Abstract
Sequence type 398 (ST398) Staphylococcus aureus, frequently carried by livestock, has caused severe human infections and often carries transmissible antibiotic resistance genes. Among methicillin-susceptible S. aureus isolates colonizing Dallas County Jail detainees, 13.2% were ST398, spa type t571, and were genetically similar to human colonization isolates from New York, Chicago, and the Dominican Republic.
TEXT
Sequence type 398 (ST398) methicillin-resistant Staphylococcus aureus (MRSA) is often transmitted among livestock and animal handlers but is uncommonly associated with human disease in the United States (1). We unexpectedly identified ST398/t571 methicillin-sensitive S. aureus (MSSA) as the most common genetic background among MSSA isolates colonizing detainees in the Dallas County Jail. In this study, we compared the pulsed-field gel electrophoresis (PFGE) pulsotypes of isolates from the Dallas jail and from a single patient in Chicago with those of strains isolated in previous studies.
Recognized first in Europe (2–9), ST398 MRSA has been isolated in the United States (10), Canada (11), and Latin America (12). ST398 methicillin-susceptible S. aureus (MSSA) isolates have been recovered from pigs (13), humans (14–17), and retail meat (18). Asymptomatic human colonization and infection with ST398 MSSA are rare in the United States and Europe (19–21). However, in New York City in 2004 to 2007, 13 people, mostly of Dominican origin, had nasal carriage of ST398 MSSA with the unusual spa type t571 (ST398/t571) (22). In addition, patients in the Dominican Republic in 2007-2008 were infected by ST398/t571 MSSA (22), as were patients in New Jersey (23).
In January 2009, we screened the anterior nares and the hands of 928 detainees, housed in 68 divisions, called tanks, in the Dallas County Jail, for S. aureus carriage. Tanks had a capacity of 24 to 36 detainees. Detainees can interact with others within a single tank but have little interaction with those in other tanks. The study was approved by the Institutional Review Boards at the University of Chicago Medical Center and the University of Texas-Southwestern Medical Center. Recovered MSSA isolates were genotyped from subjects housed in a stratified random sample of 26 of the 68 tanks; all MRSA isolates were genotyped.
In a separate study of S. aureus colonization among patients and household contacts (24), we also identified one patient in Chicago with ST398/t571 MSSA colonization.
Multilocus sequence typing (MLST) (25), spa typing (26), PCR for the Panton-Valentine leukocidin (PVL) genes (27), and PFGE using Cfr9I digestion (22, 28) were performed. Antimicrobial susceptibilities were determined by automated testing (29).
Among the 345 subjects in the 26 selected tanks, 110 (31.9%) carried MSSA. Of 158 MSSA isolates identified, 152 were available, and 34 MLST types were identified. Twenty isolates (13%) were ST398, the most common ST; all lacked PVL genes. Of the 345 subjects, the 16 (4.6%) who carried the 20 ST398 MSSA isolates were more likely than carriers of other MSSA genetic backgrounds to be female (P = 0.0009) and older (P = 0.01) (Table 1).
Table 1.
Demographic characteristics of ST398 MSSA carriers and subjects carrying other MSSA sequence types in Dallas County Jail
| Characteristic | Detainees carrying ST398 MSSA (n = 16) | Detainees carrying other MSSA backgrounds (n = 95) | P value |
|---|---|---|---|
| Gender, no. (%) | |||
| Male | 7 (44) | 77 (81) | 0.0009 |
| Female | 9 (56) | 23 (19) | |
| Race, no. (%) | |||
| White | 7 (44) | 52 (55) | 0.5 |
| Black | 8 (50) | 41 (43) | |
| Unknown | 1 (6) | 2 (2) | |
| Ethnicity, no. (%) | |||
| Hispanic | 3 (19) | 24 (25) | 0.8 |
| Non-Hispanic | 13 (81) | 70 (74) | |
| Unknown | 0 | 1 (1) | |
| Age, yrs, mean ± SD | 40.6 ± 9.5 | 33.3 ± 10.7 | 0.01 |
| Median duration of stay in jail, days (range) | 50.5 (1–245) | 74 (1–675) | 0.3 |
Among the 20 ST398 isolates, 18 were susceptible to ciprofloxacin, gentamicin, rifampin, and trimethoprim-sulfamethoxazole and resistant to clindamycin and erythromycin. The two other isolates differed in that one was susceptible to clindamycin and the other was resistant to ciprofloxacin.
All 20 MSSA ST398 isolates shared the same pulsotype (Fig. 1) and spa type t571. All ST398/t571 MSSA carriers were housed in 4 of the 26 (15%) tanks, designated tanks A, B, C, and D. In tank A, samples from 20 (63%) of the 32 female detainees were cultured; ST398/t571 MSSA was carried by 9 of 17 (53%) S. aureus carriers. All ST398/t571 isolates from tank A shared a common antibiogram. One subject who carried ST398/t571 on the hand had an MSSA isolate obtained from her nose, not available for genotyping, that shared the same antibiogram as the ST398/t571 isolates in tank A, suggesting that it may have been ST398/t571. The culture results from tanks A, B, C, and D are shown in Table 2.
Fig 1.
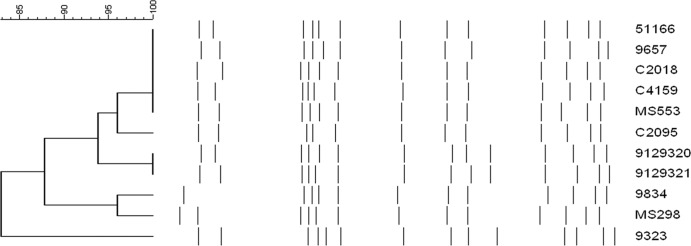
Analysis of PFGE patterns of ST398 isolates from the Dallas County Jail (C2018, C4159, and C2095; all are spa type t571), New York State Prisons (9657 [t6864], 9834 [t571], and 9323 [t6587]) (30), Chicago colonization isolates (9129320 [t571] and 9129321 [t571]), Northern Manhattan (51166 [t571] and MS553 [t6608], and MS298 [t1451]) (22). Band patterns are compared using the Dice coefficient.
Table 2.
S. aureus culture from nares and hand carriage in tank A, in which all subjects were female, and tanks B, C, and D, in which all subjects were male
| Tank | Subject | Length of stay in jail (days) | Hand culturee |
Nares culturee |
||
|---|---|---|---|---|---|---|
| Result | Genotypea | Result | Genotypea | |||
| A | 1 | 56 | MSSA | 398/pvl− | neg | NA |
| A | 2 | 8 | MSSA | 398/pvl− | neg | NA |
| A | 3 | 25 | MSSA | 15slvb/pvl− | MSSA | 959/pvl− |
| A | 4 | 91 | neg | NA | MSSA | NTc/pvl− |
| A | 5 | 1 | MSSA | 1860/pvl− | MSSA | 398/pvl− |
| A | 6 | 4 | MSSA | 1860/pvl− | MSSA | 72/pvl− |
| A | 7 | 113 | MSSA | 398/pvl− | MSSA | 398/pvl− |
| A | 8 | 101 | MSSA | 398/pvl− | MSSA | 97/pvl− |
| A | 9 | 26 | MSSA | 1860/pvl− | MSSA | 72/pvl− |
| A | 10 | 10 | neg | NA | neg | NA |
| A | 11 | 4 | MRSA | 8/IV/pvl+ | neg | NA |
| A | 12 | 66 | MSSA | 398/pvl− | MSSA | Not typedd |
| A | 13 | 2 | MSSA | 5/pvl− | neg | NA |
| A | 14 | 16 | neg | NA | neg | NA |
| A | 15 | 67 | MRSA | 8/IV/pvl+ | neg | NA |
| A | 16 | 57 | neg | NA | neg | NA |
| A | 17 | 45 | MSSA | 398/pvl− | neg | NA |
| A | 18 | 1 | MRSA | 8/IV/pvl+ | neg | NA |
| A | 19 | 21 | neg | NA | MSSA | 398/pvl− |
| A | 20 | 245 | MSSA | 398/pvl− | MSSA | 398/pvl− |
| B | 21 | 445 | neg | NA | MSSA | 432/pvl− |
| B | 22 | 21 | MSSA | 398/pvl− | MSSA | 398/pvl− |
| B | 23 | 15 | MSSA | 398/pvl− | MSSA | 8/pvl+ |
| B | 24 | 69 | MSSA | 398/pvl− | MSSA | 398/pvl− |
| B | 25 | 53 | neg | NA | neg | NA |
| B | 26 | 131 | neg | NA | MSSA | 398/pvl− |
| B | 27 | 22 | MSSA | 630/pvl− | neg | NA |
| C | 28 | 18 | neg | NA | MSSA | 8/pvl− |
| C | 29 | 7 | MSSA | 188/pvl+ | MSSA | 188/pvl− |
| C | 30 | 80 | MSSA | 434/pvl− | MSSA | 398/pvl− |
| C | 31 | 7 | neg | NA | MSSA | 45/pvl− |
| D | 32 | 197 | neg | NA | MRSA | Not typedd |
| D | 33 | 66 | MSSA | 1/pvl− | neg | NA |
| D | 34 | 45 | neg | NA | MRSA | 30/IV/pvl+ |
| D | 35 | 38 | neg | NA | neg | NA |
| D | 36 | 49 | MRSA | 8/IV/pvl+ | MRSA | 8/IV/pvl+ |
| D | 37 | 42 | neg | NA | MSSA | 398/pvl− |
| D | 38 | 22 | MSSA | 6/pvl− | neg | NA |
| D | 39 | 11 | neg | NA | neg | NA |
| D | 40 | 18 | neg | NA | neg | NA |
| D | 41 | 149 | MSSA | 6/pvl− | MSSA | 6/pvl− |
| D | 42 | 23 | neg | NA | MSSA | 398/pvl− |
| D | 43 | 23 | neg | NA | MSSA | 8/pvl+ |
| D | 44 | 78 | neg | NA | neg | NA |
| D | 45 | 184 | neg | NA | MSSA | Not typedd |
| D | 46 | 84 | neg | NA | MSSA | 8/pvl+ |
| D | 47 | 150 | MSSA | 6/pvl− | MSSA | 6/pvl− |
| D | 48 | 213 | neg | NA | neg | NA |
| D | 49 | 7 | neg | NA | neg | NA |
| D | 50 | 215 | neg | NA | MSSA | Not typedd |
| D | 51 | 71 | neg | NA | neg | NA |
| D | 52 | 16 | neg | NA | neg | NA |
| D | 53 | 222 | neg | NA | neg | NA |
Format for genotype is ST/SCCmec type (for MRSA isolates)/presence (pvl+) or absence (pvl−) of carriage of the PVL genes. NA, not applicable.
“15slv” indicates a single-locus MLST variant of ST15.
“NT” signifies a new MLST type not yet reported in the MLST database.
Isolate was not available for genotyping.
neg, negative.
For the skin and soft tissue infection (SSTI) patient from Chicago, ST398/t571 MSSA was isolated from the nose and throat. None of his 4 household contacts and no other subject in our study of 350 households was colonized with an MSSA or MRSA ST398 isolate (24).
As determined by PFGE, ST398 isolates from the Dallas County Jail, from New York State Prisons (30), from the patient in Chicago, and from northern Manhattan (22) all shared PFGE patterns with >80% identity by Dice coefficient analysis (Fig. 1). This suggests that this genetic background may be identified in geographic pockets across the United States.
The finding of 9 subjects in a single jail tank with carriage of ST398/t571 MSSA isolates with a common pulsotype suggests local spread and easy transmissibility. No other MSSA strain had a similarly wide distribution in a single tank. This finding contrasts with data from the Netherlands, where ST398 MRSA isolates were less transmissible in the hospital than other MRSA genetic backgrounds (31). Also, carriage of ST398 MRSA by animal workers is often transient and related to persistent exposure to animals (32). It is not known if the ST398/t571 MSSA isolates in our study caused only transient carriage, as shown in the study of ST398 MRSA.
Limitations of the study were that it was cross-sectional and that few demographic data were available for each subject.
ST398 MRSA strains may carry and transfer to other S. aureus backgrounds transposons encoding genes for tetracycline resistance (33, 34) or have resistance to pleuromutilin and lincosamide antimicrobials (4, 35–37). ST398 isolated from humans has been found to carry phages that encode human innate immune modulators (38). Many spa types have been associated with the ST398 background (39), and the t571 type has likely been derived on more than one occasion from strains with related spa types (40).
ST398/t571 MSSA has been found rarely among S. aureus isolates identified in studies of colonization in humans (15, 41, 42) or livestock (43, 44). However, in the United States and Europe, ST398/t571 MSSA isolates have caused fatal necrotizing pneumonia, SSTIs, and bloodstream infections (8, 16, 39, 45–48; N. van der Mee Marquet, personal communication) and after 2001 caused sterile site infections in China (49, 50).
Among ST398 MRSA, ST398/t571 MRSA isolates have been isolated rarely in studies of humans (5, 9, 51–56) or livestock (2, 3, 6, 11, 57), although they were common in an animal study from Peru (12).
The apparent ease of transmission of ST398/t571 MSSA in community settings in the absence of an animal reservoir, the frequent identification of ST398 strains among livestock, concerns about novel mechanisms of resistance in ST398 strains, and reports of invasive infections caused by ST398/t571 MSSA isolates all suggest that continued genotypic surveillance of S. aureus strains in humans and animals is warranted.
ACKNOWLEDGMENTS
This work was supported by CDC grant R01 C1000373-01 to R.S.D., M.Z.D., J.S., and S.B.-V., NIH grant 1K23 AI095361-01 from NIAID to M.Z.D., and NIH grant R01 AI077690-S1 to F.D.L.
Footnotes
Published ahead of print 8 May 2013
REFERENCES
- 1. van Cleef BA, Monnet DL, Voss A, Krziwanek K, Allerberger F, Struelens M, Zemlickova H, Skov RL, Vuopio-Varkila J, Cuny C, Friedrich AW, Spiliopoulou I, Pászti J, Hardardottir H, Rossney A, Pan A, Pantosti A, Borg M, Grundmann H, Mueller-Premru M, Olsson-Liljequist B, Widmer A, Harbarth S, Schweiger A, Unal S, Kluytmans JA. 2011. Livestock-associated methicillin-resistant Staphylococcus aureus in humans, Europe. Emerg. Infect. Dis. 17:502–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. de Neeling AJ, van den Broek MJM, Spalburg EC, van Santen-Verheuvel MG, Dam-Deisz WDC, Boshuizen HC, van de Giessen AW, van Duijkeren E, Huijsdens XW. 2007. High prevalence of methicillin resistant Staphylococcus aureus in pigs. Vet. Microbiol. 122:366–372 [DOI] [PubMed] [Google Scholar]
- 3. Espinosa-Gongora C, Larsen J, Moodley A, Nielsen JP, Skov RL, Andreasen M, Guardabassi L. 2012. Farm-specific lineages of methicillin-resistant Staphylococcus aureus clonal complex 398 in Danish pig farms. Epidemiol. Infect. 140:1794–1799 [DOI] [PubMed] [Google Scholar]
- 4. Lozano C, Aspiroz C, Ara M, Gómez-Sanz E, Zarazaga M, Torres C. 2011. Methicillin-resistant Staphylococcus aureus (MRSA) ST398 in a farmer with skin lesions and in pigs of his farm: clonal relationship and detection of lnu(A) gene. Clin. Microbiol. Infect. 17:923–927 [DOI] [PubMed] [Google Scholar]
- 5. Lozano C, Aspiroz C, Lasarte JJ, Gómez-Sanz E, Zarazaga M, Torres C. 2011. Dynamic of nasal colonization by methicillin-resistant Staphylococcus aureus ST398 and ST1 after mupirocin treatment in a family in close contact with pigs. Comp. Immunol. Microbiol. Infect. Dis. 34:e1–e7. 10.1016/j.cimid.2010.06.006 [DOI] [PubMed] [Google Scholar]
- 6. Pletinckx LJ, Verhegghe M, Crombé F, Dewulf J, De Bleecker Y, Rasschaert G, Butaye P, Goddeeris BM, De Man I. 2013. Evidence of possible methicillin-resistant Staphylococcus aureus ST398 spread between pigs and other animals and people residing on the same farm. Prev. Vet. Med. 109:293–303 [DOI] [PubMed] [Google Scholar]
- 7. Vanderhaeghen W, Cerpentier T, Adriaensen C, Vicca J, Hermans K, Butaye P. 2010. Methicillin-resistant Staphylococcus aureus (MRSA) ST398 associated with clinical and subclinical mastitis in Belgian cows. Vet. Microbiol. 144:166–171 [DOI] [PubMed] [Google Scholar]
- 8. van der Mee-Marquet N, François P, Domelier-Valentin AS, Coulomb F, Decreux C, Hombrock-Allet C, Lehiani O, Neveu C, Ratovohery D, Schrenzel J, Quentin R, Bloodstream Infection Study Group of Réseau des Hygiénistes du Centre (RHC) 2011. Emergence of unusual bloodstream infections associated with pig-borne-like Staphylococcus aureus ST398 in France. Clin. Infect. Dis. 52:152–153 [DOI] [PubMed] [Google Scholar]
- 9. Wulf MW, Sørum M, van Nes A, Skov R, Melchers WJ, Klaassen CH, Voss A. 2008. Prevalence of methicillin-resistant Staphylococcus aureus among veterinarians: an international study. Clin. Microbiol. Infect. 14:29–34 [DOI] [PubMed] [Google Scholar]
- 10. Smith TC, Male MJ, Harper AL, Kroeger JS, Tinkler GP, Moritz ED, Capuano AW, Herwaldt LA, Diekema DJ. 2009. Methicillin-resistant Staphylococcus aureus (MRSA) strain ST398 is present in midwestern U.S. swine and swine workers. PLoS One 4(1):e4258. 10.1371/journal.pone.0004258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Khanna T, Friendship R, Dewey C, Weese JS. 2008. Methicillin resistant Staphylococcus aureus colonization in pigs and pig farmers. Vet. Microbiol. 128:298–303 [DOI] [PubMed] [Google Scholar]
- 12. Arriola CS, Güere ME, Larsen J, Skov RL, Gilman RH, Gonzalez AE, Silbergeld EK. 2011. Presence of methicillin-resistant Staphylococcus aureus in pigs in Peru. PLoS One 6(12):e28529. 10.1371/journal.pone.0028529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kadlec K, Schwarz S. 2010. Identification of the novel dfrK-carrying transposon Tn559 in a porcine methicillin-susceptible Staphylococcus aureus ST398 strain. Antimicrob. Agents Chemother. 54:3475–3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fan J, Shu M, Zhang G, Zhou W, Jiang Y, Zhu Y, Chen G, Peacock SJ, Wan C, Pan W, Feil EJ. 2009. Biogeography and virulence of Staphylococcus aureus. PLoS One 4(7):e6216. 10.1371/journal.pone.0006216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moritz ED, Smith TC. 2011. Livestock-associated Staphylococcus aureus in childcare worker. Emerg. Infect. Dis. 17:742–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rasigade JP, Laurent F, Hubert P, Vandenesch F, Etienne JJ. 2010. Lethal necrotizing pneumonia caused by an ST398 Staphylococcus aureus strain. Emerg. Infect. Dis. 16:1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van Belkum A, Melles DC, Peeters JK, van Leeuwen WB, van Duijkeren E, Huijsdens XW, Spalburg E, de Neeling AJ, Verbrugh HA, Dutch Working Party on Surveillance and Research of MRSA-SOM 2008. Methicillin-resistant and -susceptible Staphylococcus aureus sequence type 398 in pigs and humans. Emerg. Infect. Dis. 14:479–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Waters AE, Contente-Cuomo T, Buchhagen J, Liu CM, Watson L, Pearce K, Foster JT, Bowers J, Driebe EM, Engelthaler DM, Keim PS, Price LB. 2011. Multidrug-resistant Staphylococcus aureus in US meat and poultry. Clin. Infect. Dis. 52:1227–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. David MZ, Boyle-Vavra S, Zychowski DL, Daum RS. 2011. Methicillin-susceptible Staphylococcus aureus as a predominantly healthcare-associated pathogen: a possible reversal of roles? PLoS One 6(4):e18217. 10.1371/journal.pone.0018217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grundmann H, Hori S, Enright MC, Webster C, Tami A, Feil EJ, Pitt T. 2002. Determining the genetic structure of the natural population of Staphylococcus aureus: a comparison of multilocus sequence typing with pulsed-field gel electrophoresis, randomly amplified polymorphic DNA analysis, and phage typing. J. Clin. Microbiol. 40:4544–4546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Monecke S, Luedicke C, Slickers P, Ehricht R. 2009. Molecular epidemiology of Staphylococcus aureus in asymptomatic carriers. Eur. J. Clin. Microbiol. Infect. Dis. 28:1159–1165 [DOI] [PubMed] [Google Scholar]
- 22. Bhat M, Dumortier C, Taylor BS, Miller M, Vasquez G, Yunen J, Brudney K, Sánchez EJ, Rodriguez-Taveras C, Rojas R, Leon P, Lowy FD. 2009. Staphylococcus aureus ST398, New York City and Dominican Republic. Emerg. Infect. Dis. 15:285–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mediavilla JR, Chen L, Uhlemann AC, Hanson BM, Rosenthal M, Stanak K, Koll B, Fries BC, Armellino D, Schilling ME, Weiss D, Smith TC, Lowy FD, Kreiswirth BN. 2012. Methicillin-susceptible Staphylococcus aureus ST398, New York and New Jersey, USA. Emerg. Infect. Dis. 18:700–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Miller LG, Eells SJ, Taylor AR, David MZ, Ortiz N, Zychowski D, Kumar N, Cruz D, Boyle-Vavra S, Daum RS. 2012. Staphylococcus aureus colonization among household contacts of patients with skin infections: risk factors, strain discordance, and complex ecology. Clin. Infect. Dis. 54:1523–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Harmsen D, Claus H, Witte W, Rothgänger J, Claus H, Turnwald D, Vogel U. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442–5448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lina G, Piémont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, Vandenesch F, Etienne J. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128–1132 [DOI] [PubMed] [Google Scholar]
- 28. Bens CC, Voss A, Klaassen CH. 2006. Presence of a novel DNA methylation enzyme in methicillin-resistant Staphylococcus aureus isolates associated with pig farming leads to uninterpretable results in standard pulsed-field gel electrophoresis analysis. J. Clin. Microbiol. 44:1875–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. NCCLS 2004. Performance standards for antimicrobial disk susceptibility testing: 14th informational supplement, M100-S14. NCCLS, Wayne, PA [Google Scholar]
- 30. Lowy FD, Aiello AE, Bhat M, Johnson-Lawrence VD, Lee MH, Burrell E, Wright LN, Vasquez G, Larson EL. 2007. Staphylococcus aureus colonization and infection in New York State prisons. J. Infect. Dis. 196:911–918 [DOI] [PubMed] [Google Scholar]
- 31. Wassenberg MWM, Bootsma MCJ, Troelstra A, Kluytmans JAJW, Bonten MJM. 2011. Transmissibility of livestock-associated methicillin-resistant Staphylococcus aureus (ST398) in Dutch hospitals. Clin. Microbiol. Infect. 17:316–319 [DOI] [PubMed] [Google Scholar]
- 32. Graveland H, Wagenaar JA, Bergs K, Heesterbeek H, Heederik D. 2011. Persistence of livestock associated MRSA CC398 in humans is dependent on intensity of animal contact. PLoS One 6(2):e16830. 10.1371/journal.pone.0016830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. de Vries LE, Christensen H, Skov RL, Aarestrup FM, Agersø Y. 2009. Diversity of the tetracycline resistance gene tet(M) and identification of Tn916- and Tn5801-like (Tn6014) transposons in Staphylococcus aureus from humans and animals. J. Antimicrob. Chemother. 64:490–500 [DOI] [PubMed] [Google Scholar]
- 34. Lozano C, Rezusta A, Gómez P, Gómez-Sanz E, Báez N, Martin-Saco G, Zarazaga M, Torres C. 2012. High prevalence of spa types associated with the clonal lineage CC398 among tetracycline-resistant methicillin-resistant Staphylococcus aureus strains in a Spanish hospital. J. Antimicrob. Chemother. 67:330–334 [DOI] [PubMed] [Google Scholar]
- 35. Hauschild T, Feßler AT, Kadlec K, Billerbeck C, Schwarz S. 2012. Detection of the novel vga(E) gene in methicillin-resistant Staphylococcus aureus CC398 isolates from cattle and poultry. J. Antimicrob. Chemother. 67:503–504 [DOI] [PubMed] [Google Scholar]
- 36. Mendes RE, Smith TC, Deshpande L, Diekema DJ, Sader HS, Jones RN. 2011. Plasmid-borne vga(A)-encoding gene in methicillin-resistant Staphylococcus aureus ST398 recovered from swine and a swine farmer in the United States. Diagn. Microbiol. Infect. Dis. 71:177–180 [DOI] [PubMed] [Google Scholar]
- 37. Schwendener S, Perreten V. 2011. New transposon Tn6133 in methicillin-resistant Staphylococcus aureus ST398 contains vga(E), a novel streptogramin A, pleuromutilin, and lincosamide resistance gene. Antimicrob. Agents Chemother. 55:4900–4904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Price LB, Stegger M, Hasman H, Aziz M, Larsen J, Andersen PS, Pearson T, Waters AE, Foster JT, Schupp J, Gillece J, Driebe E, Liu CM, Springer B, Zdovc I, Battisti A, Franco A, Zmudzki J, Schwarz S, Butaye P, Jouy E, Pomba C, Porrero MC, Ruimy R, Smith TC, Robinson DA, Weese JS, Arriola CS, Yu F, Laurent F, Keim P, Skov R, Aarestrup FM. 2012. Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. mBio 3(1):e00305-11. 10.1128/mBio.00305-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vandendriessche S, Kadlec K, Schwarz S, Denis O. 2011. Methicillin-susceptible Staphylococcus aureus ST398-t571 harbouring the macrolide-lincosamide-streptogramin B resistance gene erm(T) in Belgian hospitals. J. Antimicrob. Chemother. 66:2455–2459 [DOI] [PubMed] [Google Scholar]
- 40. van Wamel WJ, Hansenová Maňásková S, Fluit AC, Verbrugh H, de Neeling AJ, van Duijkeren E, van Belkum A. 2010. Short term micro-evolution and PCR-detection of methicillin-resistant and -susceptible Staphylococcus aureus sequence type 398. Eur. J. Clin. Microbiol. Infect. Dis. 29:119–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Donker GA, Deurenberg RH, Driessen C, Sebastian S, Nys S, Stobberingh EE. 2009. The population structure of Staphylococcus aureus among general practice patients from The Netherlands. Clin. Microbiol. Infect. 15:137–143 [DOI] [PubMed] [Google Scholar]
- 42. Lozano C, Gómez-Sanz E, Benito D, Aspiroz C, Zarazaga M, Torres C. 2011. Staphylococcus aureus nasal carriage, virulence traits, antibiotic resistance mechanisms, and genetic lineages in healthy humans in Spain, with detection of CC398 and CC97 strains. Int. J. Med. Microbiol. 301:500–505 [DOI] [PubMed] [Google Scholar]
- 43. Hasman H, Moodley A, Guardabassi L, Stegger M, Skov RL, Aarestrup FM. 2010. spa type distribution in Staphylococcus aureus originating from pigs, cattle and poultry. Vet. Microbiol. 141:326–331 [DOI] [PubMed] [Google Scholar]
- 44. Riesen A, Perreten V. 2009. Antibiotic resistance and genetic diversity in Staphylococcus aureus from slaughter pigs in Switzerland. Schweiz. Arch. Tierheilkd. 151:425–431 [DOI] [PubMed] [Google Scholar]
- 45. Grundmann H, Aanensen DM, van den Wijngaard CC, Spratt BG, Harmsen D, Friedrich AW, European Staphylococcal Reference Laboratory Working Group 2010. Geographic distribution of Staphylococcus aureus causing invasive infections in Europe: a molecular-epidemiological analysis. Plos Med. 7:e1000215. 10.1371/journal.pmed.1000215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jiménez JN, Vélez LA, Mediavilla JR, Ocampo AM, Vanegas JM, Rodríguez EA, Kreiswirth BN, Correa MM. 2011. Livestock-associated methicillin susceptible Staphylococcus aureus ST398 infection in woman, Columbia. Emerg. Infect. Dis. 17:1970–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Uhlemann AC, Dumortier C, Hafer C, Taylor BS, Sánchez JE, Rodriguez-Taveras C, Leon P, Rojas R, Olive C, Lowy FD. 2012. Molecular characterization of Staphylococcus aureus from outpatients in the Caribbean reveals the presence of pandemic clones. Eur. J. Clin. Microbiol. Infect. Dis. 31:505–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Verkade E, Bergmans AM, Budding AE, van Belkum A, Savelkoul P, Buiting AG, Kluytmans J. 2012. Recent emergence of Staphylococcus aureus clonal complex 398 in human blood cultures. PLoS One 7:e41855. 10.1371/journal.pone.0041855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen H, Liu Y, Jiang X, Chen M, Wang H. 2010. Rapid change of methicillin-resistant Staphylococcus aureus clones in a Chinese tertiary care hospital over a 15-year period. Antimicrob. Agents Chemother. 54:1842–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wu D, Wang Q, Yang Y, Geng W, Wang Q, Yu S, Yao K, Yuan L, Shen X. 2010. Epidemiology and molecular characteristics of community-associated methicillin-resistant and methicillin-susceptible Staphylococcus aureus from skin/soft tissue infections in a children's hospital in Beijing, China. Diagn. Microbiol. Infect. Dis. 67:1–8 [DOI] [PubMed] [Google Scholar]
- 51. Salmenlinna S, Lyytikäinen O, Vainio A, Myllyniemi AL, Raulo S, Kanerva M, Rantala M, Thomson K, Seppänen J, Vuopio J. 2010. Human cases of methicillin-resistant Staphylococcus aureus CC398, Finland. Emerg. Infect. Dis. 16:1626–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cuny C, Nathaus R, Layer F, Strommenger B, Altmann D, Witte W. 2009. Nasal colonization of humans with methicillin-resistant Staphylococcus aureus (MRSA) CC398 with and without exposure to pigs. PLoS One 4(8):e6800. 10.1371/journal.pone.0006800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Huijsdens XW, Bosch T, van Santen-Verheuvel MG, Spalburg E, Pluister GN, van Luit M, Heck MEOC, Haenen A, de Neeling AJ. 2009. Molecular characterisation of PFGE non-typable methicillin-resistant Staphylococcus aureus in The Netherlands, 2007. Euro Surveill. 14:pii=19335. (Erratum, 15:pii=19515, 2010.) http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19335 [DOI] [PubMed] [Google Scholar]
- 54. van Cleef BA, Verkade EJM, Wulf MW, Buiting AG, Voss A, Huijsdens XW, van Pelt W, Mulders MN, Kluytmans JA. 2010. Prevalence of livestock-associated MRSA in communities with high pig-densities in The Netherlands. PLoS One 5(2):e9385. 10.1371/journal.pone.0009385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. van Loo I, Huijsdens X, Tiemersma E, de Neeling A, van de Sande-Bruinsma N, Beaujean D, Voss A, Kluytmans J. 2007. Emergence of methicillin-resistant Staphylococcus aureus of animal origin in humans. Emerg. Infect. Dis. 13:1834–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. van Rijen MM, Bosch T, Heck MEOC, Kluytmans JAJW. 2009. Meticillin-resistant Staphylococcus aureus epidemiology and transmission in a Dutch hospital. J. Hosp. Infect. 72:299–306 [DOI] [PubMed] [Google Scholar]
- 57. Graveland H, Wagenaar JA, Heesterbeek H, Mevius D, van Duijkeren E, Heederik D. 2010. Methicillin resistant Staphylococcus aureus ST398 in veal calf farming: human MRSA carriage related with animal antimicrobial usage and farm hygiene. PLoS One 5(6):e10990. 10.1371/journal.pone.0010990 [DOI] [PMC free article] [PubMed] [Google Scholar]


