Abstract
The majority of outer membrane (OM) lipoproteins in Gram-negative bacteria are tethered to the membrane via an attached lipid moiety and oriented facing in toward the periplasmic space; a few lipoproteins have been shown to be surface exposed. The outer membrane lipoprotein P6 from the Gram-negative pathogenic bacterium nontypeable Haemophilus influenzae (NTHi) is surface exposed and a leading vaccine candidate for prevention of NTHi infections. However, we recently found that P6 is not a transmembrane protein as previously thought (L. V. Michel, B. Kalmeta, M. McCreary, J. Snyder, P. Craig, M. E. Pichichero, Vaccine 29:1624–1627, 2011). Here we pursued studies to show that P6 has a dual orientation, existing infrequently as surface exposed and predominantly as internally oriented toward the periplasmic space. Flow cytometry using three monoclonal antibodies with specificity for P6 showed surface staining of whole NTHi cells. Confocal microscopy imaging confirmed that antibodies targeted surface-exposed P6 of intact NTHi cells and not internal P6 in membrane-compromised or dead cells. Western blots of two wild-type NTHi strains and a mutant NTHi strain that does not express P6 showed that P6 antibodies do not detect a promiscuous epitope on NTHi. Depletion of targets to nonlipidated P6 significantly decreased bactericidal activity of human serum. Protease digestion of surface-exposed P6 demonstrated that P6 is predominantly internally localized in a manner similar to its homologue Pal in Escherichia coli. We conclude that P6 of NTHi is likely inserted into the OM in two distinct orientations, with the predominant orientation facing in toward the periplasm.
INTRODUCTION
The outer membrane (OM) of Gram-negative bacteria is asymmetrically structured with lipopolysaccharides in its outer leaflet and phospholipids in its inner leaflet (1). The OM is also comprised of numerous lipoproteins that are typically tethered to the inner leaflet of the OM (via their N-terminally attached lipid moieties) and oriented toward the periplasmic space of the cell (1–4). However, a subset of lipoproteins, including the outer membrane protein P6 from nontypeable Haemophilus influenzae (NTHi), are surface exposed (5–8).
Since its discovery in the mid-1980s, P6 has been a leading vaccine candidate for prevention of NTHi infections in humans (acute otitis media, sinusitis, acute exacerbations of chronic bronchitis, and pneumonia). P6 is a strong vaccine candidate because it is immunogenic in children and adults, it is surface exposed, and it is highly conserved among pathogenic strains (5, 8–16).
Previous work demonstrated noncovalent binding of P6 to the peptidoglycan layer of the cell (17–19). Therefore, P6 was thought to be a transmembrane protein, able to access both intracellular and extracellular molecules by physically spanning the OM. In 2011, we demonstrated that P6 could not be a transmembrane protein based on structural and computational studies (20) utilizing the nuclear magnetic resonance (NMR) solution structure of P6 (Protein Data Bank [PDB] identification [ID] 2AIZ) (19). That discovery led us to reexamine all previous work on P6 and to formulate a hypothesis that P6 might exhibit two distinct orientations in the OM of NTHi (20). A dual orientation would reconcile previous work and our own. While we were pursuing experiments, the dual-orientation concept was described for the first time for the Lpp lipoprotein of Escherichia coli (7). Here we describe our work demonstrating that the P6 lipoprotein likely exists in two orientations in the OM of NTHi.
MATERIALS AND METHODS
Bacterial strains and cell culture conditions.
All NTHi cultures were grown on brain heart infusion (BHI) medium (BD) supplemented with 20 μg/ml NAD (Sigma) and 10 μg/ml hemin (Sigma). Wild-type NTHi (86-028NP) was a pediatric isolate (gift from Lauren Bakaletz, The Research Institute at Nationwide Children's Hospital) (21). Wild-type NTHi strain 49P5H1 and a mutant NTHi strain that does not express P6 were gifts from Timothy Murphy (State University at Buffalo) (22). Wild-type and mutant NTHi strains were cultured on supplemented BHI medium under aerobic conditions, with shaking (200 rpm) at 37°C for 3 to 4 h until the optical density at 490 nm (OD490) reached 0.8 (log phase). Cells were pelleted gently (5,000 × g) and washed before further sample preparation.
SDS-PAGE and immunoblot assay.
For the 10% SDS-PAGE experiments, samples were prepared in nonreducing sample buffer (2× recipe; 0.12 M Tris-Cl, pH 6.8, 4% SDS, 20% glycerol, 0.01% bromphenol blue) and boiled for 10 min. Proteins were transferred to a nitrocellulose membrane (Pierce), blocked with 5% milk in Tris-buffered saline (TBS), incubated with primary antibody (4G4 and 7F3 diluted 1:120; 3B9 and anti-protein D diluted 1:4,000) in 1% milk and TBS and then secondary antibody (1:3,000) in 1% milk and TBS with 0.05% Tween 20 (TBST), and washed with TBS or TBST between antibody incubations. Anti-protein D antibody serum (a gift from Kristian Riesbeck, Lund University, Skane University Hospital) was used to detect protein D, which served as a loading control for some of the NTHi Western blots. P6 monoclonal antibodies were gifts from Timothy Murphy (7F3, 4G4) and Michael Apicella (University of Iowa) (3B9). The secondary antibody was horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Bethyl Laboratories) for the P6 monoclonal antibodies or HRP-conjugated goat anti-rabbit IgG (Invitrogen) for anti-protein D. The blot was visualized using the LumiGLO Reserve HRP chemiluminescent substrate kit (KPL) according to the manufacturer's instructions.
Flow cytometry.
Bacterial samples were cultured as described above and used for surface expression analysis by flow cytometry. Between 300 and 500 μl of NTHi cells (OD490 = ∼0.8) was washed twice with phosphate-buffered saline (PBS), pH 7.2, and then incubated with 250 μl of primary antibody (3B9, 4G4, or 7F3) in serum at a 1:10 dilution or not diluted, depending on the experiment, for 1 h at room temperature. Monoclonal antibody-bound NTHi strains were washed twice with PBS and then incubated with 250 μl of either fluorescein isothiocyanate (FITC)-labeled goat anti-mouse IgG (BioLegend) at a 1:500 dilution or Alexa Fluor 488-labeled goat anti-mouse IgG (Invitrogen) at a 1:200 or 1:350 dilution (for the permeabilization experiment). After two additional PBS washes, the samples were resuspended in 200 μl of PBS and read on an LSR II flow cytometer (BD Biosciences).
In the permeabilization experiment, samples were prepared as described above with the following exceptions. Wash steps were performed using PBS-1% bovine serum albumin (BSA)-2 mM EDTA to decrease nonspecific binding. After the initial primary (4G4) and secondary (Alexa Fluor 488) incubations, the cells were fixed using 250 μl of 4% paraformaldehyde fixative reagent (BioLegend) for 30 min at room temperature. The cells were then permeabilized with 0.1% Triton X-100 (US Biologicals) in PBS (final concentration, ∼1.7 mM Triton X-100) for 30 min at room temperature. Selected samples were also incubated with primary antibody (4G4) after permeabilization (60 min, room temperature) and then the secondary antibody Alexa Fluor 647 goat anti-mouse IgG (Life Technologies) at a 1:350 dilution (30 min, room temperature). In summary, permeabilized samples were prepared as follows: (i) control 1, no primary antibody plus Alexa Fluor 488 (no permeabilization or second round of labeling); (ii) control 2, 4G4 plus Alexa Fluor 488, permeabilization, no primary antibody plus Alexa Fluor 647; (iii) 4G4 plus Alexa Fluor 488, permeabilization, 4G4 plus Alexa Fluor 647.
Confocal microscopy.
Bacterial samples were cultured as described above with modifications for confocal microscopy. All wash steps were performed using PBS-0.1% BSA. A total of 250 μl of monoclonal antibody (7F3) diluted 1:10 in PBS-0.1% BSA was incubated with whole cells for 1 h at room temperature. After three washes, the cells were incubated with 250 μl Alexa Fluor 488-labeled goat anti-mouse IgG (Invitrogen) at a 1:200 dilution in PBS-0.1% BSA for 30 min. After three additional washes, the samples were fixed in 4% paraformaldehyde fixative reagent (BioLegend) for 30 min, washed three more times, and then resuspended in 50 μl of PBS. To identify dead or membrane-compromised cells, 1 μl of a 1.83 mM propidium iodide (Molecular Probes) solution in dimethyl sulfoxide (DMSO) was added to the 50-μl cell sample (which was washed and resuspended in 0.85% NaCl instead of PBS). The sample was quickly mixed and incubated for less than 30 min before imaging.
The confocal images were collected on a Leica TCS SP5 II AOBS filter-free tunable spectral confocal research microscope with resonant scanner and hybrid detectors (Leica Microsystems Inc., Buffalo Grove, IL) attached to a Leica DMI6000 fully automated microscope using Leica LAS system software and a 60× oil immersion objective. The Argon laser 496 laser line was used to detect Alexa Fluor 488 (excitation at 496 nm, emission at 519 nm) and propidium iodide (excitation at 496 nm, emission at 635 nm). Differential interference contrast (DIC) images, green (Alexa Fluor 488) images, and red (propidium iodide) images were merged to identify dead or compromised cells (red) and fluorescently labeled whole cells (green).
Bactericidal assay.
Recombinant nonlipidated P6 was a gift from John Orban (University of Maryland Biotechnology Institute) (19). Purified nonlipidated P6 was prepared and analyzed via NMR spectroscopy as previously described (20). The bactericidal assay was performed as previously described (16, 23) with some modifications. The CFU count was established at each particular OD. NTHi strains were cultivated, harvested, and diluted to 107 CFU ml−1 in PBS containing potassium, magnesium, and calcium salts (PCMA). In 96-well plates, 50 μl of heat-inactivated serum sample was added and then serially diluted 2-fold 11 times, leaving the last well without serum, which served as a control. Ten microliters of NTHi (105 CFU) and 500 μl of precolostral calf serum were mixed as a source of complement, followed by PCMA buffer to create a total volume of 2.5 ml. Fifty microliters of the complement and bacteria mixture was added to 50 μl of serum. After a 60-min incubation, the number of surviving bacteria was determined by plating 10 μl onto chocolate agar and counting the colonies. One adult serum pool and one pediatric serum pool with known bactericidal titers against an NTHi strain (86-028NP) were run during each experiment as controls. Experiments were done in triplicate on two separate days to confirm reproducibility of the data. The bactericidal titer of the serum was defined as the inverse of the highest dilution that led to ≥50% bacterial killing and was compared to that of the negative control (complement plus bacterium). For the depletion of anti-P6 antibodies from serum, polystyrene beads were washed extensively with borate buffer (pH 8.0). Recombinant purified nonlipidated P6 was incubated with these beads overnight at room temperature. The beads were washed extensively, incubated in BSA-borate buffer for 30 min at room temperature, and then pelleted and incubated with 200 μl of patient sera for 2 h at room temperature. The beads were centrifuged (200 × g), and the supernatant was collected. The anti-P6 absorbed serum levels were measured by an enzyme-linked immunosorbent assay (ELISA) and used for bactericidal assays. The reciprocal bactericidal titers were compared with unadsorbed sera to determine the bactericidal activity mediated by anti-P6 antibodies. To ensure the selective depletion of only anti-P6 antibodies, polystyrene beads were adsorbed with protein D (less bactericidal) or OMP26 proteins (nonbactericidal) as a control; the depletion of anti-OMP26 antibodies did not affect the levels of either protein D- or P6-specific antibodies in the serum or vice versa (24). OMP26-depleted sera were also used (as a control) to perform the bactericidal assay; bactericidal titers before and after OMP26 adsorption were the same (data not shown), confirming that (i) P6 adsorption was specific and (ii) the reduction in the bactericidal activity as a result of the depletion was attributed to anti-P6 antibodies.
Protease digestion.
The protease digestion experiment was modified from that of Shin et al. (25). NTHi bacteria were cultured as described above. Approximately 60 ml of cell culture was harvested via centrifugation (5,000 × g) for 8 min at 4°C and resuspended in 6 ml of PBS. The resuspension was split into two aliquots, 1 ml and 5 ml. The 1-ml aliquot was stored at 4°C to be used as the whole-cell sample, while the 5-ml sample was lysed via sonication (cycles of 15 s on and 30 s off for 20 cycles on ice). Fifty microliters of the whole- and lysed-cell samples was removed and placed in 2× nonreducing sample buffer (described above) supplemented with 5 mM Pefabloc SC (Sigma) and flash frozen in liquid nitrogen (“time zero” samples). One microliter of a 1:10 dilution of proteinase K (Sigma; catalog number P4850) was added to 1 ml of the whole-cell sample and to 1 ml of the lysed-cell sample (final concentration, 2.3 μg/ml). Both samples were placed in a 37°C water bath; 50-μl aliquots were removed from each sample at increasing time intervals for up to 5 h (samples were placed in a gently shaking incubator after 2 h to prevent cell aggregation). All 50-μl aliquots were immediately placed in nonreducing sample buffer supplemented with Pefabloc SC and flash frozen in liquid nitrogen. Samples were quickly thawed and boiled for 10 min immediately before being separated on a 10% SDS-PAGE gel. The Western blot was detected using the P6 monoclonal antibody 4G4 (described above). As a loading control, samples were also detected using anti-proteinase K polyclonal antibody (LifeSpan Biosciences, Inc.) at a 1:80,000 dilution in 1% milk in TBS with HRP-conjugated goat anti-rabbit IgG (Invitrogen) at a 1:3,000 dilution in 1% milk and TBST.
RESULTS
P6 surface expression is not the predominant orientation of the protein.
In previous work, anti-P6 and gold-labeled secondary antibodies visualized using electron microscopy were used to demonstrate surface exposure of P6 (5, 8). To confirm P6 surface exposure in NTHi, we utilized a similar primary-secondary antibody “sandwich” technique but employed flow cytometry to detect the fluorescently labeled secondary antibody. Flow cytometric analysis using three monoclonal antibodies (3B9, 4G4, and 7F3) with specificity for P6 demonstrated surface staining on whole NTHi cells (strain 86-028) (Fig. 1). Flow cytometric detection of a P6-null mutant NTHi strain (21) confirmed that the P6 monoclonal antibodies 4G4, 7F3, and 3B9 do not stain the cell surface of the mutant NTHi strain (Fig. 1). As shown in Fig. 2C and E, whole NTHi cells were stained with a 4G4/fluorescent secondary antibody sandwich (3.6 to 3.8% of cells). However, when NTHi cells were permeabilized with Triton X-100 and then stained with a second fluorescent antibody sandwich, we observed a large increase in labeling (84.3%) (Fig. 2F).
Fig 1.
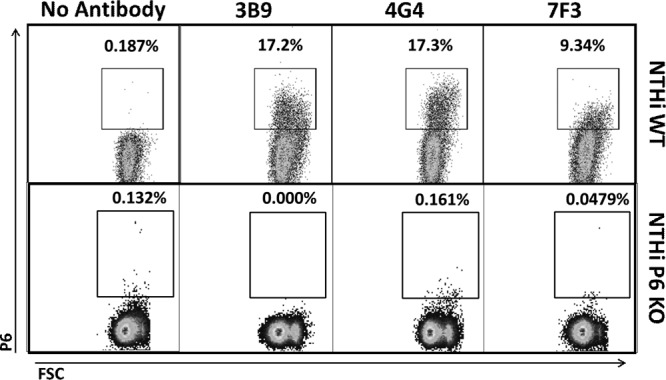
There is low surface staining of the P6 protein in NTHi cells using flow cytometric analysis. Representative data from flow cytometry experiments on NTHi wild-type (WT) whole cells (86-028) bound with no antibody or a single P6 monoclonal antibody (3B9, 4G4, or 7F3) demonstrate low levels of surface staining. Similar experiments on NTHi P6 knockout (KO) cells (no P6 expressed) demonstrate no surface staining above background. The fluorescently labeled cells are contained within the square inset (gated on the FITC-positive population), and the percentage of labeled cells is noted above the inset. FSC, forward scatter.
Fig 2.
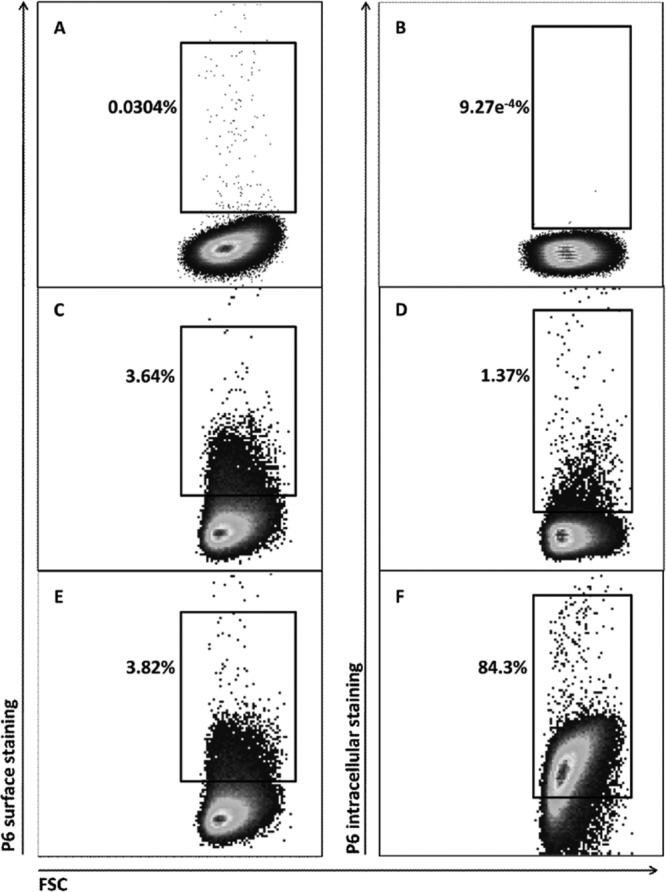
NTHi cells demonstrate high intracellular staining after permeabilization by flow cytometric analysis. Only background staining is detected in NTHi cells stained with (no primary) secondary Alexa Fluor 488 (surface staining) (A) or secondary Alexa Fluor 647 (intracellular staining) (B) after permeabilization. To quantify differences in intracellular versus surface staining of P6, NTHi cells were incubated with primary 4G4/secondary Alexa Fluor 488 (surface staining), permeabilized, and then reincubated with primary 4G4/secondary Alexa Fluor 647 (intracellular staining). Only 3.82% of cells were positive for surface staining (E), but 84.3% of cells were positive for intracellular staining (F). The shift in intracellular P6 detection is not observed in cells that were incubated with primary 4G4/secondary Alexa Fluor 488 and only the secondary Alexa Fluor 647 (no primary), as the surface-staining signal (C) and intracellular signal (D) are both low. Images are from a single set of representative flow cytometry experiments on NTHi wild-type cells (strain 86-028). The fluorescently labeled cells are contained within the square inset, with the percentage of labeled cells noted next to the inset.
NTHi strain 02-011-01 (formerly known as HH13) (26), which contains known amino acid changes in P6 (Fig. 3), was used in flow cytometric detection with 3B9, 4G4, and 7F3 monoclonal antibodies. The D59N (aspartate-to-asparagine change at amino acid 59) mutation in P6 in 02-011-01 has previously been shown to completely eliminate binding to monoclonal antibody 7F3 but does not affect binding to the monoclonal antibodies 3B9 and 4G4 (20). Results confirm that 7F3 does not bind P6 in 02-011-01 but 3B9 and 4G4 antibodies readily stain the 02-011-01 cells (Fig. 4).
Fig 3.

Comparison of P6 from NTHi wild type (WT) (86-028) and that from mutant (MT) strain 02-011-01. The P6 amino acid sequences from the NTHi wild-type strain (86-028) (top, red) and a naturally occurring P6 mutant NTHi strain (02-011-01) (bottom, blue) are aligned, and differences are highlighted in bold/italics (asterisks indicate a gap in the 02-011-01 sequence compared to the wild type). Residue 59, the site of the D59N mutation, is underlined.
Fig 4.
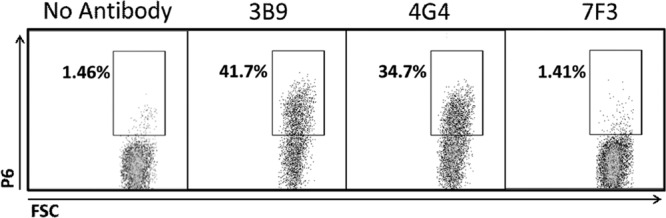
NTHi strain 02-011-01 demonstrates different amounts of P6 staining with the monoclonal antibodies 3B9, 4G4, and 7F3 using flow cytometric analysis. The representative images are from flow cytometry experiments on NTHi strain 02-011-01 whole cells (containing the P6 D59N mutant) stained with no antibody or a single P6 monoclonal antibody (3B9, 4G4, and 7F3). The fluorescently labeled cells are contained within the square inset, and the percentage of labeled cells is noted next to the inset.
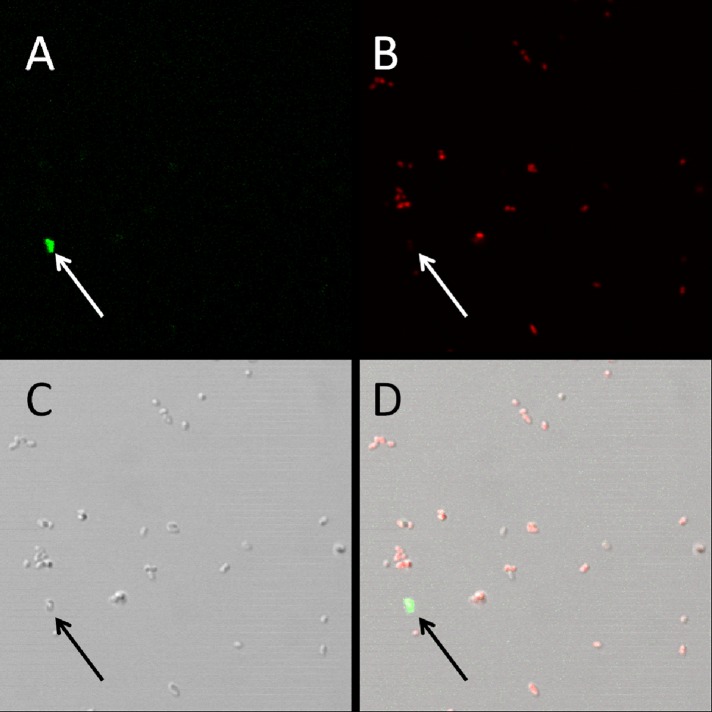
To demonstrate that P6 monoclonal antibodies target surface-exposed P6 of intact NTHi cells and not internal P6 in membrane-compromised or dead cells, we examined fluorescent staining of NTHi using confocal microscopy. A fluorophore-conjugated secondary antibody bound to the P6 monoclonal antibody 7F3 was shown to stain the surface of NTHi cells, and the cell morphologies of stained and unstained cells were similar (Fig. 5). Figure 5 also demonstrates how few NTHi cells are stained with the P6 fluorescent antibody sandwich, consistent with the flow cytometry results described above. Moreover, 7F3 stained NTHi cells that were not stained by propidium iodide, a red fluorescent dye that penetrates cells with damaged membranes (Fig. 6).
Fig 5.

A low percentage of whole NTHi cells demonstrates surface staining for P6 protein by confocal microscopy. Psuedocolor images of NTHi cells stained with primary anti-P6 antibody (7F3) and secondary Alexa Fluor 488 (green) demonstrate that only a small percentage of NTHi cells is positive for P6 surface expression.
Fig 6.
P6 antibodies bind to surface-exposed P6 protein on viable NTHi cells. The psuedocolor images are of NTHi cells stained with 7F3/secondary Alexa Fluor 488 (green) (A) and propidium iodide (red) (B). The arrow in the merged image (D) points to an NTHi cell that is positive for P6 surface staining but does not stain with propidium iodide. The differential interference contrast image (C) demonstrates normal morphology of the surface-stained NTHi cell.
Testing for a promiscuous epitope with P6 structural similarity.
To address the possibility that P6 monoclonal antibodies interacted with a promiscuous epitope on a non-P6, surface-exposed protein of NTHi (20), we performed standard Western blotting experiments on two wild-type NTHi strains (86-028 and 49P5H1) and a mutant NTHi strain that does not express P6 (constructed in strain 49P5H1) (22). Western blots of two wild-type NTHi whole-cell lysates probed using the three P6 monoclonal antibodies show that 3B9, 4G4, and 7F3 each bind to a single protein at the molecular weight of P6 and no other proteins in wild-type NTHi strains. Western blots of mutant NTHi whole-cell lysates (in which P6 is not expressed) showed that monoclonal antibodies 3B9, 4G4, and 7F3 do not bind any other NTHi proteins (Fig. 7). NTHi lipoprotein protein D, expressed in all three strains of NTHi, was also detected in the samples as a loading control.
Fig 7.
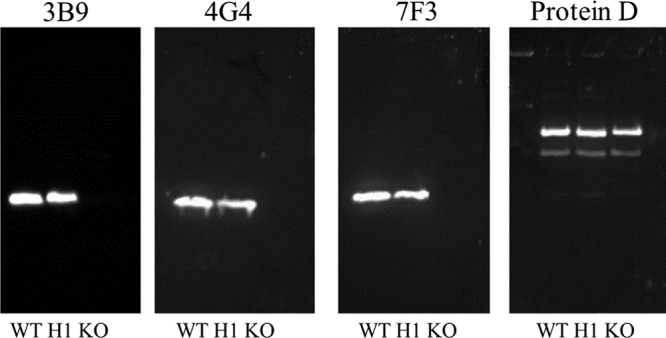
Western blot detection of P6 demonstrates specificity to the P6 antigen on whole-cell lysates from wild-type NTHi strains 86-028 (WT) and 49P5H1 (H1) and a mutant P6 knockout NTHi strain (KO). The P6 monoclonal antibodies 3B9, 4G4, and 7F3 detect only one band (at the molecular weight of P6) from wild-type whole-cell lysates and do not detect protein from a P6 knockout strain (KO). Protein D was detected using polyclonal antibody serum to demonstrate protein-loading consistency.
Bactericidal assay with folded P6.
The presence of bactericidal antibody in human serum is related to protection from disease caused by NTHi (8, 20). Importantly, the antigenic targets of bactericidal antibodies are exclusively exposed structures found on the surface of the bacteria. Prior work had shown that antibody directed to P6 produced a bactericidal effect against NTHi (22, 27). However, in those studies, the P6 protein used was either recombinant protein or native P6 extracted from NTHi. In both cases, P6 was in a lipidated form (i.e., a lipid moiety was attached to the protein's N terminus) and the conformation of the lipidated protein was not evaluated. From NMR experiments, we found that lipidated P6 protein is easily unfolded and/or aggregated (data not shown). Therefore, we sought to determine that folded P6 (as confirmed by NMR spectroscopy) was a target for human bactericidal antibody. In Fig. 8, we show that the bactericidal activity of human serum significantly decreased upon depletion of folded P6 targets, suggesting that P6 can be the target of human bactericidal (surface-exposed) antibodies.
Fig 8.
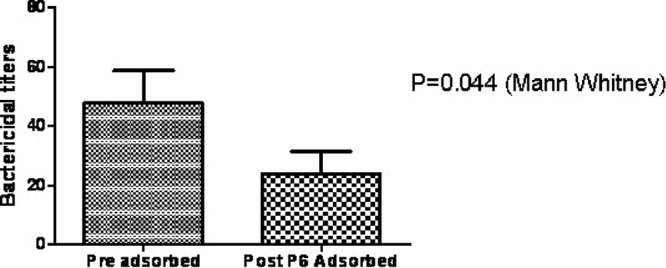
Bactericidal assay with anti-P6 adsorbed sera. Sera were depleted for anti-P6 antibodies using nonlipidated, folded P6 protein. The bactericidal assay was performed with both preadsorbed and adsorbed sera (depleted for anti-P6 antibodies) in the same plate to determine the contribution of anti-P6 antibodies to total bactericidal activity of serum. Results are expressed as the mean bactericidal activity with the standard deviation. A P value of <0.05 was considered significant.
P6 is likely expressed on the inner leaflet of NTHi.
The majority of outer membrane lipoproteins in Gram-negative bacteria are tethered to the membrane via an attached lipid moiety and oriented facing in toward the periplasmic space. To confirm that P6 is also oriented facing in toward the periplasmic space, protease digestion experiments modified from a previously described method (25) were performed using whole and lysed NTHi cells. Proteins expressed on the surface of NTHi, including P6, were removed by digestion with proteinase K. NTHi cells were then lysed to detect P6 that was not surface exposed. A Western blot using a P6 monoclonal antibody (4G4) was used to probe for P6 protein (Fig. 9). P6 was detectable in whole cells after removal of surface-exposed P6 with proteinase K. When NTHi cells were lysed prior to proteinase K digestion, P6 was completely digested between 15 min and 1 h. An immunoblot with anti-proteinase K antibody served as a loading control for the experiment, and a standard Coomassie blue stain was also performed on both the whole-cell gel and the lysed-cell gel to visualize the digested proteins (see Fig. S1 in the supplemental material).
Fig 9.
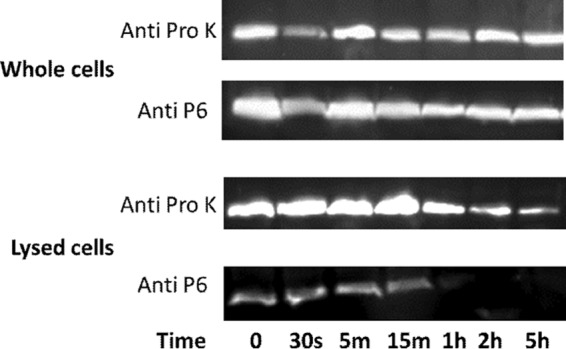
Intracellular P6 protein is protected from digestion with proteinase K. Whole or lysed cells were treated with proteinase K enzyme for 0 min, 30 s, 5 min, 15 min, 1 h, 2 h, and 5 h and then immediately mixed with sample buffer/Pefabloc and flash frozen in liquid nitrogen to stop digestion reactions. Representative Western blots probed with the P6 monoclonal antibody 4G4 (Anti P6) or anti-proteinase K antibody (Anti Pro K) demonstrate that P6 protein was protected from digestion in whole cells and degraded in lysed cells between 15 min and 1 h. Proteinase K was detected using an anti-proteinase K antibody to serve as a loading control.
DISCUSSION
In this paper, we show that lipoprotein P6 of NTHi likely exists in a dual orientation—surface exposed and oriented facing in toward the periplasmic space. We propose that P6 of NTHi is a second protein from a Gram-negative bacterium with dual orientation, similar to the recent description of Lpp of Escherichia coli (7). Interestingly, the structures of Lpp are distinct in the dual orientations (7). The proposed structure of internally localized Lpp is a homotrimer that attaches to the outer membrane via its lipid moieties, i.e., the inward orientation is achieved by three lysine residues attached to the peptidoglycan layer. Surface-exposed Lpp is also a homotrimer, but it physically spans the outer membrane of E. coli as a transmembrane protein. Lpp is a small, 56-residue lipoprotein with a single structural feature, that of an alpha helix (28). When trimerized, Lpp forms a helix bundle, a structural motif commonly seen among transmembrane proteins (29). In contrast to Lpp, P6 exhibits a more complex fold, consisting of a combination of alpha helices, loops, and a beta sheet (19). The P6 epitope for the monoclonal antibody 3B9 was mapped to be a conformational epitope consisting of nonsequential residues (residues 87 to 94, 147, 148), as shown in Fig. 10. The proposed peptidoglycan binding site (19) consists of residues overlapping the epitope for 3B9 (Fig. 10). Since the monoclonal antibody 3B9 interacts with surface-exposed P6 (as shown by flow cytometry and immunogold experiments), we know that at least part of P6 maintains its native fold in the peptidoglycan-associated and surface-exposed forms.
Fig 10.
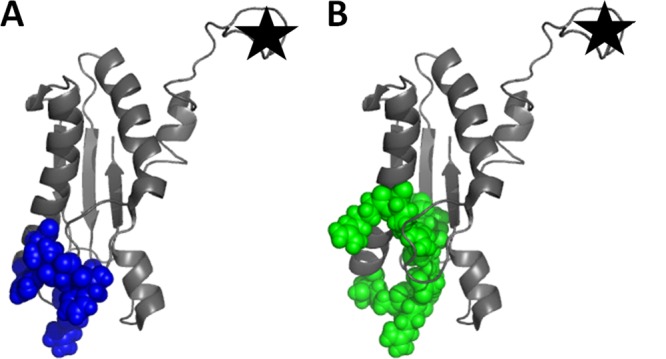
Proposed interaction sites on P6. Several residues proposed to be involved in binding peptidoglycan (blue) (A) and the monoclonal antibody 3B9 (green) (B) are the same in both cases. The N-terminal lipid moiety was not included in the NMR solution structure of P6 (PDB ID 2AIZ) but would be located at the part of the structure that is marked with a star. This structure was prepared using PyMOL Molecular Graphics System, version 1.5.0.1 (Schrödinger, LLC).
Our previous analysis of the P6 structure described it as a non-membrane-spanning protein because (i) the structure of P6 does not have the exposed hydrophobic residues necessary for it to interact favorably with the inside of the membrane and (ii) the overall length of P6 is too short to physically span the membrane and interact with the peptidoglycan layer in the periplasm and antibodies (such as 7F3) on the cell surface (20). Here we have shown that a portion of NTHi P6 is surface exposed and that the monoclonal antibodies (3B9, 4G4, and 7F3) to P6 do not bind promiscuously to a structurally similar non-P6 protein. However, our results suggest that only a small percentage of the NTHi population expresses P6 on its surface, as shown in the flow cytometric analysis and confocal microscope images. Consistently, we observed that less than 20% (and more often less than 5%) of NTHi cells expressed detectable P6 on their surface. Confocal images also confirmed that surface-exposed P6 can be detected in whole, uncompromised cells.
Because most of our experiments relied on the use of monoclonal antibodies to detect P6, it was important to ensure that the monoclonal antibodies did not bind proteins other than P6. If a “promiscuous epitope” on another protein existed (30) and that epitope was present on the surface of NTHi, our studies would have erroneously concluded that P6 was surface exposed. Although it had been shown that the P6 monoclonal antibody 7F3 does not interact with proteins other than P6 (22), a similar set of experiments was performed using 4G4 and 3B9. We tested all three monoclonal antibodies using two wild-type NTHi strains (86-028 and 49P5H1) and a mutant strain of NTHi that does not express P6 (22). If P6 monoclonal antibodies interacted with a protein other than P6, we would expect to observe multiple bands in the immunoblot of cell lysate from the wild-type strains and a single band in the Western blot of mutant NTHi. Since we observed only single bands in the wild-type immunoblots and no detectable bands in the mutant immunoblots, we conclude that the P6 monoclonal antibodies do not bind proteins other than P6. In addition, flow cytometric analysis showed that the monoclonal antibodies 4G4, 7F3, and 3B9 did not stain the cell surface of the mutant strain of NTHi that does not express P6 and that 7F3 did not stain the cell surface of NTHi strain 02-011-01 that contained a P6 variant that was incapable of binding 7F3. The monoclonal antibodies 3B9 and 4G4, however, did stain the cell surface of 02-011-01. Taken together, the results demonstrate that monoclonal antibody binding is mediated by the P6 protein and not a promiscuous epitope.
An alternative method for indirect demonstration of surface exposure is a bactericidal assay. In the past, bactericidal activity of human serum was detected using lipidated P6 that aggregates and/or unfolds due to the exposed lipid moiety (as detected by NMR spectroscopy). In order to show physiologic relevance, we repeated the bactericidal assay using nonlipidated P6 that more accurately mimics the physiological structure of the protein in vivo. Our results corroborated those of previous bactericidal studies on lipidated P6 by demonstrating that nonlipidated, folded P6 is the target of bactericidal antibodies.
Since its initial characterization (14), P6 has been known to interact noncovalently with the cell wall or peptidoglycan layer (17–19, 31, 32). Because P6 is a member of a class of proteins known as peptidoglycan-associated lipoproteins (PAL), its localization in the periplasmic space would be expected (22). In fact, strong detergents or high temperatures must be employed to separate P6 from the peptidoglycan layer during purification of P6 from native NTHi (19, 31, 32). Results from our protease digestion experiments corroborate the notion that P6 is internally localized. Incomplete digestion of P6 in whole cells suggests that some of P6 was not accessible to proteinase K activity. However, it may be that protease digestion was incomplete due to surface crowding, protein aggregation, or only partial surface exposure of P6. Therefore, another possible explanation of the results from the proteinase K experiment is that P6 is surface exposed but not susceptible to protease digestion due to surface crowding or an alternate physical/chemical reason.
Flow cytometric analysis demonstrated that while only a small fraction (3 to 4%) of whole NTHi cells displayed detectable P6 on their surface, a much larger percentage of cells (84.3%) expressed detectable P6 after they were permeabilized with 0.1% Triton X-100. Similar low levels of Triton X-100 have been shown to permeabilize cells such that antibodies are able to access intracellular antigens (33, 34). These results suggest that a much larger population of P6 is expressed on the inner leaflet of the OM in NTHi and, therefore, is accessible to antibodies only after permeabilization. However, these results are also consistent with Triton X-100 increasing access to P6 by altering the surface of NTHi.
The homologue to P6 in E. coli is also a peptidoglycan-associated lipoprotein, aptly named Pal (35). The sequence and structure of Pal (Protein Data Bank ID 1OAP) are highly similar to those of P6 (19, 36), suggesting the possibility of similar biological functions, homologous protein interactions, and/or similar localizations. It has been well documented that the majority of Pal is located in the periplasm of E. coli, with its N-terminal lipid moiety inserted into the inner leaflet of the outer membrane, allowing it to interact with the C-terminal periplasmic domain of OmpA (35), the periplasmic protein TolB (35, 37), the inner membrane protein TolA (38), and the peptidoglycan layer itself (27). Although the equivalent protein-protein interactions have not been described in NTHi, many of the proteins mentioned above have homologues in NTHi.
In summary, the experiments described here corroborate past studies on P6 which point to P6 being internally localized (5, 8). Results from the protease digestion experiment suggest that a portion of P6 protein is not accessible to proteinase K digestion. Flow cytometric analysis suggests that a significant proportion of P6 is accessible to antibodies only after permeabilization of the cells via low concentrations of Triton X-100. Confocal microscope images and flow cytometric analysis demonstrate that only a small percentage of NTHi cells exhibit P6 labeling. Native P6 has been shown to coprecipitate and/or copurify with peptidoglycan (19, 31, 32). P6's homologue in E. coli (Pal) has been shown to interact with the C-terminal periplasmic domain of OmpA (35), the periplasmic protein TolB (35, 37), the inner membrane protein TolA (27), and the peptidoglycan layer itself (27). Taken together, these results support a conclusion that P6 is also localized to the periplasmic space of NTHi, although we cannot fully exclude the alternative, though unlikely, possibilities.
Surface-exposed lipoproteins in pathogenic bacteria, in particular those proteins whose sequences/structures are conserved among strains, are often considered good protein vaccine candidates. What has not been considered is the viability of protein candidates with dual orientation, especially if the surface orientation is not the predominant population. The observation has implications for P6 as a vaccine candidate and requires further study of the mechanism responsible for infrequent expression of P6 on the surface, where bactericidal antibody might kill the organism. With increased vaccine pressure, bacterial targets often evolve to evade the protective immune system (39). As such, dual-orientation lipoprotein distributions may change after vaccine implementation.
Studies to determine the mechanisms controlling differential P6 orientation in the OM of NTHi are under way. Several integral transmembrane proteins have been shown to exhibit dual membrane topologies (40–42). In those cases, the protein exhibits two opposite orientations (one flipped upside down from the other), but both orientations maintain the same membrane-spanning regions. Studies on these dual-topology membrane proteins suggest that “flipping” occurs due to gene duplication followed by divergent topology evolution (40) or “flipping” occurs in the endoplasmic reticulum (in eukaryotic cells) shortly after protein synthesis (41). We propose that dual orientation of P6 occurs (i) during the initial insertion of the lipoprotein into the OM, most likely via the Lol system (43, 44), or (ii) after initial insertion of P6 into the OM. In the latter case, P6 would “flip” from the inner leaflet to the outer leaflet of the OM. Due to the amphipathic nature of bilayer membranes, the flipping of P6 to its external orientation would require a significant amount of energy input and most likely some sort of “flippase” enzyme to assist the process. In either case, we suggest that dual orientation of P6 has biological relevance to NTHi or it would not have been conserved through evolution.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by NIH NIDCD RO1 08671 (to M.E.P.), the Rochester Institute of Technology, and the Rochester Institute of Technology/Rochester General Hospital Research Institute Strategic Alliance Fund. Part of the work done by J. Snyder was supported by an ASBMB UAN undergraduate research award. The Confocal Microscopy Lab at the Rochester Institute of Technology is funded, in part, by a Major Research Instrumentation Grant from NSF (1126629).
We thank Cheryl Hanzlik, a technician at the Confocal Microscopy Lab at the Rochester Institute of Technology, for assistance and use of the microscope to obtain images. We thank David Veerhoven and Kathy Dermody (Rochester General Hospital Research Institute) for assistance with flow cytometric studies, Timothy Murphy (University at Buffalo, SUNY), Michael Apicella (University of Iowa), Kristian Riesbeck (Lund University, Skane University Hospital), and Lauren Bakaletz (The Research Institute at Nationwide Children's Hospital) for gifts of monoclonal antibodies and NTHi strains, John Orban (University of Maryland Biotechnology Institute) for nonlipidated P6 DNA, and Martin Pavelka and Michelle Dziejman (University of Rochester Medical Center) for thoughtful discussions.
Footnotes
Published ahead of print 17 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00185-13.
REFERENCES
- 1. Malinverni JC, Silhavy TJ. 2009. An ABC transport system that maintains lipid asymmetry in the Gram-negative outer membrane. Proc. Natl. Acad. Sci. U. S. A. 106:8009–8014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kovacs-Simon A, Titball RW, Michell SL. 2011. Lipoproteins of bacterial pathogens. Infect. Immun. 79:548–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ruiz M, Kahne D, Silhavy TJ. 2006. Advances in understanding bacterial outer-membrane biogenesis. Nat. Rev. Microbiol. 4:57–66 [DOI] [PubMed] [Google Scholar]
- 4. Tokuda H. 2009. Biogenesis of outer membranes in Gram-negative bacteria. Biosci. Biotechnol. Biochem. 73:465–473 [DOI] [PubMed] [Google Scholar]
- 5. Bogdan JA, Apicella MA. 1995. Mapping of a surface-exposed, conformational epitope of the P6 protein of Haemophilus influenzae. Infect. Immun. 63:4395–4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cornelissen CN, Kelley M, Hobbs MM, Anderson JE, Cannon JG, Cohen MS, Sparling PF. 1998. The transferrin receptor expressed by gonococcal strain FA1090 is required for the experimental infection of human male volunteers. Mol. Microbiol. 27:611–616 [DOI] [PubMed] [Google Scholar]
- 7. Cowles CE, Li Y, Semmelhack MF, Cristea IM, Silhavy TJ. 2011. The free and bound forms of Lpp occupy distinct subcellular locations in Escherichia coli. Mol. Microbiol. 79:1168–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nelson MB, Murphy TF, van Keulen H, Rekosh D, Apicella MA. 1988. Studies on P6, an important outer membrane protein antigen of Haemophilus influenzae. Rev. Infect. Dis. 10(Suppl 2):S331–S336 [DOI] [PubMed] [Google Scholar]
- 9. DeMaria TF, Murwin DM, Leake ER. 1996. Immunization with outer membrane protein P6 from nontypeable Haemophilus influenzae induces bactericidal antibody and affords protection in the chinchilla model of otitis media. Infect. Immun. 64:5187–5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Green BA, Quinn-Dey T, Zlotnik GW. 1987. Biologic activities of antibody to a peptidoglycan-associated lipoprotein of Haemophilus influenzae against multiple clinical isolates of H. influenzae type b. Infect. Immun. 55:2878–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kyd JM, Dunkley ML, Cripps AW. 1995. Enhanced respiratory clearance of nontypeable Haemophilus influenzae following mucosal immunization with P6 in a rat model. Infect. Immun. 63:2931–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Murphy TF, Bakaletz LO, Kyd JM, Watson B, Klein DL. 2005. Vaccines for otitis media: proposals for overcoming obstacles to progress. Vaccine 23:2696–2702 [DOI] [PubMed] [Google Scholar]
- 13. Murphy TF, Bartos LC, Rice PA, Nelson MB, Dudas KC, Apicella MA. 1986. Identification of a 16,000-dalton outer membrane protein on nontypeable Haemophilus influenzae as a target for human serum bactericidal antibody. J. Clin. Invest. 78:1020–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Murphy TF, Nelson MB, Dudas KC, Mylotte JM, Apicella MA. 1985. Identification of a specific epitope of Haemophilus influenzae on a 16,600-dalton outer membrane protein. J. Infect. Dis. 152:1300–1307 [DOI] [PubMed] [Google Scholar]
- 15. Murphy TF. 2005. Vaccine development for non-typeable Haemophilus influenzae and Moraxella catarrhalis: progress and challenges. Expert Rev. Vaccines 4:843–853 [DOI] [PubMed] [Google Scholar]
- 16. Sabirov A, Casey JR, Murphy TF, Pichichero ME. 2009. Breast feeding is associated with a reduced frequency of acute otitis media and high serum antibody levels against NTHi and outer membrane protein vaccine antigen candidate P6. Pediatr. Res. 66:565–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Deich RA, Metcalf BJ, Finn CW, Farley JE, Green BA. 1988. Cloning of the genes encoding a 15,000-dalton peptidoglycan-associated outer membrane protein from Haemophilus influenzae. J. Bacteriol. 170:489–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Green BA, Metcalf BJ, Quinn-Dey T, Kirkley DH, Quataert SA, Deich RA. 1990. A recombinant non-fatty acylated form of the Hi-PAL (P6) protein of Haemophilus influenzae elicits biologically active antibody against both nontypeable and type b H. influenzae. Infect. Immun. 58:3272–3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Parsons LM, Lin F, Orban J. 2006. Peptidoglycan recognition by Pal, an outer membrane lipoprotein. Biochemistry 45:2122–2128 [DOI] [PubMed] [Google Scholar]
- 20. Michel LV, Kalmeta B, McCreary M, Snyder J, Craig P, Pichichero ME. 2011. Vaccine candidate P6 of nontypeable Haemophilus influenzae is not a transmembrane protein based on protein structural analysis. Vaccine 29:1624–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bakaletz LO, Tallan BM, Hoepf T, DeMaria TF, Birck HG, Lim DJ. 1988. Frequency of fimbriation of nontypeable Haemophilus influenzae and its ability to adhere to chinchilla and human respiratory epithelium. Infect. Immun. 56:331–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Murphy TF, Kirkham C, Lesse AJ. 2006. Construction of a mutant and characterization of the role of the vaccine antigen P6 in outer membrane integrity of nontypeable Haemophilus influenzae. Infect. Immun. 74:5169–5176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Neary JM, Murphy TF. 2006. Antibodies directed at a conserved motif in loop 6 of outer membrane protein P2 of nontypeable Haemophilus influenzae recognize multiple strains in immunoassays. FEMS Immunol. Med. Microbiol. 46:251–261 [DOI] [PubMed] [Google Scholar]
- 24. Khan MN, Kaur R, Pichichero ME. 2012. Bactericidal antibody response against P6, protein D, and OMP26 of nontypeable Haemophilus influenzae after acute otitis media in otitis-prone children. FEMS Immunol. Med. Microbiol. 65:439–447 [DOI] [PubMed] [Google Scholar]
- 25. Shin JJ, Bryksin AV, Godfrey HP, Cabello FC. 2004. Localization of BmpA on the exposed outer membrane of Borrelia burgdorferi by monospecific anti-recombinant BmpA rabbit antibodies. Infect. Immun. 72:2280–2287 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26. Chang A, Kaur R, Michel LV, Casey JR, Pichichero ME. 2011. Haemophilus influenzae vaccine candidate outer membrane protein P6 is not conserved in all strains. Hum. Vaccin. 7:102–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lugtenberg B, Bronstein H, Van Selm N, Peters R. 1977. Peptidoglycan-associated outer membrane proteins of gram negative bacteria. Biochim. Biophys. Acta 465:571–578 [DOI] [PubMed] [Google Scholar]
- 28. Shu W, Liu J, Ji H, Lu M. 2000. Core structure of the outer membrane lipoprotein from Escherichia coli at 1.9 A resolution. J. Mol. Biol. 299:1101–1112 [DOI] [PubMed] [Google Scholar]
- 29. Bowie JU. 1999. Helix-bundle membrane protein fold templates. Protein Sci. 8:2711–2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kramer A, Keitel T, Winkler K, Stöcklein W, Höhne W, Schneider-Mergener J. 1997. Molecular basis for the binding promiscuity of an anti-p24 (HIV-1) monoclonal antibody. Cell 91:799–809 [DOI] [PubMed] [Google Scholar]
- 31. Munson RS, Granoff DM. 1985. Purification and partial characterization of outer membrane proteins P5 and P6 from Haemophilus influenzae type b. Infect. Immun. 49:544–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Murphy TF, Bartos LC, Campagnari AA, Nelson MB, Apicella MA. 1986. Antigenic characterization of the P6 protein of nontypable Haemophilus influenzae. Infect. Immun. 54:774–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jamur MC, Oliver C. 2010. Permeabilization of cell membranes. Methods Mol. Biol. 588:63–66 [DOI] [PubMed] [Google Scholar]
- 34. van de Ven AL, Adler-Storthz K, Richards-Kortum R. 2009. Delivery of optical contrast agents using Triton-X100, part 1. reversible permeabilization of live cells for intracellular labeling. J. Biomed. Opt. 14:021012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Clavel T, Germon P, Vianney A, Portalier R, Lazzaroni JC. 1998. TolB protein of Escherichia coli K-12 interacts with the outer membrane peptidoglycan-associated proteins Pal, Lpp and OmpA. Mol. Microbiol. 29:359–367 [DOI] [PubMed] [Google Scholar]
- 36. Godlewska R, Wiśniewska K, Pietras Z, Jagusztyn-Krynicka EK. 2009. Peptidoglycan-associated lipoprotein (Pal) of Gram-negative bacteria: function, structure, role in pathogenesis and potential application in immunoprophylaxis. FEMS Microbiol. Lett. 298:1–11 [DOI] [PubMed] [Google Scholar]
- 37. Bouveret E, Derouiche R, Rigal A, Lloubés R, Lazdunski C, Bénédetti H. 1995. Peptidoglycan-associated lipoprotein-TolB interaction. J. Biol. Chem. 270:11071–11077 [DOI] [PubMed] [Google Scholar]
- 38. Cascales E, Gavioli M, Sturgis JN, Lloubés R. 2000. Proton motive force drives the interaction of the inner membrane TolA and outer membrane Pal proteins in Escherichia coli. Mol. Microbiol. 38:904–915 [DOI] [PubMed] [Google Scholar]
- 39. Golubchik T, Brueggemann AB, Street T, Gertz RE, Jr, Spencer CCA, Ho T, Giannoulatou E, Link-Gelles R, Harding RM, Beall B, Peto TEA, Moore MR, Donnelly P, Crook DW, Bowden R. 2012. Pneumococcal genome sequencing tracks a vaccine escape variant formed through a multi-fragment recombination event. Nat. Genet. 44:352–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rapp M, Granseth E, Seppala S, von Heijne G. 2006. Identification and evolution of dual-topology membrane proteins. Nat. Struct. Mol. Biol. 13:112–116 [DOI] [PubMed] [Google Scholar]
- 41. Sebag J, Hinkle PM. 2009. Regions of melanocortin 2 (MC2) receptor accessory protein necessary for dual topology and MC2 receptor trafficking and signaling. J. Biol. Chem. 284:610–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wurie HR, Buckett L, Zammit VA. 2011. Evidence that diacylglycerol acyltransferase 1 (DGAT1) has dual membrane topology in the endoplasmic reticulum of HepG2 cells. J. Biol. Chem. 286:36238–36247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Narita S, Matsuyama S, Tokuda H. 2004. Lipoprotein trafficking in Escherichia coli. Arch. Microbiol. 182:1–6 [DOI] [PubMed] [Google Scholar]
- 44. Narita S, Tokuda H. 2010. In Economou A. (ed), Protein secretion, p 117–129 Methods in molecular biology, vol 619 Springer Science + Business Media, LLC, New York, NY [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.