Abstract
Pneumocystis jirovecii is an opportunistic pathogen in immunocompromised and AIDS patients. Detection by quantitative PCR is faster and more sensitive than microscopic diagnosis yet requires specific infrastructure. We adapted a real-time PCR amplifying the major surface glycoprotein (MSG) target from Pneumocystis jirovecii for use on the new BD MAX platform. The assay allowed fully automated DNA extraction and multiplex real-time PCR. The BD MAX assay was evaluated against manual DNA extraction and conventional real-time PCR. The BD MAX was used in the research mode running a multiplex PCR (MSG, internal control, and sample process control). The assay had a detection limit of 10 copies of an MSG-encoding plasmid per PCR that equated to 500 copies/ml in respiratory specimens. We observed accurate quantification of MSG targets over a 7- to 8-log range. Prealiquoting and sealing of the complete PCR reagents in conical tubes allowed easy and convenient handling of the BD MAX PCR. In a retrospective analysis of 54 positive samples, the BD MAX assay showed good quantitative correlation with the reference PCR method (R2 = 0.82). Cross-contamination was not observed. Prospectively, 278 respiratory samples were analyzed by both molecular assays. The positivity rate overall was 18.3%. The BD MAX assay identified 46 positive samples, compared to 40 by the reference PCR. The BD MAX assay required liquefaction of highly viscous samples with dithiothreitol as the only manual step, thus offering advantages for timely availability of molecular-based detection assays.
INTRODUCTION
Pneumocystis jirovecii is an opportunistic pathogen causing pneumonia in people suffering from AIDS (1). Moreover, it plays an increasing role as a pathogen in immunosuppressed patients and organ transplant recipients (2–4). Whereas detection of the fungus by immunofluorescent staining has long been considered the major diagnostic tool, it has now been documented consistently that molecular detection by PCR is superior to microscopic evaluation (1). Consequently, a number of PCR protocols, including nested PCR and real-time PCR, have been published and evaluated (5–19). Moreover, molecular detection of P. jirovecii in less-invasive patient specimens has also been shown to be cost-effective (20). The interpretation of PCR test results requires quantification because carriage of low numbers of P. jirovecii has been reported to occur without disease (1, 21). As the detection of a P. jirovecii infection has direct therapeutic impact for the choice of proper antibiotics, timely diagnosis is desirable. Yet availability of rapid molecular assays in routine laboratories, especially outside normal working hours, is still limited. Fully automated molecular platforms might facilitate timely and widespread application of molecular tests. The new BD MAX system (BD Diagnostics, Sparks, MD) allows fully automated DNA extraction, PCR setup, and multiplex real-time PCR for in vitro diagnostic (IVD) (22–24) as well as in-house-developed tests. The platform extracts DNA or RNA using specific extraction reagents, followed by real-time PCR amplification and detection of fluorescence in up to five channels. The system can be run in an open mode that allows adding any user-specific primers and PCR reagents. The eluted nucleic acid is then combined with those reagents and is transferred into a PCR microfluidic cartridge. Fully automated PCR without time-consuming and laborious hands-on work would be beneficial for rapid and easy detection of P. jirovecii. In this study, we aimed to adopt a published real-time PCR for detection of P. jirovecii (7) into the new BD MAX system. This is the first report that describes the usage of the BD MAX in the open system with user-specific PCR reagents.
MATERIALS AND METHODS
Samples.
The BD MAX PCR was validated with the 2011 external quality assessment quality control for molecular diagnostics (QCMD) panel (Qnostics, Glasgow, Scotland). For the retrospective analysis, extracted DNA from 54 patient samples which had previously (<2 years) tested positive for P. jirovecii by the routine real-time PCR were used. The prospective study was done from March to October 2012 at the 2,000-bed tertiary care University Hospital Heidelberg. A total of 278 samples from 188 patients with requests for P. jirovecii detection were analyzed by the reference PCR as well as the new BD MAX real-time PCR. For discrepant results, the assays were repeated once and patients were evaluated for P. jirovecii infection based on clinical signs (nonproductive cough, progressive hypoxia), radiological signs, including ground glass opacity and parenchyma consolidation, and initiation of a full course of treatment with high-dose trimethoprim-sulfamethoxazole.
Primers, probes, and plasmids.
Primers and dually labeled probes were ordered from Eurofins MWG Operon (Ebersberg, Germany). Primer stock solutions were prepared at 50 pmol/μl, except the sample process control (SPC), which was prepared at 10 pmol/μl. All probes were dissolved at 10 pmol/μl and stored at −20°C. The primer sequences (5′-3′) used were as follows for the P. jirovecii target gene MSG: MSG-fw, GAA TGC AAA TCC TTA CAG ACA ACA G, and MSG-rv, AAA TCA TGA ACG AAA TAA CCA TTG C (7). For detection, we used a dually labeled hydrolysis probe: MSG probe, FAM-AGA CAT CGA CAC ACA CAA GCA CGT CT-BHQ1. The internal control probe was TET or Texas Red-CTA GCA GCA CGC CAT AGT GAC TGG C-BHQ2. The sample process control primers were SPC-fw, GGA TCT AGC CGT GTG CCC GCT, and SPC-rv, GGC ATG GAG GTT GTC CCA TTT GTG, with the SPC probe Cy5-TTG ATG CCT CTT ACA TTG CTC CAC CTT TCC T-BHQ2. A positive-control plasmid containing a P. jirovecii HuMSG14 major surface glycoprotein gene (GenBank accession number AF033209) cloned into pCR2.1 was used and quantitated by NanoDrop (Peqlab, Erlangen, Germany) spectrophotometry. As an internal control, a tetracycline resistance gene of pBR322 encompassed by MSG primer sequences was prepared. The internal control was added to the PCR mix to control for PCR inhibition. The MSG primers amplify a 250-bp product from the target gene and a 295-bp product from the internal control that can be differentiated using the MSG probe and the internal control probe, respectively. The plasmids were obtained from H. H. Larsen (7). The sample process control is included in the BD MAX ExK DNA-2 kit (BD Diagnostics) and carries the Drosophila melanogaster scaffold protein gene (GenBank accession number AC246497.1) cloned in a pUC119 vector sequence (GenBank accession number U07650).
DNA extraction.
For routine diagnostics, DNA was extracted from 200 μl of the patient's sputum or tracheal secretion sample using the QIAamp DNA blood minikit (Qiagen, Hilden, Germany) without any preincubation. DNA was eluted in 100 μl of the kit's AE buffer. For bronchoalveolar lavage (BAL) fluid and high-volume samples, 1,000 μl was centrifuged at 8,000 rpm for 5 min, 800 μl supernatant was removed, and the sample was resuspended prior to DNA extraction. The 5-fold concentration by this pretreatment was considered when calculating target concentrations.
Reference real-time PCR.
The real-time PCR (7) was done as follows. The 25-μl PCR mixtures contained 2× quantitative PCR (qPCR) master mix (ABsolute qPCR mix [no ROX]; Thermo Fisher, Schwerte, Germany), 1 μM each MSG-fw and MSG-rv primer, 150 nM 6-carboxyfluorescein (FAM)-labeled MSG probe, 150 nM TET-labeled internal control probe, 2 × 104 copies of internal control plasmid, and 2 μl of the sample DNA extract. The PCR was run on a Chromo4 cycler (Bio-Rad, Munich, Germany) at 95°C for 15 min with 45 cycles of 95°C for 15 s and 60°C for 40 s. With each run, 102, 104, and 106 copies/PCR of a plasmid encoding the MSG target were run in parallel for quantification.
Preparation of BD MAX PCR reagents.
A 2-fold concentrated master mix in 12 μl was prepared as follows: 4.8 μl of real-time ready DNA probe master 5× (number 05502381001; Roche Applied Science, Mannheim, Germany), 0.25 μl each of MSG-fw and MSG-rv primers (1 μM in the 2-fold concentrated master mix) and SPC-fw and SPC-rv primers (0.2 μM), 1 μl each of FAM-labeled MSG probe, Texas Red-labeled internal control probe, and Cy5-labeled SPC probe (0.8 μM), 1 μl (equaling 1,000 copies) of internal control plasmid, and 2.2 μl of PCR-grade water. The master mix was prepared as a stock solution, aliquoted, and stored at −20°C for up to 3 months. Additionally, master mix was aliquoted at 12 μl directly into conical snap-in tubes fitting the BD MAX extraction strip and sealed using adhesive cover foil and a PlateMax sealer (Axygen; 2 times for 8 s each at 180°C).
BD MAX PCR.
For high-volume specimens, 2 ml was centrifuged (8,000 rpm for 5 min) and resuspended in 400 μl. Other specimens were filled up to a 400-μl volume with phosphate-buffered saline (PBS). Samples were split, and 200 μl each was analyzed by either manual DNA extraction and reference real-time PCR or the BD MAX system. Samples were analyzed within 3 days after receipt. Viscous samples were pretreated by mixing 200 μl of the sample with 200 μl of dithiothreitol (Sputasol, 1:1; Oxoid, Wesel, Germany), followed by incubation at 37°C for 30 min. A total of 200 μl of the specimen was pipetted into the sample buffer tube (SBT) of the BD MAX ExK DNA-2 kit. The SBTs were covered with a septum cap, vortexed briefly, and placed into the sample rack. Unitized reaction strips of the BD MAX ExK DNA-2 kit were supplemented with the 2-fold concentrated BD MAX PCR master mix. After extraction, the BD MAX combines the 12 μl of nucleic acid eluate with the user-provided PCR master mix and 12 μl of the mixture is then loaded into the PCR cartridge. Approximately 4 μl of the reaction mixture will be in the amplification chamber. The assay was run in the research mode using default settings for the extraction and the following PCR cycling protocol: 95°C for 60 s and 45 cycles of 98°C for 8 s and 58°C for 16.3 s. Fluorescence gains and thresholds were set for FAM (475/520 nm) at 40 and 50 (where the first value is the setting for the fluorescence gain and the second value is the setting for the threshold), VIC (530/565 nm) at 80 and 50, Texas Red (585/630 nm) at 80 and 50, Cy5 (630/665 nm) at 80 and 75, and Cy5.5 (680/715 nm) at 80 and 10. A color compensation matrix was programmed with a FAM/VIC of 0.05 and a Texas Red/Cy5 of 0.023. The Roche PCR mix has a fluorescent dye as pipetting control signaling in the Cy5.5 channel. Quantitation was done using a plasmid standard that was run once and saved as an external standard. For analysis in the PCR-only mode, 8 μl of the 2-fold concentrated mix was combined with 4 μl DNA or plasmid standard and 4 μl of 20 mM NaOH to mimic the extraction process. The cartridge was loaded with 10 μl of this mixture.
Statistical analysis.
Calculations, including a determination of linear regression, were done with Prism5 (GraphPad Software). Statistical differences between groups were analyzed by a two-tailed unpaired t test or Mann-Whitney test as indicated.
RESULTS
Technical adoption of a conventional real-time PCR on the BD MAX.
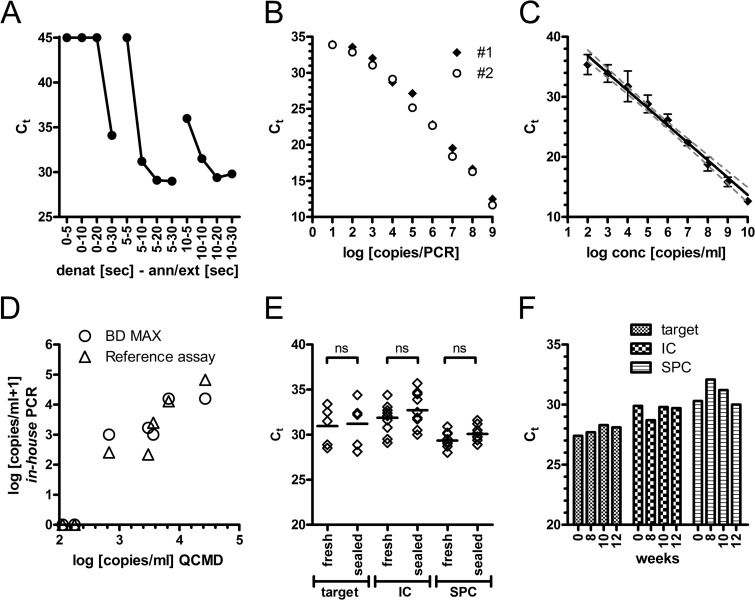
For initial optimization procedures, DNA from P. jirovecii-positive samples was added to the BD MAX PCR mixture and then 10 μl was manually loaded into the PCR cartridges and the PCR was run in the PCR-only mode. We observed that BD MAX delivered better results (i.e., lower threshold cycles [CT] values) when increasing the denaturing temperature to 98°C (data not shown). No major differences were observed when changing the annealing temperature up or down 6°C from the 60°C used on the Chromo4 cycler. As the BD MAX elutes DNA in NaOH, it is recommended to use a neutralizing buffer when preparing the PCR mix. We observed that for the particular enzyme used in this study, the neutralization step was able to be omitted since amplification worked equally well when we simulated extraction by adding the DNA in water (CT, 28.6 ± 0.54), the DNA in NaOH (CT, 29.0 ± 0.05), and the DNA in NaOH and neutralization buffer (CT, 29.6 ± 0.17). Next, we optimized amplification times. We observed the best amplification when the denaturing time exceeded 5 s and annealing/extension was above 10 s (Fig. 1A). Then we assessed the detection limit of the PCR by applying a 10-fold serial dilution of a plasmid carrying the MSG target gene (Fig. 1B). We observed that 10 copies added with the supplemented PCR mix to the PCR cartridge (equaling 4 copies within the reaction chamber) were able to be detected and that the PCR had an amplification range of 7 logs. Ten copies in the PCR equal approximately 230 copies/ml in the original material if 200 μl of specimen is added to 1,500 μl of buffer in the SBT.
Fig 1.
Technical evaluation of the BD MAX PCR. (A) The BD MAX PCR was run in the PCR-only mode, and the indicated combinations of denaturation and annealing/extension times (two-step protocol) were tested with 6 log10 copies of a plasmid standard. (B) Serial dilutions of an MSG-encoding plasmid standard were evaluated in the PCR-only mode in duplicates. CT values of the MSG target gene amplification (FAM) are plotted against the plasmid copy numbers within the PCR. (C) A total of 200 μl of serial dilutions of the plasmid standard in three negative tested BAL fluid matrices was evaluated for automated DNA extraction and PCR amplification. Mean (± standard deviation [SD]) CT values of the MSG target gene amplification (FAM) are shown (n = 3). (D) The reference and BD MAX PCRs were used to evaluate the 2011 QCMD quality panel. Concentrations determined by use of a plasmid standard are plotted against the reference data. (E) PCR reagents were either tested immediately (fresh) or aliquoted, sealed, and stored overnight at −20°C. Ten patient samples (5 positive, 5 negative) were tested with both preparations. Median and individual CT data for the target MSG gene (FAM), internal control (Texas Red), and sample process control (Cy5) are shown. An unpaired t test showed no significant differences (ns). (F) A 5 log10 plasmid standard was tested repeatedly over 12 weeks with stored PCR reagents (target, MSG; IC, internal control; SPC, sample process control).
Fully automated extraction and real-time PCR.
We next evaluated the sensitivity of the full process on the BD MAX encompassing DNA extraction, PCR setup, and real-time PCR. We used a multiplex PCR that amplified the MSG target gene (FAM), an internal control (IC) (Texas Red), and a sample process control (SPC) (Cy5). The internal control plasmid was added at a low copy number to avoid competition with amplification of the natural MSG target.
Valid negative results were reported for target gene-negative samples when the SPC was positive with any CT value and the internal control showed a CT value of <36.5. For target gene-positive samples, the SPC and IC were not evaluated but the sample was quantitated according to a recorded external standard.
Using serial dilutions of the MSG plasmid, we observed a detection limit of 200 to 400 copies/ml (data not shown), indicating high efficacy of the extraction process compared to the PCR-only mode. Next we spiked negative BAL material with an MSG-encoding plasmid (Fig. 1C). The detection limit was about 500 copies/ml (2.70 log10/ml), with linear amplification over at least a 7- to 8-log range, and similar results were obtained for tracheal secretions (data not shown). One thousand copies/ml were consistently detected (100%), whereas 100 copies/ml in matrix gave a positive signal in 33% of the tested samples. We did not observe cross-contamination between neighboring samples when alternating positive (9 log10) and negative samples were examined. Using an external quality assessment panel, both PCRs, the reference real-time PCR and the BD MAX PCR, missed two infrequently detected samples but identified five positive samples with sufficient quantitative accuracy (Fig. 1D) (R2 = 0.93 for the reference procedure; R2 = 0.85 for the BD MAX PCR).
To further simplify handling, we prepared ready-to-use PCR reagents that were aliquoted, sealed, and stored at −20°C. This process did not affect the activity of the PCR (Fig. 1E). Further analysis testing a 5-log10 plasmid standard over 3 months with stored master mix proved sufficient stability of the complemented PCR reagents (Fig. 1F).
Quantitative correlation of conventional real-time PCR and BD MAX PCR.
In a retrospective analysis, 54 samples which previously tested positive for P. jirovecii by the reference real-time PCR and from which extracted DNA had been stored at −20°C were tested with the BD MAX PCR. A total of 46 samples were identified as positive in the BD MAX PCR. The remaining 8 discrepant samples were reevaluated by repeating the original reference real-time PCR and turned out to be negative in repeat testing. These eight samples had low-positive results at the time of initial testing and most probably did not give reproducible results due to long-term storage. Excluding those nonreproducible samples, quantitative comparison of original reference results and BD MAX real-time PCR results showed correlations of 0.76 for all 46 positive samples and 0.82 when one outlier was removed (Fig. 2).
Fig 2.
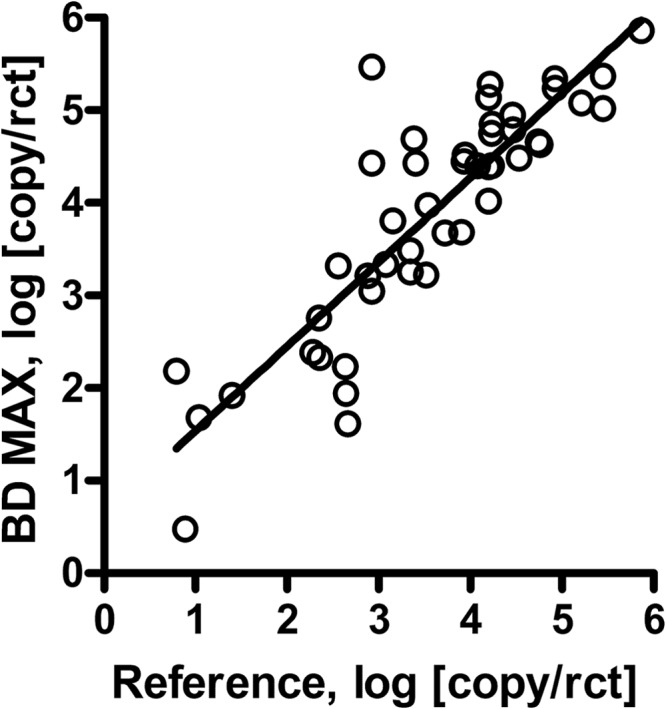
Comparison of MSG quantifications by the BD MAX and the reference PCR. Extracted DNA from 46 samples that had tested positive by the reference procedure was evaluated by the BD MAX PCR in the PCR-only mode. Quantification of the MSG target was based upon the CT values and an external standard of serial dilutions of a plasmid and is indicated as copy numbers/PCR. Linear regression was determined, and the correlation graph is plotted.
Evaluation of the BD MAX PCR in a prospective study.
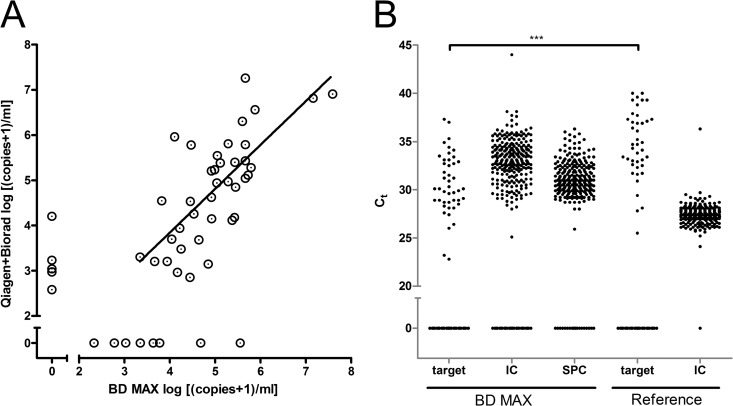
In a prospective study, 278 respiratory samples were analyzed by both assays. Of those, 18.3% (51) were positive by at least one molecular assay. The BD MAX PCR yielded 213 negative and 44 positive results; 10 samples were unresolved due to failure in detection of the internal control, and 11 samples were not able to be evaluated due to technical failures (accounting for 7.5% of the samples with no result upon first-run analysis) (Table 1). Unresolved tests on the BD MAX were repeated once with only 3 samples, giving no final result (Table 1). The reference procedure consisting of manual DNA extraction and conventional real-time PCR gave 238 negative and 40 positive results; none of the samples were unresolved. Direct comparison showed that both PCRs were concordant for 35 positive and 227 negative samples. Eleven samples were positive only by the BD MAX PCR, and 5 samples were positive only by the reference PCR (Table 2). The quantitative results showed a sufficient correlation given the heterogeneity of the materials included (Fig. 3A).
Table 1.
Results of BD MAX and reference PCRsa
| Result | No. of samples with each result by: |
|
|---|---|---|
| BD MAX PCR | Reference procedure | |
| First result | ||
| Negative | 213 | 238 |
| Positive | 44 | 40 |
| Unresolved | 10 | 0 |
| Technical failure | 11 | 0 |
| Final result | ||
| Negative | 229 | 238 |
| Positive | 46 | 40 |
| Unresolved | 3 | 0 |
| Total | 278 | 278 |
Indicated are the interpreted results of the BD MAX and the reference PCRs after the first run and upon single retesting of failed assays.
Table 2.
BD MAX and reference PCRs in comparison
| Result by reference procedure | No. of samples with each BD MAX PCR result |
|
|---|---|---|
| Positive | Negative/unresolved | |
| Positive | 35 | 5 |
| Negative | 11 | 227 |
Fig 3.
Prospective evaluation of the automated BD MAX PCR. (A) Clinical samples were tested for presence of the Pneumocystis jirovecii MSG target gene and quantified according to an external plasmid standard. Log concentrations of positive samples are indicated (log [{copies + 1}/ml]; zero refers to absence of detection). Discrepant results were reanalyzed once. Concentrations below 3 log10 and 2.7 log10 per ml for the reference and the BD MAX method, respectively, were extrapolated. Linear regression was determined, and the correlation graph is plotted. (B) Distribution of CT values of the different amplicons as detected in the BD MAX real-time PCR and the reference procedure (target, MSG; IC, internal control; SPC, sample process control; 0, negative). Target amplicon CT values were compared by a Mann-Whitney test (***, P < 0.0001).
Among the five samples that were missed in the BD MAX PCR, four had low concentrations of P. jirovecii. One sample with a higher concentration had been analyzed only with supernatant of the BAL fluid by BD MAX PCR. In a second sample from the same patient, both PCRs were positive. Among the remaining four patients, three were judged negative and one was judged positive for Pneumocystis jirovecii infections according to clinical findings.
For the 11 samples that were positive only in the BD MAX PCR, we observed a distribution of 2.34 log10 to 5.56 log10 copies/ml, results that were well above the detection limit (Fig. 3A). Upon retesting with the reference PCR for discrepancy analysis, three were positive with low concentrations and eight remained negative. Of the 8 repeat negative samples, two were from patients that had positive results by both PCRs in additional samples whereas one patient was negative in both PCRs in a second sample. For the remaining five patients that were positive only by the BD MAX PCR, one was determined to be Pneumocystis pneumonia based on clinical findings (in a patient with systemic inflammatory disease and immunosuppression), and for another patient, trimethoprim-sulfamethoxazole therapy was initiated by the clinicians despite the negative reference PCR. Of the five patients with positive results in repeat or secondary testing, diagnosis of P. jirovecii infection was made for two patients and the other three suffered from atypical pneumonia without a confirmed microbiological diagnosis. Taken together, the BD MAX PCR missed one patient but identified an additional three patients with Pneumocystis jirovecii infection.
A comparison of the CT values of both PCRs showed that the target gene was detected with a median CT value of 30.1 for the BD MAX PCR whereas the median CT value for the combination of manual extraction and reference PCR was higher, at 34.7 (Fig. 3B). Due to the design of the BD MAX PCR (amplifying only 160 copies of the internal control plasmid within the reaction chamber), the internal control was expectedly detected at higher CT values than in the reference procedure.
DISCUSSION
Rapid detection of respiratory infections with P. jirovecii is of importance to initiate early and proper treatment of this emerging pathogen. In this report, we show the successful merging of a published real-time PCR (7) for detection of P. jirovecii on the new automated BD MAX platform. We used a quantitative real-time PCR that was initially published as touchdown PCR using fluorescence resonance energy transfer (FRET) hybridization probes. The PCR was run in our routine diagnostic using a conventional protocol and a hydrolysis probe. The limit of detection was about 1,000 copies/ml of a plasmid in BAL fluid matrix that equaled 4 copies/PCR. The detection limit was in the range that had been published (5 copies/reaction). Whereas initially a cutoff of 10 copies/PCR (referring to 3.40 log10 copies/ml in the sample) was suggested to discriminate patients suffering from Pneumocystis infection from patients with asymptomatic carriage (7), a later report suggested 1,000 copies/reaction (equaling 5.40 log10 copies/ml) as more appropriate (9). In our diagnostic laboratory, signals below 5.30 log10 copies/ml were routinely reported as borderline. Low-positive PCR results have been repeatedly observed and have been interpreted as carriage or colonization. Therefore, a most-sensitive PCR does not seem to be of utmost importance; instead, a proper quantification and establishment of an in-house cutoff are mandatory.
To facilitate the handling of this PCR and to make it available for use apart from the normal working hours, we thought to migrate this assay onto the fully automated BD MAX platform run in the open mode. We successfully established a protocol that worked well on the BD MAX. The PCR reagents were delivered by the user into the DNA extraction strip of the commercial ExK DNA-2 kit; all other steps were done by the instrument. In the case of a viscous sample, a pretreatment with dithiothreitol was the only manual step necessary. Adopting the PCR, we observed that annealing and extension times were able to be decreased substantially, possibly due to the microfluidic buildup. Modulating annealing/extension temperatures had few effects. In general, however, we observed that higher denaturing temperatures of up to 98°C were beneficial. The PCR itself had a detection limit of 4 copies/PCR chamber, well in the range of the original PCR (7) as well as other real-time PCRs (10, 12, 15).
The combined processes of automated DNA extraction and multiplex PCR had a detection limit of approximately 500 copies (2.70 log10) of an MSG-encoding plasmid per ml in BAL matrix (9 copies/PCR chamber). Although both PCRs missed two infrequently detected samples in the 2011 QCMD panel, the detection limit is below what has been published as a clinically relevant cutoff (14, 25). Both PCR assays showed good correlation in quantification to the QCMD quality panel as well as in a retrospective comparison of previously positive tested samples.
To further facilitate the PCR handling, we aliquoted fully complemented PCR reagents, sealed them, and stored them at −20°C. The sealing process itself did not affect the PCR quality, and over a time period of 3 months, only a slight reduction in amplification efficacy was observed. This protocol allows use of the whole system outside a specific PCR area, as all reagents have been prealiquoted and the amplification process is closed.
In a prospective study, the BD MAX PCR had an initial failure rate of 7.5%; however, only 3.6% were due to PCR inhibition whereas 3.9% were because of a technical problem which occurred only once during the entire study. Upon single retesting, 1.1% remained unresolved. We observed a slightly weaker quantitative correlation between the two PCRs which might be attributable to the heterogeneity of BAL fluid, tracheal secretions, and sputa. Limited reproducibility of quantitative results in these specimens has been discussed before (14). Nonetheless, discordant results were limited in number, and samples that were missed by the BD MAX protocol were mainly low-positive samples. In contrast, the BD MAX identified additional samples among which were a number of true positives. For the PCR targeting the MSG gene, two cutoffs have been suggested, 10 copies/PCR (7) and 1,000 copies/PCR (9), with the latter increasing specificity from 84.9 to 98.6%. Using the thresholds of 3.40 log10 and 5.40 log10 copies/ml that equal the published cutoffs with the protocol applied here, the BD MAX PCR would have identified 41 and 14, respectively, of 46 samples as being above the threshold whereas the reference real-time PCR would have identified 33 and 12, respectively, of 40 samples as being above the threshold. Without providing a detailed analysis of the clinical findings for all patients, the discrepancy analysis already identified five patients with clinical findings of a Pneumocystis infection that were below the 5.40 log10/ml threshold. Our patient cohort consisted mainly of solid organ transplant recipients, patients with hematological malignancies, and patients with systemic inflammatory disease, known to be at risk for Pneumocystis jirovecii infection (26), but lacked a high proportion of HIV patients. The data argue for the need of establishment of an in-house cutoff value, but a cutoff of 10 copies/PCR seems to be more appropriate (14). Ten copies/reaction were also found to result in 98% sensitivity and 96% specificity in receiver-operator curve analysis for detection of hsp70 (15), and 30 copies/reaction were needed for targeting the internal transcribed spacer 2 (ITS2) region (17). Indeed, it is known that non-HIV patients have lower fungal burden (27, 28), and differentiation of carriage versus infection in non-HIV patients might therefore be even more difficult (29). In this line, using beta-tubulin as a target gene, microscopically positive samples were reported to have a burden of 3.6 to 6.6 log10 copies/ml whereas microscopically negative samples had a lower burden of 0.9 to 3.9 log10/ml (10). A gray zone of unclear clinical significance, as opposed to infection or colonization, was also suggested within the range of 2.08 and 3.28 log10 copies of mitochondrial large-subunit rRNA gene per ml, which is in the range of the above-mentioned cutoff of 3.40 log10/ml (30), or 0 to 3.16 log10 PCR pathogens/ml (29). Besides the mere determination of Pneumocystis target gene concentrations, it has been suggested to normalize those values to host DNA to account for sample heterogeneity (14).
On the other hand, multicenter studies have shown that the negative predictive value of real-time PCR in Pneumocystis diagnostics is high (99%); thus, a rapid negative report is helpful to exclude Pneumocystis jirovecii infection (29, 31, 32).
The specific clinical use of PCR is confirmed by a recent report in which immunofluorescence assay (IFA)-negative but PCR-positive patients suffering from systemic inflammatory disease had a worse 1-year survival rate than IFA-negative/PCR-negative patients (25). However, this difference was not observed for other patient groups. In this report, IFA-positive patients had PCR loads of at least 5.6 log10/ml and IFA-negative/PCR-positive patients had a mean load of 4.1 log10/ml, and among the latter, a number of patients had diffuse radiological patterns indicative of possible Pneumocystis pneumonia.
In order to make a timely decision, it would be preferable to have radiological, laboratory, and microbiological PCR findings available at once. The new assay presented here provided an easier workflow, thus offering advantages for application of molecular-based detection assays. The assay required liquefaction of highly viscous samples but otherwise needed no further pipetting step, thus improving handling significantly. The BD MAX Pneumocystis PCR assay showed equal technical performance to and good quantitative correlation with the established reference real-time PCR.
ACKNOWLEDGMENTS
A.H.D. and S.Z. have received a speaker's honorarium from BD Diagnostics.
BD Diagnostics supported the study by providing assay kits but had no influence on design and evaluation.
Footnotes
Published ahead of print 15 May 2013
REFERENCES
- 1. Thomas CF, Jr, Limper AH. 2004. Pneumocystis pneumonia. N. Engl. J. Med. 350:2487–2498 [DOI] [PubMed] [Google Scholar]
- 2. Sowden E, Carmichael AJ. 2004. Autoimmune inflammatory disorders, systemic corticosteroids and pneumocystis pneumonia: a strategy for prevention. BMC Infect. Dis. 4:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li MC, Lee NY, Lee CC, Lee HC, Chang CM, Ko WC. 10 October 2012. Pneumocystis jiroveci pneumonia in immunocompromised patients: delayed diagnosis and poor outcomes in non-HIV-infected individuals. J. Microbiol. Immunol. Infect. 10.1016/j.jmii.2012.08.024 [DOI] [PubMed] [Google Scholar]
- 4. Roblot F, Godet C, Le Moal G, Garo B, Faouzi Souala M, Dary M, De Gentile L, Gandji JA, Guimard Y, Lacroix C, Roblot P, Becq-Giraudon B. 2002. Analysis of underlying diseases and prognosis factors associated with Pneumocystis carinii pneumonia in immunocompromised HIV-negative patients. Eur. J. Clin. Microbiol. Infect. Dis. 21:523–531 [DOI] [PubMed] [Google Scholar]
- 5. Kaiser K, Rabodonirina M, Picot S. 2001. Real time quantitative PCR and RT-PCR for analysis of Pneumocystis carinii hominis. J. Microbiol. Methods 45:113–118 [DOI] [PubMed] [Google Scholar]
- 6. Palladino S, Kay I, Fonte R, Flexman J. 2001. Use of real-time PCR and the LightCycler system for the rapid detection of Pneumocystis carinii in respiratory specimens. Diagn. Microbiol. Infect. Dis. 39:233–236 [DOI] [PubMed] [Google Scholar]
- 7. Larsen HH, Masur H, Kovacs JA, Gill VJ, Silcott VA, Kogulan P, Maenza J, Smith M, Lucey DR, Fischer SH. 2002. Development and evaluation of a quantitative, touch-down, real-time PCR assay for diagnosing Pneumocystis carinii pneumonia. J. Clin. Microbiol. 40:490–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Meliani L, Develoux M, Marteau-Miltgen M, Magne D, Barbu V, Poirot JL, Roux P. 2003. Real time quantitative PCR assay for Pneumocystis jirovecii detection. J. Eukaryot. Microbiol. 50(Suppl):651. [DOI] [PubMed] [Google Scholar]
- 9. Flori P, Bellete B, Durand F, Raberin H, Cazorla C, Hafid J, Lucht F, Sung RT. 2004. Comparison between real-time PCR, conventional PCR and different staining techniques for diagnosing Pneumocystis jiroveci pneumonia from bronchoalveolar lavage specimens. J. Med. Microbiol. 53:603–607 [DOI] [PubMed] [Google Scholar]
- 10. Brancart F, Rodriguez-Villalobos H, Fonteyne PA, Peres-Bota D, Liesnard C. 2005. Quantitative TaqMan PCR for detection of Pneumocystis jiroveci. J. Microbiol. Methods 61:381–387 [DOI] [PubMed] [Google Scholar]
- 11. Strutt M, Smith M. 2005. Development of a real-time probe-based PCR assay for the diagnosis of Pneumocystis pneumonia. Med. Mycol. 43:343–347 [DOI] [PubMed] [Google Scholar]
- 12. Arcenas RC, Uhl JR, Buckwalter SP, Limper AH, Crino D, Roberts GD, Wengenack NL. 2006. A real-time polymerase chain reaction assay for detection of Pneumocystis from bronchoalveolar lavage fluid. Diagn. Microbiol. Infect. Dis. 54:169–175 [DOI] [PubMed] [Google Scholar]
- 13. Robberts FJ, Liebowitz LD, Chalkley LJ. 2007. Polymerase chain reaction detection of Pneumocystis jiroveci: evaluation of 9 assays. Diagn. Microbiol. Infect. Dis. 58:385–392 [DOI] [PubMed] [Google Scholar]
- 14. Bandt D, Monecke S. 2007. Development and evaluation of a real-time PCR assay for detection of Pneumocystis jiroveci. Transpl. Infect. Dis. 9:196–202 [DOI] [PubMed] [Google Scholar]
- 15. Huggett JF, Taylor MS, Kocjan G, Evans HE, Morris-Jones S, Gant V, Novak T, Costello AM, Zumla A, Miller RF. 2008. Development and evaluation of a real-time PCR assay for detection of Pneumocystis jirovecii DNA in bronchoalveolar lavage fluid of HIV-infected patients. Thorax 63:154–159 [DOI] [PubMed] [Google Scholar]
- 16. Rohner P, Jacomo V, Studer R, Schrenzel J, Graf JD. 2009. Detection of Pneumocystis jirovecii by two staining methods and two quantitative PCR assays. Infection 37:261–265 [DOI] [PubMed] [Google Scholar]
- 17. Fujisawa T, Suda T, Matsuda H, Inui N, Nakamura Y, Sato J, Toyoshima M, Nakano Y, Yasuda K, Gemma H, Hayakawa H, Chida K. 2009. Real-time PCR is more specific than conventional PCR for induced sputum diagnosis of Pneumocystis pneumonia in immunocompromised patients without HIV infection. Respirology 14:203–209 [DOI] [PubMed] [Google Scholar]
- 18. Wilson JW, Limper AH, Grys TE, Karre T, Wengenack NL, Binnicker MJ. 2011. Pneumocystis jirovecii testing by real-time polymerase chain reaction and direct examination among immunocompetent and immunosuppressed patient groups and correlation to disease specificity. Diagn. Microbiol. Infect. Dis. 69:145–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Orsi CF, Gennari W, Venturelli C, La Regina A, Pecorari M, Righi E, Machetti M, Blasi E. 2012. Performance of 2 commercial real-time polymerase chain reaction assays for the detection of Aspergillus and Pneumocystis DNA in bronchoalveolar lavage fluid samples from critical care patients. Diagn. Microbiol. Infect. Dis. 73:138–143 [DOI] [PubMed] [Google Scholar]
- 20. Harris JR, Marston BJ, Sangrujee N, DuPlessis D, Park B. 2011. Cost-effectiveness analysis of diagnostic options for pneumocystis pneumonia (PCP). PLoS One 6:e23158. 10.1371/journal.pone.0023158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Peterson JC, Cushion MT. 2005. Pneumocystis: not just pneumonia. Curr. Opin. Microbiol. 8:393–398 [DOI] [PubMed] [Google Scholar]
- 22. Dalpke AH, Hofko M, Zimmermann S. 2012. Comparison of the BD Max methicillin-resistant Staphylococcus aureus (MRSA) assay and the BD GeneOhm MRSA achromopeptidase assay with direct- and enriched-culture techniques using clinical specimens for detection of MRSA. J. Clin. Microbiol. 50:3365–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Le Guern R, Herwegh S, Grandbastien B, Courcol R, Wallet F. 2012. Evaluation of a new molecular test, the BD Max Cdiff, for detection of toxigenic Clostridium difficile in fecal samples. J. Clin. Microbiol. 50:3089–3090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Riedlinger J, Beqaj SH, Milish MA, Young S, Smith R, Dodd M, Hankerd RE, Lebar WD, Newton DW. 2010. Multicenter evaluation of the BD Max GBS assay for detection of group B streptococci in prenatal vaginal and rectal screening swab specimens from pregnant women. J. Clin. Microbiol. 48:4239–4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Botterel F, Cabaret O, Foulet F, Cordonnier C, Costa JM, Bretagne S. 2012. Clinical significance of quantifying Pneumocystis jirovecii DNA by using real-time PCR in bronchoalveolar lavage fluid from immunocompromised patients. J. Clin. Microbiol. 50:227–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rodriguez M, Fishman JA. 2004. Prevention of infection due to Pneumocystis spp. in human immunodeficiency virus-negative immunocompromised patients. Clin. Microbiol. Rev. 17:770–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Limper AH, Offord KP, Smith TF, Martin WJ., II 1989. Pneumocystis carinii pneumonia. Differences in lung parasite number and inflammation in patients with and without AIDS. Am. Rev. Respir. Dis. 140:1204–1209 [DOI] [PubMed] [Google Scholar]
- 28. Fillaux J, Malvy S, Alvarez M, Fabre R, Cassaing S, Marchou B, Linas MD, Berry A. 2008. Accuracy of a routine real-time PCR assay for the diagnosis of Pneumocystis jirovecii pneumonia. J. Microbiol. Methods 75:258–261 [DOI] [PubMed] [Google Scholar]
- 29. Muhlethaler K, Bogli-Stuber K, Wasmer S, von Garnier C, Dumont P, Rauch A, Muhlemann K, Garzoni C. 2012. Quantitative PCR to diagnose Pneumocystis pneumonia in immunocompromised non-HIV patients. Eur. Respir. J. 39:971–978 [DOI] [PubMed] [Google Scholar]
- 30. Alanio A, Desoubeaux G, Sarfati C, Hamane S, Bergeron A, Azoulay E, Molina JM, Derouin F, Menotti J. 2011. Real-time PCR assay-based strategy for differentiation between active Pneumocystis jirovecii pneumonia and colonization in immunocompromised patients. Clin. Microbiol. Infect. 17:1531–1537 [DOI] [PubMed] [Google Scholar]
- 31. Hauser PM, Bille J, Lass-Florl C, Geltner C, Feldmesser M, Levi M, Patel H, Muggia V, Alexander B, Hughes M, Follett SA, Cui X, Leung F, Morgan G, Moody A, Perlin DS, Denning DW. 2011. Multicenter, prospective clinical evaluation of respiratory samples from subjects at risk for Pneumocystis jirovecii infection by use of a commercial real-time PCR assay. J. Clin. Microbiol. 49:1872–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fillaux J, Berry A. 2013. Real-time PCR assay for the diagnosis of Pneumocystis jirovecii pneumonia. Methods Mol. Biol. 943:159–170 [DOI] [PubMed] [Google Scholar]