Abstract
We report that the rates of nasal cocolonization with methicillin-susceptible Staphylococcus aureus and methicillin-resistant coagulase-negative staphylococci can vary widely between patients admitted to different wards within a single hospital. Such cocolonization can greatly influence the performance of molecular methicillin-resistant S. aureus (MRSA) screening tests depending on the methods used and targets selected.
TEXT
Staphylococci, especially methicillin-resistant Staphylococcus aureus (MRSA) strains, are major pathogens implicated in nosocomial infections, and the link between S. aureus nasal colonization and staphylococcal disease has been well established (1), with nosocomial S. aureus bacteremia being three times more frequent in S. aureus carriers than in noncarriers (2). Moreover, Safdar and Bradley underlined the importance of MRSA carrier detection by showing that patients colonized with MRSA were four times more likely to develop invasive infections than patients colonized with methicillin-sensitive S. aureus (MSSA) (3). Finally, MRSA infections are associated with a high financial burden. For example, the cost of management for patients with an orthopedic device infection due to MRSA is estimated at $100,000 per case, representing a 50% additional cost compared to that of MSSA infections (4).
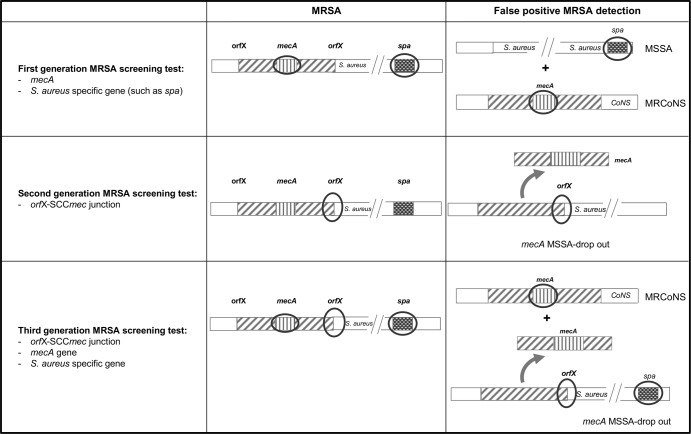
In this context, several manufacturers have developed rapid molecular MRSA screening tests as an alternative to conventional culture methods (5–8). Staphylococcal methicillin resistance is encoded by the mecA gene, located on a mobile genetic element designated the staphylococcal cassette chromosome mec (SCCmec) acquired by horizontal transfer and chromosomic insertion. Different generations of MRSA molecular tests have been successively developed using different combinations of genetic targets. The first-generation tests rely on the combined amplification of the mecA gene and of an S. aureus-specific gene such as spa (Fig. 1) (9). As the mecA gene is present in both MRSA and methicillin-resistant coagulase-negative staphylococci (MRCoNS) (10), specificity of this first generation of MRSA screening tests in clinical specimens can be altered by MSSA and MRCoNS cocolonization (11). To cope with this issue, the second-generation MRSA screening tests used a set of primers targeting the junction sequence between SCCmec and the S. aureus chromosome. Specifically, one primer targets an S. aureus-specific sequence near the SCCmec insertion site located in the orfX gene, and another primer targets an SCCmec-specific sequence. Hence, the amplification detects the presence of the SCCmec element only when it is inserted in the S. aureus genome, thus eliminating interference due to MRCoNS. Nevertheless, it had been demonstrated that this detection method can still give a false-positive result in the presence of S. aureus isolates harboring an SCC element lacking the mecA gene; such isolates are designated mecA dropout isolates (12, 13). These SCC-positive, methicillin-susceptible S. aureus (MSSA) isolates are misidentified as MRSA by commercial screening tests.
Fig 1.
Genomic targets used by different generations of MRSA screening tests and expected cases of false-positive MRSA detections. Three generations of MRSA screening tests have been successively developed to improve the specificity of MRSA detection by including new combinations of genomic targets (circled). However, each generation of MRSA screening tests still suffers from specificity issues with false detection of MRSA in the case of MSSA and MRCoNS cocolonization (first generation), presence of mecA dropout MSSA (second generation), and mecA dropout MSSA and MRCoNS cocolonization (third generation). MSSA, methicillin-susceptible S. aureus; MRSA, methicillin-resistant S. aureus; CoNS, coagulase-negative staphylococci.
The rates of false-positive MRSA results caused by mecA dropout MSSA can be as high as 12.9%, leading to unneeded expenditures linked to patient management (14, 15). To avoid obtaining these false-positive results, the detection of the mecA gene has been included along with S. aureus- and SCCmec-specific sequences in more recent MRSA screening tests. In these third-generation tests, only the simultaneous detection of mecA and the S. aureus- and SCCmec-specific sequences is interpreted as a positive result for MRSA. However, these tests can still yield false-positive MRSA results if patients are cocolonized with MRCoNS and mecA dropout MSSA. In such patients, the mecA gene is detected from the genome of MRCoNS, and the S. aureus- and SCCmec-specific sequences are detected in the MSSA genome. Therefore, the MRCoNS-MSSA cocolonization rate in a given population must be taken into account because it can negatively impact the specificity of third-generation MRSA screening tests. Becker et al. have addressed this question and reported a 3.4% rate of MRCoNS-MSSA nasal cocolonization in German cardiothoracic surgery patients upon admission (11). However, we raised the hypothesis that this rate could be higher in patients hospitalized in wards with high antibiotic pressure, such as intensive care units (ICUs), the population of which represents, in most countries, the population primarily targeted for MRSA screening. We therefore prospectively investigated the rates of nasal colonization with methicillin-resistant and methicillin-susceptible Staphylococcus spp. in patients hospitalized in ICUs and compared these rates with those observed in orthopedic surgery (OS) patients at admission; these OS patients were considered to be representative of the general inpatient population.
Double nasal swabs from both nostrils (one sample per patient) were routinely collected from June to December 2010 from ICU and OS patients at the Northern Hospital Group of the Hospices Civils de Lyon, Lyon, France. The first swab was used for routine MRSA screening, and the second swab was used to seed chromogenic ChromID S. aureus medium allowing the growth of all staphylococcal species (plate 1) (bioMérieux, Marcy l'Etoile, France) using the quadrant technique (16). Direct identification of S. aureus is based on the green appearance of colonies. After 24 h of incubation, plate 1 was replicated using Lederberg velvet onto two agar plates: one ChromID S. aureus plate (plate 2) and one ChromID MRSA plate selective for methicillin-resistant staphylococci (plate 3) (bioMérieux, France) (17). Comparison of colony positions on plates 1 and 2 allowed for the validation of accurate replication. Comparison of the replicated plates 2 and 3 allowed for the determination of the methicillin resistance status of S. aureus and CoNS colonies. All bacterial colonies were confirmed as Staphylococcus spp. using Gram staining and catalase testing. To confirm the results of replication-based methicillin resistance testing, a subset of 80 randomly selected isolates was tested for the presence of the mecA gene using PCR, as described elsewhere (18). The results of both methods were fully concordant. The differences in the colonization rates between ICU and OS patients were tested for statistical significance using Fisher's exact test with a significance threshold of 0.05. Statistical analysis was performed using R software, version 2.14.2 (The R Foundation for Statistical Computing, Vienna, Austria).
The nasal swabs of 353 patients were investigated, including those from 202 ICU patients and 151 OS patients. A total of 336 patients (95.2%) were colonized with Staphylococcus spp. (Table 1). Eighty-nine patients (25.2%) were colonized with S. aureus, and 321 patients (90.9%) were colonized with CoNS. Overall, 21% of patients were cocolonized with S. aureus and CoNS (22.5% of OS patients and 19.8% of ICU patients). Although MRSA colonization rates were comparably low in both patient groups, ICU patients were 2.2 times more likely to be colonized with MRCoNS than OS patients (P < 0.001) and 2.2 times more likely to be cocolonized with MRCoNS and MSSA (P < 0.05). Considering only patients colonized with MSSA, the cocolonization rate with MRCoNS was as high as 24.3% (9/37) in OS patients and 61.3% (27/44) in ICU patients (P < 0.01). These results indicate the following: (i) that the prevalence rates of MRSA, MSSA, MRCoNS, methicillin-sensitive coagulase-negative staphylococci (MSCoNS), and cocolonization differ markedly across different units in the same hospital; (ii) that the prevalence of MRCoNS colonization is high, even in patients representative of the community; and (iii) that cocolonization with MSSA and MRCoNS is highly prevalent, especially in ICU patients.
Table 1.
Comparison of the Staphylococcus species nasal colonization rates of ICU and OS patient populations
| Strain(s)b | No. (%) of colonized patients |
||
|---|---|---|---|
| OS patients (n = 151) | ICU patients (n = 202) | Total (n = 353) | |
| Staphylococcus spp. | 148 (98.0)a | 188 (93.1) | 336 (95.2) |
| S. aureus | 39 (25.8) | 50 (24.8) | 89 (25.2) |
| MSSA | 37 (24.5) | 44 (21.8) | 81 (22.9) |
| MRSA | 2 (1.3) | 6 (3.0) | 8 (2.3) |
| CoNS | 143 (94.7)a | 178 (88.1) | 321 (90.9) |
| MSCoNS | 128 (84.8)a | 60 (29.7) | 188 (53.3) |
| MRCoNS | 51 (33.8)a | 154 (76.2) | 205 (58.1) |
| MRCoNS and MSSA | 9 (6.0)a | 27 (13.4) | 36 (10.2) |
P < 0.05 for orthopedic surgery (OS) versus ICU patients, calculated using Fisher's exact test.
MSSA, methicillin-susceptible S. aureus; MRSA, methicillin-resistant S. aureus; CoNS, coagulase-negative staphylococci.
These findings question the relevance of mecA detection combined with the detection of S. aureus-specific and SCCmec-orfX junction sequences in third-generation MRSA tests. The detection of these three targets was designed to avoid the misidentification of mecA dropout MSSA as MRSA; only the simultaneous detection of mecA and S. aureus- and SCCmec-specific sequences is interpreted as a positive result for MRSA. Indeed, our results indicate that nearly two-thirds of ICU patients colonized with MSSA were also colonized with MRCoNS. Assuming that this proportion is similar in patients colonized with mecA dropout MSSA, this rate of cocolonization potentially leads to a substantial rate of false-positive MRSA identifications using third-generation screening tests. Although mecA detection using such tests likely improves the tests' specificity and their positive predictive values, the actual benefit of this improvement is clearly limited in settings with high MRCoNS prevalence, such as ICUs.
To conclude, we demonstrate that the rates of MRCoNS nasal carriage can vary widely between different wards of the same hospital. Local epidemiologic surveys should determine the prevalence of factors that negatively impact the accuracy of MRSA screening, such as MSSA and MRCoNS cocolonization, and the choice of MRSA screening tests should account for the results of these surveys.
ACKNOWLEDGMENTS
We are grateful to P. Tarquis and M. Chetail for expert technical assistance.
This study was supported by funds from a research grant (number DMNAFLKS1-264864) to F.L. from BD Diagnostics.
Footnotes
Published ahead of print 8 May 2013
REFERENCES
- 1. Wertheim HF, Melles DC, Vos MC, van Leeuwen W, van Belkum A, Verbrugh HA, Nouwen JL. 2005. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 5:751–762 [DOI] [PubMed] [Google Scholar]
- 2. Wertheim HF, Vos MC, Ott A, van Belkum A, Voss A, Kluytmans JA, van Keulen PH, Vandenbroucke-Grauls CM, Meester MH, Verbrugh HA. 2004. Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus non-carriers. Lancet 364:703–705 [DOI] [PubMed] [Google Scholar]
- 3. Safdar N, Bradley EA. 2008. The risk of infection after nasal colonization with Staphylococcus aureus. Am. J. Med. 121:310–315 [DOI] [PubMed] [Google Scholar]
- 4. Parvizi J, Pawasarat IM, Azzam KA, Joshi A, Hansen EN, Bozic KJ. 2010. Periprosthetic joint infection: the economic impact of methicillin-resistant infections. J. Arthroplasty 25:103–107 [DOI] [PubMed] [Google Scholar]
- 5. French GL. 2009. Methods for screening for methicillin-resistant Staphylococcus aureus carriage. Clin. Microbiol. Infect. 15(Suppl 7):10–16 [DOI] [PubMed] [Google Scholar]
- 6. Laudat P, Demondion E, Jouannet C, Charron J, Chillou C, Salaun V, Mankikian B. 2012. Detection of Staphylococcus aureus resistant to methicillin (MRSA) by molecular biology (Cepheid GeneXpert IL, GeneOhm BD, Roche LightCycler, Hyplex Evigene I2A) versus screening by culture: economic and practical strategy for the laboratory. Pathol. Biol. (Paris) 60:208–213 (In French.) [DOI] [PubMed] [Google Scholar]
- 7. Peterson LR, Liesenfeld O, Woods CW, Allen SD, Pombo D, Patel PA, Mehta MS, Nicholson B, Fuller D, Onderdonk A. 2010. Multicenter evaluation of the LightCycler methicillin-resistant Staphylococcus aureus (MRSA) advanced test as a rapid method for detection of MRSA in nasal surveillance swabs. J. Clin. Microbiol. 48:1661–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Roisin S, Laurent C, Nonhoff C, Deplano A, Hallin M, Byl B, Struelens MJ, Denis O. 2012. Positive predictive value of the Xpert MRSA assay diagnostic for universal patient screening at hospital admission: influence of the local ecology. Eur. J. Clin. Microbiol. Infect. Dis. 31:873–880 [DOI] [PubMed] [Google Scholar]
- 9. Xu B, Liu L, Li X, Wang X. 2012. A multiplex PCR assay for the rapid and sensitive detection of methicillin-resistant Staphylococcus aureus and simultaneous discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J. Food Sci. 77:M638–M642 [DOI] [PubMed] [Google Scholar]
- 10. Katayama Y, Ito T, Hiramatsu K. 2000. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 44:1549–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Becker K, Pagnier I, Schuhen B, Wenzelburger F, Friedrich AW, Kipp F, Peters G, von Eiff C. 2006. Does nasal cocolonization by methicillin-resistant coagulase-negative staphylococci and methicillin-susceptible Staphylococcus aureus strains occur frequently enough to represent a risk of false-positive methicillin-resistant S. aureus determinations by molecular methods? J. Clin. Microbiol. 44:229–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stamper PD, Louie L, Wong H, Simor AE, Farley JE, Carroll KC. 2011. Genotypic and phenotypic characterization of methicillin-susceptible Staphylococcus aureus isolates misidentified as methicillin-resistant Staphylococcus aureus by the BD GeneOhm MRSA assay. J. Clin. Microbiol. 49:1240–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Donnio PY, Oliveira DC, Faria NA, Wilhelm N, Le Coustumier A, de Lencastre H. 2005. Partial excision of the chromosomal cassette containing the methicillin resistance determinant results in methicillin-susceptible Staphylococcus aureus. J. Clin. Microbiol. 43:4191–4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Blanc DS, Basset P, Nahimana-Tessemo I, Jaton K, Greub G, Zanetti G. 2011. High proportion of wrongly identified methicillin-resistant Staphylococcus aureus carriers by use of a rapid commercial PCR assay due to presence of staphylococcal cassette chromosome element lacking the mecA gene. J. Clin. Microbiol. 49:722–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Donnio PY, Fevrier F, Bifani P, Dehem M, Kervegant C, Wilhelm N, Gautier-Lerestif AL, Lafforgue N, Cormier M, Le Coustumier A. 2007. Molecular and epidemiological evidence for spread of multiresistant methicillin-susceptible Staphylococcus aureus strains in hospitals. Antimicrob. Agents Chemother. 51:4342–4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Perry J, Rennison C, Butterworth L, Hopley A, Gould F. 2003. Evaluation of S. aureus ID, a new chromogenic agar medium for detection of Staphylococcus aureus. J. Clin. Microbiol. 41:5695–5698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lederberg J, Lederberg EM. 1952. Replica plating and indirect selection of bacterial mutants. J. Bacteriol. 63:399–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Killgore GE, Holloway B, Tenover FC. 2000. A 5′ nuclease PCR (TaqMan) high-throughput assay for detection of the mecA gene in staphylococci. J. Clin. Microbiol. 38:2516–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]