Abstract
Fully standardized reproducible and sensitive quantification assays for cytomegalovirus (CMV) are needed to better define thresholds for antiviral therapy initiation and interruption. We evaluated the newly released Abbott RealTime CMV assay for CMV quantification in whole blood (WB) that includes automated extraction and amplification (m2000 RealTime system). Sensitivity, accuracy, linearity, and intra- and interassay variability were validated in a WB matrix using Quality Control for Molecular Diagnostics (QCMD) panels and the WHO international standard (IS). The intra- and interassay coefficients of variation were 1.37% and 2.09% at 5 log10 copies/ml and 2.41% and 3.80% at 3 log10 copies/ml, respectively. According to expected values for the QCMD and Abbott RealTime CMV methods, the lower limits of quantification were 104 and <50 copies/ml, respectively. The conversion factor between international units and copies (2.18), determined from serial dilutions of the WHO IS in WB, was significantly different from the factor provided by the manufacturer (1.56) (P = 0.001). Results from 302 clinical samples were compared with those from the Qiagen artus CMV assay on the same m2000 RealTime system. The two assays provided highly concordant results (concordance correlation coefficient, 0.92), but the Abbott RealTime CMV assay detected and quantified, respectively, 20.6% and 47.8% more samples than the Qiagen/artus CMV assay. The sensitivity and reproducibility of the results, along with the automation, fulfilled the quality requirements for implementation of the Abbott RealTime CMV assay in clinical settings. Our results highlight the need for careful validation of conversion factors provided by the manufacturers for the WHO IS in WB to allow future comparison of results obtained with different assays.
INTRODUCTION
Cytomegalovirus (CMV), a member of the Herpesviridae family, establishes a life-long persistent and latent infection within its host. Reactivations occur frequently throughout the lifespan, with asymptomatic viral shedding in healthy individuals. However, in immunocompromised patients, especially in allogeneic hematopoietic stem cell or solid-organ transplant recipients, CMV may cause severe disease (1, 2). Viremia is a well-recognized risk factor for CMV disease, and viral-load kinetics has been reported to be predictive for the development of CMV disease (3, 4). Quantitative viral genome testing in blood (whole blood or plasma) is the best option for diagnosis, decision making regarding initiation of preemptive therapy, and monitoring response to therapy (5, 6). The reliability and accuracy of viral load determination are then critical points for the management of transplant patients. Due to their greater sensitivity and reduced technician hands-on time, quantitative real-time PCR assays for quantification of CMV DNA have largely replaced pp65 antigenemia testing in routine diagnostic laboratories (7, 8).
Because of the increased requirements for quality certification of laboratories, there is a trend for the use of in vitro diagnostic (IVD)/Conformité Européene (CE)-labeled or FDA-approved commercial assays. However, results of measurements of CMV DNA loads performed with commercially available assays might differ significantly, particularly according to the extraction method used, which is a source of variability with whole blood (WB) (9–11). An interlaboratory comparison of CMV viral load assays in 33 different laboratories in the United States, Canada, and Europe has shown significant variability, with >40% of results having a delta log above 0.5, which may have an impact on patient care and limit interinstitutional comparisons (12). Data from 2 other studies conducted in the United States have included interlaboratory comparisons of CMV viral load measures with both in-house and commercial assays and have shown significant differences in quantitative values ranging from 2 up to 3 log10 between two laboratories (13, 14). In order to standardize results and overcome the variability between laboratories, the World Health Organization (WHO) Expert Committee on Biological Standardization established the first international standard (IS) for human cytomegalovirus for nucleic acid amplification techniques on November 30, 2010 (15). For each assay, a conversion factor is applied in order to provide the results in international units per milliliter (IU/ml).
In this study, we performed an evaluation of the recently available IVD/CE-labeled RealTime CMV assay (Abbott RealTime CMV), which included a complete fully automated extraction and amplification of CMV DNA from WB on the m2000 RealTime system. The analytical performances have been validated in the WB matrix using quality control panels and the IS. Clinical samples were tested and the results compared to those obtained with the Qiagen artus IVD/CE-labeled CMV assay on the m2000 RealTime system. Our findings demonstrated good reliability and accuracy of results.
MATERIALS AND METHODS
Standards and clinical samples. (i) QCMD samples.
The Quality Control for Molecular Diagnostics (QCMD) 2010 CMV proficiency panel (QCMD, Scotland) consisted of 10 lyophilized samples (QCMD CMV 10-01 to QCMD CMV 10-10). Nine samples contained various concentrations of cultured CMV strain AD169 in either virus transport medium or human plasma, and one plasma sample was negative for CMV. In positive samples, CMV DNA concentrations ranged from 230 to 2,552,701 copies/ml (i.e., 2.36 to 6.41 log10 copies/ml).
(ii) WHO international standard.
The WHO international standard (IS) for human CMV for nucleic acid amplification techniques (National Institute for Biological Standards and Control [NIBSC] code: 09/162; NIBSC, Hertfordshire, Great Britain) is a lyophilized whole-virus preparation of the CMV Merlin strain (15). After reconstitution in 1 ml of water, the WHO IS has a concentration of 5 × 106 IU/ml (i.e., 6.7 log10 IU/ml).
(iii) Clinical samples.
A total of 302 blood specimens received in the laboratory for CMV load quantification from November 2010 to December 2010 were selected if at least two aliquots of 0.5 ml of WB collected in EDTA tubes were stored at −80°C. They were collected from 207 patients, including 14 hematopoietic stem cell transplant (HSCT) recipients, 94 kidney transplant recipients, 47 HIV-infected patients, 27 patients with leukemia or lymphoma, and 11 patients with inflammatory bowel disease. The clinical samples were tested in singlet with the Abbott RealTime CMV assay and with the test of record, the Qiagen artus CMV assay, in separate runs.
Quantitative real-time PCR assays. (i) Abbott RealTime CMV assay.
Quantification of CMV in WB was carried out with the Abbott RealTime CMV (Abbott Molecular Inc., Des Plaines, IL) on the m2000 RealTime platform that includes the m2000sp instrument for automated extraction of DNA and the m2000rt instrument for real-time PCR in batches of 48 tests. Extraction of DNA was done with a DNA sample preparation kit. The Abbott RealTime CMV assay uses three reagent kits, the amplification reagent kit the amplification, the control kit for external controls, and the calibrator kit for the standard curve. The amplification targets two highly conserved regions, within the UL34 and UL80.5 genes. An internal control (IC) is also supplied to check the overall process, including DNA extraction and possible PCR inhibition. Automated DNA extraction was performed from 500 μl WB (300 μl was processed and eluted in 150 μl) in the Abbott m2000sp instrument, followed by automated addition onto the PCR plate of the master mixture and DNA extracts (20 μl volume used for PCR, corresponding to 40 μl of WB). The sealed PCR plate was loaded on the m2000rt instrument for quantification of viral CMV DNA. Two controls (one positive and one negative) provided by the manufacturer were included in each run. Two calibrators (A and B) analyzed in triplicate were used to establish the standard curve and calculate the CMV DNA concentrations in samples. The results are expressed in copies/ml or log10 copies/ml, as follows: not detected, no detection of amplification signal; detected, detection of amplification signal with a value of <0.60 log10 copies/ml; quantified, absolute values for quantification between 1.60 and 8.00 log10 copies/ml; upper limit of quantification, values of >8.00 log10 copies/ml.
(ii) Qiagen artus CMV assay.
Real-time PCR with the CMV ABI Prism SDS kit (Qiagen Hamburg GmbH, Hilden, Germany) was also performed on the m2000 RealTime platform in batches of 48 tests. The Qiagen artus CMV kit contains reagents and enzymes for the specific amplification of a 105-bp region of the CMV major immediate-early gene. DNA extractions were performed using the Abbott m2000 DNA sample preparation kit. CMV DNA was extracted from 500 μl of WB (300 μl processed) including IC spiking and eluted in a final volume of 150 μl. According to the manufacturer's instructions, PCR was carried out in a 96-well plate with a reaction volume of 50 μl containing 20 μl of DNA extract and 30 μl of master mixture. Four external positive controls (CMV QS 1 to 4) were used to establish the standard curve. Controls were DNA plasmids containing either the CMV target sequence (quantitation standard [QS]) or a heterologous target sequence (IC). The results are given in copies/ml or log10 copies/ml, as follows: not detected, no detection of amplification signal; detected, detection of amplification signal with a value of <2.30 log10 copies/ml; quantified, absolute values for quantification between 2.30 and 8.00 log10 copies/ml; upper limit of quantification, values of >8.00 log10 copies/ml.
Analytical performances of the Abbott RealTime CMV assay. (i) Lower limit of quantification.
The lower limit of quantification (LLQ) was determined first by using serial dilutions of the QCMD CMV 10-08 sample (23,988 copies/ml, i.e., 4.38 log10 copies/ml) at expected values of 200, 150, 100, and 50 copies/ml. The first dilution was prepared by adding 417 μl of the QCMD CMV 10-08 sample to 50 ml of CMV DNA-negative WB. The LLQ was determined secondarily by using serial dilutions of clinical samples with expected viral loads of 150, 100, and 50 copies/ml with the Abbott RealTime CMV assay. Serial dilutions were generated starting with a 50-fold dilution of a pool of 6 WB-positive samples at 4 log10 copies/ml in CMV-negative WB. For the LLQ based on QCMD and Abbott RealTime CMV expected values, each dilution was tested 28 to 30 times.
(ii) Between-run and within-run reproducibilities.
The between-run and within-run reproducibilities were determined at 3 log10 copies/ml and 5 log10 copies/ml by serially diluting in CMV DNA-negative WB the remainder of 6 CMV DNA-positive WB samples with values around 6 log10 copies/ml. Thirty and 11 replicates were tested for intra-assay and interassay reproducibility, respectively.
(iii) Assay linearity.
The linearity of the assay was verified with dilutions of a highly CMV DNA-positive sample in CMV-negative WB. These dilutions had expected viral loads ranging from 1.7 to 5 log10 copies/ml. For each dilution, CMV DNA was quantified with the Abbott RealTime CMV assay at least three times and the mean concentration was calculated.
Concordance correlation coefficient.
To assess the agreement of the two methods, we use the concordance correlation coefficient (CCC) (16). A value of 1 denotes a perfect concordance; a value of zero denotes its complete absence. In our work, as the data were left-censored by the minimal detected value of each method, we used the maximum likelihood estimator proposed by Barnhart et al. to estimate the CCC (17); 95% confidence intervals (95% CIs) are given and were assessed using bootstrap analysis.
Conversion factor between copies/ml and IU/ml.
The conversion factor between copies/ml and UI/ml was evaluated with the Abbott RealTime assay. Serial dilutions of the WHO IS were prepared in phosphate-buffered saline (PBS) (PBS dilutions) and in WB (WB dilutions) with expected values of 5, 4, and 3 log10 IU/ml. In detail, 200 μl of the reconstituted WHO IS was added to 9.8 ml of PBS (1:50 dilution) to constitute the PBS dilution 1 at an expected concentration of 5 log10 IU/ml and the mixture was homogenized on a rotary shaker for 20 min. Two successive 10-fold dilutions were carried out to obtain PBS dilution 2 (4 log10 IU/ml) and PBS dilution 3 (3 log10 IU/ml). Each dilution was split into aliquots of 600 μl. WB dilution 1, WB dilution 2, and WB dilution 3 were prepared following the same protocol. To assess the conversion factor in PBS and WB, 11 replicates of the three PBS dilutions and the three WB dilutions were analyzed in the same run.
Statistical analysis.
Analyses were performed using the statistical R package (2.15.0) (http://www.R-project.org/). Differences were considered statistically significant at P values of <0.05.
(i) Determination of the LLQ.
Parameters of the detection-number-of-copies curve were estimated using a probit regression model. The LLQ (defined as the 95th percentile of the previous model) was then estimated.
(ii) Linearity.
Observed values were plotted against theoretical values. Parameters of the corresponding linear regression model were estimated.
(iii) Conversion factors between copies/ml and IU/ml.
Conversion factors for the number of copies were estimated using a linear regression model (observed values against theoretical values) without the intercept. Conversion factors for the log10 of copies were estimated using the mean difference between the observed and the theoretical values. The 95% CIs are given, and tests against values given by the Abbott RealTime CMV assay were performed.
RESULTS
Analytical performance of the Abbott RealTime CMV assay.
The LLQ, between-run, and within-run reproducibility and linearity of the assay were determined by using dilutions of QCMD or clinical samples positive for CMV DNA. As the assay is CE marked for whole blood, the dilutions were performed in CMV-negative WB.
(i) Lower limit of quantification.
The LLQ for WB was defined as the CMV viral load detected by the assay with a probability of 95% and tested with dilutions of QCMD samples and clinical samples. Probit analysis of the data predicted an LLQ at 104 copies/ml (95% CI, 81 to 122 copies/ml) with the QCMD dilutions (Table 1). All dilutions of clinical samples with expected values of 150 and 100 copies/ml and 96.7% with expected values of 50 copies/ml were detected.
Table 1.
Lower limit of quantification of the Abbott RealTime CMV assay for whole blood
| Sample | Expected value (copies/ml) | No. of replicates | Mean value (copies/ml) | Detection rate (%) |
|---|---|---|---|---|
| QCMDa | 200 | 30 | 91 | 100.0 |
| 150 | 30 | 67 | 100.0 | |
| 100 | 28 | 39 | 89.3 | |
| 50 | 30 | 20 | 70.0 | |
| 25 | 30 | 22 | 20.0 | |
| Clinicalb | 150 | 30 | 147 | 100.0 |
| 100 | 30 | 142 | 100.0 | |
| 50 | 29 | 72 | 96.7 |
The LLQ was determined by using dilutions of the QCMD CMV 10-08 sample in whole blood.
The LLQ was determined by using dilutions of clinical samples in whole blood.
(ii) Between-run and within-run reproducibility.
Intra-assay coefficients of variation (CV), determined on 30 replicates, were 1.37% and 2.41% at the mean values of 5.09 and 3.04 log10 copies/ml, respectively. The same two samples were tested in 11 different assays. Testing showed CVs of 2.09% and 3.80% for 5.01 and 2.95 log10 copies/ml, respectively. The variability of the detection of the IC supplied with the Abbott RealTime CMV assay to check the overall process, including DNA extraction and possible PCR inhibition, was evaluated on the clinical samples. Overall, for the 302 samples tested in 14 separate experiments, no inhibition was detected and the mean cycle threshold (CT) value (±SD) was 29.28 (±0.30). The intrarun variation coefficients ranged from 0.01% to 1.71%. The interrun variation coefficient calculated with the means of CT values of each run was 0.43%.
(iii) Linearity.
The assay was linear (r2 = 0.9682) in the range of all samples tested (1.7 to 5 log10 copies/ml) (data not shown).
Correlation with QCMD values.
CMV DNA was not detectable in the negative QCMD CMV 10-05 sample and was quantified for the other 9 positive samples (Table 2). CMV DNA loads measured by the Abbott RealTime CMV assay were lower than those expected. Differences between measured and expected values ranged from −0.13 (QCMD CMV 10-02) to −0.55 log10 copies/ml (QCMD CMV 10-08), with a mean of −0.26 log10 copies/ml When viral loads were expressed in copies/ml, the conversion factor between measured and expected values was 1.81 (95% CI, 1.78 to 1.83).
Table 2.
Quantification of CMV DNA in QCMD 2010 CMV samples
| QCMD sample no. | Expected value (log10 copies/ml) | Abbott RealTime CMV value (log10 copies/ml) |
|---|---|---|
| 10-01 | 3.27 | 2.96 |
| 10-02 | 2.36 | 2.23 |
| 10-03 | 5.44 | 5.23 |
| 10-04 | 3.74 | 3.45 |
| 10-05 | NDa | ND |
| 10-06 | 6.41 | 6.15 |
| 10-07 | 3.26 | 3.03 |
| 10-08 | 4.38 | 3.83 |
| 10-09 | 2.84 | 2.59 |
| 10-10 | 4.23 | 4.07 |
ND, CMV DNA was not detected.
Comparison of the Abbott RealTime CMV and Qiagen artus CMV assays.
The 302 clinical samples were analyzed using both analytical systems.
By using the Abbott RealTime CMV assay, we detected CMV DNA in 129 (42.7%) clinical samples. For 99 of them (32.8%), DNA was quantifiable (>1.60 log10 copies/ml). With the Qiagen artus CMV assay, CMV DNA was detected in 107 (35.4%) of samples, of which 67 (22.2%) were quantified above the LLQ value (2.30 log10 copies/ml). Thus, compared to the Qiagen artus CMV assay, the Abbott RealTime CMV assay provided increases of 20.6% and 47.8% of samples detected and quantified, respectively.
Results between the two techniques were discordant for 38 (12.6%) of samples. Discrepancies were observed only for samples with low copy numbers, mostly below the lower quantification limit. In 8 of them, CMV DNA was undetectable by using the Abbott RealTime CMV assay but detected by using the Qiagen artus CMV assay (DNA load below the LLQ for 6 samples and at 2.36 and 2.78 log10 copies/ml for the two other samples, respectively). Conversely, CMV DNA was undetectable with the Qiagen artus CMV assay in 30 samples but was detected in 23 samples (DNA load below the LLQ) or quantified in the other 7 samples (median DNA load, 2.00 log10 copies/ml; range, 1.67 to 2.18 log10 copies/ml) with the Abbott RealTime CMV assay. To further analyze the 30 Abbott RealTime CMV-positive and Qiagen artus CMV-negative samples, we determined if previous or subsequent samples collected within 2 weeks in the same patients were tested with the Qiagen artus CMV assay. For the 30 discrepant samples, 15 previous or subsequent samples were available (including 13 tested in this study) and all were positive.
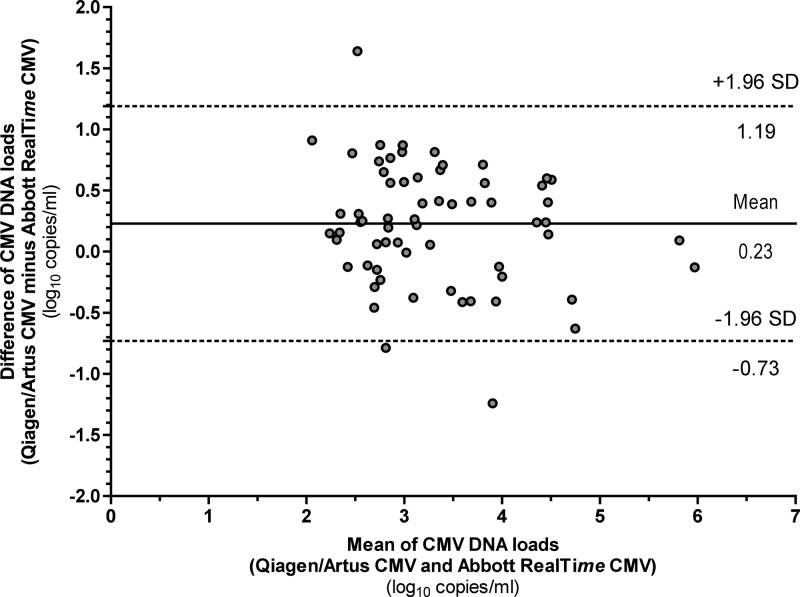
For the 65 positive samples quantified with both assays, the two assays showed good concordance, with a CCC of 0.92 (95% CI, 0.62 to 0.98). Viral load values measured with the Qiagen artus CMV assay were on average 0.23 log10 higher than those measured with the Abbott RealTime CMV assay (P = 0.0003). Bland-Altman analysis on the same 65 samples showed that 41 (63.1%) and 63 (96.9%) of them had a variation of <0.5 log10 and <1.0 log10 copies/ml, respectively (Fig. 1).
Fig 1.
Bland-Altman analysis of the CMV Qiagen artus and Abbott RealTime CMV assays for samples (n = 65) with a CMV DNA load above the lower limit of quantification (LLQ) of each assay (2.30 log10 and 1.60 log10 copies/ml for the Abbott RealTime CMV and Qiagen artus CMV assays, respectively). CMV DNA loads are expressed in log10 copies/ml.
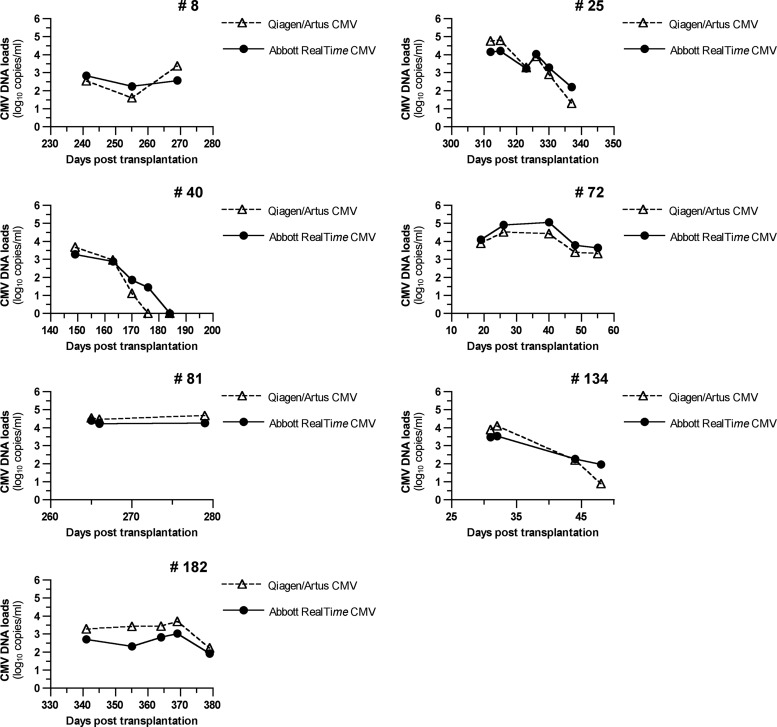
In order to further analyze the correlation between the two assays, we compared the viral-load kinetics for patients with at least three successively positive samples. As shown in Fig. 2, the profiles are very similar, with overlapping patterns in all patients. In addition, variations were always in the same direction except between the second and third samples for patient 72 (decrease of 0.08 log10 copies/ml with the Qiagen artus CMV assay and increase of 0.15 log10 with the Abbott RealTime CMV assay) and the first and second samples for patient 182 with the Abbott RealTime CMV assay.
Fig 2.
Kinetics of CMV replication in whole blood in seven patients, determined by using the Qiagen artus CMV assay and the CMV RealTime Abbott assay. Patient 72 was an HSCT recipient, and the other patients were kidney transplant recipients. CMV DNA loads are expressed in log10 copies/ml.
Determination of conversion factors between international units and copies.
Box plots of conversion factors calculated for the 11 replicates of each dilution of the WHO IS in PBS (PBS dilution 1, PBS dilution 2, and PBS dilution 3) or in WB (WB dilution 1, WB dilution 2, and WB dilution 3) are depicted in Fig. 3. In PBS, the conversion factor varied according to the dilution tested (Fig. 3A). In WB, the conversion factor was roughly constant and always higher than the manufacturer's value (Fig. 3B). The seemingly wide dispersion of conversion factor values for WB dilution 1 was due only to one extreme value (8.04), which was, however, not rejected.
Fig 3.
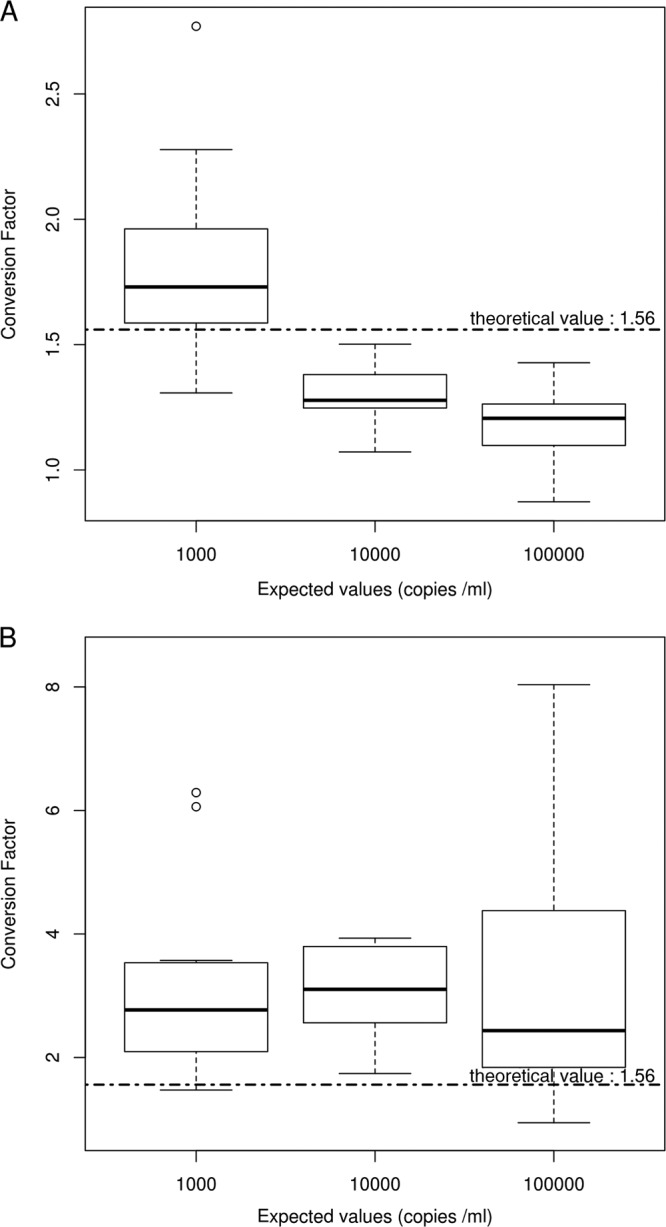
Conversion factors (copies/ml to IU/ml) in PBS and whole blood. Eleven replicates of each dilution of the WHO IS in PBS or WB (at expected values of 1,000, 10,000, and 100,000 copies/ml) were tested in the same run. Box plots of conversion factors estimated on each dilution are shown for PBS (A) and whole blood (B). Manufacturer's values are indicated by the dotted horizontal line.
The conversion factors determined using linear regression models were (in copies/ml) 1.16 (95% CI, 1.11 to 1.22, tested against the Abbott RealTime value [1.56]; P < 0.0001) for PBS and 2.18 (95% CI, 1.79 to 2.80, tested against the Abbott RealTime value [1.56]; P = 0.001) for WB and (in log10 copies/ml) 0.15 (95% CI, 0.11 to 0.18, tested against the Abbott RealTime value [0.19]; P = 0.02) for PBS and 0.45 (95% CI, 0.38 to 0.52, tested against the Abbott RealTime value [0.19]; P < 0.0001) for WB. The calculated conversion factor in WB was thus significantly different compared with the manufacturer's conversion factor.
DISCUSSION
Sensitive and reproducible quantification of CMV DNA is crucial for the initiation and the monitoring of therapy to control CMV infection in transplant recipients. In regard to the detection of CMV DNA in blood, the specimens, WB or plasma, vary. Both specimens have been shown suitable for the monitoring of CMV infection in transplant recipients (4, 18, 19). A fully automated real-time PCR assay, the IVD/CE-labeled Abbott RealTime CMV assay, provides distinct procedures adapted to each of these specimens. The use of WB as a matrix, also convenient for other viral targets of interest in the monitoring of transplant recipients (i.e., Epstein-Barr virus, human herpes 6 virus), takes advantage of laboratory workflow optimization by avoiding a centrifugation step required with plasma (20). In this study, we evaluated the Abbott RealTime CMV assay for the quantification of CMV in WB.
CMV DNAemia dynamics should be taken into consideration in the initiation of preemptive therapy, as underlined by recent studies (21, 22). Thus, the reproducibility of the assay used for monitoring CMV infection is of great concern. In this regard the Abbott RealTime CMV assay was shown to achieve a high level of both intra- and interreproducibility, with low coefficients of variation for high DNA levels and DNA levels close to the cutoffs usually proposed for therapy initiation. This low variability was further confirmed with the narrow range of CT values of the IC, supporting the effectiveness of the automated extraction procedure used in the assay. Importantly, no PCR inhibitory effect was observed with the WB samples analyzed in the present study, since the implementation of the assay in a routine (from May 21 to September 30, 2012) IC cycle threshold was suggestive of PCR inhibitor or extraction deficiency in only 16 samples out of 4,560 (0.35%) (i.e., cycle threshold value out of the range of validity as established automatically in the calibration run) (data not shown).
According to the manufacturer, the LLQ of the assay is 40 copies/ml of WB. This threshold was validated by testing QCMD panel samples and clinical samples diluted in WB to maintain extraction conditions, thus confirming the high sensitivity of this assay for CMV quantification in WB. This new commercial assay, validated and released for WB samples, reaches a higher sensitivity than the tests already commercially available, which provide a higher LLQ (23, 24).
We compared the Abbott RealTime CMV with the Qiagen artus CMV assay on 302 clinical samples by using the same platform for both extraction and amplification, thus avoiding variability due to different instruments and extraction methods. The two assays were highly concordant. Similar kinetics of CMV replication from patients with successive samples confirmed this correlation. Most of the discrepancies occurred for samples positive with the Abbott RealTime CMV assay and negative with the Qiagen artus CMV assay at low viral loads, which is in agreement with the lower limit of detection of the Abbott RealTime CMV assay validated with serial dilutions. Because the use of a third assay may have little chance to resolve the discrepancies, previous or subsequent samples collected within 2 weeks in the same patients were examined, and all confirm that patients with discrepant results had a CMV active infection. The Abbott RealTime CMV assay showed a significantly higher sensitivity, with 42.7% of samples being positive compared to 35.4% with the Qiagen artus CMV assay. In addition, the Abbott RealTime CMV assay provided a 47.8% increase of quantified samples compared to the Qiagen artus CMV assay (32.8% versus 22.2%). This high sensitivity will allow more precise assessment of the kinetics of viral replication for (i) early detection of individuals with short CMV doubling time and (ii) better determination of CMV half-life in patients receiving antiviral therapy. These measures might be useful in adjustment of the duration of treatment (3, 22, 25). Despite a good overall correlation among concordant positive results, differences above 0.5 log10 copies/ml were observed for 36.9% of these samples.
Differences in DNA quantification techniques between laboratories have led to site-specific recommendations for both initiating and monitoring antiviral treatment. The international quantitative reference standard (WHO IS for CMV) currently available will likely allow direct comparisons between different quantification techniques after conversion of copies/ml into UI/ml. For the Abbott RealTime CMV assay, the manufacturer has determined the conversion factor in plasma and provides a unique conversion factor whatever the matrix used (WB or plasma). When using the WHO IS diluted in PBS or WB, we found that the conversion factor experimentally determined in the present study was significantly different from the one proposed by the manufacturer. Its value was lower in PBS and higher in WB, meaning that less CMV DNA was quantified from WB, likely reflecting to difference in DNA extraction efficiency. An even more divergent conversion factor for WB was found by Furione et al. using the Abbott RealTime PCR assay and the WHO IS diluted in WB (26). The reasons for this discrepancy were not clear. As the same automated extraction system was used in both studies, differences in extraction efficiency were very unlikely. The dilution factors of the reconstituted WHO IS in WB differed between the studies (1/16 in the study by Furione et al. versus 1/50 in the present study) (26). As indicated in the package insert, once reconstituted, the WHO IS should be diluted in the matrix appropriate to the material being calibrated. However, the dilution factor to apply is not indicated. Thus, in order to normalize the results obtained with different commercially available and in-house assays, dilution factors and the matrix used to dilute the WHO IS as well as the statistical analysis of the results should be standardized. Then, a detailed and consensual protocol should be defined to calculate the conversion factor according to assays and matrixes used.
In conjunction with current laboratory quality requirements as defined by the International Standardization Organization (ISO) 1589 norm for laboratory techniques, commercial assays fulfill several criteria that an individual hospital laboratory will achieve with difficulty. Considering the benefits of a fully automated assay in reducing hands-on time and repetitive motion injuries and in the traceability and security of blood sample management, and considering the precision and the reproducibility of the results observed in this study, Abbott RealTime CMV assay meets several quality requirements for the CMV quantitative assay implementation in a routine laboratory.
ACKNOWLEDGMENTS
We are grateful to Ghislaine Borit and Muriel Vitu for their technical assistance and Nathalie Parquet for providing whole blood.
J. LeGoff, F. Simon, and M.-C. Mazeron have consulted for Abbott Molecular. All other authors have declared that no competing interests exist. This does not alter the authors' adherence to all of the Journal of Clinical Microbiology policies on sharing data and materials.
Footnotes
Published ahead of print 24 April 2013
REFERENCES
- 1. Beam E, Razonable RR. 2012. Cytomegalovirus in solid organ transplantation: epidemiology, prevention, and treatment. Curr. Infect. Dis. Rep. 14:633–641 [DOI] [PubMed] [Google Scholar]
- 2. Ljungman P, Hakki M, Boeckh M. 2011. Cytomegalovirus in hematopoietic stem cell transplant recipients. Hematol. Oncol. Clin. North Am. 25:151–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Emery VC, Sabin CA, Cope AV, Gor D, Hassan-Walker AF, Griffiths PD. 2000. Application of viral-load kinetics to identify patients who develop cytomegalovirus disease after transplantation. Lancet 355:2032–2036 [DOI] [PubMed] [Google Scholar]
- 4. Kotton CN, Kumar D, Caliendo AM, Asberg A, Chou S, Snydman DR, Allen U, Humar A, Transplantation Society International CMV Consensus Group 2010. International consensus guidelines on the management of cytomegalovirus in solid organ transplantation. Transplantation 89:779–795 [DOI] [PubMed] [Google Scholar]
- 5. Baldanti F, Lilleri D, Gerna G. 2008. Monitoring human cytomegalovirus infection in transplant recipients. J. Clin. Virol. 41:237–241 [DOI] [PubMed] [Google Scholar]
- 6. Gerna G, Lilleri D, Chiesa A, Zelini P, Furione M, Comolli G, Pellegrini C, Sarchi E, Migotto C, Bonora MR, Meloni F, Arbustini E. 2011. Virologic and immunologic monitoring of cytomegalovirus to guide preemptive therapy in solid-organ transplantation. Am. J. Transplant. 11:2463–2471 [DOI] [PubMed] [Google Scholar]
- 7. Cardeñoso L, Pinsky BA, Lautenschlager I, Aslam S, Cobb B, Vilchez RA, Hirsch HH. 2013. CMV antigenemia and quantitative viral load assessments in hematopoietic stem cell transplant recipients. J. Clin. Virol. 56:108–112 [DOI] [PubMed] [Google Scholar]
- 8. Piiparinen H, Hockerstedt K, Gronhagen-Riska C, Lautenschlager I. 2004. Comparison of two quantitative CMV PCR tests, Cobas Amplicor CMV Monitor and TaqMan assay, and pp65-antigenemia assay in the determination of viral loads from peripheral blood of organ transplant patients. J. Clin. Virol. 30:258–266 [DOI] [PubMed] [Google Scholar]
- 9. Lee AV, Atkinson C, Manuel RJ, Clark DA. 2011. Comparative evaluation of the QIAGEN QIAsymphony® SP system and bioMerieux NucliSens easyMAG automated extraction platforms in a clinical virology laboratory. J. Clin. Virol. 52:339–343 [DOI] [PubMed] [Google Scholar]
- 10. Mengelle C, Mansuy JM, Da Silva I, Davrinche C, Izopet J. 2011. Comparison of 2 highly automated nucleic acid extraction systems for quantitation of human cytomegalovirus in whole blood. Diagn. Microbiol. Infect. Dis. 69:161–166 [DOI] [PubMed] [Google Scholar]
- 11. Mengelle C, Mansuy JM, Saune K, Barthe C, Boineau J, Izopet J. 2012. A new highly automated extraction system for quantitative real-time PCRs from whole blood samples: routine monitoring of opportunistic infections in immunosuppressed patients. J. Clin. Virol. 53:314–319 [DOI] [PubMed] [Google Scholar]
- 12. Pang XL, Fox JD, Fenton JM, Miller GG, Caliendo AM, Preiksaitis JK. 2009. Interlaboratory comparison of cytomegalovirus viral load assays. Am. J. Transplant. 9:258–268 [DOI] [PubMed] [Google Scholar]
- 13. Hayden RT, Yan X, Wick MT, Rodriguez AB, Xiong X, Ginocchio CC, Mitchell MJ, Caliendo AM. 2012. Factors contributing to variability of quantitative viral PCR results in proficiency testing samples: a multivariate analysis. J. Clin. Microbiol. 50:337–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wolff DJ, Heaney DL, Neuwald PD, Stellrecht KA, Press RD. 2009. Multi-Site PCR-based CMV viral load assessment-assays demonstrate linearity and precision, but lack numeric standardization: a report of the association for molecular pathology. J. Mol. Diagn. 11:87–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Freyer JF, WHO Expert Committee on Biological Standardization 2010. Collaborative study to evaluate the proposed 1st WHO International Standard for human cytomegalovirus (HCMV) for nucleic acid amplification (NAT)-based assays. WHO/BS/10.2138. World Health Organization, Geneva, Switzerland [Google Scholar]
- 16. Lin LI. 1989. A concordance correlation coefficient to evaluate reproducibility. Biometrics 45:255–268 [PubMed] [Google Scholar]
- 17. Barnhart HX, Song J, Lyles RH. 2005. Assay validation for left-censored data. Stat. Med. 24:3347–3360 [DOI] [PubMed] [Google Scholar]
- 18. Andrews PA, Emery VC, Newstead C. 2011. Summary of the British Transplantation Society guidelines for the prevention and management of CMV disease after solid organ transplantation. Transplantation 92:1181–1187 [DOI] [PubMed] [Google Scholar]
- 19. Ljungman P. 2008. CMV infections after hematopoietic stem cell transplantation. Bone Marrow Transplant. 42(Suppl 1):S70–S72 [DOI] [PubMed] [Google Scholar]
- 20. Deback C, Geli J, Ait-Arkoub Z, Angleraud F, Gautheret-Dejean A, Agut H, Boutolleau D. 2009. Use of the Roche LightCycler 480 system in a routine laboratory setting for molecular diagnosis of opportunistic viral infections: evaluation on whole blood specimens and proficiency panels. J. Virol. Methods 159:291–294 [DOI] [PubMed] [Google Scholar]
- 21. Atabani SF, Smith C, Atkinson C, Aldridge RW, Rodriguez-Peralvarez M, Rolando N, Harber M, Jones G, O'Riordan A, Burroughs AK, Thorburn D, O'Beirne J, Milne RS, Emery VC, Griffiths PD. 2012. Cytomegalovirus replication kinetics in solid organ transplant recipients managed by preemptive therapy. Am. J. Transplant. 12:2457–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Muñoz-Cobo B, Solano C, Costa E, Bravo D, Clari MA, Benet I, Remigia MJ, Montoro J, Navarro D. 2011. Dynamics of cytomegalovirus (CMV) plasma DNAemia in initial and recurrent episodes of active CMV infection in the allogeneic stem cell transplantation setting: implications for designing preemptive antiviral therapy strategies. Biol. Blood Marrow Transplant. 17:1602–1611 [DOI] [PubMed] [Google Scholar]
- 23. Michelin BD, Hadzisejdic I, Bozic M, Grahovac M, Hess M, Grahovac B, Marth E, Kessler HH. 2008. Detection of cytomegalovirus (CMV) DNA in EDTA whole-blood samples: evaluation of the quantitative artus CMV LightCycler PCR kit in conjunction with automated sample preparation. J. Clin. Microbiol. 46:1241–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Raggam RB, Bozic M, Salzer HJ, Hammerschmidt S, Homberg C, Ruzicka K, Kessler HH. 2010. Rapid quantitation of cytomegalovirus DNA in whole blood by a new molecular assay based on automated sample preparation and real-time PCR. Med. Microbiol. Immunol. 199:311–316 [DOI] [PubMed] [Google Scholar]
- 25. Buyck HC, Griffiths PD, Emery VC. 2010. Human cytomegalovirus (HCMV) replication kinetics in stem cell transplant recipients following anti-HCMV therapy. J. Clin. Virol. 49:32–36 [DOI] [PubMed] [Google Scholar]
- 26. Furione M, Rognoni V, Cabano E, Baldanti F. 2012. Kinetics of human cytomegalovirus (HCMV) DNAemia in transplanted patients expressed in international units as determined with the Abbott RealTime CMV assay and an in-house assay. J. Clin. Virol. 55:317–322 [DOI] [PubMed] [Google Scholar]