Abstract
Modern pyrosequencing technology allows for a more comprehensive approach than traditional Sanger sequencing for elucidating the etiology of bovine digital dermatitis. We sought to describe the composition and diversity of treponemes in digital dermatitis lesions by using deep sequencing of the V3 and V4 hypervariable regions of the 16S rRNA gene coupled with species-level taxonomic identification. Treponema-specific 16S rRNA gene PCRs and pyrosequencing were performed on biopsy specimens originating from 10 different Catalan dairy herds (n = 36) with digital dermatitis, and this analysis yielded 75,297 sequences. We identified 20 different taxa, including a potentially novel phylotype that displayed 95% sequence identity to members of the Treponema denticola/Treponema pedis-like cluster. Species frequencies and abundances that were determined by pyrosequencing analysis were highly correlated with the results of fluorescent in situ hybridization using phylotype-specific oligonucleotide probes. In a limited number of animals from a single geographic region, we detected most of the Treponema phylotypes that were described in previous investigations of digital dermatitis. Additionally, we identified a number of phylotypes that mapped to oral treponemes of humans and dogs that had not been reported for digital dermatitis lesions. The results presented here support previous observations of a polytreponemal etiology of infections, with Treponema phagedenis-like, Treponema medium/Treponema vincentii-like, and T. denticola/T. pedis-like phylotypes being highly associated with disease. Using this new approach, it has become feasible to study large herds and their surrounding environments, which might provide a basis for a better understanding of the pathogenesis of this disease.
INTRODUCTION
Bovine digital dermatitis (DD), which causes lameness and wasting in cattle, is the most serious foot disorder of dairy cows from economic and welfare perspectives (1). The disease, which is characterized by focal proliferative to ulcerative dermatitis that is typically located on the plantar aspect of the foot, apparently is contagious (2). Currently, a bacterial etiology of DD is well documented (3, 4). Fluorescent in situ hybridization (FISH) analyses have shown that spirochetes of the genus Treponema are found mainly in the deeper parts of DD lesions, near the interface with healthy tissue (5, 6). Additionally, cows with DD demonstrate specific humoral and cell-mediated immune responses to Treponema (7). At least 18 different treponemal species have been identified by cloning of bacterial 16S rRNA genes PCR-amplified directly from DD samples (8–11). Previous works by us and others have suggested that DD has a symbiotic treponemal pathogenesis (9–12), and Treponema phagedenis-like, Treponema denticola-like, and Treponema medium-like bacteria appear to be among the most prevalent species in the lesions. The complex interplay between infecting Treponema species, their relative contributions to pathogenesis and infection reservoirs, and their transmission routes are largely unknown for this disease and must be elucidated (13).
The inherent limitations of the labor-intensive, costly approach of 16S rRNA-based clonal analysis, which utilizes the conventional Sanger capillary sequencing technique (14), has hitherto prevented large-scale in-depth analysis of the microbial ecology of DD lesions. Next-generation deep-coverage sequencing techniques now allow high-throughput sequencing of a single variable 16S rRNA region in a cost-effective manner (15). Using 454 pyrosequencing, we analyzed the treponemal biodiversity in 36 biopsy specimens from Spanish cattle with DD. Subsequently, the results of the sequencing analysis were evaluated by FISH. Presently, we are not aware of any other work that has applied next-generation sequencing technology to the investigation of DD in cattle.
MATERIALS AND METHODS
Sample collection and preparation.
Eleven Catalan dairy farms (in the provinces of Gerona and Barcelona), where DD was present endemically and no preventive herd strategies were practiced regularly, were selected for the study. Forty biopsy specimens were obtained at routine trimming, which was performed by a professional hoof trimmer who used a transportable, hydraulic, trimming chute. Only one biopsy specimen per animal was obtained. The skin was wiped clean of manure and rinsed with sterile phosphate-buffered saline before sampling. Skin biopsy specimens of 6 mm were taken from the center of each lesion using a sterile biopsy punch (UniPunch; UniPunch Products, Inc., Clear Lake, WI). Following sampling, the lesions were treated with a mixture of tetracycline and salicylic acid powder (Fagron Ibérica, S.a.u., Terrassa, Spain), and a bandage was applied. Macroscopically, more than 60% of the lesions were characterized as being in the classic ulcerative stage, i.e., the lesions had diameters of >2 cm, as illustrated in Fig. 1, left, and often were painful on palpation (score of M2, according the system described by Holzhauer et al. [16]). The remaining lesions included different stages of DD (16). The severity score (acute or chronic) and the farm location history were recorded during collection of the samples. Spirochetes were detected by dark-field microscopy or dilute carbol fuchsin (DCF) staining in tissue smears from approximately 75% of the biopsy specimens (details of the samples are provided in Table S1 in the supplemental material).
Fig 1.
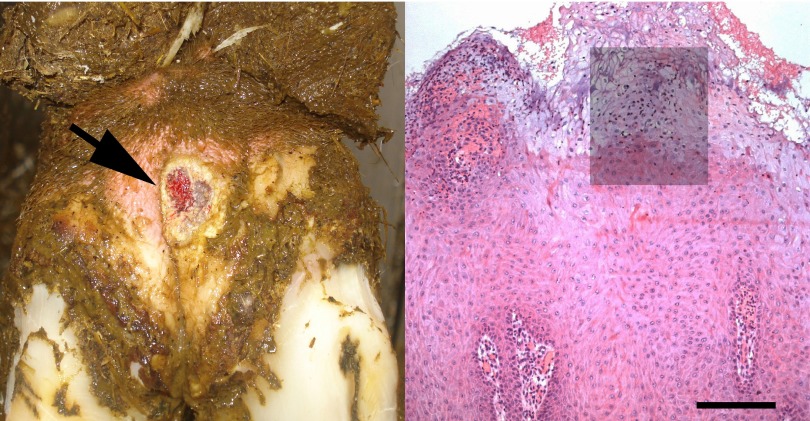
Bovine digital dermatitis appearance. (Left) Plantar aspect of the hind foot of a Catalan Holstein dairy cow affected by an ulcerative stage of the disease (arrow). (Right) Digital dermatitis lesion with a lesion score of 3, characterized by acanthotic epidermis with degenerated ballooning keratinocytes, exocytosis, ulceration of dermal papillae, and bacterial infiltration. The shading represents the area used for fluorescent in situ hybridization on parallel sections in Fig. 5. Bar, 200 μm.
For molecular analysis, one half of the biopsy specimen was cut into small pieces, suspended in 0.5 ml of sterile phosphate-buffered saline, and frozen at −20°C. For histological examination, the other half of the biopsy specimen was fixed in 10% neutral buffered formalin, dehydrated, and embedded in paraffin wax.
Bacterial DNA was extracted from tissue samples using the DNeasy tissue kit (Qiagen, Hilden, Germany). First, a piece of biopsy specimen the size of one-quarter of a pea was incubated for 1.5 h at 55°C in 180 μl lysis buffer with 20 μl proteinase K, which was provided in the kit. All subsequent steps were performed according to the instructions provided by the manufacturer. The concentrations and purity of the samples were evaluated using a Nanodrop-1000 spectrophotometer (Fisher Scientific, Wilmington, MA), and samples with 260-nm/280-nm absorbance (A260/A280) ratios of <1.8 or low concentrations of DNA were purified and concentrated according to the DNA cleanup protocol provided in the manual (Qiagen).
Histopathology.
Sections from all specimens were stained with hematoxylin and eosin for histopathological evaluation, and lesions were classified as DD, according to the system described by Rasmussen et al. (6), if they showed acanthotic epidermis (with or without parakeratotic papillomatous proliferation) with the complications of (i) bacterial colonization/infiltration of pale-staining and ballooning keratinocytes (swollen noneosinophilic cytoplasm and enlarged or condensed nuclei) and (ii) exocytosis, erosion, and/or ulceration of dermal papillae. The degree of epidermal damage in the lesions was further scored by the senior author as mild/focal (score of 1), moderate (score of 2), or extensive to diffuse (score of 3), as illustrated in Fig. 1, right. Additionally, the biopsy specimens were scored according to the inflammatory response in the dermis, i.e., mild or absent (score of 1), moderate (score of 2), or severe (score of 3), as previously described (6).
FISH analysis.
The oligonucleotide probes used in this study are listed in Table S2 in the supplemental material, and they were selected using ARB software (http://www.arb-home.de) or the Primrose program, version 2.17 (http://www.bioinformatics-toolkit.org/Primrose/index.html) (17). The oligonucleotide probes were labeled at the 5′ end with fluorescein isothiocyanate (FITC), Alexa Fluor 488, or the isothiocyanate derivative Cy3 (Eurofins MWG Operon, Ebersberg, Germany, or DNA Technology A/S, Risskov, Denmark). For FISH analysis, serial sections were cut (3 μm) and mounted on SuperFrost Plus slides (Menzel-Gläser, Braunschweig, Germany). Hybridization was carried out for 16 h at 45°C with 40 μl of hybridization buffer (100 mM Tris [pH 7.2], 0.9 M NaCl, 0.1% sodium dodecyl sulfate) and 200 ng of each probe, in a Sequenza slide rack (Thermo Shandon, Cheshire, United Kingdom). The sections then were washed three times with prewarmed (45°C) hybridization buffer for 15 min and subsequently three times with prewarmed (45°C) washing solution (100 mM Tris [pH 7.2], 0.9 M NaCl). The sections were then rinsed in water, air dried, and mounted in Vectashield (Vector Laboratories Inc., Burlingame, CA) for epifluorescence microscopy.
The hybridized sections were deidentified, and all were read and scored by the senior author. The total bacterial invasion of the epidermis within each biopsy specimen was scored from 0 to 3 (0, no invasive bacteria; 1, low number of invasive bacteria; 2, moderate number of invasive bacteria; 3, high number of invasive bacteria). For each of the Treponema phylotypes, the “within-biopsy prevalence” (phylotype prevalence score) was scored from 0 to 3 as described previously (6, 12) (0, no hybridization; 1, sparse hybridization [up to 5% of the total number of bacteria]; 2, moderate hybridization [between 5% and 10% of the total number of bacteria]; 3, strong hybridization [more than 10% of the total number of bacteria]), according to the system described by Rasmussen et al. (6). An Axio Imager M1 epifluorescence microscope equipped for epifluorescence with a 120-W HBO lamp and filter sets 43 and 38 were used to visualize the Cy3 and Alexa Fluor 488 dyes, respectively. Images were obtained using an AxioCam MRm version 3 FireWire monochrome camera and AxioVision software, version 4.5 (Carl Zeiss, Oberkochen, Germany).
Primer design.
Oligonucleotide primers targeting the V3 and V4 hypervariable regions, which spanned positions 346 to 705 in Escherichia coli K-12, were designed using the aligned 16S rRNA sequences of Treponema putidum, Treponema pedis, T. denticola, Treponema vincentii, Treponema calligyrum, and Treponema refringens (BioNumerics; Applied Maths, Sint-Martens-Latem, Belgium) with the Primer3 version 0.4.0 (http://frodo.wi.mit.edu/primer3) (18) and Primrose (http://www.bioinformatics-toolkit.org/index.html) (17) programs. According to the probe match function in the Ribosomal Database Project (RDP) database (http://rdp.cme.msu.edu), this primer set appeared to be specific for the Spirochaetaceae family, because it amplified mainly members of the genus Treponema. Of the 20 type strains that were present in the RDP database, this primer set targeted four members, i.e., T. putidum (ATCC 700334), T. denticola (ATCC 35405), T. medium (G7201), and T. pedis (T3552B).
Preparation of 16S rRNA gene amplicon libraries and sequencing.
DNA was amplified by PCR using the Treponema-specific primers that were designed for this study. Each sample was amplified with unique forward and reverse primers that included an added hexamer barcode at their 5′ ends (see Table S2 in the supplemental material). Amplification of the target region was carried out in a 50-μl reaction mixture that contained 1× AmpliTaq buffer (Applied Biosystems, Carlsbad, CA), 100 μM concentrations of each deoxynucleoside triphosphate (Amersham Biosciences, Piscataway, NJ), 0.2 pmol of each primer, 2.5 U of AmpliTaq DNA polymerase (Applied Biosystems), and 2 μl of template DNA. Thermal cycling using a T3 thermocycler (Biometram, Göttingen, Germany) was performed as follows: denaturation at 94°C for 3.5 min; 30 cycles of denaturation at 94°C for 45 s, annealing at 59°C for 45 s, and extension at 72°C for 1 min; and a final elongation step of 5 min. A positive control and a negative control (water) were included for every PCR. For samples for which no PCR product could be amplified, a DNA cleanup and concentration step was added and the PCR procedure was repeated. The DNA concentration and quality of the PCR amplicons from all 40 biopsy specimens were assessed with an Agilent 2100 bioanalyzer (Agilent Technologies Inc., Santa Clara, CA) prior to high-throughput sequencing (data not shown). Equal amounts of all 40 amplicons were pooled (final concentration, 5 μg) and purified using the Qiagen Mini Elute kit (Qiagen), according to the protocol provided by the manufacturer. Next-generation sequencing was performed by LGC Genomics (Teddington, Middlesex, United Kingdom) using a Roche/454 GS FLX+ titanium kit, according to their standard procedure (1/4 Pico-Titer plate [PTP]). The data discussed in this publication have been deposited in the NCBI Gene Expression Omnibus (19) and are accessible through accession number GSE42426 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE42426).
Sequence processing and analysis.
The 454 pyrosequencing readings were processed using the RDP Pyrosequencing Pipeline (20, 21). The raw sequences were parsed with barcodes using the Pipeline initial process step, and the amplicons were screened for the presence of forward and reverse primers, with an allowance for ≤2 mismatches to the primer sequences, no ambiguous base calls, and a minimum sequence length of 200 bases. Sequences represented by less than 10 clones were discarded from the analysis. NCBI BLAST searches (22) and the RDP sequence match process (21) were used to identify representative sequences, and queries that matched the closest database sequence with ≥98% identity over an alignment of at least 300 bp were assigned to the respective species level taxon.
For one highly prevalent sequence that shared only 95% sequence identity with any recognized Treponema species, a nearly full-length 16S rRNA gene sequence (approximately 1.4 kb) was obtained by cloning and sequencing using vector primers and the primers listed in Table S2 in the supplemental material. This procedure was described in a previous publication (12). Sequence analysis and phylogenetic tree construction were performed using the BioNumerics version 4.0 program (Applied Math).
Nucleotide sequence accession number.
The sequence of the putative Treponema phylotype discussed above has been deposited in GenBank under accession number KC161247.
RESULTS
Spirochetal diversity determined by pyrosequencing analysis.
From the 40 biopsy specimens that were included in the analysis (the clinical metrics are detailed in Table S1 in the supplemental material), 212,827 bacterial sequences were generated, of which 75,297 sequences, with an average length of 321 bp, met the quality-control criteria. In this process, all sequences that were isolated from cows 1 to 4 were discarded due to a low number of forward sequences (between 8 and 101); therefore, these animals were omitted from further analysis. Table S3 in the supplemental material presents the relative frequency distribution of the phylotypes in the remaining 36 biopsy specimens. Only spirochetes were found in the samples, which indicated a good protocol and primer specificity. The 16S rRNA sequences of treponemes differ by an average of nearly 10% (23), and, in general, the V3 and V4 hypervariable regions offered good resolution between the various taxa. Only T. medium and T. vincentii could not be reliably distinguished from each other.
Of the 75,297 sequences, 51,635 were delineated into 20 different taxa based on 16S rRNA comparisons, including 5 established and cultivable species of the genus Treponema (T. phagedenis, T. medium, T. denticola, T. pedis, and T. refringens), 9 as-yet-uncultivated phylotypes that were previously identified from DD samples (PT1 to PT4, PT8, PT9, PT12/PN20, PT13, and PT15), 5 as-yet-uncultivated phylotypes that to the best of our knowledge have not been previously described for DD lesions (Treponema clones 9T-42, 517, 356, 198, and 2.82), and 1 novel as-yet-uncultivated phylotype. The term “phylotype” was used for clusters of cloned sequences that differed from known species by approximately 2% and were ≥99% similar to members of their cluster (24). The mean number of species/phylotypes that were identified in each DD sample was 8 (range, 4 to 14 species/phylotypes). Figure S1 in the supplemental material presents the relative frequency distribution of the 9 most prevalent species/phylotypes in the 36 DD biopsy specimens. No obvious pattern in the distribution of phylotypes was observed in relation to the herd origin and/or the clinical status of individual lesions.
Based on the findings from the pyrosequencing results, we cloned and sequenced 1,442 bp of the 16S rRNA of a potential new Treponema variant, which is hereafter referred to as PT18. This sequence shared only 95% sequence identity with recognized Treponema species (T. putidum ATCC 700334, T. pedis T3552B, and T. denticola ATCC 35405). Treponema clones 9T-42 and 517 were both part of the human oral microbiome, and clone 9T-42 originated from a root canal area that displayed chronic apical periodontitis (25, 26). Clones 356 and 198 were isolated from the subgingival plaque of a dog (27). As a final curiosity, we identified a sequence displaying 100% identity with treponemal clone 2.82, which was an environmental sample from an aerogenic biogas pond in China.
Phylogenetic analysis.
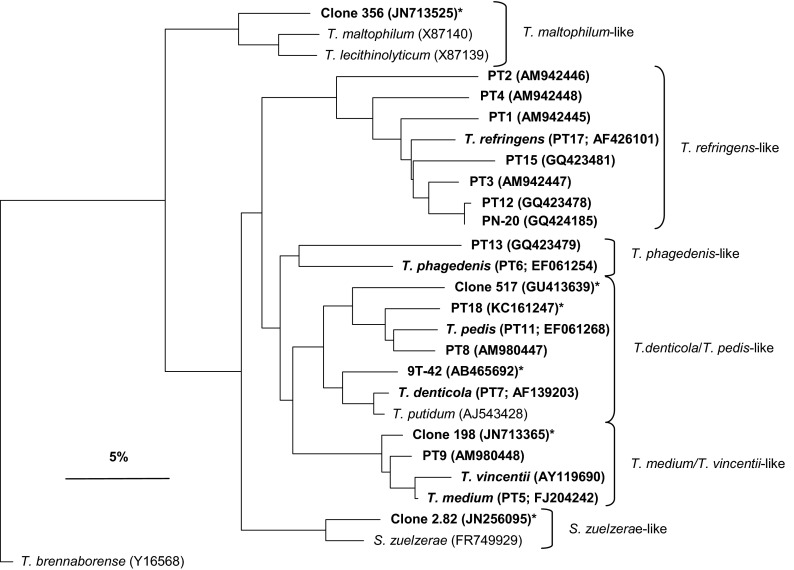
A phylogenetic tree based on comparisons of 325 bases of the V3 and V4 hypervariable regions of 16S rRNA gene sequences is presented in Fig. 2. The tree was constructed using the Jukes-Cantor correction and the neighbor-joining method. The treponemes that were detected in this study fell into 6 clusters, i.e., T. denticola/T. pedis-like, T. phagedenis-like, T. refringens-like, T. medium/T. vincentii-like, Treponema maltophilum-like, and Spirochaeta zuelzerae-like (11). PT18 clustered with the T. denticola/T. pedis-like group, which comprised treponemes that were identified in human periodontal disease and DD (9). The two isolates from the human oral cavity, i.e., 9T-42 and 517, also belonged to this group. Clone 356, the sole member of the T. maltophilum-like cluster that was identified in this study, appeared to be related only distantly to the other Treponema species isolated from DD lesions and displayed 93% sequence identity with Treponema lecithinolyticum and T. maltophilum. However, Yano et al. (11) also detected a clone from the T. maltophilum-like cluster in DD lesions.
Fig 2.
Phylogenetic tree based on 16S rRNA gene sequence comparisons (hypervariable regions V3 and V4), showing the relationship between the strains identified in this study (bold type) and related treponemes. Treponema brennaborense was included as an out-group. *, phylotypes not previously isolated from bovine digital dermatitis lesions. The scale bar represents a 5% difference in nucleotide sequences.
Based on the number of sequences per specific phylotype that were identified in the high-throughput sequencing results, we were able to estimate the relative frequency distribution of the Treponema species/phylotypes in the DD samples (see Table S3 in the supplemental material). The prevalence and densities of the T. phagedenis-like, T. medium-like, and T. denticola-like taxa were high. PT9, Treponema clone 9T-42, and PT4 were identified in more than one-half of the biopsy specimens but at relatively low densities, whereas T. pedis, PT8, the new phylotype PT18, and PT12/PN20 were identified in less than one-half of the samples but were highly abundant when present (Table 1).
Table 1.
Frequencies and densities of Treponema phylotypes in DD biopsy samples
| Phylotype | Pyrosequencing results |
FISH results |
||
|---|---|---|---|---|
| Frequency (%) (n = 36)a | Mean densityb | Frequency (%) (n = 37)c | Mean density scored | |
| T. phagedenis (PT6) | 100 | 0.19 | 95 | 2.54 |
| T. medium/T. vincentii (PT5) | 86 | 0.16 | 73 | 2.22 |
| T. denticola (PT7) | 86 | 0.13 | 63 | 1.96 |
| PT9 | 81 | 0.08 | 81 | 2.10 |
| Treponema clone 9T-42 | 67 | 0.02 | NDe | ND |
| PT4 | 61 | 0.09 | 62 | 1.70 |
| T. pedis (PT16) | 44 | 0.31 | 24 | 2.56 |
| PT18 | 42 | 0.21 | 59 | 2.55 |
| PT8 | 39 | 0.23 | 57 | 1.90 |
| Treponema clone 517 | 39 | 0.05 | 5 | 1.00 |
| T. refringens | 28 | 0.03 | 68 | 1.52 |
| PT12/PN20 | 25 | 0.21 | 3 | 2.00 |
| PT1 | 22 | 0.04 | 19 | 1.43 |
| PT15 | 22 | 0.07 | 5 | 1.50 |
| Treponema clone 356 | 17 | 0.02 | ND | ND |
| PT2 | 14 | 0.03 | 8 | 1.00 |
| PT13 | 11 | 0.04 | 11 | 1.50 |
| Treponema clone 198 | 11 | 0.03 | ND | ND |
| PT3 | 6 | 0.01 | 0 | 0 |
| Treponema clone 2.82 | 3 | 0.02 | ND | ND |
Values represent the proportions of positive biopsy samples. Findings for cows 1 to 4 were not included in the analysis.
Mean density values for positive biopsy samples.
Findings for cows 2, 7, and 8 were not included in the analysis.
Mean density scores (from 1 to 3) for positive biopsy samples.
ND, not determined.
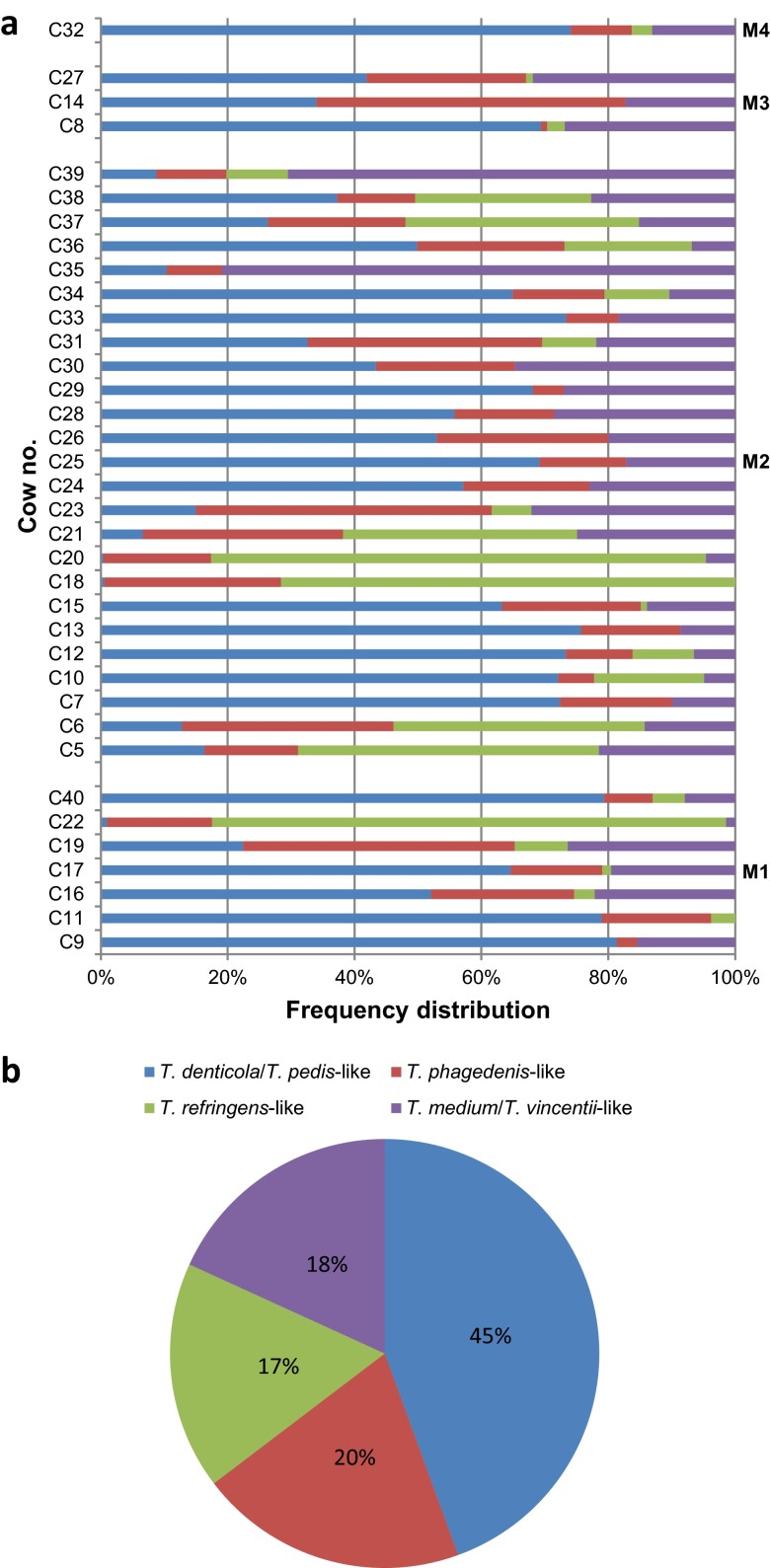
Next, we compared the relative frequency distributions of the 4 most prevalent treponemal clusters organized in Fig. 2 for the 36 DD samples (Fig. 3a). The T. maltophilum-like and S. zuelzerae-like clusters were not included in this part of the analysis because they were minor populations in the examined DD lesions. The prevalence rates for each cluster differed considerably among the DD samples (Fig. 3a). The phylotypes belonging to the T. denticola/T. pedis-like cluster were by far the most prevalent. These phylotypes were present in all of the examined samples (Fig. 3a) and represented a total of 45% of the phylotypes that were identified in the 36 biopsy specimens (Fig. 3b). In 20 biopsy specimens, the T. denticola/T. pedis-like cluster accounted for more than 50% of the phylotypes. The T. denticola/T. pedis-like group was sparsely represented only in the C18, C20, and C22 samples; in these three samples, members of T. medium/T. vincentii-like treponemes were scarce or lacking, and members of the T. refringens-like cluster made up more than two-thirds of all phylotypes. In total, the T. phagedenis-like cluster (20%), the T. medium/T. vincentii-like cluster (18%), and the T. refringens-like cluster (17%) were nearly equally prevalent (Fig. 3b). However, whereas the T. phagedenis-like and T. medium/T. vincentii-like phylotypes were present in 36 and 34 samples, respectively, T. refringens-like treponemes were identified in only 24 biopsy specimens (Fig. 3a).
Fig 3.
Relative frequencies of occurrence of the 4 most prevalent treponemal clusters (T. denticola/T. pedis-like, T. phagedenis-like, T. medium/T. vincentii-like, and T. refringens-like) among the 36 digital dermatitis biopsy specimens (a) and in the 36 biopsy specimens in total (b). The samples were grouped according to clinical lesion scores (M1 to M4).
Spirochetal diversity determined by FISH analysis.
FISH analyses were performed with separate biopsy specimens from the same lesions. With double in situ hybridization with the general probe for the bacterial domain (EUB338) and the Treponema group probe, severe extensive treponemal epidermal infiltration that constituted more than 90% of the total bacterial population (score of 3) was observed in 28 biopsy specimens (see Table S4 in the supplemental material). In the middle and deeper parts of the lesions, the presence of bacteria other than treponemes was negligible. Three biopsy specimens (C18, C33, and C34) displayed only moderate bacterial infiltration but treponemes constituted more than 90% of the total bacterial population. Biopsy specimen C6 (histopathological score of 2) was only focally affected by treponemes, and four severely ulcerated biopsy specimens (C5, C26, C27, and C39) displayed only scarce and superficial colonization of the debris by treponemes and a few other bacteria. The treponemal and bacterial colonization of the remaining three biopsy specimens (C2, C7, and C8) could not be determined due to incorrect preparation of the tissue specimens (i.e., we were unable to find the correct cutting angle, which resulted in slides with only minor surface areas).
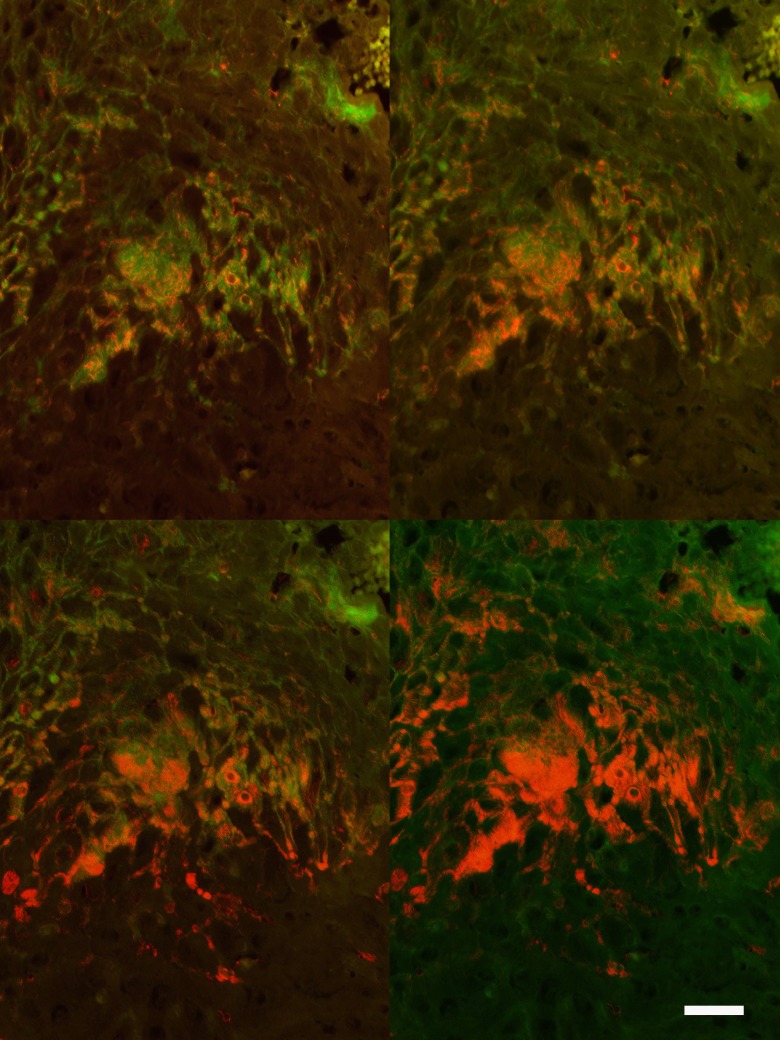
The prevalence, density, and spatial distribution of the different treponemes were visualized with the application of species/phylotype-specific FISH probes. With this approach, we were able to identify and to score 15 phylotypes of Treponema (Table 1). The probes for clones 9T-42 and 356 were not specific; therefore, the results for these probes were omitted from the study. Clones 198 and 2.82 were not included in this part of the analysis. The Treponema species/phylotypes were usually found intermingled with several other phylotypes. An example of serial hybridizations of the same biopsy section is shown in Fig. 4, illustrating the accumulated density of treponemes using 1, 2, and 3 species/phylotype-specific probes and the Treponema group probe against the EUB338 probe for bacteria in general. With an estimated cross section of less than 0.3 μm, the 3-μm-thick tissue sections might easily contain 5 to 8 layers of parallel organisms. Especially deep in the lesion, the treponemes were found to be closely grouped parallel to the epithelial growth direction and independent of cellular boundaries for cells in the epidermis. The treponemes appeared highly epitheliotropic, as the infection was restricted to the epidermis. Treponemes only infiltrated connective tissue and vessels choked by neutrophilic exudates apically in the dermal papillae. In cases of ulcers with formation of granulation tissue, the treponemes were found only superficially in the lesions. The prevalence of the new phylotype, PT18, in positive biopsy specimens varied from a few organisms to high-density infiltration, including heading spirochetes at the border between damaged and vital epidermis.
Fig 4.
Visualization of Treponema organisms in the epidermis in bovine digital dermatitis by fluorescent in situ hybridization. The skin surface is up, and deep layers of the epidermis are down (also see Fig. 1, right, for orientation). Sequential hybridization of the same section was performed by applying probes for detection of different Treponema phylotypes (Cy3-labeled probes), compared with the total bacterial load (fluorescein-labeled EUB338 probe) (green). Since treponemes are marked by both Cy3-labeled (red) and fluorescein-labeled (green) probes, the organisms appear orange. Hybridization was performed with probes for phylotype 18 (upper left), phylotypes 6 and 18 (upper right), or phylotypes 6, 8, and 18 (lower left) or the Treponema group probe (lower right) (Cy3) and the EUB338 probe (green). Bar, 50 μm.
Comparison of pyrosequencing and FISH results.
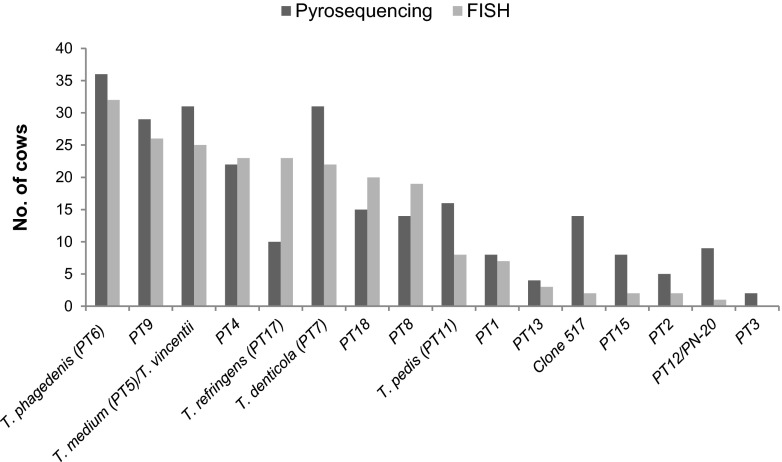
The pyrosequencing and FISH results were highly correlated with respect to prevalence (number of positive cows) [Spearman's rank ρ(14) = 0.82, P = 0.0001] (Fig. 5) and the density of the single phylotypes [Spearman's rank ρ(14) = 0.84, P = 0.00004]. Biopsy specimens from cows 1 to 4, 7, and 8, which for technical reasons were omitted from the pyrosequencing and/or FISH analyses, were not included in the correlation analysis. Of the 8 species that were most often identified in the biopsy specimens, the two methods agreed on 7, although the ranking orders were not entirely the same. According to both methods, however, T. phagedenis was the most prevalent species and, when present, T. pedis was the most highly abundant species (Table 1). T. phagedenis was usually demonstrated to be present in all layers of the lesion, including as the heading spirochetes at the border between damaged and healthy epidermis. PT9 and T. medium, which, according to both methods, were among the four most prevalent Treponema phylotypes in the examined DD biopsy specimens, were found at the interface between damaged and healthy epidermis. The density of the new phylotype, PT18, also was high according to pyrosequencing and FISH results. Comparison of pyrosequencing results and epidermal histopathological scores revealed an interesting trend. The six biopsy specimens with scores of 2 (moderate DD) were colonized by 6 different phylotypes on average, while the 24 biopsy specimens with scores of 3 (fully developed DD) were colonized by 8.5 phylotypes on average. Although the treponemal diversity was low in the ulcerated biopsy specimens in FISH analyses, an average of 8.7 different phylotypes was revealed by pyrosequencing.
Fig 5.
Proportions of positive cows for each phylotype, as determined by pyrosequencing and fluorescent in situ hybridization.
DISCUSSION
Because Treponema species are notoriously difficult to culture, they have been identified in DD lesions mainly by culture-independent methods such as costly, labor-intensive, traditional 16S rRNA gene-based, clonal analysis (8, 9, 12). For that reason, many previous studies focused on a limited number of samples or on the detection of selected Treponema species or phylotypes (9, 10, 28). With the advent of high-throughput sequencing technologies, however, it is now possible to obtain high-resolution bacterial communities and to process large numbers of samples at an affordable price (29, 30). In this study, we used 454 pyrosequencing to perform in-depth, phylogenetic analysis of 36 DD biopsy specimens that were removed from 10 different dairy herds in Spain.
Spirochetes are scarce on healthy foot skin, but this group of bacteria, mainly from the genus Treponema, thrive and become a dominant component of the bacterial population in infected DD tissue, especially in the deeper parts of the lesions (5, 6, 28). The use of universal bacterial primers may underestimate spirochetal populations, especially less-abundant species/phylotypes (25). Therefore, to obtain high levels of coverage of the Treponema species from DD biopsy specimens, we targeted our pyrosequencing analysis specifically against members of the genus Treponema, more specifically the T. pallidum, T. phagedenis, and T. denticola subgroups (23), which were composed mostly of the Treponema spp. that were identified in DD lesions previously (8, 9, 11, 31, 32).
To validate the results, FISH analysis was performed with separate biopsy specimens from the same lesions. In addition to phylogenetic identification and enumeration, this method provides information on the spatial localization of individual microorganisms at the infection site (33). Furthermore, when 16S RNA is used as the target molecule for FISH, this method also reflects the metabolic activity of the bacterial cells.
The results presented here demonstrated that the pyrosequencing approach, which targeted only the V3 and V4 variable regions of the 16S rRNA of treponemal species, was well suited for obtaining deep coverage of the spirochete taxa that were present in DD samples, including rare species. From the 36 biopsy specimens, we were able to discover most of the species/phylotypes that had been identified previously in DD lesions of dairy cows (6, 9–11). We also discovered 16S rRNA gene sequences that had not been reported previously from the DD biopsy specimens and a prevalent, potentially novel species/phylotype of Treponema, PT18. When comparing the pyrosequencing and FISH results, we observed strong correlations between the prevalence rates of species that were identified in the 10 herds and the abundance measures (Table 1). The correlation between the results for the two methods indicated that the pyrosequencing approach was suited for phylogenetic analyses and for the determination of bacterial loads in individual samples.
A polytreponemal etiology seems to be the hallmark of this disease. We observed several Treponema species that were intermingled within the DD lesions, as reported previously (6, 11). In the study by Rasmussen et al. (6), the treponemal diversity was lower in subclinical DD cases and in chronic DD cases obtained from slaughterhouses, compared with clinical DD cases. A similar trend of lower treponemal diversity in less-severe lesions was found in this study, with an average of 6.0 phylotypes in moderate DD lesions and averages of 8.5 and 8.7 phylotypes in fully developed clinical DD lesions and ulcerated lesions, respectively.
The DD-associated spirochetes form a number of distinct phylogenetic clusters (Fig. 2). Although with considerable variability in frequency, members of the T. denticola/T. pedis-like, T. phagedenis-like, and T. medium/T. vincentii-like phylogenetic groups made up 83% of the samples that were identified in the 36 biopsy specimens (Fig. 3). This result is consistent with previous reports from other areas of the world (9–11). Among the four clusters that represented most of the detected phylotypes, the T. denticola/T. pedis-like group was the main contributor and was responsible for nearly one-half of the load of treponemes in the 36 investigated samples (Fig. 3), which was interesting because this group comprises a number of taxa with close phylogenetic resemblance to oral spirochetes of human and canine origin. For example, T. denticola is considered the prime contributor to periodontal disease-associated tissue destruction (34). A parallel was drawn previously between DD and human periodontitis, because both are tissue-destructive diseases with multibacterial etiology (35). However, while human periodontitis appears to be caused by a pathogenic collection that consists of bacteria other than Treponema (36), the results of our study and other recent studies (6, 9, 11) render it likely that the etiology of DD is mainly multitreponemal, although a few other bacteria (e.g., Dichelobacter nodosus) also are involved. In this study, however, we were unable to detect D. nodosus by FISH (data not shown).
We have demonstrated that Treponema-specific pyrosequencing of the V3 and V4 variable regions of the 16S rRNA is well suited for investigation of the treponemal community structure, diversity, and abundance in DD lesions. With this method, it is possible to process multiple clinical specimens at a lower cost and possibly at a higher taxonomic resolution than with clonal libraries. The present pyrosequencing results extend and support previous studies using 16S clonal libraries to describe the treponemal communities in DD lesions, implicating T. denticola/T. pedis-like, T. phagedenis-like, and T. medium/T. vincentii-like clusters as being highly associated with DD infections. For further clarification of the pathogenesis of this disease, studies of lesion development over time and of the prevalence of DD-associated spirochetes in large herds and their surrounding environments are needed. The pyrosequencing approach presented here provides a powerful molecular epidemiological tool that might form the basis for the development of effective preventive and therapeutic strategies for this very painful and disabling disease.
Supplementary Material
ACKNOWLEDGMENTS
This project was funded by HIPRA and the National Veterinary Institute, Technical University of Denmark (DTU-VET).
We thank the participating farmers and claw trimmers from Digitrim S.L. for collaboration regarding the herds and the technical staff members at HIPRA and DTU-VET for their excellent assistance.
Footnotes
Published ahead of print 8 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00320-13.
REFERENCES
- 1. Bruijnis MR, Beerda B, Hogeveen H, Stassen EN. 2012. Assessing the welfare impact of foot disorders in dairy cattle by a modeling approach. Animal 6:962–970 [DOI] [PubMed] [Google Scholar]
- 2. Cheli R, Mortellaro C. 1974. La dermatite digitale del bovino, p 208–213 In Proceedings of the 8th International Conference on Diseases of Cattle, Milan, Italy [Google Scholar]
- 3. Demirkan I, Walker RL, Murray RD, Blowey RW, Carter SD. 1999. Serological evidence of spirochaetal infections associated with digital dermatitis in dairy cattle. Vet. J. 157:69–77 [DOI] [PubMed] [Google Scholar]
- 4. Elliott MK, Alt DP, Zuerner RL. 2007. Lesion formation and antibody response induced by papillomatous digital dermatitis-associated spirochetes in a murine abscess model. Infect. Immun. 75:4400–4408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Moter A, Leist G, Rudolph R, Schrank K, Choi BK, Wagner M, Göbel UB. 1998. Fluorescence in situ hybridization shows spatial distribution of as yet uncultured treponemes in biopsies from digital dermatitis lesions. Microbiology 144:2459–2467 [DOI] [PubMed] [Google Scholar]
- 6. Rasmussen N, Capion N, Klitgaard K, Rogdo T, Fjeldaas T, Boye M, Jensen TK. 2012. Bovine digital dermatitis: possible pathogenic consortium consisting of Dichelobacter nodosus and multiple Treponema species. Vet. Microbiol. 160:151–161 [DOI] [PubMed] [Google Scholar]
- 7. Trott DJ, Moeller MR, Zuerner RL, Goff JP, Waters WR, Alt DP, Walker RL, Wannemuehler MJ. 2003. Characterization of Treponema phagedenis-like spirochetes isolated from papillomatous digital dermatitis lesions in dairy cattle. J. Clin. Microbiol. 41:2522–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Choi BK, Nattermann H, Grund S, Haider W, Göbel UB. 1997. Spirochetes from digital dermatitis lesions in cattle are closely related to treponemes associated with human periodontitis. Int. J. Syst. Bacteriol. 47:175–181 [DOI] [PubMed] [Google Scholar]
- 9. Evans NJ, Brown JM, Demirkan I, Singh P, Getty B, Timofte D, Vink WD, Murray RD, Blowey RW, Birtles RJ, Hart CA, Carter SD. 2009. Association of unique, isolated treponemes with bovine digital dermatitis lesions. J. Clin. Microbiol. 47:689–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nordhoff M, Moter A, Schrank K, Wieler LH. 2008. High prevalence of treponemes in bovine digital dermatitis: a molecular epidemiology. Vet. Microbiol. 131:293–300 [DOI] [PubMed] [Google Scholar]
- 11. Yano T, Moe KK, Yamazaki K, Ooka T, Hayashi T, Misawa N. 2010. Identification of candidate pathogens of papillomatous digital dermatitis in dairy cattle from quantitative 16S rRNA clonal analysis. Vet. Microbiol. 143:352–362 [DOI] [PubMed] [Google Scholar]
- 12. Klitgaard K, Boye M, Capion N, Jensen TK. 2008. Evidence of multiple Treponema phylotypes involved in bovine digital dermatitis as shown by 16S rDNA analysis and fluorescent in situ hybridization. J. Clin. Microbiol. 46:3012–3020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Evans NJ, Timofte D, Isherwood DR, Brown JM, Williams JM, Sherlock K, Lehane MJ, Murray RD, Birtles RJ, Hart CA, Carter SD. 2012. Host and environmental reservoirs of infection for bovine digital dermatitis treponemes. Vet. Microbiol. 156:102–109 [DOI] [PubMed] [Google Scholar]
- 14. Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Claesson MJ, Wang Q, O'Sullivan O, Greene-Diniz R, Cole JR, Ross RP, O'Toole PW. 2010. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 38:e200. 10.1093/nar/gkq873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Holzhauer M, Bartels CJ, Döpfer D, van Schaik G. 2008. Clinical course of digital dermatitis lesions in an endemically infected herd without preventive herd strategies. Vet. J. 177:222–230 [DOI] [PubMed] [Google Scholar]
- 17. Ashelford KE, Weightman AJ, Fry JC. 2002. PRIMROSE: a computer program for generating and estimating the phylogenetic range of 16S rRNA oligonucleotide probes and primers in conjunction with the RDP-II database. Nucleic Acids Res. 30:3481–3489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rozen S, Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365–386 [DOI] [PubMed] [Google Scholar]
- 19. Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cole JR, Chai B, Farris RJ, Wang Q, Kulam SA, McGarrell DM, Garrity GM, Tiedje JM. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294–D296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic Local Alignment Search Tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 23. Paster BJ, Dewhirst FE, Weisburg WG, Tordoff LA, Fraser GJ, Hespell RB, Stanton TB, Zablen L, Mandelco L, Woese CR. 1991. Phylogenetic analysis of the spirochetes. J. Bacteriol. 173:6101–6109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770–3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sakamoto M, Siqueira JF, Jr, Rôcas IN, Benno Y. 2009. Diversity of spirochetes in endodontic infections. J. Clin. Microbiol. 47:1352–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner ACR, Yu WH, Lakshmanan A, Wade WG. 2010. The human oral microbiome. J. Bacteriol. 192:5002–5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dewhirst FE, Klein EA, Thompson EC, Blanton JM, Chen T, Milella L, Buckley CM, Davis IJ, Bennett ML, Marshall-Jones ZV. 2012. The canine oral microbiome. PLoS One 7:e36067. 10.1371/journal.pone.0036067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brandt S, Apprich V, Hackl V, Tober R, Danzer M, Kainzbauer C, Gabriel C, Stanek C, Kofler J. 2011. Prevalence of bovine papillomavirus and Treponema DNA in bovine digital dermatitis lesions. Vet. Microbiol. 148:161–167 [DOI] [PubMed] [Google Scholar]
- 29. Calvo-Bado LA, Oakley BB, Dowd SE, Green LE, Medley GF, Ul-Hassan A, Bateman V, Gaze W, Witcomb L, Grogono-Thomas R, Kaler J, Russell CL, Wellington EM. 2011. Ovine pedomics: the first study of the ovine foot 16S rRNA-based microbiome. ISME J. 5:1426–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, Podar M, Leys EJ. 2012. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 6:1176–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Collighan RJ, Woodward MJ. 1997. Spirochaetes and other bacterial species associated with bovine digital dermatitis. FEMS Microbiol. Lett. 156:37–41 [DOI] [PubMed] [Google Scholar]
- 32. Demirkan I, Carter SD, Hart CA, Woodward MJ. 1999. Isolation and cultivation of a spirochaete from bovine digital dermatitis. Vet. Rec. 145:497–498 [DOI] [PubMed] [Google Scholar]
- 33. Moter A, Göbel UB. 2000. Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms. J. Microbiol. Methods 41:85–112 [DOI] [PubMed] [Google Scholar]
- 34. Holt SC, Ebersole JL. 2005. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex,” a prototype polybacterial pathogenic consortium in periodontitis. Periodontol. 2000 38:72–122 [DOI] [PubMed] [Google Scholar]
- 35. Edwards AM, Dymock D, Woodward MJ, Jenkinson HF. 2003. Genetic relatedness and phenotypic characteristics of Treponema associated with human periodontal tissues and ruminant foot disease. Microbiology 149:1083–1093 [DOI] [PubMed] [Google Scholar]
- 36. Dashper SG, Seers CA, Tan KH, Reynolds EC. 2011. Virulence factors of the oral spirochete Treponema denticola. J. Dent. Res. 90:691–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.