Abstract
The performance of hepatitis B surface antigen (HBsAg) screening assays is continuously improved to reduce the risk of transfusion-associated hepatitis B. In this study, a semiautomated immune complex transfer chemiluminescence enzyme immunoassay (ICT-CLEIA) for the detection of HBsAg, which is as sensitive as hepatitis B virus (HBV) DNA PCR, was developed; the ICT-CLEIA assay performance was compared with the performance of the Architect HBsAg QT assay and HBV DNA PCR. The specificities in the initial assay and after retesting were 99.50% (1,988/1,998 samples) and 99.95% (1,997/1,998 samples), respectively. The analytical detection limit was determined to be 0.2 mIU/ml using the 2nd International WHO HBsAg standard, and the cutoff value (0.5 mIU/ml) of the ICT-CLEIA assay was 8.0 standard deviations (SD) above the mean of the HBsAg-negative specimens. The ICT-CLEIA assay could detect HBsAg even in the presence of anti-HBs antibodies and demonstrated a 23.6-day-shorter window period using commercially available HBsAg seroconversion panels than the Architect HBsAg QT assay. Furthermore, the monitoring of the viral kinetics by the ICT-CLEIA assay and the HBV DNA PCR produced very similarly shaped curves during both the HBsAg seroconversion and reverse seroconversion periods. Therefore, the ICT-CLEIA assay may be useful not only for an earlier detection of HBV reactivation but also for the monitoring of hepatitis B patients.
INTRODUCTION
Hepatitis B virus (HBV) infection is one of the world's most prevalent infectious diseases and a serious global health problem. According to World Health Organization (WHO) statistics, more than 240 million people in the world are estimated to be persistently infected with HBV, and approximately 600,000 people die every year due to the acute or chronic forms of hepatitis B (1). HBV is transmitted by exposure to infected blood or fluids through transfusion of unscreened infectious blood or blood products, by intravenous drug abuse, by sexual contact with infected persons, or perinatally.
Immunoassays to detect hepatitis B surface antigen (HBsAg) are routinely used for the diagnosis of HBV infection and the screening of blood from donors because of simplicity and cost-effectiveness. The number of HBsAg particles is approximately 1,000- to 10,000-fold higher than the number of complete DNA-containing virus particles (2), making HBsAg a very sensitive and useful marker for HBV infection. However, despite HBsAg measurement, there remains a residual risk of transfusion-transmitted infection with HBV through the transfusion of infected blood or blood components, due mainly to a relatively long preseroconversion window period following HBV infection or occult HBV infection (3, 4, 5, 6). Therefore, there is a continuous need to develop more sensitive HBsAg assays capable of reducing the window period and detecting occult HBV carriage.
In addition, HBV has been classified into 10 genotypes, designated A to J, on the basis of an intergroup divergence of >8% in the complete nucleotide sequences (7, 8, 9). Furthermore, a large number of amino acid substitutions were found within the central region of amino acid residues 120 to 147 of HBsAg, and some of the amino acid substitutions affect the antigenicity and immunogenicity (10, 11, 12, 13, 14, 15, 16). Therefore, the sensitivity of immunoassays for HBsAg must be continuously improved to detect all genotypes and, at least, the frequently observed escape mutants to reduce the risk of false-negative results (17).
Although the immune complex transfer (ICT) technique, which could markedly reduce the nonspecific signals by transfer of the immune complexes from the first solid phase to the second one, has been developed to increase the sensitivity of immunoassay, the assay is time-consuming and takes more than 20 h to obtain the results (18, 19).
As a gold standard, a highly sensitive multiplex (MPX) nucleic acid amplification test (NAT) for blood screening, capable of detecting HBV DNA, HCV RNA, and HIV RNA in a single tube, has been used since the 1990s. While the minipool sample MPX NAT was superior to the HBsAg assay for detecting HBV during the early stage of acute infection (20–22), the cost-effectiveness of NAT is a major concern, especially in populations with low HBV prevalence when donors are screened for HBsAg and hepatitis B virus core antibody (anti-HBc antibody). Clinically, HBV DNA quantification is useful for monitoring chronic hepatitis B patients during antiviral therapy as well as HBV-resolved patients during chemotherapy. Indeed, the highly sensitive HBV DNA assay might be useful for detecting low viral loads, but the assay was too expensive to be applied in developing countries.
In this study, a semiautomated highly sensitive chemiluminescent enzyme immunoassay for HBsAg using the ICT technique (ICT-CLEIA) was developed, and clinical usefulness was evaluated. This HBsAg assay would be more convenient and cheaper than the HBV DNA assay including NAT.
MATERIALS AND METHODS
Specimens.
HBsAg-negative sera and potentially interfering specimens were obtained from ProMedDx, Inc. (Norton, MA). A total of 14 human seroconversion panels for HBV were purchased from ZeptoMetrix (Buffalo, NY) and Boston Biomedica, Inc. (West Bridgewater, MA). The 2nd International WHO HBsAg standard was obtained from the National Institute for Biological Standards and Control (NIBSC code number 00/588; Hertfordshire, United Kingdom).
HBsAg mutants.
Plasmids expressing wild-type small HBsAg (genotype C, serotype adr, GenBank accession number AB033550) and 9 types of recombinant small HBsAg mutants (I126S, Q129H, M133L, D144A, D144E, G145R, G145K, I126S D145R, D144A G145R mutants) were transiently expressed in COS7 cells. For the expression, the expression vector pcDNA3.1 (Life Technologies, Carlsbad, CA) and PolyMag neo (OZ Biosciences, France) as a transfection reagent were used according to the manufacturers' recommended protocol. The culture supernatant was collected 2 days after transfection for analysis.
Alkaline phosphatase-conjugated anti-HBsAg monoclonal antibodies.
A murine monoclonal anti-HBsAg Fab was conjugated with alkaline phosphatase (Oriental Yeast Co., Ltd., Osaka, Japan) using N-(ε-maleimidocaproyloxy)succinimide ester (Dojindo Laboratories, Kumamoto, Japan). The conjugate was then purified using size exclusion chromatography. Hybridoma clones producing anti-HBsAg monoclonal antibody were prepared in our laboratory using a conventional method.
2,4-Dinitrophenyl-biotinyl bovine serum albumin-conjugated anti-HBsAg monoclonal antibodies.
A murine monoclonal anti-HBsAg Fab was labeled with 2,4-dinitrophenol (2,4-DNP) and biotin via bovine serum albumin (BSA) using succinimidyl-6-(2,4-dinitrophenyl)aminohexanoate (AnaSpec, Fremont, CA) and sulfosuccinimidyl-6-(biotin-amido)hexanoate (Thermo Fisher Scientific, Rockford, IL) as described previously (18, 19). The labeled anti-HBsAg Fab was purified using size exclusion chromatography. Hybridoma clones producing the anti-HBsAg monoclonal antibody were established in our laboratory using a conventional method.
Magnetic microparticles coated with anti-2,4-DNP monoclonal antibodies.
The magnetic microparticles having amino groups on the surface (Micromod Partikeltechnologie GmbH, Rostock-Warnemuende, Germany) were activated with sulfosuccinimidyl-4-(N-maleimidomethyl)cyclohexane-1-carboxylate (Sulfo-SMCC; Pierce, Rockford, IL) and coupled with a monoclonal murine anti-2,4-DNP monoclonal antibody according to the manufacturer's technical note. The particles were washed and then blocked with BSA. Hybridoma clones producing the anti-2,4-DNP monoclonal antibody were established in our laboratory using a conventional method.
ICT-CLEIA for HBsAg.
The HBsAg assay is a one-step chemiluminescent enzyme immunoassay coupled with an immune complex transfer method (ICT-CLEIA). The assay detects HBsAg in the absence or presence of anti-HBs antibodies (HBsAb) by performing sample pretreatment. To dissociate the HBsAg-HBsAb immune complexes for the detection of HBsAg in samples, an aliquot (120 μl) of specimens was incubated with 80 μl of pretreatment reagent 1 (15 mM phosphate buffer containing 0.15 N NaOH, 1.2 M urea, and 0.4% Brij35) at room temperature for 7 min and subsequently neutralized with pretreatment reagent 2 (0.05 M citric acid, 0.03 M 2-mercaptoethylamine, and 0.6 M NaCl) before being assayed.
The pretreated specimens were incubated with 100 μl of alkaline phosphatase-conjugated anti-HBsAg monoclonal antibodies for 9 min, incubated with 100 μl of 2,4-dinitrophenol-biotinyl-BSA-conjugated anti-HBsAg monoclonal antibodies for 9 min, and subsequently incubated with 75 μl of 0.5% (wt/vol) magnetic microparticles coated with anti-2,4-dinitorphenol monoclonal antibodies for 10 min at 37°C. After being washed with a washing buffer (Tris-buffered saline containing 0.05% Tween 20) three times, magnetic microparticles were incubated with 35 μl of 5 mM epsilon-DNP-l-lysine hydrochloride (Tokyo Chemical Industry, Tokyo, Japan) to elute the immune complex, and the supernatants containing immune complexes were prepared by magnetic separation. The reaction, including the pretreatment of the specimens as described above, was automatically performed using a 12GC PLUS Magtration System (Precision System Science, Matsudo, Japan). Of the supernatant, 30 μl was aspirated and dispensed into a reaction vessel of HISCL-2000i, a fully automated chemiluminescent enzyme immunoassay analyzer (Sysmex, Kobe, Japan), and reacted with 60 μl of 0.25% (wt/vol) streptavidin-coated microparticles (Sysmex, Kobe, Japan) for 7 min at 42°C. After being washed, the bound alkaline phosphatase activity was measured using a chemiluminescent substrate, CDP-Star with Sapphire II (Life Technologies, Carlsbad, CA), as relative light units. The results were expressed in IU values, which were determined according to an in-house standard curve using the 2nd International WHO HBsAg standard (NIBSC code number 00/588; Hertfordshire, United Kingdom). In each of the assays, a quantitative value greater than or equal to 0.5 mIU/ml was considered a reactive result.
ICT-CLEIA confirmatory assay for HBsAg.
The ICT-CLEIA confirmatory assay was performed to confirm the presence of HBsAg in specimens by means of specific antibody neutralization using Abbott Architect HBsAg QT confirmatory reagents 1 and 2. Confirmatory reagent 1 is human plasma containing anti-HBs antibodies, and reagent 2 is human plasma negative for anti-HBs antibodies. In the ICT-CLEIA confirmatory assay, pretreated samples, which were incubated with reagent 1 or 2 for 15 min at room temperature, were used in the ICT-CLEIA assay. If the signal for the neutralized sample was reduced by at least 50% compared to the nonneutralized sample, the sample was judged as HBsAg positive.
Reproducibility.
The reproducibility over 10 days was assessed using three serum samples (7.2, 13.9, and 453.7 mIU/ml). Each sample was assayed in triplicate daily for 10 days (n = 30 for each sample). Within-run reproducibility (n = 10) was assessed using two serum samples (11.7 mIU/ml and 577.7 mIU/ml).
Dilution linearity.
The HBsAg-positive samples were diluted with HBsAg-negative serum, and the dilution linearity was evaluated. On the basis of the in-house standard curve, the linearity was defined relative to the calculated amount of HBsAg.
RESULTS
Reproducibility.
The between-day reproducibility of the ICT-CLEIA assay was assessed using three HBsAg-positive serum samples (7.2, 13.9, and 453.7 mIU/ml). Each sample was tested in triplicate daily for 10 days (n = 30 tests for each sample). The between-day coefficients of variation (CVs) for 7.2, 13.9, and 453.7 mIU/ml were 2.9%, 2.7%, and 1.9%, respectively (Fig. 1). The within-run CVs for 11.7 and 577.7 mIU/ml were 1.7% and 1.5%, respectively (data not shown).
Fig 1.
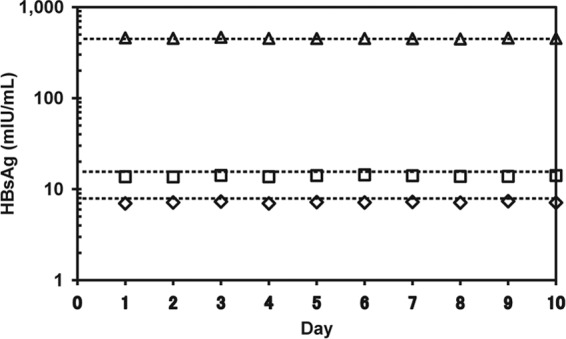
Reproducibility of HBsAg ICT-CLEIA. Three HBsAg-positive serum samples (♢, 7.2 mIU/ml; □, 13.9 mIU/ml; △, 453.7 mIU/ml) were assayed in triplicate daily for 10 days. The results are the means from triplicate assays.
Analytical sensitivity.
To determine the analytical sensitivity of the ICT-CLEIA assay, a calibration curve was obtained using the 2nd International WHO HBsAg standard serially diluted with HBsAg-negative serum (Fig. 2). The analytical sensitivity of the ICT-CLEIA assay was 0.2 mIU per ml. At this concentration, the mean minus 2 standard deviations (SD) for the relative light units (RLUs) did not overlap the mean plus 2 SD for the zero calibrator (n = 10 tests for each sample). The concentration-related CVs for 0 to 0.8 mIU/ml ranged from 1.8 to 3.6%.
Fig 2.
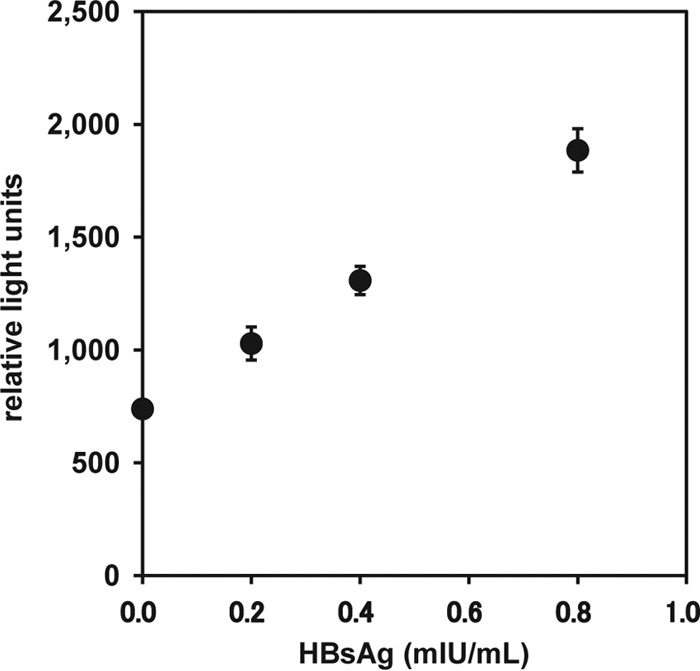
Analytical sensitivity of HBsAg ICT-CLEIA. The assay reactivity is shown as relative light units. The results are the means from 10 assays, and the error bars show 2 SD.
Dilution linearity.
To evaluate the dilution linearity of the ICT-CLEIA assay, four individual HBsAg-positive samples diluted with HBsAg-negative serum were used. As shown in Table 1, the dilution linearity of the ICT-CLEIA assay ranged from 96.9 to 107.3% relative to the expected amount of HBsAg.
Table 1.
Dilution linearity test for HBsAg-positive samplesa
| HBsAg-positive sample | Dilution factor (DF) | Observed value (mIU/ml) × DF | Expected value (mIU/ml) × DF | Recovery rate (%) |
|---|---|---|---|---|
| 1 | Neat | 30.3 | 30.3 | 100.0 |
| 5 | 32.0 | 105.6 | ||
| 25 | 32.2 | 106.3 | ||
| 2 | Neat | 66.1 | 66.1 | 100.0 |
| 5 | 66.0 | 99.8 | ||
| 25 | 70.9 | 107.3 | ||
| 3 | Neat | 392.9 | 392.9 | 100.0 |
| 5 | 401.2 | 102.1 | ||
| 25 | 407.1 | 103.6 | ||
| 4 | Neat | 799.5 | 799.5 | 100.0 |
| 5 | 774.4 | 96.9 | ||
| 25 | 814.3 | 101.9 |
Four HBsAg-positive samples were diluted with HBsAg-negative serum and assayed in duplicate.
Assay specificity.
To evaluate the specificity of the ICT-CLEIA assay, 1,842 HBsAg-negative specimens and 160 potentially interfering specimens were tested (Table 2). The initial and repeatedly reactive rates were 0.70% (14/2,002) and 0.25% (5/2,002), respectively. While one repeatedly reactive specimen out of 1,842 HBsAg-negative specimens was confirmed negative, four repeatedly reactive (1 anti-HIV antibody-positive and 3 from HBV vaccine recipients) specimens out of 160 potentially interfering specimens were confirmed positive for HBsAg by the confirmatory assay using anti-HBs neutralizing antibodies. An anti-HIV antibody-positive specimen reactive according to the ICT-CLEIA assay (43.1 mIU/ml) was also reactive with the Architect HBsAg QT assay (50 mIU/ml). Three specimens (0.5, 1.8, and 6.7 mIU/ml) from the HBV vaccine recipients were bled within approximately 1 month from the date of vaccination. The specificities in the initial assay and after retesting were 99.50% (1,988/1,998) and 99.95% (1,997/1,998), respectively. The cutoff value (0.5 mIU/ml) of the ICT-CLEIA assay was 8.0 SD above the mean of the HBsAg-negative specimens (Fig. 3).
Table 2.
Specificity of ICT-CLEIA for HBsAg
| Specimen | No. of samples tested | No. of samples |
||
|---|---|---|---|---|
| Initially reactive (% of total) | Repeatedly reactive (% of total) | Confirmatory assay reactive (% of repeatedly reactive) | ||
| HBsAg-negative specimens | 1,842 | 10 (0.54) | 1 (0.05) | 0 (0.00) |
| Potentially interfering specimensa | 160 | 4 (2.50) | 4 (2.50)b | 4 (100.00) |
| Total | 2,002 | 14 (0.70) | 5 (0.25) | 4 (80.00) |
Potentially interfering specimens included samples that were anti-HCV antibody positive (10), anti-CMV antibody positive (10), anti-EBV antibody positive (10), anti-HAV antibody positive (10), anti-HSV antibody positive (10), anti-HIV antibody positive (10), anti-rubella antibody positive (10), anti-syphilis antibody positive (10), anti-toxoplasma antibody positive (10), anti-nuclear antigen antibody positive (10), anti-mouse antibody positive (10), and rheumatoid factor positive (10), from patients with alcoholic cirrhosis (10), from HBV vaccine recipients (10), from pregnant women (10), and from patients with candidiasis (10).
Four samples repeatedly reactive when using the ICT-CLEIA assay consist of 1 anti-HIV antibody-positive specimen and 3 specimens from the HBV-vaccinated individuals.
Fig 3.
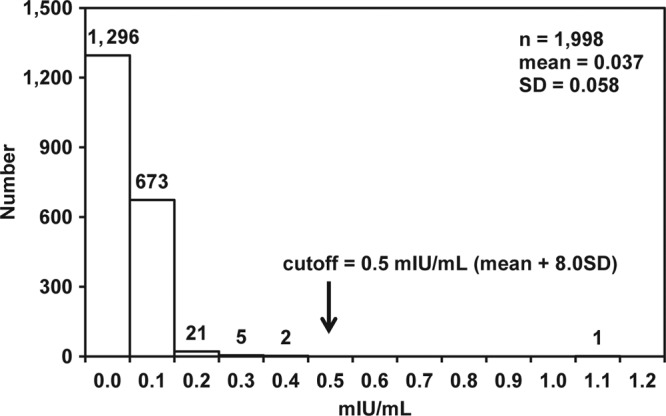
Distribution of HBsAg-negative specimens. A total of 2,002 samples were tested, but 4 samples that were repeatedly reactive with the ICT-CLEIA assay and confirmed for the presence of HBsAg with a confirmatory assay were excluded from this calculation.
Mutant sensitivity.
All 10 recombinant HBsAg samples, consisting of the wild type and 9 mutants, were utilized to evaluate the ability of the ICT-CLEIA assay to detect HBsAg mutants. The recombinant mutants consisted of recombinant HBsAg with seven single-amino-acid substitutions at position 126, 129, 133, 144, and 145 and two double-amino-acid substitutions at position 126 or 144 and 145. The results of the ICT-CLEIA and the Architect HBsAg QT assays, which were expressed as IU/ml, were converted to cutoff index (COI) by dividing the quantitative values with the respective cutoff values. As shown in Fig. 4, the ICT-CLEIA assay could detect all types of HBsAg mutants tested, and the COI of the ICT-CLEIA assay was 23.2- to 121.5-fold higher than those of the Architect HBsAg QT assay.
Fig 4.
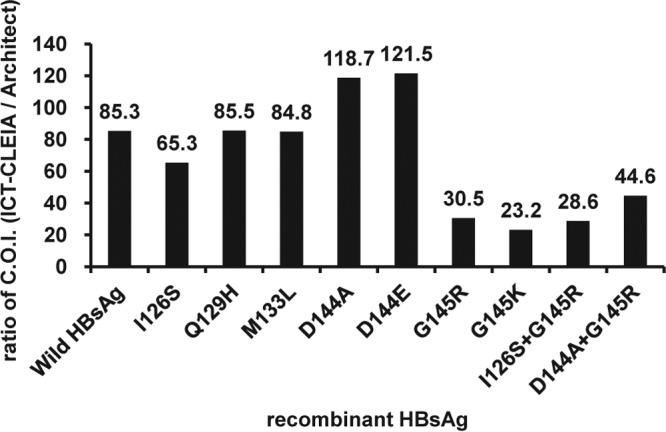
Reactivities of ICT-CLEIA for HBsAg and Architect HBsAg QT assay of HBV mutants. Recombinant HBsAg mutants were assayed with the ICT-CLEIA and Architect HBsAg QT assay. The results are expressed as a ratio of COI (ICT-CLEIA/Architect HBsAg QT).
Sensitivity for HBsAg seroconversion.
To evaluate the ability of the ICT-CLEIA assay to shorten the window period for HBV infection, 12 commercially available preseroconversion panels, which become HBsAg positive during the collection period, were tested. The results of the ICT-CLEIA assay were compared with those of the Architect HBsAg QT assay and HBV DNA PCR. The ICT-CLEIA assay revealed a higher sensitivity than the Architect HBsAg QT assay for the early detection of HBsAg, as shown in Table 3. The mean window closure time from the first bleed of the ICT-CLEIA assay was 23.6 days shorter than the Architect HBsAg QT assay. In addition, the window periods for the ICT-CLEIA assay and HBV DNA PCR were almost equal. For HBV6279, PCR detected HBV DNA 12 days earlier than the ICT-CLEIA assay detected HBsAg, whereas HBsAg detection by the ICT-CLEIA assay occurred 7 days earlier than HBV DNA detection by PCR with HBV6290. For the remaining 10 panels, HBsAg detection by the ICT-CLEIA assay and HBV DNA detection by PCR occurred simultaneously.
Table 3.
Comparison of the assay performance for detection of HBV in seroconversion panels
| Panel | No. of days to detection from the first bleed |
||
|---|---|---|---|
| Architect HBsAga | ICT-CLEIA | HBV-DNAb | |
| PHM909 | 9 | 0 | 0 |
| PHM921 | 0 | 0 | 0 |
| PHM922 | 16 | 0 | 0 |
| PHM923 | 15 | 0 | 0 |
| PHM929 | 14 | 0 | 0 |
| PHM931 | 19 | 0 | 0 |
| HBV6272 | 94 | 0 | 0 |
| HBV6279 | 26 | 12 | 0 |
| HBV6281 | 19 | 0 | 0 |
| HBV6290 | 21 | 0 | 7 |
| HBV11012 | 18 | 0 | 0 |
| HBV11048 | 44 | 0 | 0 |
| Mean | 24.6 | 1.0 | 0.6 |
Assay with the Architect HBsAg QT assay kit was performed by Mitsubishi Chemical Medience Corporation.
Results of HBV DNA assay, except the PHM929, PHM931, and HBV6272 panels were based on the vendor's data sheet. HBV DNA assay results for PHM929 and PHM931 were based on the results published previously (22). HBV DNA for HBV6272 was detected using the Roche Cobas TaqMan HBV version 2.0 by SRL, Inc.
In addition, three panels (PHM935B, HBV6281, and HBV11000) were tested to evaluate the ability of the ICT-CLEIA assay to detect HBsAg in the presence of anti-HBs antibodies. PHM935B and HBV6281 were composed of serial specimens collected during a seroconversion period, whereas HBV11000 was composed of serial specimens collected during a reverse seroconversion period. In PHM935B (Fig. 5A), the HBsAg levels gradually decreased, but HBV DNA could not be detected between days 203 and 273. Although the Architect HBsAg assay could not detect HBsAg between days 262 and 273, the ICT-CLEIA assay could detect HBsAg with all panels, even in the presence of anti-HBsAb antibody during the late stage of infection. With the HBV6281 panel (Fig. 5B), which was an acute and recovered hepatitis B seroconversion panel, the ICT-CLEIA assay presented a similar profile change in viral load to that of the HBV DNA PCR during the course of HBV infection. The ICT-CLEIA assay could detect HBsAg even in the presence of anti-HBs antibodies, as shown in PHM935B, whereas both the Architect HBsAg QT assay and HBV DNA PCR (the limit of detection in the provided data was 100 copies/ml) could not detect HBV infection during the recovered phase of infection between days 50 and 54. In addition, the Architect HBsAg QT assay could not detect HBsAg between days 0 and 13 during the acute phase of infection. With the HBV11000 panel (Fig. 5C), which was a case of reverse seroconversion of anti-HBsAb antibody to HBsAg, the ICT-CLEIA assay demonstrated a similar profile change in viral load to that of the HBV DNA PCR, while the Architect HBsAg QT assay could not detect HBsAg in the presence of anti-HBsAb antibody between days 0 and 18.
Fig 5.
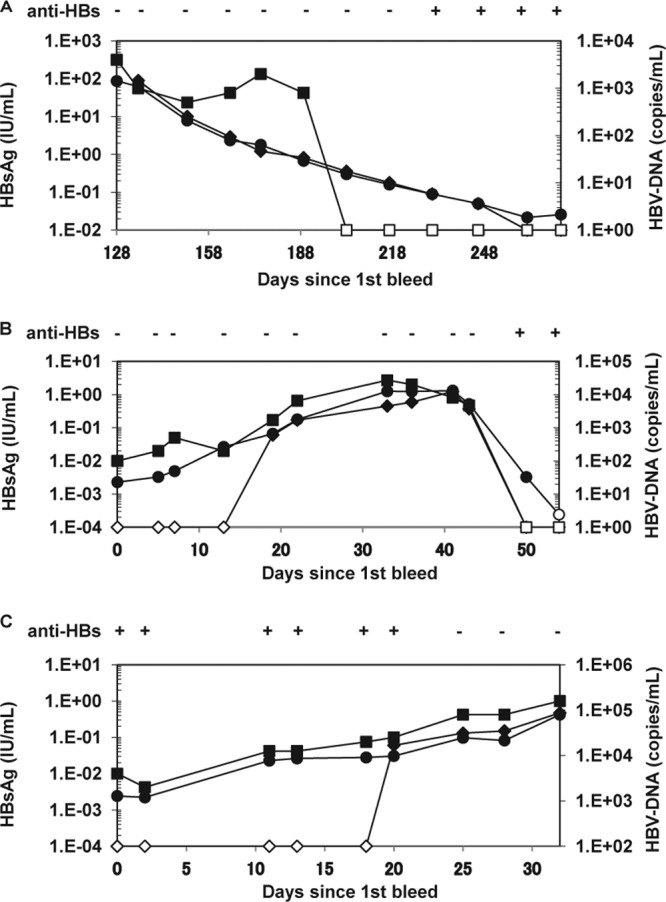
Comparison of HBsAg and HBV DNA profiles in seroconversion panels, PHM935B (A), HBV6281 (B), and a reverse seroconversion panel, HBV11000 (C). The anti-HBs data (−, negative; +, positive) and HBV DNA data were obtained from the supplier's data sheet. HBV DNA detection for HBV11000 was performed using the Roche Cobas TaqMan HBV version 2.0 by SRL, Inc. The results of the ICT-CLEIA (●, ○) were compared with those of the Architect HBsAg QT assay (◆, ♢) and HBV DNA PCR (■, □). Closed symbols, positive; open symbols, negative.
DISCUSSION
The HBsAg assay is the first-line screen for HBV in blood donations, and therefore a highly sensitive screening assay is very much needed to reduce the risk of transfusion-associated HBV infection. In this study, a semiautomated ICT-CLEIA assay was developed and evaluated for the detection of HBsAg. Compared with the Abbott Architect HBsAg QT assay, which has a clinical detection limit of 50 mIU/ml, the ICT-CLEIA assay has a 100-fold-higher sensitivity, with a detection limit of 0.5 mIU/ml.
To improve the sensitivity of the assay, the ICT technique (18, 19) was coupled with a magnetic microparticle enzyme immunoassay using a chemiluminescent substrate. By using this technique, it was possible to utilize the alkaline phosphatase conjugates at a high concentration without increasing background noise. As a result, the signal-to-noise ratio was increased and the sensitivity of the assay (0.5 mIU/ml) was improved 100-fold higher than that of the Architect HBsAg QT assay (50 mIU/ml). The total assay time could be shortened 10-fold compared to the two-site immune complex transfer enzyme immunoassay described previously (18, 19). In addition, the specimens were treated with pretreatment reagents to detect HBsAg even in the presence of anti-HBs antibodies in this assay. The ingredients in the pretreatment reagents were optimized for sensitive HBsAg detection by releasing HBsAg from immune complexes consisting of HBsAg and anti-HBs antibody.
The sensitivity improvement of the ICT-CLEIA assay did not compromise assay specificity (99.95%, n = 1,998), and no false-positive results were observed when testing potentially interfering specimens unrelated to HBV infection. Although 4 out of 160 potentially interfering specimens (1 anti-HIV antibody-positive specimen and 3 specimens from the individuals with HBV vaccination) were positive according to the ICT-CLEIA assay, all four specimens were positive according to the ICT-CLEIA confirmatory assay, indicating that the ICT-CLEIA assay specifically detected HBsAg in the samples. The specimen positive for anti-HIV antibody was also reactive using the Architect HBsAg QT assay. This specimen might have been collected from an individual dually infected with HBV and HIV, although HBV DNA could not be detected in the specimen using the Roche Cobas TaqMan HBV version 2.0 test (data not shown). Indeed, it has been reported that chronic HBV infection is found in approximately 10% of the HIV-infected population (23, 24). Alternatively, another possibility is that the specimen might have been collected from an HIV-infected individual who had received HBV vaccination. This specimen was negative for anti-HBc antibody (data not shown).
For the three specimens from the HBV vaccine recipients that were reactive with the ICT-CLEIA assay, the patients were bled within approximately 1 month after vaccination according to the vendor's information. There are several reports describing the HBs antigenemia postvaccination (25–32). Lunn et al. reported a prolonged hepatitis B surface antigenemia case due to vaccination (30). A positive HBsAg was found when the individual donated blood 18 days after vaccination. Köksal et al. reported that the longest duration of confirmed antigenemia was 21 days (29). Because of the higher sensitivity of the ICT-CLEIA assay, it might be possible to detect vaccine-derived HBsAg for a longer period than reported previously. In addition, all three specimens were negative for anti-HBc antibodies (data not shown), which might support the interpretation that the HBsAg detected with the ICT-CLEIA assay was derived from vaccines.
During the evaluation of the detection of HBsAg mutants, nine recombinant HBsAg mutants all tested positive using the ICT-CLEIA assay. The sensitivity improvement of the ICT-CLEIA assay resulted in 23.2- to 121.5-fold enhanced detection of the HBsAg mutants compared to the Architect HBsAg QT assay, although mutants with amino acid substitutions at position 145 appeared to be underquantified by the ICT-CLEIA assay. The ICT-CLEIA assay was established utilizing several monoclonal antibodies that recognize different epitopes to detect HBsAg mutants. Indeed, the ICT-CLEIA assay could detect the most frequently reported HBsAg mutant, G145R, with or without additional mutation at position 126 or 144, while commercial assays established utilizing a monoclonal capture antibody and monoclonal detection antibody cannot detect these mutants (10, 12). Moerman et al. reported that the assays using a monoclonal antibody against the amino acids outside the “a” determinant (amino acid residues 124 to 147) were obviously less prone to creating false-negative results (13). The fact that one of the monoclonal antibodies used in the ICT-CLEIA assay is directed against an epitope located outside the “a” determinant (data not shown) may confer HBsAg mutant detection reliability.
To evaluate the early detection of HBV infection using the ICT-CLEIA assay, commercially available seroconversion panels were tested. The ICT-CLEIA assay reduced the window period by 23.6 days compared with the Architect HBsAg QT assay. In addition, in 10 out of 12 panels tested, HBsAg detection by the ICT-CLEIA assay and HBV DNA detection by PCR occurred simultaneously. We also examined the ability of the ICT-CLEIA assay to detect HBsAg in anti-HBs-positive samples using three types of seroconversion panels (PHM935B, HBV6281, and HBV11000). The PHM935B and HBV6281 panels were cases of seroconversion of HBsAg to anti-HBs antibody, while the HBV11000 was a case of reverse seroconversion of anti-HBs antibody to HBsAg.
When using the PHM935B panels, HBV DNA was negative between days 203 and 273 based on the vendor's data sheet (the Roche Amplicor HBV monitor test; limit of detection was 400 copies/ml), but Kimura et al. (33) demonstrated the presence of HBV DNA during this period by using in-house real-time detection PCR (limit of detection was 20 copies/ml) and nested PCR and revealed that HBV DNA levels gradually decrease, as seen in the ICT-CLEIA assay (Fig. 5A). Together with the result for the HBV6281 panel, the monitoring of the viral kinetics by the ICT-CLEIA assay and the HBV DNA PCR resulted in very similar-shaped curves both in the early and late phases of HBV infection despite the presence or the absence of anti-HBs antibody. Therefore, highly sensitive HBsAg detection by the ICT-CLEIA assay would be useful for not only an earlier detection of acute hepatitis B infection but also in the monitoring of hepatitis B patients during antiviral therapy.
HBV reactivation is a well-recognized complication of solid organ or hematopoietic stem cell transplantation (34, 35), anticancer therapy with or without rituximab (36, 37, 38, 39), treatment with biologic agents (40), or human immunodeficiency virus infection (41). HBV reactivation occurs in both HBsAg-positive patients and HBsAg-negative patients. In particular, the appearance of HBsAg in patients previously positive for anti-HBs antibody is called reverse seroconversion, as seen in the HBV11000 panel (Fig. 5C). Recently, it was reported that the reduction or the loss of anti-HBs antibody during or after the treatment was followed by HBV reactivation (42, 43). Therefore, the ICT-CLEIA assay, which is able to detect HBsAg before the loss of anti-HBs antibody, might be useful for the earlier diagnosis of HBV reactivation.
ACKNOWLEDGMENTS
We gratefully acknowledge the helpful advice and comments of Yasuhito Tanaka (Nagoya City University Graduate School of Medical Sciences).
Footnotes
Published ahead of print 8 May 2013
REFERENCES
- 1. World Health Organization 2012. Hepatitis B. WHO, Geneva, Switzerland: http://www.who.int/mediacentre/factsheets/fs204/en/ [Google Scholar]
- 2. Ganem D, Prince AM. 2004. Hepatitis B virus infection—natural history and clinical consequences. N. Engl. J. Med. 350:1118–1129 [DOI] [PubMed] [Google Scholar]
- 3. Allain J-P, Candotti D, Soldan K, Phelps B, Giachetti C, Shyamala V, Yeboah F, Ankowa M, Owusu-Ofori S, Opare-Sem O. 2003. The risk of hepatitis B virus infection by transfusion in Kumai, Ghana. Blood 101:2419–2425 [DOI] [PubMed] [Google Scholar]
- 4. Biswas R, Tabor E, Hsia CC, Wright DJ, Laycock ME, Fiebig EW, Peddada L, Smith R, Schreiber GB, Epstein JS, Nemo GJ, Busch MP. 2003. Comparative sensitivity of HBV NATs and HBsAg assays for detection of acute HBV infection. Transfusion 43:788–798 [DOI] [PubMed] [Google Scholar]
- 5. Linauts S, Saldanha J, Strong DM. 2008. PRISM hepatitis B surface antigen detection of hepatitis B virus minipool nucleic acid testing yield samples. Transfusion 48:1376–1382 [DOI] [PubMed] [Google Scholar]
- 6. Vermeulen M, Lelie N, Sykes W, Crookes R, Swanevelder J, Gaggia L, Roux ML, Kuun E, Gulube S, Reddy R. 2009. Impact of individual-donation nucleic acid testing on risk of human immunodeficiency virus, hepatitis B virus, and hepatitis C virus transmission by blood transfusion in South Africa. Transfusion 49:1115–1125 [DOI] [PubMed] [Google Scholar]
- 7. Norder H, Couroucé Coursaget A-MP, Echevarria JM, Lee Mushahwar S-DIK, Robertson BH, Locarnini S, Magnius LO. 2004. Genetic diversity of hepatitis B virus strains derived worldwide; genotypes, subgenotypes, and HBsAg subtypes. Intervirology 47:289–309 [DOI] [PubMed] [Google Scholar]
- 8. Olinger CH, Jutavijittum P, Hübschen JM, Yousukh A, Samountry B, Thammavong T, Toriyama K, Muller CP. 2008. Possible new hepatitis B virus genotype, southeast Asia. Emerg. Infect. Dis. 14:1777–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tatematsu K, Tanaka Y, Kurbanov F, Sugauchi F, Mano S, Maeshiro T, Nakayoshi T, Wakuta M, Miyakawa Y, Mizokami M. 2009. A genetic variant of hepatitis B virus divergent from known human and ape genotypes isolated from a Japanese patient and provisionally assigned to new genotype J. J. Virol. 83:10538–10547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Coleman PF, Chen YCJ, Mushahwar IK. 1999. Immunoassay detection of hepatitis B surface antigen mutants. J. Med. Virol. 59:19–24 [DOI] [PubMed] [Google Scholar]
- 11. Ge J-H, Liu H-M, Sun J, Zhang L-Z, He J, Li Y-L, Liu H, Xu Y, Yu H-Y, Hu Y-P. 2004. Antigenic and immunogenic changes due to mutation of s gene of HBV. World J. Gastroenterol. 10:3137–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ly TD, Servant-Delmas A, Bagot S, Gonzalo S, Férey Ebel M-PA, Dussaix E, Laperche S, Roque-Afonso A-M. 2006. Sensitivity of four new commercial hepatitis B virus surface antigen (HBsAg) assays in detection of HBsAg mutant forms. J. Clin. Microbiol. 44:2321–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moerman B, Moons V, Sommer H, Schmitt Y, Stetter M. 2004. Evaluation of sensitivity for wild type and mutant forms of hepatitis B surface antigen by four commercial HBsAg assays. Clin. Lab. 50:159–162 [PubMed] [Google Scholar]
- 14. Scheiblauer H, Soboll H, Nick S. 2006. Evaluation of 17 CE-marked HBsAg assays with respect to clinical sensitivity, analytical sensitivity, and hepatitis B virus mutant detection. J. Med. Virol. 78:S66–S70 [DOI] [PubMed] [Google Scholar]
- 15. Seddigh-Tonekaboni S, Waters JA, Jeffers S, Geheke R, Ofenloch B, Horsch A, Hess G, Thomas HC, Karayiannis P. 2000. Effect of variation in the common “a” determinant on the antigenicity of hepatitis B surface antigen. J. Med. Virol. 60:113–121 [DOI] [PubMed] [Google Scholar]
- 16. Tian Y, Xu Y, Zhang Z, Meng Z, Qin L, Lu M, Yang D. 2007. The amino acid residues at position 120 to 123 are crucial for the antigenicity of hepatitis B surface antigen. J. Clin. Microbiol. 45:2971–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gerlich WH. 2004. Diagnostic problems caused by HBsAg mutants—a consensus report of an expert meeting. Intervirology 47:310–313 [DOI] [PubMed] [Google Scholar]
- 18. Hashida S, Hashinaka K, Nishikata I, Oka S, Shmada K, Saitoh A, Takamizawa A, Shinagawa H, Ishkawa E. 1995. Measurement of human immunodeficiency virus type 1 p24 in serum by an ultrasensitive enzyme immunoassay, the two-site immune complex transfer enzyme immunoassay. J. Clin. Microbiol. 33:298–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hashida S, Ishikawa S, Hashinaka K, Nishikata I, Oka S, Ishikawa E. 2000. Earlier detection of human immunodeficiency virus type 1 p24 antigen and immunoglobulin G and M antibodies to p17 antigen in seroconversion serum panels by immune complex transfer enzyme immunoassay. Clin. Diagn. Lab. Immunol. 7:872–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Meng Q, Wong C, Rangachari A, Tamatsukuri S, Sasaki M, Fiss E, Cheng L, Ramankutty T, Clarke D, Yawata H, Sasakura Y, Hirose T, Impraim C. 2001. Automated multiplex assay system for simultaneous detection of hepatitis B virus DNA, hepatitis C virus RNA, and human immunodeficiency virus type 1 RNA. J. Clin. Microbiol. 39:2937–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Minegishi K, Yoshikawa A, Kishimoto S, Yugi H, Yokoya N, Sakurada M, Kiyokawa H, Nishioka K, the Japanese Red Cross Screening Research Group NAT 2003. Superiority of minipool nucleic acid amplification technology for hepatitis B virus over chemiluminescence immunoassay for hepatitis B surface antigen screening. Vox Sang. 84:287–291 [DOI] [PubMed] [Google Scholar]
- 22. Wiedmann M, Kluwick S, Walter M, Fauchald G, Howe J, Bronold M, Zauke M. 2007. HIV-1, HCV and HBV seronegative window reduction by the new Roche cobas TaqScreen MPX test in seroconversion donors. J. Clin. Microbiol. 39:282–287 [DOI] [PubMed] [Google Scholar]
- 23. Alter MJ. 2006. Epidemiology of viral hepatitis and HIV co-infection. J. Hepatol. 44(1 Suppl):S6–S9 [DOI] [PubMed] [Google Scholar]
- 24. Thio CL. 2009. Hepatitis B and human immunodeficiency virus coinfection. Hepatology 49:S138–S145 [DOI] [PubMed] [Google Scholar]
- 25. Brodersen H-P, Beckers B, Köhler H, Dahlmanns C, Kruska L, Larbig D. 1997. The test for hepatitis B surface antigen is transiently positive after vaccination with recombinant vaccine. Nephrol. Diagn. Transplant. 12:2756–2757 [DOI] [PubMed] [Google Scholar]
- 26. Dow BC, Yates P, Galea G, Munro H, Buchanan I, Ferguson K. 2002. Hepatitis B vaccines may be mistaken for confirmed hepatitis B surface antigen-positive blood donors. Vox Sang. 82:15–17 [DOI] [PubMed] [Google Scholar]
- 27. Janzen L, Minuk GY, Fast M, Bernstein KN. 1996. Vaccine-induced hepatitis B surface antigen positivity in adult hemodialysis patients: incidental and surveillance data. J. Am. Soc. Nephrol. 7:1228–1234 [DOI] [PubMed] [Google Scholar]
- 28. Kloster B, Kramer R, Eastlund T, Grossman B, Zarvan B. 1995. Hepatitis B surface antigenemia in blood donors following vaccination. Transfusion 35:475–477 [DOI] [PubMed] [Google Scholar]
- 29. Köksal N, Altinkaya N, Perk Y. 1996. Transient hepatitis B surface antigenemia after neonatal hepatitis B immunization. Acta Pediatr. 85:1501–1502 [DOI] [PubMed] [Google Scholar]
- 30. Lunn ER, Hoggarth BJ, Cook WJ. 2000. Prolonged hepatitis B surface antigenemia after vaccination. Pediatrics 105:e81. [DOI] [PubMed] [Google Scholar]
- 31. Mantadakis E, Thomaidis S, Efraimidou Ramatani E-NA, Chatzimichael A. 2010. Transient hepatitis B surface antigen circulation after Infanrix-Hexa: a case report and review of the literature. Eur. J. Pediatr. 169:1139–1141 [DOI] [PubMed] [Google Scholar]
- 32. Otağ F. 2003. False positive HBsAg result in blood donors due to administration of three different recombinant DNA hepatitis B vaccines. Vaccine 21:3734–3737 [DOI] [PubMed] [Google Scholar]
- 33. Kimura T, Rokuhara A, Sakamoto Y, Yagi S, Tanaka E, Kiyosawa K, Maki N. 2002. Sensitive immunoassay for hepatitis B virus core-related antigens and their correlation to virus load. J. Clin. Microbiol. 40:439–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Blanpain C, Knoop C, Delforge ML, Antoine M, Peny MO, Liesnard C, Vereerstraeten P, Cogan E, Adler M, Abramowicz D. 1998. Reactivation of hepatitis B after transplantation in patients with preexisting anti-hepatitis B surface antigen antibodies: report on three cases and review of the literature. Transplantation 66:883–886 [DOI] [PubMed] [Google Scholar]
- 35. Kempinska A, Kwak EJ, Angel JB. 2005. Reactivation of hepatitis B infection following allogeneic bone marrow transplantation in a hepatitis B-immune patient: case report and review of the literature. Clin. Infect. Dis. 41:1277–1282 [DOI] [PubMed] [Google Scholar]
- 36. Ide Y, Ito Y, Takahashi S, Tokudome N, Kobayashi K, Sugihara T, Hattori M, Yokoyama M, Uchiyama A, Inoue K, Sakurai N, Hatake K. 24 July 2010. Hepatitis B virus reactivation in adjuvant chemotherapy for breast cancer. Breast Cancer [Epub ahead of print.] 10.1007/s12282-010-0213-x [DOI] [PubMed] [Google Scholar]
- 37. Dervite I, Hober D, Morel P. 2001. Acute hepatitis B in a patient with antibodies to hepatitis B surface antigen who was receiving rituximab. N. Engl. J. Med. 344:68–69 [DOI] [PubMed] [Google Scholar]
- 38. Kim EB, Kim DS, Park SJ, Park Y, Rho KH, Kim SJ. 2008. Hepatitis B virus reactivation in a surface antigen-negative and antibody-positive patient after rituximab plus CHOP chemotherapy. Cancer Res. Treat. 40:36–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ozaras R, Ar C, Ongoren S, Mete B, Tabak F, Mert A, Ozturk R. 2010. Acute hepatitis B despite a previous high titer of anti-HBs. Hepatol. Int. 4:530–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Urata Y, Uesato R, Tanaka D, Kowatari K, Nitobe T, Nakamura Y, Motomura S. 2011. Prevalence of reactivation of hepatitis B virus replication in rheumatoid arthritis patients. Mod. Rheumatol. 21:16–23 [DOI] [PubMed] [Google Scholar]
- 41. Manegold C, Hannoun C, Wywiol A, Dietrich M, Polywka S, Chiwakata CB, Günther S. 2001. Reactivation of hepatitis B virus replication accompanied by acute hepatitis in patients receiving highly active antiviral therapy. 32:144–148 [DOI] [PubMed] [Google Scholar]
- 42. Pei S-N, Ma M-C, Wang M-C, Kuo C-Y, Rau K-M, Su C-Y, Chen C-H. 2012. Analysis of hepatitis B surface antibody titers in B cell lymphoma patients after rituximab therapy. Ann. Hematol. 91:1007–1012 [DOI] [PubMed] [Google Scholar]
- 43. Yoshida T, Kusumoto S, Inagaki A, Mori F, Ito A, Ri M, Ishida T, Komatsu H, Iida S, Sugauchi F, Tanaka Y, Mizokami M, Ueda R. 2010. Reactivation of hepatitis B virus in HBsAg-negative patients with multiple myeloma: two case reports. Int. J. Hematol. 91:844–849 [DOI] [PubMed] [Google Scholar]


