Abstract
Degradation of poly(3-hydroxybutyrate) (PHB) by the thiolytic activity of the PHB depolymerase PhaZ1 from Ralstonia eutropha H16 was analyzed in the presence of different phasins. An Escherichia coli strain was constructed that harbored the genes for PHB synthesis (phaCAB), the phasin PhaP1, and the PHB depolymerase PhaZ1. PHB was isolated in the native form (nPHB) from this recombinant E. coli strain, and the in vitro degradation of the polyester was examined. Degradation resulted in the formation of the expected 3-hydroxybutyryl coenzyme A (3HB-CoA) and in the formation of a second product, which occurred in significantly higher concentrations than 3HB-CoA. This second product was identified by liquid chromatography mass spectrometry (LC-MS) as crotonyl-CoA. Replacement of PhaP1 by PhaP2 or PhaP4 resulted in a lower degradation rate, whereas the absence of the phasins prevented the degradation of nPHB by the PHB depolymerase PhaZ1 almost completely. In addition, the in vitro degradation of nPHB granules isolated from R. eutropha H16 (wild type) and from the R. eutropha ΔphaP1 and ΔphaP1-4 deletion mutants was examined. In contrast to the results obtained with nPHB granules isolated from E. coli, degradation of nPHB granules isolated from the wild type of R. eutropha yielded high concentrations of 3HB-CoA and low concentrations of crotonyl-CoA. The degradation of nPHB granules isolated from the ΔphaP1 and ΔphaP1-4 deletion mutants of R. eutropha was significantly reduced in comparison to that of nPHB granules isolated from wild-type R. eutropha. Stereochemical analyses of 3HB-CoA revealed that the (R) stereoisomer was collected after degradation of granules isolated from E. coli, whereas the (S) stereoisomer was collected after degradation of granules isolated from R. eutropha. Based on these results, a newly observed mechanism in the degradation pathway for PHB in R. eutropha is proposed which is connected by crotonyl-CoA to the β-oxidation cycle. According to this model, the NADPH-dependent synthesis of PHB with (R)-3HB-CoA as the intermediate and the PHB degradation yielding (S)-3HB-CoA, which is further converted in an NAD-dependent reaction, are separated.
INTRODUCTION
Poly(3-hydroxybutyrate) (PHB) is a short-carbon-chain-length polyester belonging to the bacterial polyhydroxyalkanoates (PHAs), which are formed by a wide range of Bacteria and Archaea and comprise a large variety of constituents (1–3). PHAs are stored as granules in the cytoplasm when one nutrient is limited and a carbon source is provided in excess. They serve as carbon and energy storage compounds and are degraded if no carbon source is available and if the limiting element is provided again (4). Due to their thermoplastic properties and biodegradability, PHAs are of great interest for industry (5–7).
The model organism for PHB metabolism is the Gram-negative facultative chemolithoautotrophic bacterium Ralstonia eutropha H16, which was isolated in 1961 (8–10). About 50 years of research unraveled many aspects of the synthesis and composition of PHB. Three enzymes catalyze the formation of PHB in R. eutropha H16: a β-ketothiolase (PhaA), an acetoacetyl coenzyme A (acetoacetyl-CoA) reductase (PhaB), and the PHA synthase (PhaC). The genes of these enzymes are located in an operon and are constitutively expressed (11–13). PHB granules are insoluble and coated with phospholipids and proteins. The first protein besides PhaC that was found to be attached to the granule surface was the phasin PhaP1 (originally called PhaP before homologous genes of phaP1 were found) (14). This small protein provides the majority of the protein layer. Later on, six other phasins (PhaP2 to PhaP7) were detected, and all of them were detected at the surface of PHB granules during in vivo or in vitro binding studies. However, the amount of PhaP1 at the surface of PHB granules is by far the largest in comparison to the amounts of other phasins (15–17). The phasins coat the PHB granules, but a catalytic function remains unknown so far. R. eutropha possesses seven different PHB depolymerases (PhaZ1 to PhaZ7) and two oligomer hydrolases (PhaY1 and PhaY2) for the intracellular degradation of PHB (18–21). The binding of PhaZ1 to the granule surface was shown in vivo (22).
Whereas the degradation process of extracellular PHB depolymerases is well understood, the elucidation of the reaction mechanism of the intracellular PHB depolymerases is still in progress. PHB granules are sensitive to chemical and physical stress, whereby the protein layer is destroyed and the native granules become partially crystalline (23, 24). Granules consisting of crystalline PHB are persistent to degradation by intracellular PHB depolymerases but are sensitive to extracellular PHB depolymerases. Only methods exclusively including centrifugation steps employing glycerol or sucrose gradients for the isolation of PHB provide conditions to obtain native granules (nPHB). As a result, enzyme assays performed in the past did often not reflect the natural degradation of nPHB by intracellular PHB depolymerases (25). To simulate natural conditions, such enzymes that are involved in the degradation process have to be attached to the surface of the granules during cell growth and PHB accumulation.
The first degradation assays carried out with such isolated nPHB granules revealed the release of 3-hydroxybutyrate (3HB) in a hydrolytic reaction, although only small amounts could be detected (47). In contradiction to that study, the same laboratory revealed in 2007 a thiolytic activity of the PHB depolymerase PhaZ1. nPHB granules isolated from R. eutropha were not hydrolyzed by PHB depolymerase PhaZ1 to 3HB but degraded to 3-hydroxybutyryl coenzyme A (3HB-CoA) (22). It was assumed that the previously observed release of 3HB in degradation assays was not related to a specific PHB depolymerase activity. Moreover, degradation to 3HB would not be energetically feasible, because a simultaneous synthesis and mobilization of PHB (26, 27) would constantly require the activation of released 3HB to 3HB-CoA in an energy-consuming reaction. Based on the results of these studies, the authors proposed an alternative PHB metabolic pathway for R. eutropha in which the degradation represents the reverse reaction of the synthesis, with (R)-3HB-CoA being released.
In past years, many efforts were made to enhance the production of PHAs in different microorganisms by metabolic engineering to provide more 3-hydroxyacyl–CoA precursors derived from the β-oxidation cycle (28–31). In bacterial β-oxidation, fatty acids are successively shortened by two carbon atoms during each cycle, with acetyl-CoA being released and with one molecule of NAD and FAD being reduced. First, an acyl-CoA synthetase activates the fatty acid by the addition of CoA, and the resulting acyl-CoA is oxidized to enoyl-CoA by an acyl-CoA dehydrogenase. The addition of H2O to the double bond by an enoyl-CoA hydratase leads to the formation of 3-(S)-hydroxyacyl–CoA, which is further oxidized to ketoacyl-CoA by a 3-hydroxyacyl–CoA dehydrogenase. In the last step, acetyl-CoA is released by a 3-ketoacyl–CoA thiolase, yielding a truncated acyl-CoA molecule that reenters the cycle without the need for further activation (32). The metabolic engineering often targets the conversion of enoyl-CoA to 3-(R)-hydroxyacyl–CoA by heterologous expression of (R)-specific enoyl-CoA hydratases to provide an excess of precursors for PHA synthesis (28, 30).
In our study, we examined the in vitro degradation of nPHB granules by PHB depolymerase PhaZ1 of R. eutropha and the influence of the presence of different phasins on the degradation rate. Although the degradation of nPHB granules isolated from a recombinant Escherichia coli strain harboring the phaP1 and phaZ1 genes of R. eutropha was analyzed previously, the degradation of nPHB granules in the presence of phasins other than PhaP1 was not yet investigated (22). This study aimed to reveal a catalytic activity or a regulatory influence of the different phasins of R. eutropha on PHB degradation. During our studies, two degradation products appeared when nPHB granules were incubated with CoA. One of these products was the expected 3HB-CoA, and the other was crotonyl-CoA. A closer look was taken at the degradation products, their stereoisomeric forms, and their putative role in the metabolic context of R. eutropha.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. R. eutropha was grown in nutrient broth (NB) or in mineral salts medium (MSM) with 1% (wt/vol) sodium gluconate (9) at 30°C for 30 h in 2-liter Erlenmeyer flasks with baffles on a rotary shaker with an agitation of 120 rpm. To provide conditions permissive for PHB accumulation, the ammonium chloride concentration was reduced to 0.05% (wt/vol). E. coli strains were grown in Luria-Bertani medium (LB) at 37°C (33) or, for PHB accumulation, in ammonium chloride-reduced MSM containing 1% (wt/vol) glucose at 30°C for 30 h in 2-liter Erlenmeyer flasks with baffles on a rotary shaker at an agitation of 120 rpm. Expression of heterologous genes was induced by the addition of 200 μM isopropyl-β-d-thiogalactopyranoside (IPTG) in the early exponential growth phase. To maintain the utilized plasmids, antibiotics were added to the medium at the following concentrations: 100 μg/ml ampicillin and 50 μg/ml kanamycin.
Table 1.
Bacterial strains, plasmids, and oligonucleotides
| Strain, plasmid, or oligonucleotide | Description or sequence | Source, reference, or location |
|---|---|---|
| Strains | ||
| R. eutropha H16 | Wild type; Gmr | DSM 428 |
| R. eutropha Re1052 (ΔphaP1) | phaP1 precise deletion gene replacement strain | 44 |
| R. eutropha ΔphaP1 ΔphaP2ΩKm ΔphaP3 ΔphaP4 (ΔphaP1-4) | phaP1 and phaP2 negative; Kmr; phaP3 and phaP4 precise deletion gene replacement strain | 45 |
| E. coli Top10 | Cloning strain, F− mcrA Δ(mrr-hsdRMS-mcrBC) galK rpsL Φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU (Strr) endA1 nupG | Invitrogen |
| E. coli BL21(DE3) | Expression strain, E. coli B, F− ompT hsdSB(rB− mB−) gal dcm (DE3) | Stratagene |
| Plasmids | ||
| pCR2.1-TOPO | Cloning vector; Apr Kmr | Invitrogen |
| pBBR1MCS-2 | Broad-host-range vector; Kmr lacPOZ′mobRP4 | Invitrogen |
| pETDuet-1 | Expression vector with two MCS, each preceded by a T7 promoter/lac operator and rbs; ColE1 orilacI Apr | Novagen |
| pBHR68 | pBluescript SK(−) harboring the phaCABRe operon, including the promoter; Apr | 46 |
| pBBR1MCS-2::phaCAB | pBBR1MCS-2 carrying the BamHI-EcoRI fragment of pBHR68 harboring the phaCAB operon, including the promoter; Kmr | This study |
| pETDuet-1::phaP1 | pETDuet carrying phaP1Re in MCS-1; Apr | This study |
| pETDuet-1::phaP2 | pETDuet carrying phaP2Re in MCS-1; Apr | This study |
| pETDuet-1::phaP4 | pETDuet carrying phaP4Re in MCS-1; Apr | This study |
| pETDuet-1::phaZ1 | pETDuet carrying phaZ1Re in MCS-2; Apr | This study |
| pETDuet-1::phaP1::phaZ1 | pETDuet carrying phaP1Re in MCS-1 and phaZ1Re in MCS-2; Apr | This study |
| pETDuet-1::phaP2::phaZ1 | pETDuet carrying phaP2Re in MCS-1 and phaZ1Re in MCS-2; Apr | This study |
| pETDuet-1::phaP4::phaZ1 | pETDuet carrying phaP4Re in MCS-1 and phaZ1Re in MCS-2; Apr | This study |
| Oligonucleotides | ||
| phaP1_EcoRV | 5′-GGAGACCAGCAATGATATCCTCACCCCG-3′ | 5′ region of phaP1 |
| phaP1_EcoRI | 5′-CAACGCAGGCAGGAATTCTTATCAGGCAGCCGTCG-3′ | 3′ region of phaP1 |
| phaP2_ScaI | 5′-GGAGATGCAAGAGTACTCAGTGGAC-3′ | 5′ region of phaP2 |
| phaP2_EcoRI | 5′-AAAAAAGAATTCGGTATAACCGCATGGATGAGGCGGGCCC-3′ | 3′ region of phaP2 |
| phaP4_ScaI | 5′-GGAGACGCAAGAGTACTCAGTGG-3′ | 5′ region of phaP4 |
| phaP4_EcoRI | 5′-GGCCTCCCAAGAGCCAAAGCCGACGAATTCTTAATT-3′ | 3′ region of phaP4 |
| phaZ1_NdeI | 5′-CCAGGCAGAAAAGGCCATATGCTCTACCAATTGC-3′ | 5′ region of phaZ1 |
| phaZ1_KpnI | 5′-CCCGCAATCGCGGGCGTTTTCGGTACCTTACCTGGTGGC-3′ | 3′ region of phaZ1 |
Construction of recombinant E. coli strains.
All plasmids and oligonucleotides used in this study are listed in Table 1. Plasmid pBBR1MCS-2::phaCAB was constructed by digesting the vector pBHR68 with EcoRI and BamHI to obtain the phaCAB operon of R. eutropha, including the natural promoter region; the fragment was then ligated into the vector pBBR1MCS-2. Genes encoding the phasins PhaP1, PhaP2, and PhaP4 and the PHB depolymerase PhaZ1 were amplified from genomic DNA of R. eutropha with the oligonucleotides listed in Table 1 and subcloned in pCR2.1-TOPO (Invitrogen, Darmstadt, Germany) according to the manufacturer's instructions. For construction of the expression plasmids, the vector pETDuet-1 was digested with NcoI, and blunt ends were obtained by a fill-in reaction employing T7 polymerase (Thermo Scientific, Dreieich, Germany). After inactivation of the T7 polymerase, the vector was digested with EcoRI, and the phasin genes were inserted in multiple cloning site 1 (MCS-1) of the vector. phaZ1 was cut with NdeI and KpnI and ligated with pETDuet-1 (MCS-2) or with those plasmids which already contained one of the three phasins. The resulting plasmids were used for the recombinant expression of the phasin and the PHB depolymerase genes, expressed as a single gene alone or in different combinations, in E. coli BL21.
Isolation of nPHB.
For isolation of native PHB granules (nPHB) (modified from reference 34), cells from the cultivations permissive for PHB accumulation were suspended in 50 mM potassium phosphate buffer (pH 7.0) containing 1 mM dithiothreitol (DTT). After four passages through a French press, the resulting lysate was loaded onto the top of a glycerol step gradient. To separate the nPHB granules from the cell debris, the first discontinuous gradient consisted of 4.5 ml each of 90% and 60% (vol/vol) glycerol solutions. After 1 h of centrifugation at 100,000 × g and 4°C, the interphase was diluted with an equal volume of potassium phosphate buffer and was loaded onto the top of a second discontinuous gradient consisting of 87%, 75%, and 55% (vol/vol) glycerol solutions, each with a volume of 2.5 ml. The resulting interphase between 55% and 75% (vol/vol) glycerol contained the nPHB granules. If it was necessary to concentrate the granules, the resulting fraction was diluted again with an equal volume of buffer and was centrifuged with the first-step gradient. If necessary, the granule fractions were stored at −20°C.
Degradation of nPHB by thiolysis.
The degradation assay to monitor thiolysis of nPHB granules was done as described elsewhere (22). In brief, 300 μl of nPHB granules (100 to 200 mg/ml) was incubated with 50 mM potassium phosphate buffer (pH 7.0) containing 1 mM DTT in a total volume of 600 μl. To start the reaction, 1 mM CoA was added. After appropriate periods of time, samples were withdrawn and acidified with 0.1 N HCl or with 0.05% (wt/vol) trichloroacetic acid (TCA) in cases in which LC-MS analysis followed. To prepare the samples for high-performance liquid chromatography (HPLC) analysis, they were centrifuged for 10 min at 16,000 × g and filtered with a TITAN syringe filter made of regenerated cellulose (0.20 μm; Sun Sri, Rockwood, TN).
HPLC and LC-MS analyses.
The conditions for the HPLC analyses for the enzyme assay were modified from those used by Uchino et al. (22). In brief, samples of 20 μl were injected onto an Eclipse XDB-C18 HPLC column (5 μm; 4.6 by 150 mm; Agilent Technologies, Böblingen, Germany). The products of the enzyme reaction were eluted by employing a mixture of 50 mM ammonium acetate buffer at pH 4.7 (solution A) and methanol (solution B). A multistep gradient was applied at a flow rate of 0.8 ml/min from 5% (vol/vol) solution B to 50% (vol/vol) solution B within 25 min, up to 80% (vol/vol) solution B within 5 min (total run time, 30 min), followed by a decrease of solution B to 5% (vol/vol) within 8 min (total run time, 38 min). During the last 4 min, an isocratic flow of 5% (vol/vol) solution B (total run time, 42 min) was applied to equilibrate for the next sample injection. Standards for coenzyme A, 3-hydroxybutyryl–CoA (3HB-CoA), and crotonyl-CoA were purchased from Sigma-Aldrich.
In addition to the comparison of the retention times with those of the standards during HPLC analysis, LC-MS analysis was performed to clearly identify the resulting products of the enzyme reaction. A Nucleosil RP C18 column (5 μm; Knauer GmbH, Berlin, Germany) was used, and the multistep gradient of solution A and B was optimized for LC-MS performance. The flow rate was reduced to 0.5 ml/min, and the fraction of solution B, initially 5% (vol/vol), was increased within 45 min to 45% (vol/vol). To equilibrate the column for the next sample, solution B was then reduced in 10 min (total run time, 55 min) to 5% (vol/vol), followed by an isocratic flow for the next 10 min (total run time, 65 min). After detection of the CoA esters with a photodiode array detector at 259 nm, the eluate was directly injected to an LXQ Finnigan (Thermo Scientific, Dreieich, Germany) mass spectrometer. The tuning parameters optimized in advance for CoA were as follows: a capillary temperature of 300°C, a sheath gas flow of 12 liters/h, an auxiliary gas flow of 6 liters/h, and a sweep gas flow of 1 liter/h. The mass range was set to m/z 50 to 1,000 when run in the scan mode. The collision energy in the MS mode was set to 30 V (35).
Sample preparation of 3HB-CoA and identification of stereoisomers.
To identify the stereoisomeric form of 3HB-CoA, the products of the degradation assay were separated in an HPLC run, and the resulting peak fraction of 3HB-CoA was collected. The sample was diluted with 2 vol of 0.01 M HCl and purified by a passage through a Sep-Pak Classic C18 cartridge (Waters, Milford, MA). The cartridge was conditioned with 2.5 ml of 80% (vol/vol) methanol in 0.01 M HCl followed by equilibration with two washes with 2.5 ml 0.01 M HCl. Loading of the sample and two washes with 2.5 ml 0.01 M HCl were performed by gravity flow; 3HB-CoA was eluted with 2.5 ml of 80% (vol/vol) methanol and 0.01 M HCl, dried at 40°C in a vacuum furnace, and dissolved in 30 μl H2O.
For the identification of the stereoisomers of 3HB-CoA by HPLC, removal of CoA and linkage to a fluorescent agent were necessary. Therefore, 3HB-CoA was hydrolyzed by the addition of 30 μl 1 N NaOH and subsequently neutralized with 1 N HCl. The following steps, including sample analysis by HPLC, were performed as described elsewhere (36, 37), with some modifications. A sample of 50 μl after hydrolysis and neutralization was mixed with 150 μl of pure ethanol, 100 μl of 280 mM triphenylphosphine (TPP; Sigma-Aldrich, Seelze, Germany) in acetonitrile, 100 μl of 280 mM 2,2′-dipyridyl disulfide (DPDS; Sigma-Aldrich, Seelze, Germany) in acetonitrile, and 200 μl of 2 mM 4-nitro-7-piperazino-2,1,3-benzoxadiazole (NBD-PZ; Tokyo Chemical Industry, Tokyo, Japan) in acetonitrile. After incubation for 4 h at 30°C, the reaction was stopped by acidification with 60 μl of 0.5% (wt/vol) TCA. To remove the excess of NBD-PZ, Chromabond PS/DVB cation-exchange cartridges (Macherey-Nagel, Düren, Germany) were used.
After equilibration with 200 μl eluting solution, the cartridges were loaded with 600 μl reaction mixture and centrifuged in 15-ml tubes at 100 × g for 1 min. Without the tube being exchanged, 300 μl of eluting solution was added to the cartridges, and the centrifugation step was repeated. This eluate was subsequently used for the identification of stereoisomers. A Shimadzu HPLC system (Shimadzu, Duisburg, Germany), equipped with Nucleocel DELTA-RP S (5 μm, 250 mm in length; Macherey-Nagel, Düren, Germany), was used for the identification of the stereoisomers. Samples of 2 μl were injected into the column, an isocratic flow rate of acetonitrile-water (40/60, vol/vol) at 0.3 ml/min and 17°C for 70 min was performed, and the fluorescent detection occurred at 532 nm using an excitation wavelength of 485 nm. (R)-3HB and (S)-3HB (Sigma-Aldrich, Seelze, Germany) were used as standards and processed like the collected 3HB-CoA, starting from the evaporation step in the 40°C vacuum furnace.
RESULTS
Degradation of nPHB granules by heterologously expressed PHB depolymerase PhaZ1 of R. eutropha in E. coli.
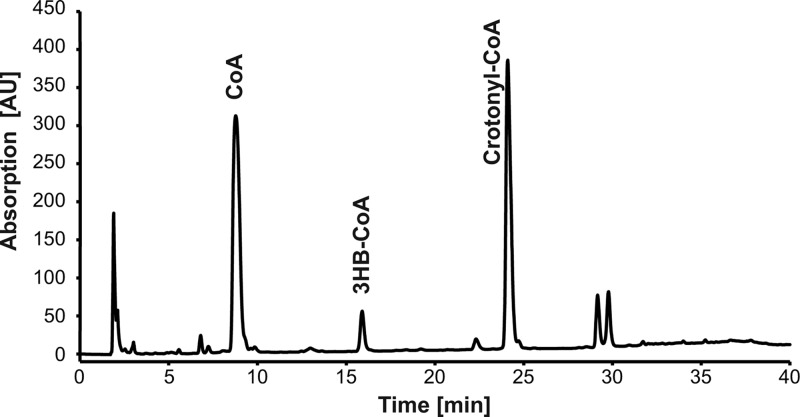
An enzyme assay was performed to analyze the influence of the PHB depolymerase PhaZ1 of R. eutropha on PHB degradation in the presence of different phasins of this bacterium. For this, phasin genes phaP1, phaP2, or phaP4 and the PHB depolymerase gene phaZ1 were heterologously expressed in an E. coli BL21(DE3) strain harboring additionally the phaCAB operon of R. eutropha to allow PHB synthesis. Granules were isolated in the native form as described in Materials and Methods, and thiolysis of the polyester was started by the addition of CoA. The first assay was performed with nPHB granules in the presence of phasin PhaP1 and PHB depolymerase PhaZ1. After 90 min of incubation, the HPLC analysis was performed as described by Uchino et al. (22) without modification. The assay revealed the presence of three products (Fig. 1). The first peak corresponded to CoA, which was consumed during the thiolysis reaction. The compound of the second, minor peak was identified as 3HB-CoA, as expected for the degradation of PHB. In addition, crotonyl-CoA was identified as a compound of the third, major peak according to its retention time and the spectrum of a standard.
Fig 1.
HPLC analysis of the degradation products formed during enzyme assay with nPHB granules incubated with 1 mM CoA. nPHB granules were accumulated in E. coli during simultaneous expression of phaP1 and phaZ1 and were isolated and subjected to degradation assay as described in Materials and Methods. The obtained supernatant was filtered after 30 min of incubation, and 20 μl was analyzed by HPLC. Peaks were identified by comparison with standards, as indicated.
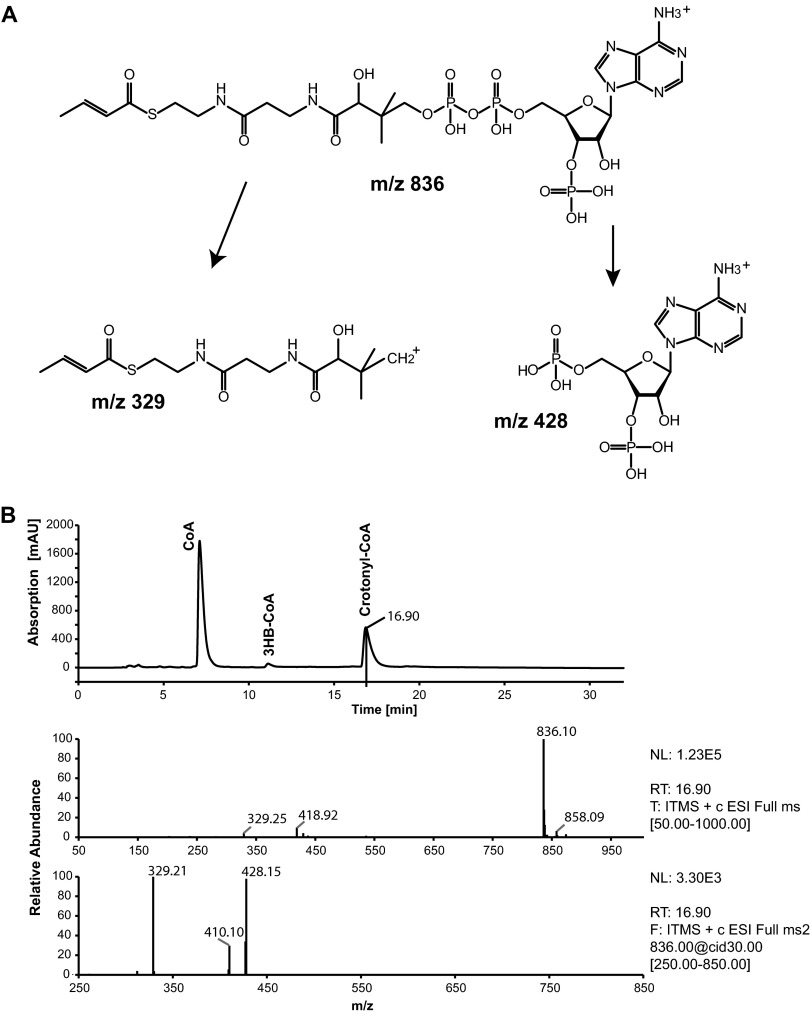
To confirm the identity of crotonyl-CoA, it was also analyzed by LC-MS. The full MS (MS 1) showed a parental ion with an m/z 836, which corresponded to the expected mass of crotonyl-CoA. Fragmentation of this ion (MS 2) led to two main daughter ions (m/z 329 and 428), thereby confirming the identity of crotonyl-CoA. Chromatograms and the chemical structure of the fragmentation products are shown in Fig. 2. In the study by Uchino et al. (22), the occurrence of crotonyl-CoA was not described. The comparison of the chromatogram in Fig. 2 to the chromatograms shown by Uchino et al. (22) revealed a shift of the retention time for CoA and 3HB-CoA of about 6 min. When the observed shift is conferred to the retention time of crotonyl-CoA obtained in this study, it should occur at about 32 min in the chromatogram of the previous study. However, all provided chromatograms terminated after 26 min of total run time. The putatively formed crotonyl-CoA may therefore have been missed. For further experiments of this study, the method for separation of the CoA esters was modified as described in Materials and Methods in order to ensure that all putatively occurring products could be monitored.
Fig 2.
LC-MS analyses of degradation products formed during enzyme assay with nPHB granules isolated from E. coli. nPHB granules were accumulated in E. coli during simultaneous expression of phaP1 and phaZ1 and were isolated and incubated with 1 mM CoA. The reaction was stopped by acidification, and 10 μl of the filtered sample was analyzed by LC-MS. (A) Structural formula of crotonyl-CoA and resulting mass fragments; (B) LC chromatogram and mass spectrometric data at the 16.9-min run time point. Top, LC chromatogram of the sample. The peaks were identified by comparison with standards, as indicated. Middle, MS spectrum at the 16.9-min run time point, which revealed one parental ion of m/z 836. Bottom, MS2 spectrum of the parental ion (m/z 836) and two daughter ions (m/z 329 and 428).
Crotonyl-CoA is an intermediate of the β-oxidation pathway and is formed from 3HB-CoA by enoyl-CoA hydratases. During the isolation process, small amounts of soluble enzymes may remain as impurities in the native granule fraction and may affect the range of compounds formed. To fully exclude this artifact, the following controls were made by starting the reaction by the addition of CoA or a racemic mixture of 3HB-CoA: reaction mixture (i) without any granule fraction, (ii) with granules lacking phasin and PHB depolymerase, or (iii) with granules and with phasin PhaP1 but lacking PHB depolymerase (Table 2). The control reaction without granule fraction (i) showed no spontaneous decomposition of CoA or (R/S)-3HB-CoA (Table 2, experiment 1). In reactions with granules lacking phasin and PHB depolymerase (ii), CoA did not induce any degradation of PHB (Table 2, experiment 2). Addition of CoA to reactions with granule-bound PhaP1 but lacking PHB depolymerase (iii) did also not cause any degradation of PHB. On the other hand, (R/S)-3HB-CoA as the starting reagent led to increasing concentrations of CoA, with a concomitant decrease of the concentration of 3HB-CoA and no other CoA ester appearing (Table 2, experiment 3). Such a change of the concentration of 3HB-CoA and the appearance of CoA was not observed in experiment 1 without granules. In control reaction (iii) [Table 2, experiment 3; induction with (R/S)-3HB-CoA], the availability of 3HB-CoA most likely induced PHB synthesis by the PHB synthase PhaC, which is also attached to the granule surface. 3HB-CoA is used for the elongation of the polyester chain, with CoA being released. If enoyl-CoA hydratases were present in the granule fraction, they could not convert the provided (R/S)-3HB-CoA; otherwise, crotonyl-CoA should have been monitored. Therefore, the crotonyl-CoA formed during the degradation assays of nPHB in the presence of phasin PhaP1 and PHB depolymerase PhaZ1 should not originate from the enzymatic conversion of 3HB-CoA by enoyl-CoA hydratases.
Table 2.
Assay of in vitro degradation of nPHB granules isolated from E. colia
| Condition | Detected substance | Amt ±SD (mM) by time in min after addition of 1 mM of: |
||||||
|---|---|---|---|---|---|---|---|---|
| CoA |
3HB-CoA |
|||||||
| 5 | 10 | 30 | 60 | 90 | 30 | 90 | ||
| 1. Negative control,b no nPHB | CoA | 0.93 | 0.93 | 1.03 | 1.08 | 1.05 | <0.02 | <0.02 |
| 3HB-CoA | <0.02 | <0.02 | <0.02 | <0.02 | <0.02 | 1.00 | 1.02 | |
| Crot-CoA | <0.02 | <0.02 | <0.02 | <0.02 | <0.02 | <0.02 | <0.02 | |
| 2. E. coli (phaCAB) nPHB | CoA | 0.88 ± 0.15 | 1.03 ± 0.30 | 1.03 ± 0.25 | 1.08 ± 0.31 | 1.29 ± 0.35 | ND | ND |
| 3HB-CoA | <0.02 | <0.02 | <0.02 | <0.02 | <0.02 | ND | ND | |
| Crot-CoA | <0.02 | <0.02 | <0.02 | <0.02 | <0.02 | ND | ND | |
| 3. E. coli (phaCAB phaP1) nPHB | CoA | 0.82 ± 0.07 | 0.84 ± 0.15 | 0.95 ± 0.21 | 1.07 ± 0.27 | 1.11 ± 0.42 | 0.57b | 0.66b |
| 3HB-CoA | <0.02 | <0.02 | <0.02 | <0.02 | <0.02 | 0.49b | 0.43b | |
| Crot-CoA | <0.02 | <0.02 | <0.02 | <0.02 | <0.02 | <0.02 | <0.02 | |
| 4. E. coli (phaCAB phaZ1) nPHB | CoA | 0.80 ± 0.08 | 0.81 ± 0.10 | 0.81 ± 0.11 | 0.80 ± 0.14 | 0.77 ± 0.15 | 0.29 ± 0.05 | 0.33 ± 0.05 |
| 3HB-CoA | <0.02 | <0.02 | <0.02 | <0.02 | <0.02 | 0.72 ± 0.03 | 0.52 ± 0.05 | |
| Crot-CoA | <0.02 | <0.02 | <0.02 | 0.03 ± 0.04 | 0.03 ± 0.04 | <0.02 | <0.02 | |
| 5. E. coli (phaCAB phaP1 phaZ1) nPHB | CoA | 0.63 ± 0.06 | 0.48 ± 0.04 | 0.27 ± 0.03 | 0.20 ± 0.04 | 0.20 ± 0.02 | 0.32 ± 0.02 | 0.24 ± 0.02 |
| 3HB-CoA | <0.02 | 0.03 ± 0.03 | 0.06 ± 0.00 | 0.07 ± 0.00 | 0.08 ± 0.00 | 0.37 ± 0.07 | 0.31 ± 0.07 | |
| Crot-CoA | 0.22 ± 0.02 | 0.54 ± 0.00 | 0.85 ± 0.08 | 0.78 ± 0.20 | 0.82 ± 0.06 | 0.26 ± 0.10 | 0.44 ± 0.12 | |
| 6. E. coli (phaCAB phaP2 phaZ1) nPHB | CoA | 0.79 ± 0.08 | 0.75 ± 0.10 | 0.63 ± 0.08 | 0.56 ± 0.13 | 0.50 ± 0.06 | 0.32 ± 0.00 | 0.28 ± 0.00 |
| 3HB-CoA | <0.02 | <0.02 | 0.03 ± 0.03 | 0.03 ± 0.03 | 0.03 ± 0.03 | 0.46 ± 0.03 | 0.38 ± 0.01 | |
| Crot-CoA | 0.08 ± 0.08 | 0.13 ± 0.10 | 0.30 ± 0.06 | 0.40 ± 0.06 | 0.37 ± 0.05 | 0.06 ± 0.06 | 0.08 ± 0.08 | |
| 7. E. coli (phaCAB phaP4 phaZ1) nPHB | CoA | 0.91 ± 0.02 | 0.92 ± 0.00 | 0.76 ± 0.02 | 0.58 ± 0.00 | 0.52 ± 0.00 | 0.36 ± 0.02 | 0.37 ± 0.04 |
| 3HB-CoA | <0.02 | <0.02 | 0.03 ± 0.03 | 0.03 ± 0.03 | 0.03 ± 0.03 | 0.54 ± 0.02 | 0.48 ± 0.00 | |
| Crot-CoA | 0.15 ± 0.00 | 0.19 ± 0.00 | 0.40 ± 0.03 | 0.35 ± 0.13 | 0.40 ± 0.18 | <0.02 | 0.12 ± 0.03 | |
Degradation assays were performed as described in Materials and Methods. The amounts of coenzyme A (CoA), 3-hydroxybutyryl–CoA (3HB-CoA) and crotonyl-CoA (Crot-CoA) were calculated from HPLC peak areas. The values represent the averages from up to 4 assay repetitions and are presented in mM. ND, not determined.
Single determination.
When phaZ1 but no phasin gene was expressed during PHB accumulation, and when these granules were used for the enzyme assay, the concentration of crotonyl-CoA was only slightly above the detection limit (Table 2, experiment 4). Only in those reactions in which PhaP1 and PhaZ1 were present in the granule fraction were a significant reduction of CoA and simultaneous formation of 3HB-CoA and crotonyl-CoA observed (Table 2, experiment 5). Within 90 min, the concentration of CoA decreased from the provided 1.00 mM to 0.20 mM, and the concentrations of 3HB-CoA and crotonyl-CoA increased from initially 0.00 mM to 0.08 and 0.82 mM, respectively. When the reactions, which led to degradation of PHB granules, were induced by (R/S)-3HB-CoA, HPLC analysis also revealed formation of crotonyl-CoA. Control reaction (iii), with phasin PhaP1 but lacking PHB depolymerase, indicated no enoyl-CoA hydratase activity but probably pointed to the PHB synthesis reaction, releasing CoA. This CoA could then in turn be used to induce the degradation of PHB. In these reactions, the concentration of crotonyl-CoA was lower than in those reaction mixtures containing the same granule fractions that were started with CoA as the inductor.
Phasins PhaP2 and PhaP4 were also used in addition to PhaP1 for the degradation assay in combination with PHB depolymerase PhaZ1. After 90 min of incubation, the concentration of CoA decreased from the provided 1.00 mM to 0.56 mM, and the concentration of crotonyl-CoA increased from initially 0.00 mM to 0.40 mM. 3HB-CoA could not be detected in concentrations higher than 0.03 mM. Despite the considerably lower degradation rate (Table 2, experiments 6 and 7), the replacement of PhaP1 by PhaP2 or PhaP4 did not influence the nPHB degradation by PHB depolymerase PhaZ1 and the formation of the main degradation product crotonyl-CoA.
Degradation of nPHB granules isolated from R. eutropha (wild type) or from the R. eutropha ΔphaP1 and R. eutropha ΔphaP1-4 deletion mutants.
Enzyme assays were also performed with granules from the wild type or the single and quadruple phasin mutants (ΔphaP1 and ΔphaP1-4 mutants) of R. eutropha to explore the degradation process and to identify the resulting products in the original PHB-producing strain.
HPLC analysis of the enzyme products obtained with nPHB granules isolated from the wild type of R. eutropha revealed increasing concentrations of 3HB-CoA (from 0.00 mM to 0.21 mM) and crotonyl-CoA (from 0.00 mM to 0.06 mM) after 90 min of incubation, as also observed for granules isolated from E. coli. During the degradation of granules isolated from R. eutropha, 3HB-CoA occurred as the main degradation product, whereas crotonyl-CoA was detected only in low concentrations (Table 3, experiment 1). In addition, enzyme assays with CoA as the inductor using granules from R. eutropha mutants lacking phaP1 or even phasin genes phaP1 to phaP4 (R. eutropha ΔphaP1or ΔphaP1-4 mutants, respectively) were also performed. In both cases, the absence of a single phasin gene (phaP1) or of four phasin genes (phaP1 to phaP4) reduced the degradation of nPHB dramatically to rates which were almost below the detection limit of the assay. The concentrations of 3HB-CoA and crotonyl-CoA increased within 90 min from 0.00 mM to 0.06 mM and 0.03 mM, respectively. However, in contrast to the observations obtained with granules isolated from recombinant E. coli, reactions in which (R/S)-3HB-CoA instead of CoA was provided yielded higher concentrations of crotonyl-CoA (from 0.00 mM up to 0.16 mM) than CoA-induced reactions (Table 3, experiments 1 to 3).
Table 3.
Assay of in vitro degradation of nPHB granules isolated from R. eutrophaa
| Condition | Detected substance | Amt ±SD (mM) by time in min after addition of 1 mM of: |
||||||
|---|---|---|---|---|---|---|---|---|
| CoA |
3HB-CoA |
|||||||
| 5 | 10 | 30 | 60 | 90 | 30 | 90 | ||
| 1. H16 nPHB | CoA | 0.84 ± 0.12 | 0.87 ± 0.06 | 0.79 ± 0.06 | 0.68 ± 0.13 | 0.66 ± 0.06 | 0.53 ± 0.14 | 0.49 ± 0.11 |
| 3HB-CoA | 0.03 ± 0.03 | 0.08 ± 0.05 | 0.14 ± 0.04 | 0.20 ± 0.06 | 0.21 ± 0.05 | 0.45 ± 0.12 | 0.40 ± 0.13 | |
| Crot-CoA | <0.02 | 0.04 ± 0.02 | 0.05 ± 0.02 | 0.06 ± 0.02 | 0.06 ± 0.02 | 0.11 ± 0.05 | 0.11 ± 0.05 | |
| 2. H16 ΔphaP1 nPHB | CoA | ND | 0.87 ± 0.12 | 0.88 ± 0.12 | 0.89 ± 0.10 | 0.89 ± 0.10 | 0.60 ± 0.05 | 0.52 ± 0.11 |
| 3HB-CoA | ND | <0.02 | <0.02 | 0.02 ± 0.02 | 0.02 ± 0.02 | 0.50 ± 0.04 | 0.43 ± 0.02 | |
| Crot-CoA | ND | <0.02 | <0.02 | <0.02 | <0.02 | 0.16 ± 0.01 | 0.16 ± 0.01 | |
| 3. H16 ΔphaP1 ΔphaP2 ΩKm ΔphaP3 ΔphaP4 nPHB | CoA | 0.74 ± 0.11 | 0.69 ± 0.15 | 0.70 ± 0.09 | 0.67 ± 0.10 | 0.63 ± 0.11 | 0.49 ± 0.03 | 0.40 ± 0.08 |
| 3HB-CoA | <0.02 | <0.02 | 0.04 ± 0.01 | 0.05 ± 0.02 | 0.06 ± 0.02 | 0.52 ± 0.01 | 0.43 ± 0.00 | |
| Crot-CoA | <0.02 | <0.02 | <0.02 | 0.03 ± 0.02 | 0.03 ± 0.02 | 0.17 ± 0.00 | 0.16 ± 0.00 | |
Degradation assays were performed as described in Materials and Methods. The amounts of coenzyme A (CoA), 3-hydroxybutyryl–CoA (3HB-CoA), and crotonyl-CoA (Crot-CoA) were calculated from HPLC peak areas. The values represent the averages from up to 4 assay repetitions and are presented in mM. ND, not determined.
Analysis of the chirality of the nPHB degradation product 3HB-CoA.
Control reactions comparable to those performed with nPHB granules isolated from E. coli are not feasible for nPHB granules isolated from R. eutropha. It is known that R. eutropha possesses the genes for seven phasins and also seven PHB depolymerases (15–18, 20, 21). To obtain nPHB granules isolated from R. eutropha without those enzymes, a multideletion mutant of R. eutropha would be necessary. Even if such a mutant was available, it could not be excluded that a putative, so-far-unknown phasin or PHB depolymerase would affect the control reaction. Hence, we can only assume that the shift of the main degradation product from crotonyl-CoA (in the case of E. coli) to 3HB-CoA (in the case of R. eutropha) derived from a further conversion of the product by enoyl-CoA hydratases. R. eutropha possesses several enoyl-CoA hydratases (13, 15) which possibly catalyze the conversion of crotonyl-CoA to (S)-3HB-CoA. Hence, a chiral analysis of 3HB-CoA obtained from enzyme assays with nPHB granules isolated from E. coli and from R. eutropha was necessary to determine the stereoisomeric form.
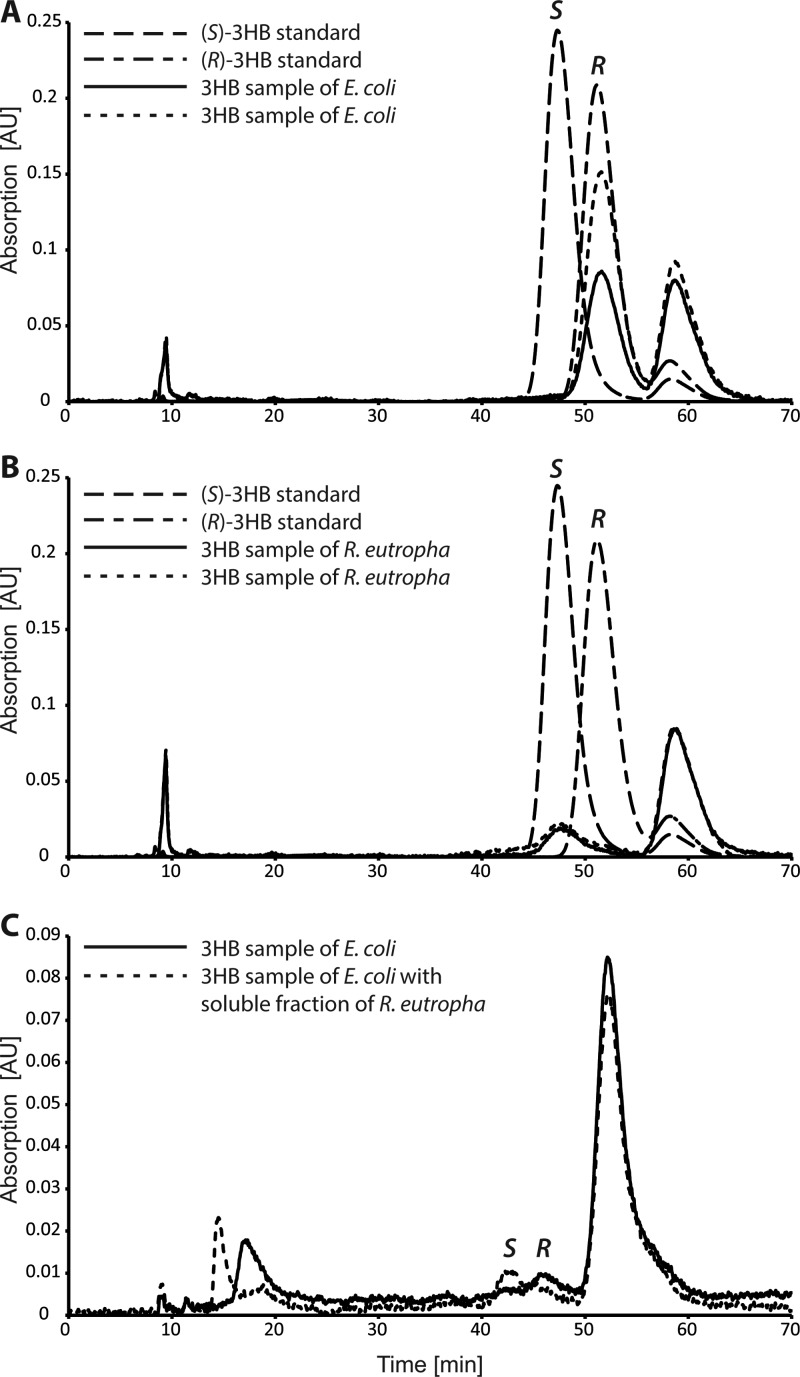
Granules isolated from E. coli with heterologously expressed phaP1 and phaZ1 and from the wild-type cells of R. eutropha were employed in enzyme assays performed as described above. The reaction mixture was incubated for 2 h and stopped with HCl. During separation of the CoA ester by HPLC, 3HB-CoA was collected and prepared for the identification of the stereoisomers as described in Materials and Methods. The analysis of the stereoisomers revealed the (R) stereoisomer for all collected fractions of 3HB-CoA from enzyme assays with granules isolated from E. coli (Fig. 3A), whereas the resulting 3HB-CoA collected after degradation of granules isolated from R. eutropha consisted mainly of the (S) stereoisomer (Fig. 3B). In the case of 3HB-CoA collected from degradation reactions with nPHB granules isolated from R. eutropha, also a little peak corresponding to (R)-3HB-CoA was detected. The precise amount of (R)-3HB-CoA could not be determined due to the separation limits of the used HPLC system, but the observed content of (R)-3HB-CoA was in all cases less than 5% in comparison to that of the (S) stereoisomer.
Fig 3.
Chiral analyses of the degradation product 3HB-CoA. The obtained 3HB-CoA from the degradation assays performed with nPHB granules isolated from E. coli (A) or R. eutropha (B) were prepared for stereoisomeric identification as described in Materials and Methods. Samples of 2 μl after removal of CoA and linkage to the fluorescent dye NBD-PZ were analyzed by chiral HPLC. Standards of (R)-3HB and (S)-3HB are shown with dashed lines, and the relevant peaks are indicated with R and S, respectively. The solid and the dotted lines represent two samples of isolated 3HB-CoA after the degradation assay. The peak at 59 min resulted from unlinked NBD-PZ. (C) 3HB-CoA obtained from the assay with nPHB granules isolated from E. coli and degraded in the absence (solid line) or presence (dotted line) of a soluble fraction derived from nPHB granules of R. eutropha. The peak at 54 min resulted from unlinked NBD-PZ.
The above-mentioned enoyl-CoA hydratases are possibly responsible for the conversion of crotonyl-CoA to (S)-3HB-CoA in R. eutropha. To determine if enzymes, which were present in the soluble part of the granule fraction of R. eutropha, are able to catalyze this reaction, another enzyme test was performed. Granules isolated from E. coli with heterologously expressed phaP1 and phaZ1 were used in addition to 120 μl of the soluble fraction of granules isolated from R. eutropha. After 2 h of incubation, HPLC analysis of this reaction mixture showed a reduced concentration of crotonyl-CoA and an increased concentration of 3HB-CoA in comparison to the assay without the addition of the soluble granule fraction from R. eutropha (data not shown).
The identification of the stereoisomers of 3HB-CoA collected from reactions with or without the addition of the soluble granule fraction of R. eutropha is shown in Fig. 3C. A clear shift from (R)-3HB-CoA to (S)-3HB-CoA was observed. Thus, the soluble part of the granule fraction isolated from R. eutropha contained enzymes, which catalyzed the reaction from crotonyl-CoA to (S)-3HB-CoA and which were presumably absent or nonfunctional in the granule fractions isolated from E. coli.
DISCUSSION
In past years, the in vivo and in vitro degradation of PHB by intracellular PHB depolymerases was investigated by several laboratories (18, 25–27, 38). Recently, a study showed that the degradation of nPHB granules by PHB depolymerase PhaZ1 of R. eutropha led to increasing concentrations of 3HB-CoA after the start of the reaction by the addition of CoA (22). Thiolysis of PHB fits well with the model of continuous accumulation and degradation of PHB during cell growth by overcoming the problem of permanent required activation of 3HB monomers with CoA in an energy-consuming reaction (26, 27). Hence, the observed mobilization of PHB no longer represents an energetically futile cycle. However, several aspects remain to be elucidated. Uchino et al. (22) suggest a putative metabolic pathway for PHB in which the polyester is degraded to (R)-3HB-CoA and further to acetyl-CoA via acetoacetyl-CoA in the reverse direction and by the same enzymes of the synthesis pathway. The regulation of the mobilization on demand regarding the environmental conditions remains unclear, especially when the degradation represents the reverse reaction of the synthesis and when the degradation products are identical to the synthesis products. Furthermore, experiments in which different cofactors were added to the reaction mixture monitored a reprocessing of 3HB-CoA to acetyl-CoA when NAD was added but not, as expected for the reverse reaction, when NADP was added as a cofactor (22).
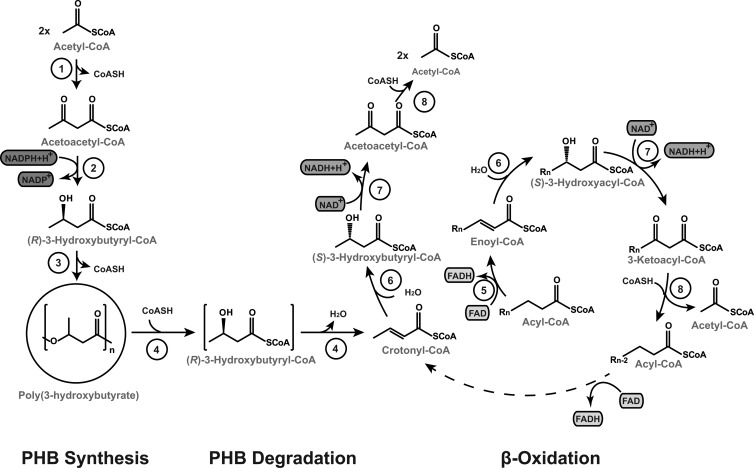
In conclusion of the results of this study, we postulate a different degradation pathway for PHB in R. eutropha, which is directly connected to the β-oxidation pathway (Fig. 4). The degradation of PHB by the intracellular PHB depolymerase PhaZ1 leads to (R)-3HB-CoA, which is probably not released (Fig. 4, square brackets) but is directly converted by the PHB depolymerase to crotonyl-CoA. Crotonyl-CoA represents the linkage between the degradation of PHB and the β-oxidation pathway. It is converted by the addition of H2O by an enoyl-CoA hydratase to (S)-3HB-CoA, which is further converted to acetoacetyl-CoA by a 3-hydroxyacyl–CoA dehydrogenase in an NAD-dependent reaction. Acetoacetyl-CoA is then cleaved by a 3-ketoacyl–CoA thiolase to two molecules of acetyl-CoA, which could enter the central metabolism in terms of limited availability of a carbon source.
Fig 4.
Model of PHB synthesis and degradation in R. eutropha. PHB is synthesized by the condensation of two molecules of acetyl-CoA to acetoacetyl-CoA by a β-ketothiolase (PhaA) (1) and reduction by acetoacetyl-CoA reductase (PhaB) (2) in an NADPH-dependent reaction from acetoacetyl-CoA to (R)–3-hydroxybutyryl–CoA. The PHA synthase (PhaC) (3) incorporates the monomers to poly(R)–3-hydroxybutyrate. PHB is degraded to crotonyl-CoA via (R)–3-hydroxybutyryl–CoA in a thiolytic reaction by intracellular PHB depolymerases (PhaZ) (4), probably without release of the intermediate (square brackets). Crotonyl-CoA combines the degradation of PHB with β-oxidation, where a double bond is added to an acyl-CoA by an acyl-CoA dehydrogenase (FadE) (5), and an enoyl-CoA is formed. Enoyl-CoA is stereospecifically converted to (S)–3-hydroxyacyl–CoA by an enoyl-CoA hydratase (multifunctional FadB) (6), and 3-ketoacyl–CoA is formed in an NAD-dependent reaction by a 3-hydroxyacyl–CoA dehydrogenase (multifunctional enzyme FadB) (7). A 3-ketoacyl–CoA thiolase (FadA) (8) cleaves the ketoacyl-CoA, and one molecule of acetyl-CoA is formed, yielding a truncated acyl-CoA. The β-oxidation cycle starts again with the addition of a double bond to the truncated acyl-CoA.
In this study, we determined the so-far-unknown PHB degradation product crotonyl-CoA. The spontaneous or enzyme-induced conversion of provided (R/S)-3HB-CoA could be excluded by control reactions with granules isolated from E. coli lacking PHB depolymerase PhaZ1. Thus, only PhaZ1 and no other enzyme present in the granule fraction seems to be capable of converting 3HB-CoA to crotonyl-CoA. As mentioned before, the thiolytic activity of PHB depolymerase PhaZ1 of R. eutropha was already shown in 2007, but no occurrence of crotonyl-CoA after incubation of nPHB granules with CoA was described (22). The comparison of the HPLC chromatograms of our study here and those of the former study suggests a putative retention time of about 32 min for crotonyl-CoA, which was not monitored anymore in the analyses by Uchino et al. (22). Although the method of the HPLC analyses comprises a total run time of 39 min, the end of detection is not specified in the manuscript. If the detection ended after 26 min (as shown in the figures of the HPLC chromatograms), the putatively occurred peak of crotonyl-CoA may have been missed.
Naturally, crotonyl-CoA is found, inter alia, in the butanoate pathway of bacteria as an intermediate during the conversion of (R)-3HB-CoA to (S)-3HB-CoA. Indeed, in this current study, both stereoisomers of 3HB-CoA collected during HPLC analyses were identified. The (R) stereoisomer of 3HB-CoA was obtained when the PHB depolymerase PhaZ1 degraded nPHB granules isolated from E. coli, while the (S) stereoisomer of 3HB-CoA was obtained when nPHB granules were degraded that were isolated from R. eutropha (Fig. 3A and B). In the enzyme assay performed with native granules isolated from E. coli, 3HB-CoA always appeared in low concentrations, whereas up to 90% of the product consisted of crotonyl-CoA (Table 2, experiment 5). Reprocessing of crotonyl-CoA to (S)-3HB-CoA, as it was observed in experiments with granules isolated from R. eutropha, requires enzymatic activity of enoyl-CoA hydratases. E. coli possesses the genes for these hydratases, but these are expressed only during growth on long-chain fatty acids with 14 or more carbon atoms, like oleic or palmitic acid (39). In addition, the presence of glucose, as in this study, inhibits the expression of fatty acid degradation genes (4, 39, 40). We tried to induce the enoyl-CoA hydratases in E. coli during cell growth by using MSM without glucose and with 0.1% (wt/vol) oleate as the carbon source. Although sufficient cell mass could be obtained, the accumulated PHB reached only up to 4% (wt/wt) of cell dry weight, which is much too low for isolation of nPHB granules and the subsequent degradation assay (data not shown). Thus, we did not further proceed with this approach.
R. eutropha possesses several enoyl-CoA hydratases, which are also induced during growth on fatty acids but are probably not strictly repressed during growth on gluconate (13, 21). A hint of the presence of these enoyl-CoA hydratases in the granule fractions is given by the results of the degradation assays with nPHB granules isolated from the single and the quadruple mutant of R. eutropha (the ΔphaP1 and ΔphaP1-4 mutants). In these reactions, CoA induced almost no degradation of nPHB granules, and therefore only very low concentrations of crotonyl-CoA were detected after 90 min of incubation (0.03 mM; Table 3, experiment 3). On the other hand, higher concentrations of crotonyl-CoA (up to 0.17 mM) were obtained after 90 min, after the reactions were started by the addition of (R/S)-3HB-CoA instead of CoA (Table 3, experiments 2 and 3). Thus, the conversion of 3HB-CoA to crotonyl-CoA was most probably catalyzed by an enoyl-CoA hydratase that was present in the soluble fraction of the granules instead of the PHB depolymerases of R. eutropha. Furthermore, the addition of the soluble granule fraction isolated from R. eutropha to granule fractions of E. coli resulted in higher concentrations of 3HB-CoA during PHB degradation than without the addition thereof (data not shown). Chiral HPLC analysis identified the collected 3HB-CoA of this reaction as the (S) stereoisomer. Therefore, the soluble fraction of granules isolated from R. eutropha must have contained enzymes possessing the catalytic function to convert crotonyl-CoA to (S)-3HB-CoA. Although no 3-hydroxybutyryl–CoA epimerases are described for R. eutropha H16 so far, their existence cannot be fully excluded. These enzymes catalyze the conversion of (R)-3HB-CoA to (S)-3HB-CoA and vice versa. If these enzymes were present and active in the used granule fractions, the monitored (S)-3HB-CoA could derive from the conversion of (R)-3HB-CoA instead of crotonyl-CoA after degradation of nPHB by the PHB depolymerase. This would mean, in turn, that the PHB depolymerase may not be responsible for the formation of crotonyl-CoA. These considerations are not supported by the findings of the experiments with granules isolated from E. coli. Enzymes involved in the β-oxidation of E. coli (including enoyl-CoA hydratases and 3-hydroxybutyryl–CoA epimerases) are strictly repressed during growth on glucose. Hence, the observed crotonyl-CoA must originate from the activity of the PHB depolymerase PhaZ1.
To monitor the putatively involved enoyl-CoA hydratases, a protein profile of purified PHB granules via SDS-PAGE was performed. In the case of PHB granules isolated from R. eutropha H16, minor copurified bands that have molecular weights similar to those of enoyl-CoA hydratases were obtained. In the protein profile of granules isolated from E. coli, the respective protein bands were missing (data not shown). However, matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) analysis failed to unambiguously identify enoyl-CoA hydratases.
Transcriptome analyses of former studies revealed that transcripts of enoyl-CoA hydratases occur during all investigated growth conditions: (i) growth on fructose with a nitrogen source, (ii) growth on fructose lacking a nitrogen source, (iii) growth on PHB (lacking an external carbon source) with a nitrogen source (41, 42). An upregulation of the expression of an enoyl-CoA hydratase (H16_A3311; log2 expression value of 3.1) in the late stationary phase under conditions permissive for PHB accumulation was shown in the study by Peplinski et al. (41). As PHB is most likely continuously accumulated and degraded during cell growth (26, 27), enoyl-CoA hydratases may already be required in the late stationary growth phase. Further utilization of PHB was not analyzed in this study. The study by Brigham et al. (42) showed that two enoyl-CoA hydratases were slightly upregulated (H16_B0382 and H16_B0657; log2 expression values of 1.2 and 1.3, respectively) during growth on PHB in comparison to growth on fructose (with a nitrogen source). However, no considerable upregulation of the expression of an enoyl-CoA hydratase could be confirmed during the conversion from conditions permissive for PHB accumulation to conditions permissive for PHB degradation.
Due to the fact that we observed only very low concentrations of (R)-3HB-CoA but significantly higher concentrations of (S)-3HB-CoA, we suppose that the (R) stereoisomer of 3HB-CoA is not released during PHB degradation. The detected amounts of (R)-3HB-CoA are presumably a consequence of the interruption of the degradation process by acidification of the reaction mixture, and therefore a release of the intermediate occurred. Our model is also in accordance with the interesting results of former studies in which the formation of acetyl-CoA during thiolysis of nPHB granules was strongly enhanced after the addition of NAD instead of NADP (22).
Initially, the aim of this study was to unravel a catalytic activity or a regulatory influence of the different phasins of R. eutropha on the degradation of PHB. As shown before, the highest in vitro degradation rate of the PHB depolymerase PhaZ1 was achieved in the presence of PhaP1 (22). Replacements of the phasin PhaP1 by PhaP2 or PhaP4 resulted in slightly reduced degradation rates of nPHB granules by PHB depolymerase PhaZ1. This fits with previous results where the lack of PhaP1 caused a reduced degradation of nPHB in R. eutropha (22). Furthermore, the obtained concentrations of 3HB-CoA and crotonyl-CoA in the enzyme assay in the presence of PhaP2 were almost equal to those in the presence of PhaP4. The lack of all phasins considerably reduced the degradation of nPHB by PhaZ1, but degradation was not fully prevented. Thus, under the conditions used in this study, the degradation of nPHB granules by PHB depolymerase PhaZ1 is most likely not strictly regulated or even inhibited in the presence of phasin PhaP2 or PhaP4 compared to PHB degradation in the presence of PhaP1. The catalytic activity of the seven so-far-known phasins and their putative regulatory influence on PHB degradation remain unknown. Although this issue could not be elucidated, the experiments of this study revealed the formation of crotonyl-CoA and both stereoisomers of 3HB-CoA during PHB degradation, depending on which microorganism, E. coli or R. eutropha, was applied. These results led us to adapt the degradation pathway of PHB in R. eutropha.
The proposed alternative pathway for the degradation of PHB in R. eutropha (Fig. 4) provides the opportunity of simultaneous synthesis and degradation of PHB without the loss of energy. In addition, due to the stereoselectivity of the synthesis and degradation reaction and due to the requirement of different cofactors for these two reactions, the regulation of the PHB metabolism might be controlled in the cell by varying the ratio of NAD to NADPH. Further work is necessary to fully understand the regulation mechanism and the involved key metabolites, like CoA and acetyl-CoA, which were already determined to influence the PHB metabolism in vitro (22, 43).
ACKNOWLEDGMENTS
We thank G. Barbenheim and S. Frech from Macherey-Nagel for kind support during the development of an appropriate application of separation of 3HB stereoisomers and the pretest with chiral HPLC columns.
Footnotes
Published ahead of print 10 May 2013
REFERENCES
- 1. Steinbüchel A, Valentin HE. 1995. Diversity of microbial polyhydroxyalkanoic acids. FEMS Microbiol. Lett. 128:219–228 [Google Scholar]
- 2. Steinbüchel A, Füchtenbusch B. 1998. Bacterial and other biological systems for polyester production. Trends Biotechnol. 16:419–427 [DOI] [PubMed] [Google Scholar]
- 3. Madison LL, Huisman GW. 1999. Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic. Microbiol. Mol. Biol. Rev. 63:21–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anderson AJ, Dawes EA. 1990. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Rev. 54:450–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reinecke F, Steinbüchel A. 2009. Ralstonia eutropha strain H16 as model organism for PHA metabolism and for biotechnological production of technically interesting biopolymers. J. Mol. Microbiol. Biotechnol. 16:91–108 [DOI] [PubMed] [Google Scholar]
- 6. Chanprateep S. 2010. Current trends in biodegradable polyhydroxyalkanoates. J. Biosci. Bioeng. 110:621–632 [DOI] [PubMed] [Google Scholar]
- 7. Gao X, Chen JC, Wu Q, Chen GQ. 2011. Polyhydroxyalkanoates as a source of chemicals, polymers, and biofuels. Curr. Opin. Biotechnol. 22:768–774 [DOI] [PubMed] [Google Scholar]
- 8. Schlegel HG, Gottschalk G, von Bartha R. 1961. Formation and utilization of poly-β-hydroxybutyric acid by Knallgas bacteria (Hydrogenomonas). Nature 191:463–465 [DOI] [PubMed] [Google Scholar]
- 9. Schlegel HG, Kaltwasser H, Gottschalk G. 1961. Ein Submersverfahren zur Kultur wasserstoffoxidierender Bakterien: Wachstumsphysiologische Untersuchungen. Arch. Mikrobiol. 38:209–222 [PubMed] [Google Scholar]
- 10. Wilde E. 1962. Untersuchungen über Wachstum und Speicherstoffsynthese von Hydrogenomonas eutropha. Arch. Mikrobiol. 43:109–13714042378 [Google Scholar]
- 11. Haywood G, Anderson A, Dawes E. 1989. The importance of PHB synthase substrate specificity in polyhydroxyalkanoate synthesis by Alcaligenes eutrophus. FEMS Microbiol. Lett. 57:1–6 [Google Scholar]
- 12. Lawrence AG, Schoenheit J, He A, Tian J, Liu P, Stubbe J, Sinskey AJ. 2005. Transcriptional analysis of Ralstonia eutropha genes related to poly-(R)-3-hydroxybutyrate homeostasis during batch fermentation. Appl. Microbiol. Biotechnol. 68:663–672 [DOI] [PubMed] [Google Scholar]
- 13. Brigham CJ, Budde CF, Holder JW, Zeng Q, Mahan AE, Rha C, Sinskey AJ. 2010. Elucidation of beta-oxidation pathways in Ralstonia eutropha H16 by examination of global gene expression. J. Bacteriol. 192:5454–5464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wieczorek R, Pries A, Steinbüchel A, Mayer F. 1995. Analysis of a 24-kilodalton protein associated with the polyhydroxyalkanoic acid granules in Alcaligenes eutrophus. J. Bacteriol. 177:2425–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pötter M, Müller H, Reinecke F, Wieczorek R, Fricke F, Bowien B, Friedrich B, Steinbüchel A. 2004. The complex structure of polyhydroxybutyrate (PHB) granules: four orthologous and paralogous phasins occur in Ralstonia eutropha. Microbiology 150:2301–2311 [DOI] [PubMed] [Google Scholar]
- 16. Pfeiffer D, Jendrossek D. 2011. Interaction between poly(3-hydroxybutyrate) granule-associated proteins as revealed by two-hybrid analysis and identification of a new phasin in Ralstonia eutropha H16. Microbiology 157:2795–2807 [DOI] [PubMed] [Google Scholar]
- 17. Pfeiffer D, Jendrossek D. 2012. Localization of poly(3-hydroxybutyrate) (PHB) granule-associated proteins during PHB granule formation and identification of two new phasins, PhaP6 and PhaP7, in Ralstonia eutropha H16. J. Bacteriol. 194:5909–5921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Saegusa H, Shiraki M, Kanai C, Saito T. 2001. Cloning of an intracellular poly[d(−)-3-hydroxybutyrate] depolymerase gene from Ralstonia eutropha H16 and characterization of the gene product. J. Bacteriol. 183:94–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Saegusa H, Shiraki M, Saito T. 2002. Cloning of an intracellular d(-)-3-hydroxybutyrate-oligomer hydrolase gene from Ralstonia eutropha H16 and identification of the active site serine residue by site-directed mutagenesis. J. Biosci. Bioeng. 94:106–112 [DOI] [PubMed] [Google Scholar]
- 20. York GM, Lupberger J, Tian J, Lawrence AG, Stubbe J, Sinskey AJ. 2003. Ralstonia eutropha H16 encodes two and possibly three intracellular poly[d-(−)-3-hydroxybutyrate] depolymerase genes. J. Bacteriol. 185:3788–3794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pohlmann A, Fricke WF, Reinecke F, Kusian B, Liesegang H, Cramm R, Eitinger T, Ewering C, Pötter M, Schwartz E, Strittmatter A, Voss I, Gottschalk G, Steinbüchel A, Friedrich B, Bowien B. 2006. Genome sequence of the bioplastic-producing “Knallgas” bacterium Ralstonia eutropha H16. Nat. Biotechnol. 24:1257–1262 [DOI] [PubMed] [Google Scholar]
- 22. Uchino K, Saito T, Gebauer B, Jendrossek D. 2007. Isolated poly(3-hydroxybutyrate) (PHB) granules are complex bacterial organelles catalyzing formation of PHB from acetyl coenzyme A (CoA) and degradation of PHB to acetyl-CoA. J. Bacteriol. 189:8250–8256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Merrick JM, Doudoroff M. 1964. Depolymerization of poly-beta-hydroxybutyrate by intracellular enzyme system. J. Bacteriol. 88:60–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cornibert J, Marchessault RH. 1972. Physical properties of poly-β-hydroxybutyrate. IV. Conformational analysis and crystalline structure. J. Mol. Biol. 28:735–756 [DOI] [PubMed] [Google Scholar]
- 25. Jendrossek D. 2007. Peculiarities of PHA granules preparation and PHA depolymerase activity determination. Appl. Microbiol. Biotechnol. 74:1186–1196 [DOI] [PubMed] [Google Scholar]
- 26. Doi Y, Segawa A, Kawaguchi Y, Kunioka M. 1990. Cyclic nature of poly(3-hydroxyalkanoate) metabolism in Alcaligenes eutrophus. FEMS Microbiol. Lett. 55:165–169 [DOI] [PubMed] [Google Scholar]
- 27. Taidi B, Mansfield D, Anderson AJ. 1995. Turnover of poly(3-hydroxybutyrate) (PHB) and its influence on the molecular mass of the polymer accumulated by Alcaligenes eutrophus during batch culture. FEMS Microbiol. Lett. 129:201–206 [Google Scholar]
- 28. Tsuge T, Fukui T, Matsusaki H, Taguchi S, Kobayashi G, Ishizaki A, Doi Y. 2000. Molecular cloning of two (R)-specific enoyl-CoA hydratase genes from Pseudomonas aeruginosa and their use for polyhydroxyalkanoate synthesis. FEMS Microbiol. Lett. 184:193–198 [DOI] [PubMed] [Google Scholar]
- 29. Park SJ, Lee SY. 2003. Identification and characterization of a new enoyl coenzyme A hydratase involved in biosynthesis of medium-chain-length polyhydroxyalkanoates in recombinant Escherichia coli. J. Bacteriol. 185:5391–5397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Davis R, Anilkumar PK, Chandrashekar A, Shamala TR. 2008. Biosynthesis of polyhydroxyalkanoates co-polymer in E. coli using genes from Pseudomonas and Bacillus. Antonie Van Leeuwenhoek 94:207–216 [DOI] [PubMed] [Google Scholar]
- 31. Kawashima Y, Cheng W, Mifune J, Orita I, Nakamura S, Fukui T. 2012. Characterization and functional analyses of R-specific enoyl coenzyme A hydratases in polyhydroxyalkanoate-producing Ralstonia eutropha. Appl. Environ. Microbiol. 78:493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schulz H. 1991. Beta oxidation of fatty acids. Biochim. Biophys. Acta 1081:109–120 [DOI] [PubMed] [Google Scholar]
- 33. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 34. Preusting H, Kingma J, Huisman G, Steinbüchel A, Witholt B. 1993. Formation of polyester blends by a recombinant strain of Pseudomonas oleovorans: different poly(3-hydroxyalkanoates) are stored in separate granules. J. Environ. Polym. Degrad. 1:11–21 [Google Scholar]
- 35. Schürmann M, Wübbeler JH, Grote J, Steinbüchel A. 2011. Novel reaction of succinyl coenzyme A (succinyl-CoA) synthetase: activation of 3-sulfinopropionate to 3-sulfinopropionyl-CoA in Advenella mimigardefordensis strain DPN7T during degradation of 3,3′-dithiodipropionic acid. J. Bacteriol. 193:3078–3089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tsai YC, Liao TH, Lee JA. 2003. Identification of l-3-hydroxybutyrate as an original ketone body in rat serum by column-switching high-performance liquid chromatography and fluorescence derivatization. Anal. Biochem. 319:34–41 [DOI] [PubMed] [Google Scholar]
- 37. Tsai YC, Chou YC, Wu AB, Hu CM, Chen CY, Chen FA, Lee JA. 2006. Stereoselective effects of 3-hydroxybutyrate on glucose utilization of rat cardiomyocytes. Life Sci. 78:1385–1391 [DOI] [PubMed] [Google Scholar]
- 38. Abe T, Kobayashi T, Saito T. 2005. Properties of a novel intracellular poly(3-hydroxybutyrate) depolymerase with high specific activity (PhaZd) in Wautersia eutropha H16. J. Bacteriol. 187:6982–6990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Clark D. 1981. Regulation of fatty acid degradation in Escherichia coli: analysis by operon fusion. J. Bacteriol. 148:521–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sato S, Nomura CT, Abe H, Doi Y, Tsuge T. 2007. Poly[(R)-3-hydroxybutyrate] formation in Escherichia coli from glucose through an enoyl-CoA hydratase-mediated pathway. J. Biosci. Bioeng. 103:38–44 [DOI] [PubMed] [Google Scholar]
- 41. Peplinski K, Ehrenreich A, Döring C, Bömeke M, Reinecke F, Hutmacher C, Steinbüchel A. 2010. Genome-wide transcriptome analyses of the ‘Knallgas’ bacterium Ralstonia eutropha H16 with regard to polyhydroxyalkanoate metabolism. Microbiology 156:2136–2152 [DOI] [PubMed] [Google Scholar]
- 42. Brigham CJ, Speth DR, Rha C, Sinskey AJ. 2012. Whole-genome microarray and gene deletion studies reveal regulation of the polyhydroxyalkanoate production cycle by the stringent response in Ralstonia eutropha H16. Appl. Environ. Microbiol. 78:8033–8044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ren Q, de Roo G, Ruth K, Witholt B, Zinn M, Thöny-Meyer L. 2009. Simultaneous accumulation and degradation of polyhydroxyalkanoates: futile cycle or clever regulation? Biomacromolecules 10:916–922 [DOI] [PubMed] [Google Scholar]
- 44. York GM, Stubbe J, Sinskey AJ. 2001. New insight into the role of the PhaP phasin of Ralstonia eutropha in promoting synthesis of polyhydroxybutyrate. J. Bacteriol. 183:2394–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pötter M, Müller H, Steinbüchel A. 2005. Influence of homologous phasins (PhaP) on PHA accumulation and regulation of their expression by the transcriptional repressor PhaR in Ralstonia eutropha H16. Microbiology 151:825–833 [DOI] [PubMed] [Google Scholar]
- 46. Spiekermann P, Rehm BHA, Kalscheuer R, Baumeister D, Steinbüchel A. 1999. A sensitive, viable-colony staining method using Nile red for direct screening of bacteria that accumulate polyhydroxyalkanoic acids and other lipid storage compounds. Arch. Microbiol. 171:73–80 [DOI] [PubMed] [Google Scholar]
- 47. Gebauer B, Jendrossek D. 2006. Assay of poly(3-hydroxybutyrate) depolymerase activity and product determination. Appl. Environ. Microbiol. 72:6094–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]