Abstract
An assay to accurately quantitate cytomegalovirus (CMV) load in finger-stick-collected dried blood spots (DBS) could potentially be useful for field studies or for analyzing patient self-collected specimens. We therefore assessed CMV DNA load in paired venipuncture-collected plasma samples and finger-stick DBS, using a previously validated quantitative PCR assay. Assay variability, sensitivity, and changes in viral load during antiviral therapy in finger-stick DBS were compared to the reference plasma quantitative PCR assay, using 106 prospectively collected pairs of finger-stick DBS and plasma samples from 35 solid-organ transplant (SOT) patients. The DBS assay showed good agreement with the reference plasma viral load assay on the log10 scale (Pearson correlation coefficient, 0.92; P < 0.001). The 95% limit of detection of the DBS assay was estimated at 2,700 plasma copies/ml (675 plasma IU/ml). In 94% (76/81) of paired DBS and plasma samples above the limit of detection, the difference in CMV load was <1 log10. CMV viral load changes during antiviral treatment were comparable in plasma and DBS. We conclude that finger-stick DBS provides a convenient sample type for quantitation of CMV load that correlates well with plasma levels. Future studies to optimize and evaluate this methodology for patient self-collected samples are warranted.
INTRODUCTION
Assessment of cytomegalovirus (CMV) load in blood has been shown to be clinically useful in various patient populations (1). In transplant recipients, quantitation of CMV load in blood (plasma, whole blood, peripheral blood mononuclear cells, or lymphocytes) is commonly used for diagnosis of CMV disease, as a marker for initiating antiviral therapy in patients receiving preemptive therapy, and for monitoring response to therapy (1). Typically, CMV quantitation is performed using blood collected by phlebotomy (venipuncture). However, this approach requires that patients have access to facilities and personnel where phlebotomy can be performed. This can be a limitation in situations where such access is not readily available (field studies in resource-limited settings or for patients who live long distances from medical facilities).
Several previous studies described the use of dried blood spots (DBS) for detection of CMV (2–12). However, most of these prior studies using DBS for detection of CMV focused on children with congenital CMV infection, where quantitation and monitoring of viral load over time are not routinely performed. Other limitations of prior studies have included relatively small numbers of patients, use of systematically spotted cards with known pipetted venipuncture-collected blood volumes (rather than utilizing actual finger-stick blood, in which precise volumes are not known and homogeneity of blood on cards is not ensured), lack of comparison with concomitantly collected blood samples, or lack of serially collected specimens for assessment of viral load changes during treatment. To our knowledge, there are no prior published studies that have specifically focused on quantitation of CMV in finger-stick-collected DBS in transplant patients. If DBS are shown to be useful for accurate quantitation of CMV load, they might be a potentially useful tool to facilitate patient self-collected samples that could ultimately be used for long-term natural history studies of viremia, adapted for use in field studies in resource-limited settings, or used to facilitate monitoring for transplant patients who do not have convenient access to phlebotomy facilities or who are unwilling to undergo venipuncture. Also, unlike plasma samples, in which processing procedures and storage conditions are potential concerns (13), dried blood spots are stable for months (14–16) and pose a low biohazard risk (17), and thus DBS samples are suitable for routine transport through the existing United States mail system.
The goal of the present study was to prospectively assess CMV viral load in concomitantly collected finger-stick dried blood spots and standard venipuncture-collected blood samples using a previously published and validated CMV quantitative PCR assay.
MATERIALS AND METHODS
Patients and samples.
This was a prospective, institutional review board-approved study performed at the University of Washington. Written informed consent was obtained from all participants. At the time of clinically performed phlebotomy (venipuncture), blood was obtained by finger stick by study personnel as follows. The finger pad of a digit was first disinfected with an alcohol gauze pad saturated with 70% isopropyl alcohol for 2 s and then wiped dry with gauze. An Accu-Chek Safe-T-Pro lancet (Roche Diagnostics, Indianapolis, IN) was used to puncture the pad of the digit, and the resulting blood was blotted directly onto FTA-Elute cards (Whatman International Ltd., United Kingdom), and allowed to dry at room temperature for at least 24 h. The cards containing the dried blood spots were stored at room temperature in multibarrier pouches (Whatman International Ltd., United Kingdom) containing desiccant pouches (MiniPax Sorbent; Multisorb Technologies, Inc., Buffalo, NY) until processed for PCR. Venipuncture was performed according to standard procedures using 5.0-ml (K2EDTA) tubes (PPT plasma preparation tube; BD Vacutainer, Franklin Lakes, NJ). Plasma was separated by centrifuging the tube at 2,500 rpm for 10 min and then stored at −70C until processed for PCR.
Extraction and PCR from plasma and dried-blood-spot samples.
For plasma samples, DNA was extracted from 200 μl of plasma using a Qiagen 96 blood kit (Qiagen Inc.) and eluted into 100 μl of Qiagen AE buffer. Blood spot samples were collected on FTA Elute microcards (1-cm diameter, with a maximum blood volume of 40 μl, according to the manufacturer). A 3-mm paper puncher was used to punch 4 spots from each blood card. The puncher was decontaminated with dry flame between different cards. DNA was extracted from four 3-mm-diameter punches and eluted into 100 μl autoclave-sterilized 5% Chelex (Bio-Rad Inc.) as follows. The 4 spots were transferred into a 1.7-ml screw-cap vial and washed twice using 1 ml of molecular grade water, vortexing, and then removing the water using a 1-ml pipette. After washing, 100 μl of 5% Chelex was added, and the vials were incubated at 95°C for 30 min, per the manufacturer's instructions, to elute the DNA from the paper.
Twenty microliters (i.e., 1/5 of the total) eluted DNA was used for real-time TaqMan CMV PCR as described previously (18). Briefly, the 50-μl PCR mixture contained 25 μl of 2× QuantiTect multiplex PCR master mix (Qiagen), a 415 nM concentration of each primer (gB and IE-Ex4), and 100 nM concentrations of the probes (gB and IE-Ex4), and the mixture was spiked with an internal control to monitor PCR interference. To compare the CMV levels measured on plasma samples and on blood card samples, CMV results were normalized to copies of CMV per 1 ml of plasma, using following formulas: CMV level on plasma sample = (CMV copies per reaction × 1,000)/40 (each PCR used DNA from 40 μl of plasma) and CMV level on blood spot sample = (CMV copies per reaction × 1,000)/2.88/0.6 (each 1-cm blood spot contains about 40 μl of blood; each PCR used DNA from about 2.88 μl of blood as noted above; in blood approximately 60% of the volume is plasma). For conversion to IU/ml, we used a conversion factor of 4 copies/IU, as determined by internal laboratory quality control studies.
Statistical analyses.
To assess the association between quantitative levels of the two measurement methods, linear regression was used. To assess the association between plasma quantity CMV detected and proportion positive by DBS, logistic, probit, and complementary log-log regression analyses were performed using DBS as the outcome (19). The predicted proportions positive for DBS were compared using each of these models and quantifying plasma CMV in 1/2-log increments, in 1/4-log increments, and continuously. Findings for all models were similar, with probit models providing the best match to actual data.
RESULTS
Correlation of venipuncture and finger stick blood viral loads.
Table 1 shows the characteristics of the 35 patients in our study population, which reflected the demographics of our solid organ transplant service. Participants provided a median of 2 samples per person (range, 1 to 12). We first compared the CMV load as assessed in 106 paired dried blood spot and plasma samples from these patients (Fig. 1). The DBS levels showed a linear correlation with plasma levels throughout the measuring range of the assay (slope of the regression line = 1.0; r = 0.92). Twenty-five (23.6%) of the 106 sample pairs were positive by plasma PCR but negative by the DBS assay. The plasma levels tended to be low for these samples, with a median (range) of 320 (109 to 2826) copies/ml compared to the overall median of 4,060 copies/ml. The ratio of viral loads between the two assays for the remaining sample pairs was assessed using the Bland-Altman analysis (Fig. 2). The cumulative percentages of samples that differed according to specified log differences in viral load between the two assays are shown in Table 2.
Table 1.
Characteristics of the study population
| Characteristic | n (%) |
|---|---|
| No. of patients | 35 |
| No. of samples | 106 |
| Mean | 3.02 |
| Median | 2 |
| Range | 1–12 |
| Age (yr) | |
| Mean ± SD | 57.1 ± 11 |
| Range | 30–75 |
| Gender | |
| Female | 13 (37.1) |
| Male | 22 (62.9) |
| Transplant type | |
| Heart | 4 (11.4) |
| Kidney | 7 (20.0) |
| K/Pa | 2 (5.7) |
| Liver | 3 (8.6) |
| Lung | 18 (51.4) |
| Other | 1 (2.9) |
| Time posttransplantation (mo) | |
| Mean ± SD | 20.5 ± 40 |
| Range | 1.9–186.0 |
K/P, kidney and pancreas.
Fig 1.
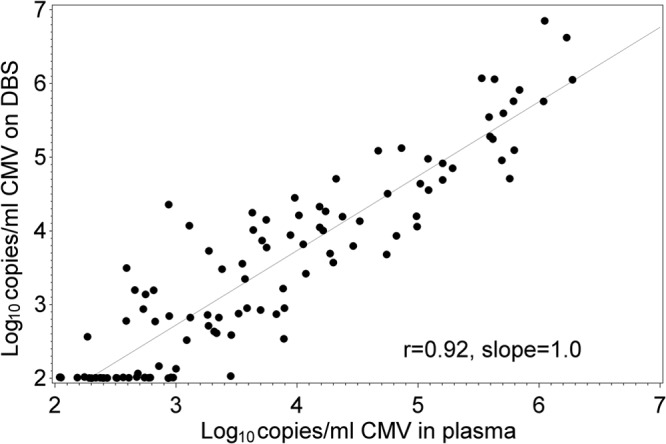
Comparison of CMV viral load in dried blood spot and plasma samples.
Fig 2.
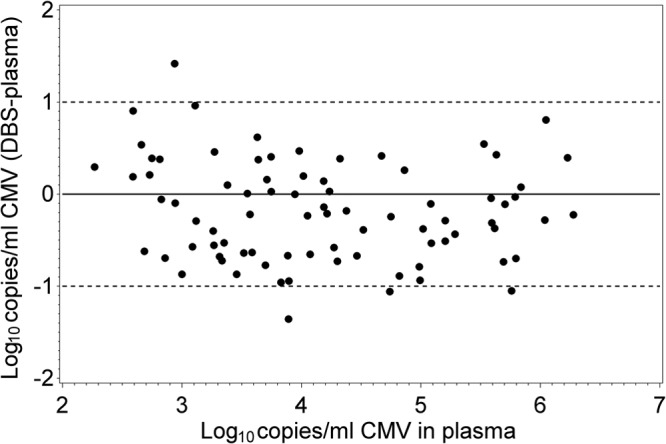
Bland-Altman plot showing the difference in CMV viral load between dried blood spots and plasma as a function of the plasma viral load.
Table 2.
Cumulative percentages of samples that differed according to specified log differences in viral load between the DBS and plasma PCR assays
| Absolute log difference in CMV load between plasma and DBS | No. of values (n = 81) | Cumulative % |
|---|---|---|
| 0.25 | 26 | 32 |
| 0.3 | 32 | 40 |
| 0.5 | 47 | 58 |
| 0.75 | 70 | 86 |
| 1 | 76 | 94 |
| 1.5 | 80 | 99 |
| 2 | 81 | 100 |
Sensitivity of DBS versus plasma assay.
The sensitivity of clinical PCR assays is typically determined by evaluating multiple replicates near the limit of detection using probit analysis and is then expressed as the lowest viral concentration expected to test positive in 95% of replicates. By this definition the limit of detection of our plasma PCR assay was found to be 80 copies/ml (20 IU/ml). Since obtaining multiple replicate DBS from patients was not within the scope of our protocol, we used the data from Fig. 1 to estimate the plasma CMV concentration at which 95% of DBS specimens would be expected to be positive. By probit modeling (Fig. 3), we predict that the DBS assay will be 95% sensitive at a plasma CMV concentration of 2,700 copies/ml (675 IU/ml), suggesting that as currently performed, the DBS assay is less sensitive than plasma PCR.
Fig 3.
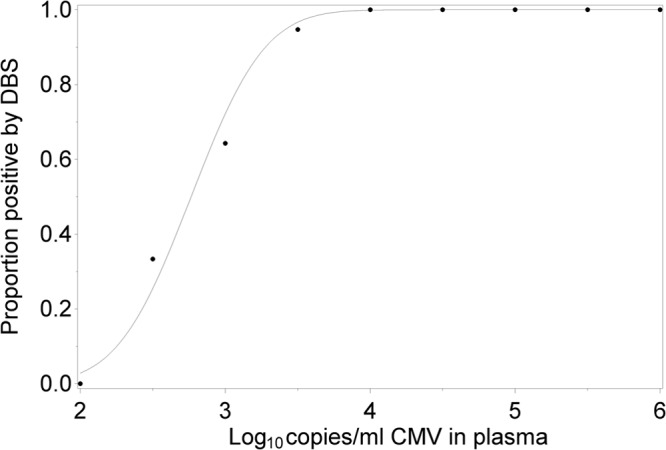
Probit modeling of the likelihood of DBS positivity as a function of plasma CMV load.
Assay variability.
To evaluate the variability of the DBS and plasma quantitative PCR assays, we evaluated 8 separate DBS and 5 aliquots of plasma each from one patient with low viral load and from another patient with a high viral load. At high viral loads, the DBS and plasma assays showed similar sample-to-sample variability (mean ± SD of 5.0 ± 0.10 log10 copies/ml versus 5.9 ± 0.07). However, at low viral loads, the CMV levels determined by DBS were more variable than those determined by plasma testing (mean ± SD of 3.6 ± 0.54 log10 copies/ml versus 4.0 ± 0.10), likely due to the copy number of the low-viral-load sample being close to the limit of detection of the DBS assay.
Changes in CMV load in DBS versus plasma samples during therapy.
To evaluate the potential clinical utility of DBS testing in patients, we measured the CMV load in serially collected paired DBS and plasma samples from three individual patients receiving the antiviral drug ganciclovir (Fig. 4A to C). For all three patients, the DBS and plasma measurements correlated well, and both tests clearly demonstrated the drop in viral load following effective antiviral therapy.
Fig 4.
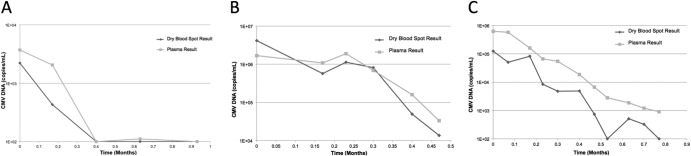
Kinetics of CMV load in plasma and dried blood samples during ganciclovir therapy in three individual patients.
DISCUSSION
We describe a convenient and relatively sensitive quantitative PCR assay for assessment of CMV load in dried blood spots obtained via finger stick. The potential applications of this technology are broad and include field studies in resource-limited settings and the use of patient-self collected specimens that could facilitate monitoring for patients who do not have convenient access to phlebotomy facilities or who are unwilling to undergo venipuncture. This could be particularly relevant for transplant patients, since DBS have also been shown to be suitable for assessing levels of immunosuppressant medications (20). Thus, theoretically, two of the most commonly monitored analytes in transplant patients (immunosuppressant-drug levels and viral load) could potentially be assessed in patient self-collected DBS samples that are sent to central laboratory facilities using the standard United States mail system. Future studies to assess patient preference, acceptability, and adherence to monitoring programs incorporating self-collected DBS specimens should be performed. Studies to independently validate this method for other frequently monitored viruses in the transplant setting (Epstein-Barr virus [EBV], BK virus, and human herpesvirus 6 [HHV-6]) will be required. Facilitating monitoring could have a significant clinical impact in transplant patients, since failure to comply with recommended CMV monitoring has been shown to be an important reason for breakthrough CMV disease (21, 22).
The estimated sensitivity of the present CMV PCR assay using dried blood spots was approximately 2,700 copies/ml (675 IU/ml) in plasma, which, although lower than the sensitivity of the current plasma PCR assay, approaches the sensitivity of a widely used commercial assay, the COBAS Amplicor CMV monitor test (23) (although not that of a more recently published commercial assay, the Artus CMV Rotor-Gene (RG) PCR [24]) and is significantly better than that of a previously published assay for CMV using DBS (3). The sensitivity appears to approach levels adequate for monitoring at least a subset of low-risk SOT patients based on previously defined clinically significant CMV viral load thresholds (1, 25). The appropriate viral load threshold for initiating antiviral therapy in hematopoietic cell transplant (HCT) recipients appears to be lower (26) than that for SOT recipients. However, in HCT patients, who typically have long-term central venous catheters (CVC), the utility of finger-stick DBS (versus blood drawn through the CVC) is less clear. It is important to emphasize that assays in the pre-WHO standard era often showed poor agreement, making it difficult to directly extrapolate between previously defined clinically relevant threshold values and those in the current assay (27, 28). Prospective trials to evaluate the use of DBS for patient monitoring will be required. Modifications to the present assay, including a greater input of dried blood spot sample, might increase the sensitivity but will need to be specifically studied. Viral loads in DBS and plasma samples were generally comparable at clinically significant levels of viremia, and changes in CMV load during treatment were also similar between the two specimen types, suggesting that DBS could be used not only for screening and diagnosis but also for monitoring response to therapy. The variability for the DBS assay was greater than that for the plasma CMV PCR assay and that reported for other DBS viral PCR assays, especially at low viral loads. However, in prior studies, known measured volumes of whole blood were systematically spotted (pipetted) onto blood cards rather than spotted directly from finger-stick blood. We hypothesize that this may have contributed to the increased assay variability seen with directly spotted blood compared to plasma samples. Given that an important proportion of samples differed by >0.5 log between the plasma and DBS assays, the same assay should be used to monitor viral loads within an individual over time. Since direct finger-stick-collected samples would offer the greatest flexibility (field sample collection, patient self-collection), further studies should focus on minimizing the variability of the assay. By assessing different extraction methods, we were able to develop a relatively simple method (boiling) that did not require expensive extraction reagents or equipment and that could be done readily in laboratories that perform clinical PCR testing. Future studies should assess the feasibility of automated DNA extraction from DBS.
The strengths of our study include the prospective study design with simultaneously collected plasma and finger stick samples, inclusion of a reasonable number of patients/samples, sequential monitoring to define viral load changes over time during therapy, and assessment of a broad range of solid-organ transplant patients. We also acknowledge potential limitations. Since only solid-organ transplant patients were included, it will be important to verify these results in other populations in whom CMV infection is a clinical problem. However, there is no biologic reason to suggest that the type of patient population would influence the results. Also, in the present study, finger-stick specimens were obtained by research personnel, not by self-collection by patients. Thus, future studies will need to verify that patient self-collected specimens are comparable to those collected by trained staff. Also, it would be important to determine whether patients consider finger stick to be more acceptable than standard venipuncture if methods using self-collected specimens (“home testing”) are to be developed.
In summary, we have demonstrated that CMV DNA load can be accurately assessed in dried blood spots collected by finger stick and that the results are generally comparable to those obtained with venipuncture-collected plasma specimens. Future studies should define specific situations in which the use of DBS for CMV load assessment might have advantages over the use of standard venipuncture-collected specimens.
ACKNOWLEDGMENTS
We are grateful to the patients who participated in the study and to Mujalin Thuntarug, Sarah Johnson, and Kristin Salmi for excellent clinical coordination of the study.
This study was supported in part by NIH grants AI NO1 272201100041C-0-0-1 and HL102547 to A.P.L. and HL093294 to M.B.
M.B. has received consulting fees from Genentech, a member of the Roche group.
Footnotes
Published ahead of print 15 May 2013
REFERENCES
- 1. Boeckh M, Boivin G. 1998. Quantitation of cytomegalovirus: methodologic aspects and clinical applications. Clin. Microbiol. Rev. 11:533–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barbi M, Binda S, Caroppo S, Calvario A, Germinario C, Bozzi A, Tanzi ML, Veronesi L, Mura I, Piana A, Solinas G, Pugni L, Bevilaqua G, Mosca F. 2006. Multicity Italian study of congenital cytomegalovirus infection. Pediatr. Infect. Dis. J. 25:156–159 [DOI] [PubMed] [Google Scholar]
- 3. Barbi M, MacKay WG, Binda S, van Loon AM. 2008. External quality assessment of cytomegalovirus DNA detection on dried blood spots. BMC Microbiol. 8:2. 10.1186/1471-2180-8-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. de Vries JJ, Vesseur A, Rotteveel LJ, Korver AM, Rusman LG, Wessels E, Kroes AC, Mylanus EA, Oudesluys-Murphy AM, Frijns JH, Vossen AC. 2013. Cytomegalovirus DNA detection in dried blood spots and perilymphatic fluids from pediatric and adult cochlear implant recipients with prelingual deafness. J. Clin. Virol. 56:113–117 [DOI] [PubMed] [Google Scholar]
- 5. Johansson PJ, Jonsson M, Ahlfors K, Ivarsson SA, Svanberg L, Guthenberg C. 1997. Retrospective diagnostics of congenital cytomegalovirus infection performed by polymerase chain reaction in blood stored on filter paper. Scand. J. Infect. Dis. 29:465–468 [DOI] [PubMed] [Google Scholar]
- 6. Scanga L, Chaing S, Powell C, Aylsworth AS, Harrell LJ, Henshaw NG, Civalier CJ, Thorne LB, Weck K, Booker J, Gulley ML. 2006. Diagnosis of human congenital cytomegalovirus infection by amplification of viral DNA from dried blood spots on perinatal cards. J. Mol. Diagn. 8:240–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shibata M, Takano H, Hironaka T, Hirai K. 1994. Detection of human cytomegalovirus DNA in dried newborn blood filter paper. J. Virol. Methods 46:279–285 [DOI] [PubMed] [Google Scholar]
- 8. Soetens O, Vauloup-Fellous C, Foulon I, Dubreuil P, De Saeger B, Grangeot-Keros L, Naessens A. 2008. Evaluation of different cytomegalovirus (CMV) DNA PCR protocols for analysis of dried blood spots from consecutive cases of neonates with congenital CMV infections. J. Clin. Microbiol. 46:943–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vauloup-Fellous C, Ducroux A, Couloigner V, Marlin S, Picone O, Galimand J, Loundon N, Denoyelle F, Grangeot-Keros L, Leruez-Ville M. 2007. Evaluation of cytomegalovirus (CMV) DNA quantification in dried blood spots: retrospective study of CMV congenital infection. J. Clin. Microbiol. 45:3804–3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Walter S, Atkinson C, Sharland M, Rice P, Raglan E, Emery VC, Griffiths PD. 2008. Congenital cytomegalovirus: association between dried blood spot viral load and hearing loss. Arch. Dis. Child. 93:F280–F285 [DOI] [PubMed] [Google Scholar]
- 11. Yamagishi Y, Miyagawa H, Wada K, Matsumoto S, Arahori H, Tamura A, Taniguchi H, Kanekiyo T, Sashihara J, Yoda T, Kitagawa M, Ozono K. 2006. CMV DNA detection in dried blood spots for diagnosing congenital CMV infection in Japan. J. Med. Virol. 78:923–925 [DOI] [PubMed] [Google Scholar]
- 12. Yamamoto AY, Mussi-Pinhata MM, Pinto PC, Figueiredo LT, Jorge SM. 2001. Usefulness of blood and urine samples collected on filter paper in detecting cytomegalovirus by the polymerase chain reaction technique. J. Virol. Methods 97:159–164 [DOI] [PubMed] [Google Scholar]
- 13. Nesbitt SE, Cook L, Jerome KR. 2004. Cytomegalovirus quantitation by real-time PCR is unaffected by delayed separation of plasma from whole blood. J. Clin. Microbiol. 42:1296–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cassol S, Salas T, Gill MJ, Montpetit M, Rudnik J, Sy CT, O'Shaughnessy MV. 1992. Stability of dried blood spot specimens for detection of human immunodeficiency virus DNA by polymerase chain reaction. J. Clin. Microbiol. 30:3039–3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chaisomchit S, Wichajarn R, Janejai N, Chareonsiriwatana W. 2005. Stability of genomic DNA in dried blood spots stored on filter paper. Southeast Asian J. Trop. Med. Public Health 36:270–273 [PubMed] [Google Scholar]
- 16. Leelawiwat W, Young NL, Chaowanachan T, Ou CY, Culnane M, Vanprapa N, Waranawat N, Wasinrapee P, Mock PA, Tappero J, McNicholl JM. 2009. Dried blood spots for the diagnosis and quantitation of HIV-1: stability studies and evaluation of sensitivity and specificity for the diagnosis of infant HIV-1 infection in Thailand. J. Virol. Methods 155:109–117 [DOI] [PubMed] [Google Scholar]
- 17. Cassol S, Weniger BG, Babu PG, Salminen MO, Zheng X, Htoon MT, Delaney A, O'Shaughnessy M, Ou CY. 1996. Detection of HIV type 1 env subtypes A, B, C, and E in Asia using dried blood spots: a new surveillance tool for molecular epidemiology. AIDS Res. Hum. Retroviruses 12:1435–1441 [DOI] [PubMed] [Google Scholar]
- 18. Boeckh M, Huang M, Ferrenberg J, Stevens-Ayers T, Stensland L, Nichols WG, Corey L. 2004. Optimization of quantitative detection of cytomegalovirus DNA in plasma by real-time PCR. J. Clin. Microbiol. 42:1142–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Agresti A. 2007. An introduction to categorical data analysis, 2nd ed Wiley, Edison, NJ [Google Scholar]
- 20. Hoogtanders K, van der Heijden J, Christiaans M, van de Plas A, van Hooff J, Stolk L. 2007. Dried blood spot measurement of tacrolimus is promising for patient monitoring. Transplantation 83:237–238 [DOI] [PubMed] [Google Scholar]
- 21. Kunzle N, Petignat C, Francioli P, Vogel G, Seydoux C, Corpataux JM, Sahli R, Meylan PR. 2000. Preemptive treatment approach to cytomegalovirus (CMV) infection in solid organ transplant patients: relationship between compliance with the guidelines and prevention of CMV morbidity. Transpl. Infect. Dis. 2:118–126 [DOI] [PubMed] [Google Scholar]
- 22. Kusne S, Grossi P, Irish W, St George K, Rinaldo C, Rakela J, Fung J. 1999. Cytomegalovirus PP65 antigenemia monitoring as a guide for preemptive therapy: a cost effective strategy for prevention of cytomegalovirus disease in adult liver transplant recipients. Transplantation 68:1125–1131 [DOI] [PubMed] [Google Scholar]
- 23. Caliendo AM, Schuurman R, Yen-Lieberman B, Spector SA, Andersen J, Manjiry R, Crumpacker C, Lurain NS, Erice A. 2001. Comparison of quantitative and qualitative PCR assays for cytomegalovirus DNA in plasma. J. Clin. Microbiol. 39:1334–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Waggoner J, Ho DY, Libiran P, Pinsky BA. 2012. Clinical significance of low cytomegalovirus DNA levels in human plasma. J. Clin. Microbiol. 50:2378–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Humar A, Gregson D, Caliendo AM, McGeer A, Malkan G, Krajden M, Corey P, Greig P, Walmsley S, Levy G, Mazzulli T. 1999. Clinical utility of quantitative cytomegalovirus viral load determination for predicting cytomegalovirus disease in liver transplant recipients. Transplantation 68:1305–1311 [DOI] [PubMed] [Google Scholar]
- 26. Pollack M, Heugel J, Xie H, Leisenring W, Storek J, Young JA, Kukreja M, Gress R, Tomblyn M, Boeckh M. 2011. An international comparison of current strategies to prevent herpesvirus and fungal infections in hematopoietic cell transplant recipients. Biol. Blood Marrow Transplant. 17:664–673. 10.1016/j.bbmt.2010.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Caliendo AM. 2013. Editorial commentary: the long road toward standardization of viral load testing for cytomegalovirus. Clin. Infect. Dis. 56:374–375. 10.1093/cid/cis905 [DOI] [PubMed] [Google Scholar]
- 28. Kraft CS, Armstrong WS, Caliendo AM. 2012. Interpreting quantitative cytomegalovirus DNA testing: understanding the laboratory perspective. Clin. Infect. Dis. 54:1793–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]


