Abstract
Miniature inverted repeat transposable elements (MITEs) have been identified flanking class 1 integrons. We have identified and characterized a 439-bp MITE-like structure in seven Acinetobacter species isolates from Portugal and Brazil. The complete sequence similarity of the elements and flanking regions suggests that MITEs may act as mobilizable vectors for the dissemination of integrons.
TEXT
Small DNA repeat sequences are present in a variety of bacterial genomes and display a wide range of structures and functions. Some of these sequences are defined as miniature inverted repeat transposable elements (MITEs) (1). MITEs are nonautonomous mobile genetic elements that require a transposase provided in trans for transposition and have been found in Eubacteria, Archaea, and Eukaryota (2, 3). It has been suggested that they derive from insertion sequences (ISs), as they have terminal inverted repeats (TIRs) and are flanked by target site duplications (TSDs) (1, 3). The transposases of ISs may be able to mobilize MITEs with similar TIRs (1).
A few studies have found MITE-like structures flanking class 1 integrons (4–6). Remarkably, an identical MITE of 439 bp was identified in a clinical Acinetobacter baumannii isolate collected in Portugal and in a prawn-associated Acinetobacter johnsonii isolate from Australia (5–7). In both cases, transposition was suggested as the mechanism of acquisition, as TSDs flanked the MITE-integron complex (5, 6). We hypothesized that this element might be more disseminated in Acinetobacter spp. because of the different origins and geographic locations of the previously identified strains. The presence of the 439-bp MITE-like structure was screened for by PCR in 28 nonrepetitive clinical Acinetobacter isolates collected between 1992 and 1999 in two regions of Portugal. The amplification protocol used was 5 min at 94°C; 30 cycles of 1 min at 94°C, 1 min at 52°C, and 1.5 min at 72°C; and a final step of 10 min at 72°C. Primers MITE1 (5′-TGTGACTGACCATTAAAG-3′) and MITE2 (5′-TGTCTTTGCACATTAAAG-3′) were used; A. baumannii 65FFC was used as the positive control (6). MITEs were detected in only one isolate, Acinetobacter sp. strain 118FFC, and sequenced. The nucleotide sequence of the MITE of this strain was identical to the one previously identified in A. baumannii strain 65FFC and A. johnsonii strain NFM2. In silico analysis of the MITE sequence revealed a partial alignment with the class 1 integron 5′-CS (conserved segment) flanking region from several clinical Acinetobacter isolates (A. baumannii 694, 695, 696, and 9043 and Acinetobacter sp. strains 5227 and 5248) collected in Brazil in 2001 and 2002 (8). PCR detection of the 439-bp MITE was positive for the six Brazilian isolates, and the sequence also confirmed 100% identity to the 439-bp MITE mentioned before. Acinetobacter isolates 118FFC, 5227, and 5248 were identified to the species level by direct genomic DNA sequencing of the rpoB gene (9). Sequencing reactions were performed with the BigDye 3.1 cycle sequencing terminator reactions (Applied Biosystems) as previously described (10). Acinetobacter sp. strain 118FFC was identified as A. bereziniae (previously Acinetobacter genomic species 10 [11]), while Brazilian isolates 5227 and 5248 were A. baumannii.
PCR with specific primers (12) amplified a class 1 integron of 2.2 kb in the uncommon nosocomial pathogen A. bereziniae 118FFC in which sequencing revealed three gene cassettes, the aacA7 gene (coding sequence [CDS] of 459 bp), the blaVIM-2 gene (CDS of 801 bp), and the aacC1 gene (CDS of 465 bp), coding for aminoglycoside (aac-type) and carbapenem (VIM-2) resistance. This is a new gene cassette array, which was designated In796 by the INTEGRALL database of class 1 integrons (13). Also, to our knowledge, VIM-2 is the first reported metallo-β-lactamase in an A. bereziniae strain. The gene cassette array of In796 is similar to the array of In58 (14). The latter contains an additional aacA4 cassette after the aacC1 cassette. The nucleotide sequences of both integrons are 100% identical over stretches of 3,361 and 1,650 bp that include the intI1, aacA7, blaVIM-2, and aacC1 genes and the qacEΔ1, sul1, and orf5 genes, respectively. A. bereziniae 118FFC containing In796 is a clinical isolate collected in Portugal in 1998, and In58 was first described in clinical strain P. aeruginosa RON-1, which was collected in France, also in 1998 (14). The similar nucleotide sequences and the array of gene cassettes suggest that In58 could be the precursor of In796 because of the loss of the last cassette, which can occur in the absence of antibiotic pressure (15, 16). In796 is flanked by two MITE-like structures. Whether the sequences surrounding In58 contain MITEs, and if these have any role in the dissemination of the integron, is not known. So far, this 439-bp MITE has been found only in Acinetobacter species.
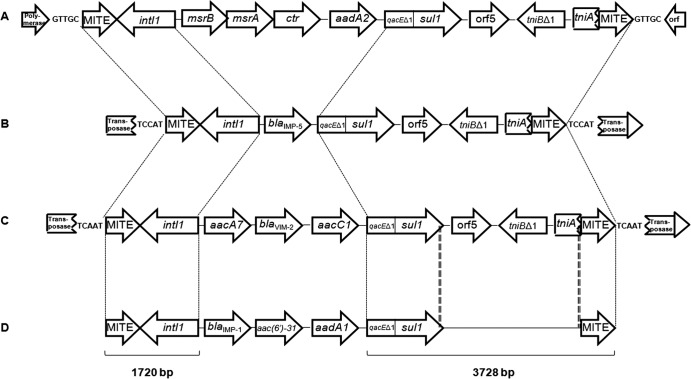
The positions of the MITEs in relation to the integron were confirmed in the seven MITE-containing strains by using primer pairs MITE1-CS3 and MITE2-CS2 (CS3 and CS2 bind in the conserved regions of the class 1 integron in an outward direction [12]) and the same PCR program previously described but with an extension time of 2 min and the addition of 5% dimethyl sulfoxide. The sizes of the PCR products showed that the MITEs were in the same positions relative to the class 1 integron as in the previously reported isolates. Sequencing of the conserved and flanking regions of the class 1 integron of A. bereziniae 118FFC confirmed that the nucleotide composition was 100% identical to the published sequences. In addition, a 5-bp TSD (TCAAT) was detected on each side of the MITEs and one gene was interrupted by the MITE-integron-MITE structure, suggesting acquisition by transposition, as also seen in A. baumannii 65FFC (6) and A. johnsonii NFM2 (5). Figure 1 shows the integron cassette arrays and the identical regions found between the MITEs in the Acinetobacter sp. strains.
Fig 1.
Schematic representation of the class 1 integrons and flanking regions in A. johnsonii strain NFM2 (A) (5); A. baumannii strain 65FFC (B) (6); A. bereziniae strain 118FFC (C) (this study); and A. baumannii strains 694, 695, 696, 5227, 5248, and 9043 (D) (8 and this study). The 1,720- and 3,728-bp segments between the dashed lines are 100% identical in panels A, B, and C; the sequence of the region between the double dashed lines in panel D was not determined, but it was PCR amplified and has the same size as that of the sequenced strains (panels A, B, and C).
Overall, we report the presence of an identical 439-bp MITE surrounding different class 1 integrons in seven clinical strains of Acinetobacter spp. in addition to the two previously reported. It is noteworthy that all of the strains of clinical origin carried different metallo-β-lactamase determinants (IMP-5, IMP-1, VIM-2); thus, MITE mobilizable integrons may be important in the dissemination of diverse metallo-β-lactamase determinants and therefore of major clinical importance. TSDs, interrupted genes, and the presence of identical MITE-like structures in three Acinetobacter species from three continents and different sources strongly suggest that MITEs may be disseminated horizontally and act as mobilizable vectors for integron dissemination.
Nucleotide sequence accession number.
The nucleotide sequence obtained from A. bereziniae 118FFC has been deposited in the GenBank database under accession number JX235356.
ACKNOWLEDGMENTS
This work received financial support from the Center of Pharmaceutical Studies, University of Coimbra, Coimbra, Portugal, and the University of Tromsø, Tromsø, Norway. S.D. is supported by grant SFRH/BD/49061/2008 from the Fundação para a Ciência e a Tecnologia, Lisbon, Portugal.
We have no conflict of interest to declare.
Footnotes
Published ahead of print 17 April 2013
REFERENCES
- 1. Delihas N. 2011. Impact of small repeat sequences on bacterial genome evolution. Genome Biol. Evol. 3:959–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Delihas N. 2008. Small mobile sequences in bacteria display diverse structure/function motifs. Mol. Microbiol. 67:475–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Siguier P, Filee J, Chandler M. 2006. Insertion sequences in prokaryotic genomes. Curr. Opin. Microbiol. 9:526–531 [DOI] [PubMed] [Google Scholar]
- 4. Poirel L, Carrer A, Pitout JD, Nordmann P. 2009. Integron mobilization unit as a source of mobility of antibiotic resistance genes. Antimicrob. Agents Chemother. 53:2492–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gillings MR, Labbate M, Sajjad A, Giguere NJ, Holley MP, Stokes HW. 2009. Mobilization of a Tn402-like class 1 integron with a novel cassette array via flanking miniature inverted-repeat transposable element-like structures. Appl. Environ. Microbiol. 75:6002–6004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Domingues S, Nielsen KM, da Silva GJ. 2011. The blaIMP-5-carrying integron in a clinical Acinetobacter baumannii strain is flanked by miniature inverted-repeat transposable elements (MITEs). J. Antimicrob. Chemother. 66:2667–2668 [DOI] [PubMed] [Google Scholar]
- 7. Da Silva GJ, Correia M, Vital C, Ribeiro G, Sousa JC, Leitão R, Peixe L, Duarte A. 2002. Molecular characterization of blaIMP-5, a new integron-borne metallo-β-lactamase gene from Acinetobacter baumannii nosocomial isolate in Portugal. FEMS Microbiol. Lett. 215:33–39 [DOI] [PubMed] [Google Scholar]
- 8. Mendes RE, Castanheira M, Toleman MA, Sader HS, Jones RN, Walsh TR. 2007. Characterization of an integron carrying blaIMP-1 and a new aminoglycoside resistance gene, aac(6′)-31, and its dissemination among genetically unrelated clinical isolates in a Brazilian hospital. Antimicrob. Agents Chemother. 51:2611–2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. La Scola B, Gundi VA, Khamis A, Raoult D. 2006. Sequencing of the rpoB gene and flanking spacers for molecular identification of Acinetobacter species. J. Clin. Microbiol. 44:827–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ray JL, Harms K, Wikmark OG, Starikova I, Johnsen PJ, Nielsen KM. 2009. Sexual isolation in Acinetobacter baylyi is locus-specific and varies 10,000-fold over the genome. Genetics 182:1165–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nemec A, Musilek M, Sedo O, De Baere T, Maixnerova M, van der Reijden TJ, Zdrahal Z, Vaneechoutte M, Dijkshoorn L. 2010. Acinetobacter bereziniae sp. nov. and Acinetobacter guillouiae sp. nov., to accommodate Acinetobacter genomic species 10 and 11, respectively. Int. J. Syst. Evol. Microbiol. 60:896–903 [DOI] [PubMed] [Google Scholar]
- 12. Domingues S, Harms K, Fricke WF, Johnsen PJ, da Silva GJ, Nielsen KM. 2012. Natural transformation facilitates transfer of transposons, integrons and gene cassettes between bacterial species. PLoS Pathog. 8:e1002837. 10.1371/journal.ppat.1002837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moura A, Soares M, Pereira C, Leitao N, Henriques I, Correia A. 2009. INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25:1096–1098 [DOI] [PubMed] [Google Scholar]
- 14. Poirel L, Lambert T, Turkoglu S, Ronco E, Gaillard J, Nordmann P. 2001. Characterization of class 1 integrons from Pseudomonas aeruginosa that contain the blaVIM-2 carbapenem-hydrolyzing beta-lactamase gene and of two novel aminoglycoside resistance gene cassettes. Antimicrob. Agents Chemother. 45:546–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rosser SJ, Young HK. 1999. Identification and characterization of class 1 integrons in bacteria from an aquatic environment. J. Antimicrob. Chemother. 44:11–18 [DOI] [PubMed] [Google Scholar]
- 16. Collis CM, Hall RM. 1992. Site-specific deletion and rearrangement of integron insert genes catalyzed by the integron DNA integrase. J. Bacteriol. 174:1574–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]