in THB containing 500 μg ml−1 spectinomycin at 37°C for plasmid maintenance. The overnight cultures were diluted 1:100 in sterile THB containing spectinomycin and cultured to reach an optical density at 600 nm (OD600) of 0.5. At this point, cells were induced with lysozyme at a concentration of 1 mg/ml for 30 min. The cultures were then processed to evaluate β-galactosidase activity according to the modified Miller assay as described previously (17). To establish a dose response curve to induction by lysozyme, SV11 [V583(pSV14)] was exposed to increasing concentrations of lysozyme (0, 1, 10, 100, and 1,000 μg/ml) for 30 min prior to assaying for β-galactosidase activity. All assays were repeated three times, and statistical significance was determined using 1-way analysis of variance (ANOVA).
Temperature, ethanol, and acid challenge assays.
In order to determine the survival of V583, SV03, SV14, WM02, and SV07 under different stress conditions, the protocol described by Benachour et al. (4) was followed. The strains were grown in 5 ml THB medium at 37°C to an OD600 of 0.5 (mid-exponential growth phase). Bacteria were harvested by centrifugation, resuspended in 5 ml of fresh medium, and then exposed to stresses as follows: (i) for high-temperature heat shock, the cultures were transferred to 62°C; (ii) for ethanol shock, ethanol was added to a final concentration of 22% (vol/vol); and (iii) for acid shock, the pH was adjusted to 3.2 with 85% lactic acid. Cells were exposed to stress conditions, and the numbers of surviving bacteria were quantified by plate counting at 0, 1, and 2 h after stress initiation. Assays were repeated three times, and statistical significance was established using 2-way ANOVA in the GraphPad 5 software package (Prism, San Diego, CA). The percent survival shown in the graphs represents the ratio of the number of viable cells after exposure to stress to the number of cells at time zero prior to challenge.
CBB staining of whole-cell lysates.
Whole-cell extracts of V583, SV03, and SV07 were prepared by growing strains in THB to an OD600 of 0.7 to 0.8. Lysozyme at a concentration of 1 mg/ml was added to the cultures for 2 h, the cultures were centrifuged, and the pellet was washed twice with 1 ml of Tris-EDTA (TE) buffer (pH 8.0). These 1-ml suspensions were lysed using a mini-BeadBeater (BioSpec Products, Bartlesville, OK) with a 500-μl volume of 0.1-mm zirconia beads and a speed setting of 4,800 rpm for 1 min. After brief centrifugation to settle the beads, 5× SDS loading dye (0.25% bromophenol blue; 50% glycerol; 10% SDS in 0.3 M Tris-Cl, pH 6.8) was added to the whole-cell lysate, and samples were boiled at 100°C for 10 min. To normalize the amount of protein loaded onto the polyacrylamide gel, the whole-cell lysates were also subjected to Bradford protein assay to determine the protein amounts in each sample. The same amount of protein was loaded into each well, and the samples were run at 200 V for an hour and were then subjected to Coomassie brilliant blue (CBB) staining. Whole-cell lysates from cultures without lysozyme added and purified lysozyme protein were used as controls.
Immunoblotting to detect RsiV degradation.
Plasmid pSV23 expresses the green fluorescent protein (GFP)-RsiV fusion protein and was constructed in the following manner. The rsiV gene was amplified from the V583 genome using primers RsiV5′ and RsiV3′, which contained an NdeI and an SphI site, respectively. In a second round of PCR, this product was amplified with an additional primer designated FLAGTAG and with RsiV3′; the resulting product contained an introduced BamHI site and a Flag tag-encoding sequence at the 5′ end. This BamHI- and SphI-digested product was cloned into the similarly digested pML28 (a pAT28 derivative containing an aph promoter) (18) to create pSV17. We next swapped the Flag tag for a GFP tag by digesting pSV17 with BamHI and NdeI. The GFP-encoding region was amplified from pMV158GFP (19) using primers GFP5′ and GFP3′. This PCR product was digested with BamHI and AseI and ligated to BamHI- and NdeI-digested pSV17 to obtain pSV23. E. faecalis strains V583 and SV03 were transformed with pSV23 to create SV09 and SV10, respectively. Cultures of SV09 [V583(pSV23)] and SV10 [SV03(pSV23)] were grown overnight in THB at 37°C and diluted 1:100 into 10 ml of fresh THB. Cultures were grown to an OD600 of 0.7 to 0.8, at which point lysozyme was added to a concentration of 1 mg/ml and the cultures were incubated for an additional 2 h. SV09 and SV10 cultures to which no lysozyme was added were used as negative controls. All four cultures were centrifuged, and the pellets were resuspended in 1 ml TE buffer (pH 8.0). Protease inhibitors and EDTA were added to these suspensions, and the mixtures subjected to bead beating in the mini-BeadBeater as described above. The lysates were analyzed on a 10% SDS–PAGE gel. Following electrophoresis, samples were electrotransferred to a polyvinylidene difluoride (PVDF) membrane and blotted with rabbit anti-GFP primary antibody (Cell Signaling Technology, Inc.) and anti-rabbit horseradish peroxidase (HRP)-conjugated secondary antibody (Sigma-Aldrich).
RESULTS
Deletion of eep renders E. faecalis more susceptible to lysozyme.
To test whether an eep deletion mutant showed increased susceptibility toward lysozyme compared to the susceptibility of the wild-type strain, we determined the lysozyme MICs for V583, SV03, and SV08 (eep complement). The results of this assay are shown in Table 4. The increased susceptibility of the eep mutant paralleled that of the sigV mutant, as both displayed MIC values at 5 mg/ml, compared to the >62 mg/ml for V583. When the eep gene was complemented back into the eep deletion mutant using a low-copy-number plasmid, the complement strain behaved similarly to the wild type, suggesting that the phenotype we observed was Eep dependent. Consistent with previous observations, the pgdA deletion mutant did not show any change in lysozyme susceptibility compared to that of the wild type (20), even though it is known to be regulated by SigV at the transcriptional level (4, 5). FA2-2 and mutants lacking eep and sigV in this genetic background were used as controls in this study to eliminate any strain bias in the lysozyme resistance mechanism. We observed a similar reduction in the MIC for the eep and sigV mutants in the FA2-2 strain background compared to the MIC of FA2-2. Finally, as a proof of principle that Eep acts through RsiV in the activation of SigV, we deleted rsiV in the eep mutant background and showed that the lysozyme resistance level of this double mutant paralleled that of the wild type (>62.5 mg/ml).
Table 4.
Lysozyme MIC assay
| Strain | Lysozyme MIC (mg/ml) |
|---|---|
| V583 | >62.5 |
| FA2-2 | >62.5 |
| SV03 (V583 Δeep) | 5.0 |
| SV07 (V583ΔsigV) | 5.0 |
| VI50 (V583 ΔpgdA) | >62.5 |
| SV05 (FA2-2 Δeep) | 5.0 |
| WM01 (FA2-2 ΔsigV) | 5.0 |
| SV08 [SV03(pSV24)] | >62.5 |
| SV17 [SV03(pSV04)] | 5.0 |
| WM02 (V583 Δeep ΔrsiV) | >62.5 |
| SV14 (V583 ΔrsiV) | >62.5 |
Eep is essential for the expression of sigV at the transcriptional level.
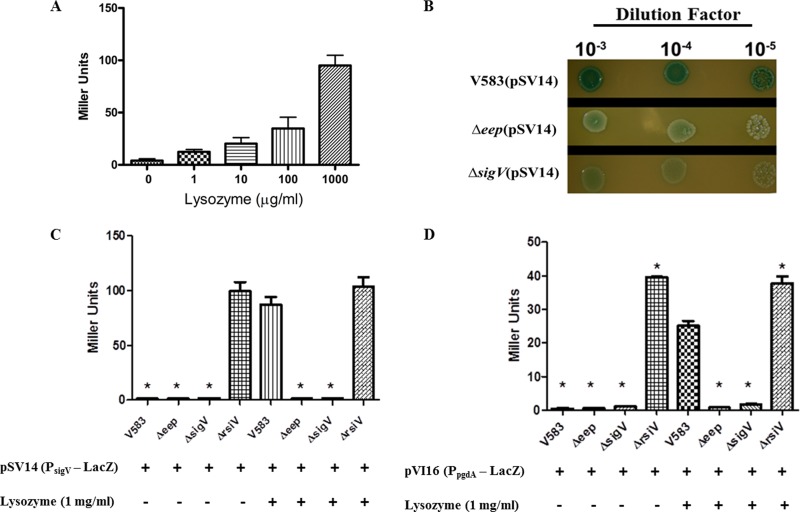
It is known that SigV autoregulates its own expression (4). We constructed a sigV promoter fusion to lacZ to confirm these observations and to ascertain the effects of both sigV and eep deletion on promoter activity. We confirmed that, in wild-type cells containing the reporter plasmid (SV11), the activity of the sigV promoter is induced by lysozyme in a dose-dependent manner (Fig. 1A). In the experiment whose results are shown in Figure 1B, we also demonstrated that promoter activity is dependent on a functional SigV, as well as Eep, for the response to lysozyme, as mutants with mutations in eep and sigV remain white in the presence of lysozyme, whereas the wild-type reporter strain turned blue on X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)-containing agar. Miller assays were performed with the reporter strains to quantify the amount of β-galactosidase protein produced in the wild type compared to the mutants. SV11 produces ∼80-fold more β-galactosidase activity than the eep and sigV mutants upon lysozyme induction (Fig. 1C). We also analyzed reporter strains with the pgdA promoter fusion to lacZ, as the expression of pgdA was previously shown to be SigV dependent (4). Similar results were observed in these reporter strains with the Miller assay (Fig. 1D), as both the eep and sigV mutants displayed reduced promoter activity compared to that of the parental strain. Consistent with its known role as an anti-sigma factor, the deletion of rsiV resulted in constitutive expression of both SigV-dependent promoters in the absence of lysozyme induction (Fig. 1C and D).
Fig 1.
Qualitative and quantitative measurement of sigV promoter and pgdA promoter activity. (A) sigV-lacZ reporter strain SV11 was subjected to Miller assay analysis after treatment with increasing concentrations of lysozyme. (B) Spot assay of sigV-lacZ reporter strains SV11, SV12 (Δeep), and SV13 (ΔsigV) on THB plates containing 1 mg/ml lysozyme and 80 μg/ml X-Gal. (C) Miller assay using LacZ reporter strains SV11, SV12, SV13, and SV15 (ΔrsiV) treated with and without 1 mg/ml of lysozyme. (D) Miller assay using pgdA-lacZ reporter strains VI60, VI61 (Δeep), VI62 (ΔsigV), and SV16 (ΔrsiV) treated with and without 1 mg/ml of lysozyme. *, significant difference (P < 0.001) relative to wild-type V583 in the presence of lysozyme. Experiments were performed in triplicate, and error bars represent standard errors of the means (SEM).
Eep confers resistance to other biological stressors.
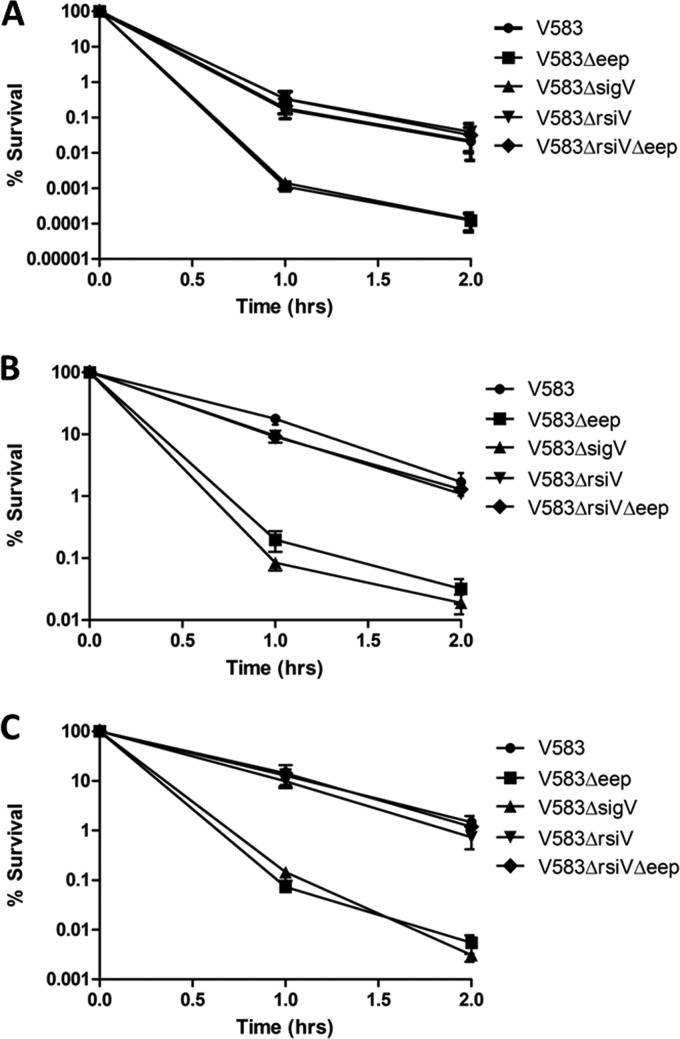
Benachour et al. (4) showed that a sigV deletion mutant is attenuated compared to a wild type when subjected to high-temperature, low-pH, and ethanol stress conditions. Having established a link between Eep and SigV phenotypes for lysozyme resistance, we reasoned that other biological stresses known to impact a sigV mutant might also negatively affect an eep mutant compared to their effects on its isogenic parent. We therefore tested the eep and sigV mutants against a variety of biological stresses (heat, low-pH, and ethanol stress) and found that the eep mutant phenocopied a sigV mutant in these biological aspects. Both mutants displayed nearly 2-log reductions against these stresses compared to the growth of V583 (Fig. 2). To confirm that the deletion of rsiV in the eep deletion strain restores the stress tolerance to wild-type levels, the strain harboring a double deletion of rsiV and eep was also tested against the aforementioned biological stresses. This strain showed tolerance to these stresses similar to that of the wild type. As a control, an rsiV deletion mutant was also used in this study. This suggests that the deletion of Eep has a direct effect on the ability of cells to adapt to stress and that this phenotype could be rescued by the constitutive expression of SigV.
Fig 2.
V583, SV03 (V583 Δeep), SV14 (V583 ΔrsiV), WM02 (V583 ΔrsiV Δeep), and SV07 (V583 ΔsigV) strains were subjected to different stresses. (A) Strains were subjected to a temperature of 62°C. (B) Strains were subjected to a pH of 3.2 using 85% lactic acid. (C) Strains were subjected to 22% ethanol treatment. Strains subjected to lactic acid stress and ethanol stress were incubated at 37°C. Samples were drawn every hour for 2 h, were serially diluted in sterile PBS (pH 7.4), and plated onto THB agar. V583 Δeep and V583 ΔsigV were significantly attenuated compared to the growth of the wild-type (P < 0.001). Experiments were performed in triplicate, and error bars represent SEM.
Eep is essential for complete processing of RsiV.
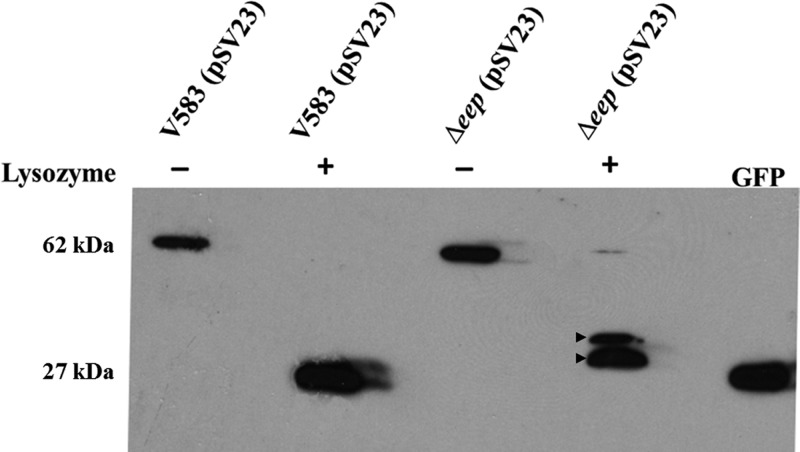
Since the strain that lacked eep phenocopied the strain that lacked sigV in all the phenotypes tested, our next goal was to determine the exact role of Eep in the regulated intramembrane proteolysis pathway leading to the release of SigV. To confirm that Eep is indeed the site 2 protease, we constructed a plasmid system wherein GFP was fused to RsiV and introduced into either the parental strain V583 or its isogenic eep mutant (SV03). The image in Figure 3 demonstrate the results of this experiment, wherein the plasmid-bearing strains SV09 and SV10 (Δeep) expressed the ∼62-kDa GFP-RsiV fusion protein in the absence of lysozyme stress. When lysozyme was added, RsiV in the V583 background was completely degraded by membrane and cytosolic proteases involved in RIP, leaving only the ∼27-kDa GFP protein. The presence of the GFP protein after SigV activation is likely attributable to the fact that GFP lacks the recognition domain that is required for Clp protease degradation of target substrates (21). However, accumulation of two partially processed GFP-RsiV products predicted to migrate between 34 and 38 kDa was observed when SV10 was treated with lysozyme.
Fig 3.
Immunoblot analysis. V583 and V583 Δeep strains containing pSV23 (expresses GFP-RsiV full-length fusion protein under the constitutive aph promoter) were subjected to 1 mg/ml lysozyme treatment for 30 min after they were grown to an OD600 of 0.8. Untreated cultures were used as controls. The whole-cell lysates were transferred to a PVDF membrane and immunoblotted with rabbit anti-GFP antibodies (Cell Signaling Technology, Inc.). A native GFP cell lysate was used as a control. The unprocessed GFP-RsiV translational fusion is predicted to migrate at 62 kDa, whereas the fully processed GFP-RsiV migrates at the native GFP position (27 kDa). Arrows indicate incompletely processed GFP-RsiV in the eep mutant exposed to lysozyme.
eep and sigV mutants bind more lysozyme than the wild type.
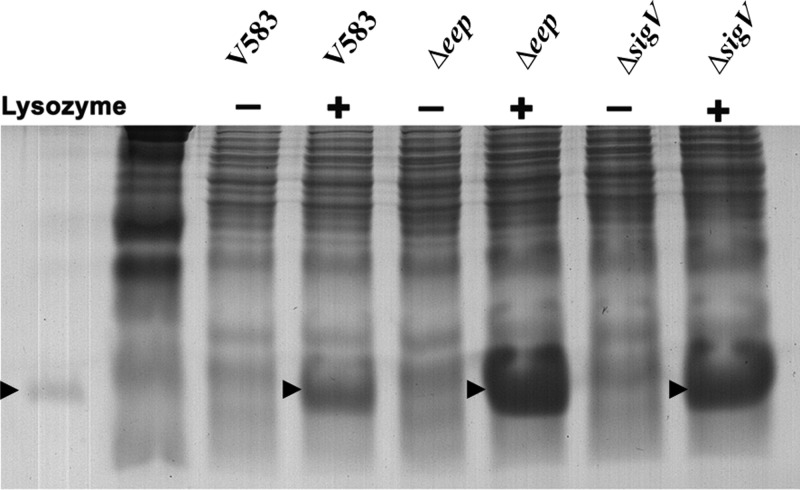
The enhanced susceptibility of SV03 (Δeep) and SV07 (ΔsigV) toward lysozyme led us to believe that these strains might be lacking some crucial mechanism that reduces the accumulation of lysozyme on the cell surface. The results depicted in Figure 4 demonstrate that the sigV and eep mutants bind more lysozyme than the wild type. Mass spectrometry analysis was performed to confirm that the protein band that was visualized consisted primarily of lysozyme (data not shown). These data suggest that the hypersusceptibility of SV03 and SV07 toward lysozyme is likely due to the enhanced accumulation of lysozyme on the cell surface of these mutants.
Fig 4.
SDS-PAGE and Coomassie brilliant blue staining assay. V583, SV03 (Δeep), and SV07 (ΔsigV) were grown to an OD600 of 0.8 in the presence and absence of 1 mg/ml lysozyme. Normalized lysates were loaded onto a 12% polyacrylamide gel and stained with Coomassie brilliant blue dye. Purified lysozyme (1 μg) was added to the left of the marker lane as a control. Arrows indicate bands corresponding to lysozyme.
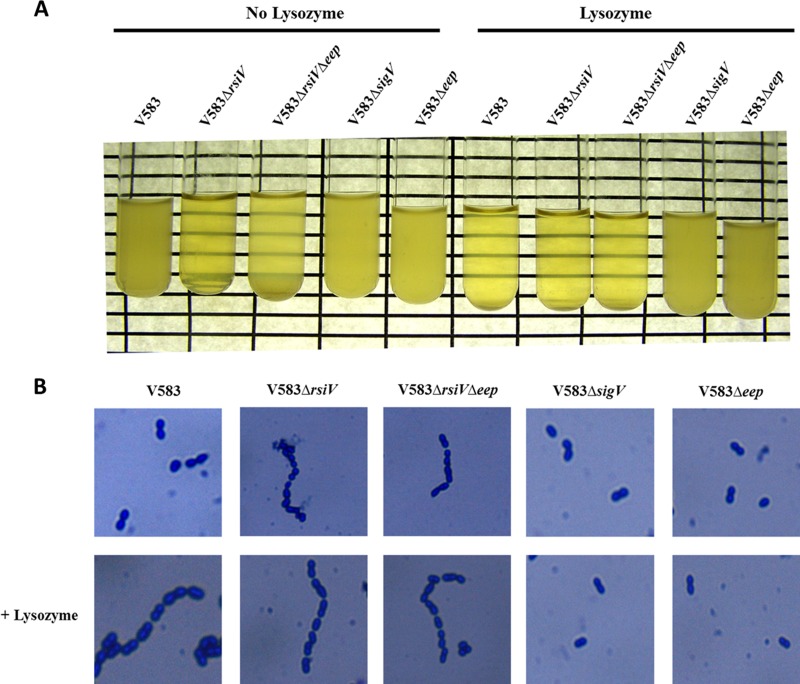
Constitutive expression of SigV results in a chaining phenotype.
Interestingly, when the gene encoding RsiV was deleted in the wild-type and V583 Δeep genetic backgrounds, a settling phenotype of the overnight culture was observed in both strains. The image in Figure 5 demonstrates this settling phenotype. Gram staining revealed significant chaining in strains lacking RsiV compared to the chaining in the wild type, suggesting that appropriate cell wall remodeling is compromised by constitutive expression of SigV. The chaining phenotype of the rsiV mutant is similar to that observed for an atlA mutant (22), suggesting that the cell wall modifications that render E. faecalis more resistant to lysozyme also perturb the activity of the major autolysin.
Fig 5.
Chaining and settling phenotypes. (A) Indicated strains were grown in the presence or absence of 1 mg/ml lysozyme in THB overnight at 37°C and photographed to depict the settling of the culture on the bottom of the tube. (B) Gram staining was performed on each culture to observe cell chaining.
Lysozyme induction results in a chaining phenotype.
Because of the chaining phenotype observed in the rsiV mutant, we reasoned that exposure to lysozyme would induce a chaining phenotype that was SigV dependent. To test this prediction, we exposed V583, SV03 (Δeep), SV07 (ΔsigV), SV14 (ΔrsiV), and WM02 (Δeep ΔrsiV) to 1 mg/ml lysozyme and examined the cultures for a settling phenotype.
As predicted, a settling phenotype was observed in the wild type when the cells were induced with lysozyme, and chaining was confirmed by Gram staining (Fig. 5). In contrast, the eep and sigV mutants did not settle or chain in the presence of lysozyme. The strains containing an rsiV deletion, SV14 (V583ΔrsiV) and WM02 (V583ΔrsiV Δeep), continued to chain and settle under lysozyme exposure. This suggests that the activation of SigV leads to the chaining phenotype and that genes under SigV control are also likely regulating the activity of the endogenous autolysins of E. faecalis.
DISCUSSION
In this study, we have shown that Eep is essential for the activation of SigV, which is an ECF sigma factor that contributes to lysozyme resistance in E. faecalis via the degradation of the anti-sigma factor RsiV (5). Eep belongs to a family of membrane-embedded zinc metalloproteases (23). Previous studies have shown that Eep is involved in the processing of peptide pheromones (24–26). To the best of our knowledge, this is the first study that demonstrates an additional role for Eep in the regulated intramembrane proteolysis of the anti-sigma factor RsiV leading to the activation of SigV. An eep deletion mutant phenocopied a sigV deletion mutant and was more than 10-fold more susceptible to lysozyme than the wild type. Promoter fusion studies demonstrate that the sigV promoter is inactive in the absence of Eep. Our hypothesis that Eep is directly involved in the degradation of RsiV was strengthened by the observation that the resistance to lysozyme was restored to wild-type levels in the double mutant (ΔrsiV Δeep) strain.
In addition, Western blot analysis revealed that Eep is required for the complete degradation of RsiV. It has been shown that the activation of other ECF sigma factors (6) in the presence of a specific stress requires the action of membrane proteases, and the data from this current study confirm the rationale that Eep is one such membrane protease. Of note, we were able to detect two intermediate processed forms of GFP-RsiV in the eep mutant background. This observation is consistent with the requirement for multiple RsiV-processing events prior to Eep cleavage. In the activation of the B. subtilis SigW (an ECF sigma factor), PrsW, a known site 1 protease, cleaves RsiW (9, 27). Upon cleavage by PrsW, further processing of RsiW occurs by an as-yet-unidentified trimming protease activity (28), and this cleavage event prepares RsiW for further processing by the known site 2 protease RasP.
The data from a Coomassie-stained SDS-PAGE gel indicate that mutants lacking eep and sigV bind more lysozyme than the wild type. Previous research has shown that two cell wall-modifying enzymes, O-acetyltransferase (OatA) and the d-alanylation complex (DltA to DltD [DltA-D]), confer resistance to lysozyme in B. subtilis and that these genes are directly regulated by SigV (29). However, in E. faecalis, it has been shown previously that individual deletion of these genes does not significantly affect the lysozyme susceptibility of E. faecalis, nor were the oatA and dltA-D genes shown to be regulated in a SigV-dependent fashion (5). In our hands, we found that single deletions of both oatA and dltA-D in the V583 background did not alter the lysozyme resistance of these strains, as these mutants still showed growth at up to 62.5 mg/ml lysozyme (data not shown). We also examined a double deletion of both oatA and dltA-D in the V583 background and found that this strain was only marginally reduced in lysozyme resistance, to ∼32 mg/ml (data not shown), which is in contrast to what was observed by Le Jeune et al. (5), where a double mutant of oatA and dlt was as sensitive to lysozyme as the sigV mutant in the JH2-2 genetic background. Whether this difference in lysozyme susceptibility can be accounted for by strain differences awaits additional study. It is, however, noteworthy that strain JH2-2 was derived from the parent strain JH2 by nitrosoguanidine mutagenesis (15), and the exposure to the mutagenizing agent may account for these aberrant strain differences. In contrast, the deletion of sigV rendered E. faecalis much more sensitive to lysozyme, suggesting that SigV-dependent gene products are involved in modifying the enterococcal cell wall in a manner independent of peptidoglycan O-acetylation or teichoic acid d-alanylation. Furthermore, a study by Hébert et al. (30) showed that a mutation in the gene encoding peptidoglycan deacetylase (pgdA) did not contribute to lysozyme resistance, despite the fact that this gene has been shown to be regulated in a SigV-dependent manner in E. faecalis (5). Our present studies also confirmed the absence of a link between PgdA and lysozyme resistance in E. faecalis V583. PgdA does, however, contribute to lysozyme susceptibility in Streptococcus pneumoniae and virulence in animal models of infection (31–33), and recent evidence suggests that PgdA is linked to virulence in E. faecalis, as a mutant in pgdA was attenuated in a Galleria mellonella infection model (20). What role PgdA might be playing in a mammalian host during infection remains to be elucidated. What is also clear from the present study and the additional cited literature is that SigV contributes to lysozyme resistance in a unique manner, and identifying the SigV regulon will be of paramount importance in understanding the unusual lysozyme resistance strategies employed by E. faecalis.
Lysozyme is a naturally secreted antimicrobial agent and is considered to be part of the innate immune system. It is secreted by a wide array of organisms in body fluids such as tears, mucus, and saliva (34–37). A study by Frank et al. (11) showed that Eep is essential for the pathogenesis of E. faecalis in a rabbit endocarditis model. It has also been shown that the concentration of lactic acid is particularly high in the heart (38). One possible explanation as to why the virulence of an eep deletion mutant was highly attenuated in the endocarditis model is that strains that lack eep are more susceptible to lactic acid stress, as shown in the present study. Another plausible explanation is that strains lacking eep are more susceptible to lysozyme than wild-type controls and, hence, are cleared more efficiently by components of the innate immune system (39, 40). The same idea could potentially be applied to a urinary tract infection (UTI) model due to the fact that it has been shown that the body responds to urinary tract infection by secreting an increased amount of lysozyme (41–43). It is noteworthy that a sigV mutant of E. faecalis JH2-2 was shown to be attenuated by ∼1.5 to 2 log in bladder and kidney colonization compared to the amount of JH2-2, and an eep mutant of E. faecalis OG1RF is attenuated in a catheter-associated UTI model (5, 44). Collectively, the available information to date suggests that the virulence associated with Eep is likely due to its effect on SigV activation. It will be of interest to establish a link with SigV in endocarditis. It is also important to note that lysozyme has been shown to be an important component of the innate immune defense system in the gastrointestinal tract (45). Since E. faecalis is a commensal bacterium which predominantly resides in the gastrointestinal tract of mammals, it is possible that this bacterium has unique mechanisms that confer high levels of lysozyme resistance, giving it a competitive edge in the gastrointestinal tract consortium.
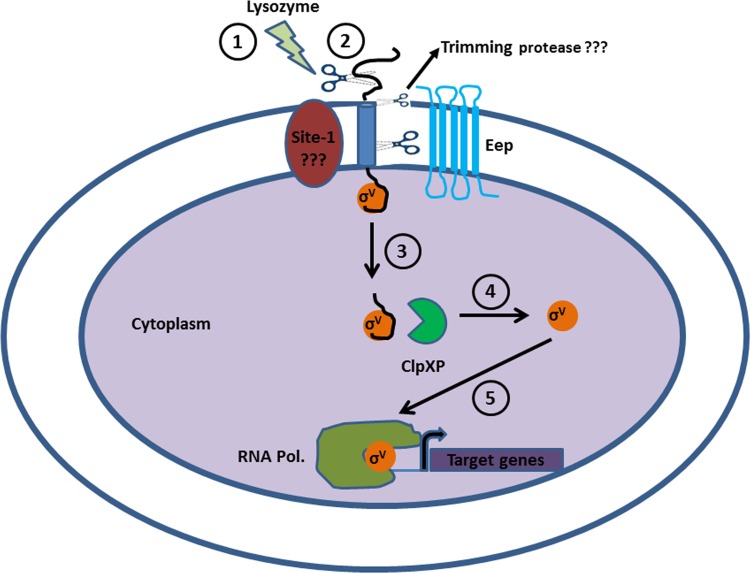
Figure 6 depicts the pathway by which SigV is released from the anti-sigma factor in the presence of lysozyme. Having evidence that supports the rationale that Eep is a site 2 protease in the regulated intramembrane proteolysis pathway, we postulate that both a site 1 and trimming protease act upstream of Eep cleavage to initiate the response to lysozyme. The biochemical analysis of the GFP-RsiV fusion in an eep background exposed to lysozyme suggests the existence of such proteases, as noted by the intermediate forms of GFP-RsiV that accumulate in the immunoblot (Fig. 3). Numerous studies have shown a requirement for site 1 proteases in initiating the activation of ECF sigma factors (6), and future studies will be aimed at the identification of such a protease. Following Eep cleavage, the model predicts that RsiV is further degraded by the cytosolic protease complex ClpXP, as this has been shown to be essential in the activation of SigW, a known ECF sigma factor in Bacillus subtilis (46).
Fig 6.
A model for the regulated intramembrane proteolysis (RIP) of RsiV. A series of proteolytic events leads to the release and activation of SigV from its anti-sigma factor RsiV. (1) E. faecalis perceives a stress signal, which in this case is lysozyme. (2) This leads to the cleavage of RsiV by the putative site 1 protease. Based on data from the experiment whose results are shown in Figure 3 and from reference 28, E. faecalis and other Gram-positive bacteria, such as B. subtilis, possess an additional putative trimming protease activity that prepares the processed RsiV for further proteolytic targeting by Eep. (3) Eep degrades the site 1 protease-processed and -trimmed RsiV, leading to the release of SigV into the cytoplasm. (4) ClpXP cytoplasmic protease further degrades RsiV to release active SigV. (5) SigV initiates the binding of the RNA polymerase upstream from specific genes that confer lysozyme resistance.
Recent work by Ellermeier's group established that SigV activation in Bacillus appears to uniquely recognize lysozyme cues (47, 49), as other cell wall- and membrane-acting agents failed to induce SigV activation. Work by Le Jeune et al. (5) showed that nisin does not induce SigV activation in E. faecalis and that the sigV mutant displays wild-type resistance to nisin, consistent with lysozyme being a key driver in SigV activation. An unusual finding from our present study is that both SigV and Eep also contribute to heat, low-pH, and ethanol stress tolerance. Attempts to use those conditions to induce SigV activation did not result in detectable β-galactosidase activity (data not shown), likely because the reporter protein might have been denatured under the conditions tested. Conversely, data presented by Benachour et al. (4) corroborate the fact that heat, low-pH and ethanol stress fail to induce SigV activation, as Northern blots failed to show an increase in sigV transcript levels following exposure to these stress inducers. Western immunoblot analysis of SV09 [V583(pSV23)] lysates subjected to heat, low-pH and ethanol stresses failed to induce the degradation of RsiV (data not shown). This could be attributed to the fact that the aforementioned stresses do not strongly induce the sigV regulon but resistance toward these stresses could still be SigV dependent. We speculate that a basal level of SigV is required for activating genes required for tolerance to these stress conditions and that Eep must also contribute to basal levels of SigV activation. It will be of interest to determine whether the transcriptional profile in the wild type and an isogenic sigV mutant differ even in the absence of lysozyme induction.
ACKNOWLEDGMENTS
We thank Craig Ellermeier (Carver College of Medicine, University of Iowa) for helpful discussions on the Western immunoblotting. We also thank Ian Huck (Kansas State University) for his help with the Miller assays and Yasuaki Hiromasa and John M. Tomich (Kansas State University Biotechnology Core Facility) for mass spectrometry analysis.
This work was supported by National Institutes of Health grant 1R01 AI 77782 (L.E.H.).
Footnotes
Published ahead of print 3 May 2013
REFERENCES
- 1. Hidron AI, Edwards JR, Patel J, Horan TC, Sievert DM, Pollock DA, Fridkin SK. 2008. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006-2007. Infect. Control Hosp. Epidemiol. 29:996–1011 [DOI] [PubMed] [Google Scholar]
- 2. Sievert DM, Ricks P, Edwards JR, Schneider A, Patel J, Srinivasan A, Kallen A, Limbago B, Fridkin S. 2013. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009-2010. Infect. Control Hosp. Epidemiol. 34:1–14 [DOI] [PubMed] [Google Scholar]
- 3. Hegstad K, Mikalsen T, Coque TM, Werner G, Sundsfjord A. 2010. Mobile genetic elements and their contribution to the emergence of antimicrobial resistant Enterococcus faecalis and Enterococcus faecium. Clin. Microbiol. Infect. 16:541–554 [DOI] [PubMed] [Google Scholar]
- 4. Benachour A, Muller C, Dabrowski-Coton M, Le Breton Y, Giard JC, Rince A, Auffray Y, Hartke A. 2005. The Enterococcus faecalis SigV protein is an extracytoplasmic function sigma factor contributing to survival following heat, acid, and ethanol treatments. J. Bacteriol. 187:1022–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Le Jeune A, Torelli R, Sanguinetti M, Giard JC, Hartke A, Auffray Y, Benachour A. 2010. The extracytoplasmic function sigma factor SigV plays a key role in the original model of lysozyme resistance and virulence of Enterococcus faecalis. PLoS One 5:e9658. 10.1371/journal.pone.0009658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ho TD, Ellermeier CD. 2012. Extra cytoplasmic function sigma factor activation. Curr. Opin. Microbiol. 15:182–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Heinrich J, Wiegert T. 2009. Regulated intramembrane proteolysis in the control of extracytoplasmic function sigma factors. Res. Microbiol. 160:696–703 [DOI] [PubMed] [Google Scholar]
- 8. Li X, Wang B, Feng L, Kang H, Qi Y, Wang J, Shi Y. 2009. Cleavage of RseA by RseP requires a carboxyl-terminal hydrophobic amino acid following DegS cleavage. Proc. Natl. Acad. Sci. U. S. A. 106:14837–14842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ellermeier CD, Losick R. 2006. Evidence for a novel protease governing regulated intramembrane proteolysis and resistance to antimicrobial peptides in Bacillus subtilis. Genes Dev. 20:1911–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Makinoshima H, Glickman MS. 2006. Site-2 proteases in prokaryotes: regulated intramembrane proteolysis expands to microbial pathogenesis. Microbes Infect. 8:1882–1888 [DOI] [PubMed] [Google Scholar]
- 11. Frank KL, Barnes AM, Grindle SM, Manias DA, Schlievert PM, Dunny GM. 2012. Use of recombinase-based in vivo expression technology to characterize Enterococcus faecalis gene expression during infection identifies in vivo-expressed antisense RNAs and implicates the protease Eep in pathogenesis. Infect. Immun. 80:539–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cruz-Rodz AL, Gilmore MS. 1990. High efficiency introduction of plasmid DNA into glycine treated Enterococcus faecalis by electroporation. Mol. Gen. Genet. 224:152–154 [DOI] [PubMed] [Google Scholar]
- 13. Thurlow LR, Thomas VC, Hancock LE. 2009. Capsular polysaccharide production in Enterococcus faecalis and contribution of CpsF to capsule serospecificity. J. Bacteriol. 191:6203–6210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Clewell DB, Tomich PK, Gawron-Burke MC, Franke AE, Yagi Y, An FY. 1982. Mapping of Streptococcus faecalis plasmids pAD1 and pAD2 and studies relating to transposition of Tn917. J. Bacteriol. 152:1220–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jacob AE, Hobbs SJ. 1974. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J. Bacteriol. 117:360–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Del Papa MF, Perego M. 2008. Ethanolamine activates a sensor histidine kinase regulating its utilization in Enterococcus faecalis. J. Bacteriol. 190:7147–7156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hancock LE, Shepard BD, Gilmore MS. 2003. Molecular analysis of the Enterococcus faecalis serotype 2 polysaccharide determinant. J. Bacteriol. 185:4393–4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hancock LE, Perego M. 2004. The Enterococcus faecalis fsr two-component system controls biofilm development through production of gelatinase. J. Bacteriol. 186:5629–5639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nieto C, Espinosa M. 2003. Construction of the mobilizable plasmid pMV158GFP, a derivative of pMV158 that carries the gene encoding the green fluorescent protein. Plasmid 49:281–285 [DOI] [PubMed] [Google Scholar]
- 20. Benachour A, Ladjouzi R, Le Jeune A, Hebert L, Thorpe S, Courtin P, Chapot-Chartier MP, Prajsnar TK, Foster SJ, Mesnage S. 2012. The lysozyme-induced peptidoglycan N-acetylglucosamine deacetylase PgdA (EF1843) is required for Enterococcus faecalis virulence. J. Bacteriol. 194:6066–6073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hoskins JR, Yanagihara K, Mizuuchi K, Wickner S. 2002. ClpAP and ClpXP degrade proteins with tags located in the interior of the primary sequence. Proc. Natl. Acad. Sci. U. S. A. 99:11037–11042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Qin X, Singh KV, Xu Y, Weinstock GM, Murray BE. 1998. Effect of disruption of a gene encoding an autolysin of Enterococcus faecalis OG1RF. Antimicrob. Agents Chemother. 42:2883–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brown MS, Ye J, Rawson RB, Goldstein JL. 2000. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100:391–398 [DOI] [PubMed] [Google Scholar]
- 24. Chandler JR, Dunny GM. 2008. Characterization of the sequence specificity determinants required for processing and control of sex pheromone by the intramembrane protease Eep and the plasmid-encoded protein PrgY. J. Bacteriol. 190:1172–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. An FY, Sulavik MC, Clewell DB. 1999. Identification and characterization of a determinant (eep) on the Enterococcus faecalis chromosome that is involved in production of the peptide sex pheromone cAD1. J. Bacteriol. 181:5915–5921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. An FY, Clewell DB. 2002. Identification of the cAD1 sex pheromone precursor in Enterococcus faecalis. J. Bacteriol. 184:1880–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Heinrich J, Wiegert T. 2006. YpdC determines site 1 degradation in regulated intramembrane proteolysis of the RsiW anti-sigma factor of Bacillus subtilis. Mol. Microbiol. 62:566–579 [DOI] [PubMed] [Google Scholar]
- 28. Heinrich J, Hein K, Wiegert T. 2009. Two proteolytic modules are involved in regulated intramembrane proteolysis of Bacillus subtilis RsiW. Mol. Microbiol. 74:1412–1426 [DOI] [PubMed] [Google Scholar]
- 29. Guariglia-Oropeza V, Helmann JD. 2011. Bacillus subtilis sigma(V) confers lysozyme resistance by activation of two cell wall modification pathways, peptidoglycan O-acetylation and d-alanylation of teichoic acids. J. Bacteriol. 193:6223–6232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hébert L, Courtin P, Torelli R, Sanguinetti M, Chapot-Chartier MP, Auffray Y, Benachour A. 2007. Enterococcus faecalis constitutes an unusual bacterial model in lysozyme resistance. Infect. Immun. 75:5390–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vollmer W, Tomasz A. 2002. Peptidoglycan N-acetylglucosamine deacetylase, a putative virulence factor in Streptococcus pneumoniae. Infect. Immun. 70:7176–7178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vollmer W, Tomasz A. 2000. The pgdA gene encodes for a peptidoglycan N-acetylglucosamine deacetylase in Streptococcus pneumoniae. J. Biol. Chem. 275:20496–20501 [DOI] [PubMed] [Google Scholar]
- 33. Blair DE, Schuttelkopf AW, MacRae JI, van Aalten DM. 2005. Structure and metal-dependent mechanism of peptidoglycan deacetylase, a streptococcal virulence factor. Proc. Natl. Acad. Sci. U. S. A. 102:15429–15434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Audran R. 1972. Bactericidal and bacteriolytic immune reactions. Respective roles of complement and lysozyme. Their value in defense mechanisms against infection. Rev. Fr. Transfus. 15:81–137 (In French.) [DOI] [PubMed] [Google Scholar]
- 35. Glynn AA. 1968. Lysozyme: antigen, enzyme and antibacterial agent. Sci. Basis Med. Annu. Rev. 1968:31–52 [PubMed] [Google Scholar]
- 36. Fabian TK, Hermann P, Beck A, Fejerdy P, Fabian G. 2012. Salivary defense proteins: their network and role in innate and acquired oral immunity. Int. J. Mol. Sci. 13:4295–4320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wiesner J, Vilcinskas A. 2010. Antimicrobial peptides: the ancient arm of the human immune system. Virulence 1:440–464 [DOI] [PubMed] [Google Scholar]
- 38. Fuller JR, Vitko NP, Perkowski EF, Scott E, Khatri D, Spontak JS, Thurlow LR, Richardson AR. 2011. Identification of a lactate-quinone oxidoreductase in Staphylococcus aureus that is essential for virulence. Front. Cell. Infect. Microbiol. 1:19. 10.3389/fcimb.2011.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Davis KM, Nakamura S, Weiser JN. 2011. Nod2 sensing of lysozyme-digested peptidoglycan promotes macrophage recruitment and clearance of S. pneumoniae colonization in mice. J. Clin. Invest. 121:3666–3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cole AM, Thapa DR, Gabayan V, Liao HI, Liu L, Ganz T. 2005. Decreased clearance of Pseudomonas aeruginosa from airways of mice deficient in lysozyme M. J. Leukoc. Biol. 78:1081–1085 [DOI] [PubMed] [Google Scholar]
- 41. Eudy WW, Burrous SE. 1971. Renal lysozyme levels in animals developing Proteus mirabilis-induced pyelonephritis. Appl. Microbiol. 21:300–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Eudy WW, Burrous SE, Sigler FW. 1971. Renal lysozyme levels in animals developing “sterile pyelonephritis”. Infect. Immun. 4:269–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dick W, Theilmann L. 1980. Urinary levels of lysozyme in children with acute chronic recurrent urinary tract infection. Padiatr Padol. 15:345–350 (In German.) [PubMed] [Google Scholar]
- 44. Frank KL, Guiton PS, Barnes AM, Manias DA, Chuang-Smith ON, Kohler PL, Spaulding AR, Hultgren SJ, Schlievert PM, Dunny GM. 2013. AhrC and Eep are biofilm infection-associated virulence factors in Enterococcus faecalis. Infect. Immun. 81:1696–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dommett R, Zilbauer M, George JT, Bajaj-Elliott M. 2005. Innate immune defence in the human gastrointestinal tract. Mol. Immunol. 42:903–912 [DOI] [PubMed] [Google Scholar]
- 46. Zellmeier S, Schumann W, Wiegert T. 2006. Involvement of Clp protease activity in modulating the Bacillus subtilis sigmaW stress response. Mol. Microbiol. 61:1569–1582 [DOI] [PubMed] [Google Scholar]
- 47. Ho TD, Hastie JL, Intile PJ, Ellermeier CD. 2011. The Bacillus subtilis extracytoplasmic function sigma factor sigma(V) is induced by lysozyme and provides resistance to lysozyme. J. Bacteriol. 193:6215–6222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sahm DF, Kissinger J, Gilmore MS, Murray PR, Mulder R, Solliday J, Clarke B. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 33:1588–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hastie JL, Williams KB, Ellermeier CD. 2013. The activity of σV, an extracytoplasmic function σ factor of Bacillus subtilis, is controlled by regulated proteolysis of the anti-σ factor RsiV. J. Bacteriol. 195:3135–3144 [DOI] [PMC free article] [PubMed] [Google Scholar]