Abstract
We previously showed that culture of samples obtained by prosthesis vortexing and sonication was more sensitive than tissue culture for prosthetic joint infection (PJI) diagnosis. Despite improved sensitivity, culture-negative cases remained; furthermore, culture has a long turnaround time. We designed a genus-/group-specific rapid PCR assay panel targeting PJI bacteria and applied it to samples obtained by vortexing and sonicating explanted hip and knee prostheses, and we compared the results to those with sonicate fluid and periprosthetic tissue culture obtained at revision or resection arthroplasty. We studied 434 subjects with knee (n = 272) or hip (n = 162) prostheses; using a standardized definition, 144 had PJI. Sensitivities of tissue culture, of sonicate fluid culture, and of PCR were 70.1, 72.9, and 77.1%, respectively. Specificities were 97.9, 98.3, and 97.9%, respectively. Sonicate fluid PCR was more sensitive than tissue culture (P = 0.04). PCR of prosthesis sonication samples is more sensitive than tissue culture for the microbiologic diagnosis of prosthetic hip and knee infection and provides same-day PJI diagnosis with definition of microbiology. The high assay specificity suggests that typical PJI bacteria may not cause aseptic implant failure.
INTRODUCTION
Accurate prosthetic joint infection (PJI) diagnosis is important, since revision arthroplasty management depends on the presence or absence of infection (1). Diagnosis involves assessment for purulence, sinus tract, histopathology, inflammatory markers, synovial fluid cells/differential, and/or isolation of the microorganism(s) from multiple tissue cultures or from sonicate fluid culture (1). Only cultures define microbiology, which is important for directing antimicrobial therapy.
The sensitivity of tissue culture is 61 to 65% (2, 3). PJI microorganisms are in biofilms on the prosthesis surface. We previously demonstrated that culture of material dislodged from the implant surface after vortexing and sonication is more sensitive than tissue culture (3–5). Despite improved sensitivity, culture-negative cases remained, and culture is slow (up to 14 days).
Compared to culture, PCR is theoretically more sensitive, faster, and not as affected by treatment. Several investigators have preliminarily evaluated PCR for PJI diagnosis (2, 6–14), and most have used 16S rRNA gene (rRNA) PCR (7–14). This method has limitations since the bacteria detected are not identified (unless sequenced), results may be nonspecific (due to background bacterial DNA in specimens/reagents), and polymicrobial infection may be missed by sequencing or yield uninterpretable sequences that demand extra analysis (e.g., use of RipSeq; Isentio AS, Paradis, Norway) before an interpretation can be provided. On the other hand, an advantage of 16S rRNA PCR is that it is not limited to certain types of bacteria.
A few studies have evaluated synovial fluid or tissue 16S rRNA PCR (9, 11–13). Panousis et al. evaluated synovial fluid PCR for samples from 91 subjects (12 with PJI) undergoing revision hip or knee arthroplasty; sensitivity and specificity were 92 and 74%, respectively (9). Fihman et al. evaluated synovial fluid and/or tissue PCR on samples from 20 subjects with arthroplasties (13 with PJI); sensitivity and specificity were 54 and 86%, respectively (12). DeMan et al. analyzed tissue from 26 subjects with arthroplasties (12 with PJI); PCR sensitivity was 50% (11). Vandercam et al. analyzed tissue from 69 subjects (34 with PJI); PCR sensitivity and specificity were 91 and 97%, respectively (13). Simultaneously achieving high sensitivity and specificity may be challenging when using broad-range PCR of synovial fluid or tissue (9, 11, 12).
Testing material dislodged from prosthesis surfaces may improve performance. Dora et al. sonicated resected arthroplasties from 69 subjects (14 with PJI); sonicate fluid broad-range PCR sensitivity was 86%, but bacteria typically associated with water were detected in those without PJI, possibly due to contamination associated with sonication in bags (8). We previously reported culture contamination associated with bag sonication, and we counsel against this (15). Recently, we evaluated 16S rRNA PCR results for sonicated knee and hip arthroplasties from 366 subjects (135 with PJI); PCR sensitivity and specificity were 70 and 98%, respectively (equivalent to sonicate fluid culture) (14).
Achermann et al. sonicated a small number of resected implants in containers and tested sonicate fluid by using SeptiFast (Roche Diagnostics, Basel, Switzerland), a commercial multiplex real-time PCR assay designed for testing blood and that does not target Propionibacterium or Corynebacterium species (2). Culture and PCR sensitivity were not statistically different (62 and 78%, respectively). Also, only 37 PJI cases were analyzed, limiting the study's power. Portillo et al. recently published a similar study, but they only studied 24 infected resected arthroplasties (16).
We developed a genus-/group-specific rapid real-time closed system PCR assay panel that targets bacteria typically associated with PJI and applied it to vortexed and sonicated implants, hypothesizing that this would sensitively and rapidly detect PJI. We evaluated a large cohort of subjects and compared this approach with tissue and sonicate fluid culture.
(Presented in part at the 110th General Meeting of the American Society for Microbiology, 23 to 27 May 2010, San Diego, CA, and at the 51st Interscience Conference on Antimicrobial Agents and Chemotherapy, 17 to 20 September 2011, Chicago, IL.)
MATERIALS AND METHODS
Study population.
A total of 449 hip or knee prostheses (complete or partial) removed between 20 April 2006 and 14 May 2011 from 438 Mayo Clinic subjects were studied. All implants and ≥2 tissue samples/joint were cultured. The study was approved by the Mayo Clinic Institutional Review Board. Only the first implant culture per subject was studied. Polyethylene liner exchanges alone were excluded.
Diagnosis of prosthetic joint infection.
PJI was diagnosed if ≥1 of the following was present: acute inflammation on periprosthetic tissue histopathology, joint space purulence, or a sinus tract (1, 17). Aseptic failure was defined as prosthesis failure without these criteria. Previous antimicrobial therapy was defined as treatment during the 28 days preceding surgery. An infectious disease physician with expertise in bone and joint infection (J. M. Steckelberg), unaware of PCR results, reviewed select cases. An infectious diseases pathologist (B. S. Pritt) blindly reviewed specimens not clearly classified as showing or not showing acute inflammation.
Specimen collection.
Intraoperatively, tissues were collected for microbiology and histopathology. Prosthesis components were placed in a 1-liter, sterilized, straight-sided, wide-mouthed polypropylene jar (Nalgene, Rochester, NY) and processed within 6 h. Results for blood leukocyte count within one preoperative week and erythrocyte sedimentation rate (ESR), C-reactive protein (CRP), and synovial fluid leukocyte count, differential, and culture within six preoperative months were recorded.
Conventional microbiologic methods.
Tissue was homogenized in brain heart infusion broth for 1 min and inoculated onto aerobic sheep blood, chocolate, and anaerobic blood agar and into thioglycolate broth (BD Diagnostic Systems, Sparks, MD). Aerobic plates were incubated at 35 to 37°C in 5 to 7% CO2 for 2 to 4 days. Anaerobic plates were anaerobically incubated at 35 to 37°C for 7 days through 18 April 2011, and 14 days thereafter. Thioglycolate broth, incubated for 7 days through 18 April 2011 and 14 days thereafter, was subcultured if cloudy. Two definitions of tissue culture positivity were evaluated: any growth, and ≥2 positive tissues with the same organism.
Synovial fluid volumes of ≥1 ml were inoculated into a Bactec Peds Plus/F bottle and incubated on a Bactec 9240 instrument (BD Diagnostic Systems, Sparks, MD) for 5 days (18). Smaller volumes were inoculated onto aerobic sheep blood and chocolate agar and aerobically incubated for 2 to 4 days and onto anaerobic blood agar and into thioglycolate broth and anaerobically incubated for 7 days through 18 April 2011 and 14 days thereafter. Any growth was considered positive.
Prosthesis vortexing and sonication.
We used a previously described implant processing technique (3) with a modification to concentrate sonicate fluid (4–5, 19). After sonication (3), sonicate fluid was centrifuged in 50-ml aliquots at 4,000 rpm (3,150 × g) for 5 min, and all but 0.5 ml in the bottom was discarded. Concentrated fluid was plated in 0.1-ml aliquots onto aerobic sheep blood, chocolate, and anaerobic sheep blood agar plates. Aerobic and anaerobic plates were incubated aerobically for 2 to 4 days and anaerobically for 14 days, respectively. Microorganism numbers were classified and reported as <20, 20 to 50, 51 to 100, or >100 CFU/plate. Growth of ≥20 CFU/plate was considered significant (19).
Ten-assay real-time PCR panel.
A panel of real-time fluorescence resonance energy transfer probe PCR assays that detects multiple bacterial pathogens typically considered causes of PJI (staphylococci, streptococci, Enterobacteriaceae, anaerobic Gram-positive cocci, Enterococcus/Granulicatella/Abiotrophia spp., Propionibacterium/Actinomyces spp., Pseudomonas aeruginosa, Corynebacterium spp., and Bacteroides fragilis group) with use of the LightCycler 1.0 instrument (Roche Applied Science, Indianapolis, IN) was designed using a previously reported strategy (Table 1) (20). Cross-reactivity/inclusivity were evaluated using DNA from 354 isolates from biofilm-associated diseases (Table 2). The Staphylococcus and P. aeruginosa assays were adapted from that described by Sakai and Qin, respectively (21, 22), and the Corynebacterium assay was from our previous work (20).
Table 1.
Real-time PCR assay panel design
| Target organisma | Target gene | Fragment size (bp) | Forward/reverse primer sequences (5′–3′) | Probe sequences (5′–3′)b | Mg2+ concn (mM) | Primer concns (forward/reverse, in μM) | Cycling parameters |
Limit of detection (CFU/ml of sonicate fluid) | % inclusivityc | % cross-reactivityc | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Anneal temp (°C) | Anneal time (s) | No. of cycles | ||||||||||
| Staphylococcus (22) | tufd | 447 | CAATGCCACAAACTCG/GCTTCAGCGTAGTCTA | ACGGCCTGTAGCAACAGTAC-FL,e LC640-CGACCAGTGATTGAGAATACGTCCe, GGCGATGCTCAATACGAAGAAAAAATC-FL,f LC705-AGAATTAATGGAAGCTGTAGATACf | 3 | 1/1 | 55 | 8 | 36 | 550 SCN IDRL-7,371, 325 S. aureus ATCC 43300 | 100 (201/201) | 0 (0/153) |
| Streptococcus | rpoBg | 137 | CCGGHCGTCACGGWAA/CCATACCAAGRTGAAGYTCCATA | GGAACGCCAGTTGATATCATGYTKAAYCC-FL, LC640-CTTGGGGTGCCATCACGKATGAA | 4 | 0.4/4 | 57 | 15 | 35 | 110 S. agalactiae, 115 S. pneumoniae | 100 (15/15)h | 0 (0/339) |
| Enterococcus/Granulicatella/Abiotrophia | rpoB | 143 | CGYGAAGCYGGCGATGAAT/AWGGCATRTCTTCTTCYGGC | CAYGAAGGRGATAARATGGCSGG-FL, LC640-GTCAYGGWAATAAAGGGGTYGTWTC | 4 | 0.4/4 | 55 | 8 | 35 | 200 E. faecalis | 100 (14/14)i | 0 (0/340) |
| Enterobacteriaceae other than Proteus | rpoB | 185 | TCTGCWATYGAAGAAGGCAACT/ATCAGGGAHGCACCRAC | GCTTGTTCAGCCGCGACC-FL, LC640-GTTGACTACATGGACGTATCCACCCAGC | 3.5 | 0.4/2 | 55 | 8 | 35 | 190 E. coli | 83.3 (20/24)j | 0 (0/330) |
| Gram-positive anaerobic cocci | 16S rRNA | 162 | TCGCGTCYSATTAGCTAGT/GCTGCATCAGRGTTYCC | CACATTGGRACTGAGAHACGGY-FL, LC640-ARACTCCTACGGGAGGCAGCAG | 2.5 | 0.4/2 | 57 | 8 | 35 | 10 F. magna | 100 (12/12) | 0 (0/342) |
| Propionibacterium/Actinomyces | 16S rRNA | 243 | CGGATTTATTGGGCGTAAAGR/AGGGTATCTAAGCCTGTTCG | GGCGAAGGCGGTTCTCTG-FL, LC640-CCTTTCCTGACGCTGAGRAGCG | 2.5 | 0.4/2 | 55 | 8 | 35 | 200 P. acnes | 100 (52/52)k | 0.3 (1/302)l |
| P. aeruginosa (21) | gyrBm | 222 | CCTGACCATCCGTCGCCACAAC/CGCAGCAGGATGCCGACGCC | GGCGAGACCGATGGCT-FL, LC640-GGCACCGAAGTTCACTTCAAGCCG | 2.5 | 0.4/1 | 55 | 8 | 35 | 200 | 100 (10/10) | 0 (0/344) |
| Corynebacteriumn (20) | Kinasen,o | 143 (forward/reverse 1), 149 (forward/reverse2) | CGRTTGTACCARGARCGGT/GCACCTSAAYCCSCGT (reverse 1), CAACGAGCACCTSAACCC (reverse 2) | TAGCGCTGGAAGTACCASGAGGT-FL, LC640-GACTCRCGCGGCGACGG, LC705-GACTCGCGCTCGGAAGG | 4 | 0.4/2 | 57 | 10 | 35 | 200 C. tuberculo-stearicum, 440 C. jeikeium | 80 (4/5)p | 0 (0/349) |
| Proteus | rpoB | 183 | CTGTCWGCAATTGAAGAAGGTAACT/GGATCAGTGAWGCACCGA | CTTGCCGTCATARAGGYG-FL, LC705-TCAAGYTTATTTAGTCGTGAT | 2.5 | 0.4/2 | 55 | 8 | 35 | 52 P. vulgaris | 100 (4/4) | 0 (0/350) |
| B. fragilis group | leuBq | 185 | CGTCCTGAGATAGAACG/GCAGCATTATCCACAAACATATAA | CTTGCTTCCAGTCGTCTATGGA-FL, LC640-CAGATTGCACAAGAAATGGCGCCGC | 2.5 | 0.4/2 | 55 | 8 | 35 | 150 B. fragilis | 92.3 (12/13)r | 0 (0/341) |
References are given in parentheses.
FL, fluorescein; LC640, LightCycler Red 640; LC705, LightCycler Red 705.
Calculated based on 354 isolates.
tuf, elongation factor Tu.
PanStaphHP1 and HP2, probes that detect Staphylococcus aureus and SCN.
SAtufHP1 and HP2, S. aureus-specific probes.
rpoB encodes the β-subunit of RNA polymerase.
S. agalactiae positive, with a Tm of 63°C, compared to Tms of <60°C for other Streptococcus spp.
The G. adiacens (n = 3) and Abiotrophia species isolates had Tms of 58°C and 50°C, respectively, compared to 55°C for the Enterococcus sp. isolate.
The four negative isolates were Morganella morganii, Pantoea agglomerans, Providencia rettgeri, and Serratia liquefaciens.
The Actinomyces spp. had Tms of ≤56°C, compared to 61 to 63°C for Propionibacterium spp.
Corynebacterium amycolatum positive, with a Tm of 58°C.
gyrB encodes DNA gyrase B.
This assay uses one forward and two reverse primers and three probes in the same reaction, primer reverse 1 and probe LC705-GACTCGCGCTCGGAAGG, specific for the resistant species (C. jeikeium and C. urealyticum).
Polyphosphate kinase gene, corresponding to pvdS2 of C. jeikeium K411 (NC_007164).
Corynebacterium group F1 negative.
leuB encodes β-isopropylmalate dehydrogenase.
B. caccae negative.
Table 2.
Isolates, from patients with biofilm-associated disease, that were used for analytical assay validationa
| Group and species | No. of isolates (total n = 354) |
|---|---|
| Staphylococcus spp. | 201 |
| S. aureus | 85 |
| Coagulase-negative Staphylococcus spp. | 116 |
| S. epidermidis | 80 |
| S. lugdunensis | 16 |
| S. warneri | 8 |
| S. capitis | 3 |
| S. caprae | 3 |
| S. simulans | 3 |
| Otherb | 3 |
| Propionibacterium spp. | 41 |
| P. acnes | 29 |
| P. avidum | 10 |
| P. granulosum | 2 |
| Enterobacteriaceae | 28 |
| Escherichia coli | 5 |
| Enterobacter cloacae | 4 |
| Proteus mirabilis | 3 |
| Klebsiella pneumoniae | 3 |
| Klebsiella oxytoca | 2 |
| Otherc | 11 |
| Streptococcus spp. | 15 |
| S. agalactiae | 3 |
| S. dysgalactiae | 3 |
| S. pyogenes | 2 |
| S. pneumoniae | 2 |
| S. salivarius | 2 |
| Otherd | 3 |
| Bacteroides fragilis group | 13 |
| B. fragilis | 8 |
| B. thetaiotaomicron | 2 |
| Othere | 3 |
| Gram-positive anaerobic cocci | 12 |
| Finegoldia magna | 8 |
| Otherf | 4 |
| Actinomyces spp.g | 11 |
| Pseudomonas aeruginosa | 10 |
| Enterococcus faecalis | 9 |
| Corynebacterium spp.h | 5 |
| Granulicatella adiacens | 3 |
| Abiotrophia defectiva | 2 |
| Otheri | 4 |
Categories shown in boldface were targeted with specific assays.
Includes one S. haemolyticus, one S. hominis, and one S. saprophyticus isolate.
Includes one Citrobacter freundii, one C. koseri, one E. aerogenes, one Morganella morganii, one Pantoea agglomerans, one P. vulgaris, one Providencia rettgeri, one Salmonella sp., one Serratia liquifaciens, one S. marcescens, and one Shigella flexneri isolate.
Includes one S. anginosus, one S. mitis, and one S. mutans isolate.
Includes one B. caccae, one B. distasonis, and one B. ovatus isolate.
Includes one Peptoniphilus asaccharolyticus, one P. harei, one Parvimonas micra, and one Anaerococcus prevotii isolate.
Includes three A. neuii, three A. odonolyticus, two A. naeslundii, one A. meyeri, one A. viscosus, and one A. radingae isolate.
Includes one C. amycolatum, one C. simulans, one C. propinquum, one Corynebacterium sp. group F1, and one C. afermentans.
Includes one Candida parapsilosis, one Dermabacter hominis, one P. fluorescens, and one S. saccharolyticus isolate.
Nucleic acid extraction from sonicate fluid.
One milliliter of concentrated sonicate fluid was extracted using the DNA-free QIAamp UCP pathogen minikit (Qiagen, Valencia, CA) and eluted in a volume of 100 μl. A human β-globin control (LightCycler control kit DNA; Roche Molecular Biochemicals, Indianapolis, IN) was run for each patient specimen to assess PCR inhibition. Briefly, a 110-bp fragment of the β-globin gene was amplified and detected by fluorescence using SYBR green I reaction mix (Roche Applied Science, Indianapolis, IN). Turnaround time was ∼4.5 h, including 0.5, 2.5, and 1.5 h for vortexing/sonication, extraction, and PCR, respectively.
Statistical analysis.
Baseline characteristics of the study groups were summarized as frequencies and percentages or medians and compared using the Wilcoxon rank-sum test or chi-square test, as appropriate. Sensitivities and specificities were compared using McNemar's test of paired proportions. Sensitivity, specificity, and positive and negative values were estimated along with 95% exact binomial confidence intervals. P values of <0.05 were considered statistically significant. Analyses were performed using the SAS statistical software package, version 9.1 (SAS Institute Inc., Cary, NC).
RESULTS
Study population.
Eleven implants were excluded because only the first implant culture per subject was studied, and four were excluded due to polyethylene liner exchange alone. Of the remaining 434 subjects, 272 had knee and 162 hip prostheses studied; 290 had aseptic failure and 144 had PJI; 244 underwent first revision and 190 a second (or greater) revision; 414 had complete and 20 had partial hardware removal (Table 3). The groups were similar in age, gender, reason for arthroplasty, and joint site. Second (or greater) revision and diabetes were more frequent in the PJI group (P = 0.004 and 0.003, respectively). A blood leukocyte count of >10 × 109/liter, ESR of >30 mm/h, and CRP of >10 mg/liter were more common with PJI versus aseptic failure (P = 0.005, <0.001, and <0.001, respectively). A synovial fluid leukocyte count of >1,700/μl and differential of >65% neutrophils were more common with PJI than aseptic failure (P < 0.001).
Table 3.
Characteristics of the 434 patients with or without PJI
| Characteristic | Value for patient group |
P value | |
|---|---|---|---|
| Patients with aseptic failure (total n = 290) | Patients with prosthetic joint infection (n = 144) | ||
| Age (yrs) | 0.53 | ||
| Median | 68 | 66 | |
| Range | 24–91 | 31–92 | |
| Gender [n (%)] | 0.14 | ||
| Male | 131 (45.2) | 76 (52.8) | |
| Female | 159 (54.8) | 68 (4..2) | |
| Reason for primary arthroplasty [n (%)] | 0.43 | ||
| Osteoarthritis | 208 (71.7) | 108 (75) | |
| Bone fracture or trauma | 44 (15.2) | 19 (13.2) | |
| Inflammatory joint disordera | 20 (6.9) | 8 (5.5) | |
| Avascular necrosis | 12 (4.1) | 3 (2.1) | |
| Congenital abnormality | 2 (0.7) | 4 (2.8) | |
| Otherb | 4 (1.4) | 2 (1.4) | |
| Second (or more) revision [n (%)] | 0.004 | ||
| No | 177 (61.0) | 67 (46.5) | |
| Yes | 113 (39.0) | 77 (53.5) | |
| Hardware removal [n (%)] | 1.00 | ||
| Partial | 14 (4.8) | 6 (4.2) | |
| Complete | 276 (95.2) | 138 (95.8) | |
| Risk factor for PJI [n (%)] | 0.002 | ||
| Diabetes mellitus | 35 (12.1) | 33 (22.8) | 0.003 |
| Long-term use of immunosuppressive therapyc | 2 (0.7) | 4 (2.8) | 0.10 |
| Site of arthroplasty [n (%)] | 0.87 | ||
| Knee | 181 (62.4) | 91 (63.2) | |
| Hip | 109 (37.6) | 53 (36.8) | |
| Antecedent antimicrobial therapy [n/total no. evaluated (%)] | 24/284 (8.4) | 58/134 (43.3) | <0.0001 |
| Diagnostic criterion for PJI | |||
| Acute inflammation [n/total no. evaluated (%)] | 0/290 (0) | 90/112 (80.4) | |
| Purulence [n (%)] | 0 | 112 (77.8) | |
| Sinus tract [n (%)] | 0 | 39 (27.1) | |
| Preoperative laboratory findings [n/total no. evaluated (%)] | |||
| Blood leukocyte count > 10 × 109/literd,e | 17/231 (7.4) | 21/123 (17.1) | 0.005 |
| Erythrocyte sedimentation rate > 30 mm/hd,f | 44/266 (16.5) | 83/135 (61.5) | <0.0001 |
| Serum C-reactive protein > 10 mg/literd,f | 53/266 (19.9) | 109/136 (81.2) | <0.0001 |
| Synovial fluid leukocyte count > 1,700/μlf,g | 33/140 (23.6) | 63/71 (88.7) | <0.0001 |
| Synovial fluid differential > 65% neutrophilsf,g | 15/137 (10.9) | 61/71 (85.9) | <0.0001 |
Includes rheumatoid arthritis, ankylosing spondylitis, and systemic lupus erythematosus.
Includes bone neoplasia, arthrofibrosis, polymyalgia rheumatica, reflex sympathetic dystrophy, and acromegaly.
Includes corticosteroids, methotrexate, and tumor necrosis factor inhibitors.
The cutoff was taken from Bernard et al. (23).
Within one preoperative week.
Within six preoperative months.
The cutoff was taken from Trampuz et al. (24), a study that excluded patients with underlying inflammatory joint diseases or connective tissue diseases and evaluated only knee arthroplasties.
Comparison of microbiologic tests.
The sensitivities of sonicate fluid PCR and culture to detect PJI were 77.1 and 72.9%, respectively (P = 0.13). Tissue culture with positive defined as any growth had the highest sensitivity (82.6%) but the lowest specificity (84.5%). Tissue culture when defined as ≥2 tissues with the same organism had a lower sensitivity (70.1%), but improved specificity (97.9%); this definition was applied for subsequent comparisons. Sensitivity of tissue culture was lower than that for sonicate fluid PCR (P = 0.04) and equivalent to that for sonicate fluid culture (P = 0.41) and synovial fluid culture (66.3%; P = 0.29) (Table 4). The specificities of PCR, sonicate fluid culture, and synovial fluid culture were similar, 97.9, 98.3, and 96.9%, respectively (Table 4).
Table 4.
Comparison of microbiologic tests for diagnosis of PJI
| Test | No. of patients with positive specimens and: |
Sensitivity (95% CI) | Specificity (95% CI) | Positive predictive value (95% CI) | Negative predictive value (95% CI) | |
|---|---|---|---|---|---|---|
| Aseptic failure (n = 290) | PJI (n = 144) | |||||
| Synovial fluid culturea | 5/161 | 59/89 | 66.3 (55.5–76.0) | 96.9 (92.9–99.0) | 92.2 (82.7–97.4) | 83.9 (77.8–88.8) |
| Tissue culture | ||||||
| Any growth | 45 | 119 | 82.6 (75.4–88.4) | 84.5 (79.8–88.5) | 72.6 (65.1–79.2) | 90.7 (86.6–93.9) |
| ≥2 positive tissues (same organism) | 6 | 101 | 70.1 (62.0–77.5) | 97.9 (95.6–99.2) | 94.4 (88.2–97.9) | 86.9 (82.7–90.3) |
| Sonicate fluid culture | 5 | 105 | 72.9 (64.9–80.0) | 98.3 (96.0–99.4) | 95.5 (89.7–98.5) | 88.0 (83.9–91.3) |
| Sonicate fluid PCR (10-assay panel) | ||||||
| Any positive result | 6 | 111 | 77.1 (69.3–83.7) | 97.9 (95.6–99.2) | 94.9 (89.2–98.1) | 89.6 (85.7–92.7) |
| Staphylococcus species | 2 | 75 | ||||
| S. aureus | 0 | 28 | ||||
| Coagulase-negative staphylococci | 2 | 47 | ||||
| Streptococcus species | 3 | 11 | ||||
| Enterococcus/Granulicatella/Abiotrophia species | 0 | 11 | ||||
| Enterobacteriaceae | 1 | 8 | ||||
| Gram-positive anaerobic cocci | 0 | 8 | ||||
| Propionibacterium species | 0 | 8 | ||||
| P. aeruginosa | 0 | 5 | ||||
| Corynebacterium species | 0 | 4 | ||||
| C. jeikeium/C. urealyticum | 0 | 0 | ||||
| Non-C. jeikeium species | 0 | 4 | ||||
| Proteus species | 0 | 1 | ||||
| B. fragilis group | 0 | 0 | ||||
Denominators are smaller for synovial fluid cultures because samples from fewer patients were submitted for this test.
Conventional microbiologic results.
Using a definition of positivity of ≥2 tissues with the same organism, 101/144 and 6/290 subjects with and without PJI, respectively, had positive tissue cultures (Fig. 1; Table 4). The six aseptic failures with positive tissue cultures included Propionibacterium spp. (n = 4) and coagulase-negative staphylococci (SCN; n = 2). Synovial fluid culture was performed for 250 subjects; 59/89 and 5/161 with and without PJI, respectively, were positive (Table 4). Finally, 105/144 and 5/290 with or without PJI had positive sonicate fluid cultures (Fig. 1; Table 4); the aseptic failures with positive cultures included SCN (n = 2), viridans group streptococcus (VGS; n = 1), Propionibacterium spp. plus Clostridium perfringens (n = 1), and Staphylococcus aureus (n = 1), and these cases were distinct from tissue culture-positive aseptic failure cases.
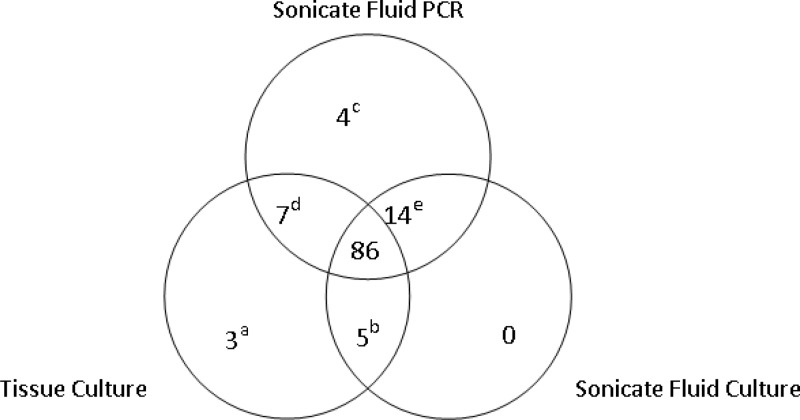
Fig 1.
Positive microbiologic results (albeit not necessarily concordant with respect to the organism detected) for patients with prosthetic joint infection. Superscript symbol definitions: a, cases 200 (Mycobacterium abscessus), 298 (Staphylococcus aureus), and 369 (polymicrobial, including S. aureus); b, cases 164 (Capnocytophaga canimorsus), 236 (Candida albicans), and 211, 259, and 425 (all three S. aureus); c, cases 214, 250, 280, and 379; d, cases 74, 110, 217, 226, 286, 352, and 430; e, cases 70, 91, 153, 203, 207, 212, 269, 300, 342, 343, 381, 396, 412, and 429.
Sonicate fluid PCR.
The β-globin gene was detected in all sonicate fluids. A total of 111 (77.1%) subjects with PJI and 6 (2.1%) with aseptic failure had a positive sonicate fluid PCR result (Fig. 1; Table 4). Among those with PJI, the Staphylococcus assay was most frequently positive, followed by the Streptococcus and Enterococcus assays. The Streptococcus assay differentiates S. agalactiae from other species by melting temperature (Tm) analysis. Three of 11 streptococcal PCR-positive sonicate fluids were consistent with S. agalactiae, concordant with culture. Similarly, one sonicate fluid was Enterococcus/Granulicatella/Abiotrophia PCR positive with a Tm consistent with Granulicatella adiacens (58°C); culture revealed G. adiacens. The Propionibacterium/Actinomyces assay was positive for three subjects, with a Tm consistent with Actinomyces spp. (<58°C); cultures from all three subjects grew Actinomyces spp.
The panel detected 11 polymicrobial infections, including a positive staphylococcal result in 10. Eight of 11 had a sinus tract. For five, two assays were positive (Staphylococcus plus Propionibacterium assays, n = 2; Staphylococcus plus Enterococcus, n = 2; Staphylococcus plus Enterobacteriaceae, n = 1). For the other six, three (n = 2) or four (n = 4) assays were positive.
Among the PJI subjects, 33 (22.9%) had negative PCR results (see Table S1 in the supplemental material). Four were infected with an organism not detectable by PCR (two fungal, one Capnocytophaga canimorsus, and one Mycobacterium sp.). Tissue and sonicate fluid cultures were negative in 24. Five had positive tissue cultures for S. aureus, of which three had S. aureus cultured from sonicate fluid.
Six (2.1%) subjects with aseptic failure had positive PCR results (SCN [n = 2], streptococci [n = 3], and Enterobacteriaceae [n = 1]). For the two subjects PCR positive for SCN (cases 89 and 252), sonicate fluid cultures and single tissue cultures grew SCN. For the three subjects PCR positive for Streptococcus, one sonicate fluid culture was positive for VGS (case 73), one subject (case 397) had a prior S. agalactiae infection in the same joint, and for the last subject (case 299) all the cultures were negative and vancomycin (given during 18 days before surgery) was stopped after surgery. For the subject PCR positive for Enterobacteriaceae in sonicate fluid (case 321), sonicate fluid culture and tissue culture were negative.
Discordant results between sonicate fluid PCR and culture.
There were 24 discordant results between sonicate fluid PCR and culture (see Table S2 in the supplemental material). Eleven PJIs were detected by PCR alone. For 6 out of 11, the Staphylococcus PCR was positive. Eight had received antibiotics. There were 8 out of 11 PJI subjects who had positive tissue and/or synovial fluid cultures; cultures were concordant with PCR results (except for case 110, where tissue culture revealed SCN and PCR indicated S. aureus). Review of the records of the other three showed that in two (cases 250 and 280), the same microorganism identified by PCR had been identified by antecedent culture.
For two subjects with PJI (cases 175 and 245), the Enterobacteriaceae assay was positive; sonicate fluid cultures did not grow Enterobacteriaceae. Case 245 had one tissue culture positive for Klebsiella pneumoniae. For two subjects with PJI (cases 236 and 164), sonicate fluid cultures were positive for organisms not targeted by the PCR panel (Candida albicans and Capnocytophaga canimorsus, respectively) and PCR was negative. For the remaining six (cases 38, 211, 259, 342, 425, and 428), sonicate fluid cultures were positive for staphylococci; one had Enterococcus spp. additionally cultured and a positive Enterococcus PCR. Staphylococcus PCR was positive for three of the six on repeat testing. One sonicate fluid culture with S. aureus (case 38) was possibly contaminated during processing; the subject underwent a one-stage exchange, received 2 weeks of oral antimicrobial therapy, and was well 1 year later.
There were three aseptic failures with positive PCR result and negative sonicate fluid cultures. For two (cases 299 and 321), no prolonged antimicrobial therapy was administered after surgery, and they were well at the 2-year follow-up. The third (case 397) had a prior beta-hemolytic Streptococcus infection and was prescribed antibiotics for 6 weeks; the Streptococcus PCR assay results were compatible with S. agalactiae.
Previous antimicrobial therapy.
Of subjects with PJI, 58 (43.3%) and 33 (24.6%) had received antimicrobial therapy within 28 and 14 days prior to surgery, respectively (Table 3). Among subjects receiving antibiotics within 14 days of surgery, 69.7% had positive tissue and sonicate fluid cultures and 87.9% had positive sonicate fluid by PCR (P = 0.01). Among subjects receiving antibiotics within 28 days of surgery, 86.2, 77.6, and 75.9% had a positive PCR, sonicate fluid culture, and tissue culture, respectively.
DISCUSSION
A molecular approach, applied to biofilms dislodged from an implant surface of a resected hip or knee arthroplasty, can sensitively and rapidly detect PJI. The described PCR panel, applied to implant-derived sonicate fluid, is more sensitive than tissue culture and, for those patients receiving antibiotics within 14 days of surgery, more sensitive than sonicate fluid culture.
PCR had similar specificity to culture of sonicate fluid, tissue, or synovial fluid, with only six aseptic failures with positive results. Based on medical record review, and also using a culture-based PJI definition (25), the two Staphylococcus PCR-positive subjects were considered to have PJI. One Streptococcus PCR-positive subject, classified as having aseptic failure, had had a history of Streptococcus PJI and was managed accordingly. This positive PCR result may reflect killed rather than live organisms, since the patient had received antibiotics for 17 days before surgery. It is therefore likely that three of the six false-positive results were misclassified cases.
PCR offers definitive results in half a day, compared with 1 to 14 days for culture. Preoperative administration of antimicrobial agents (including those administered for suppression of PJI and discontinued within the month before surgery) may affect culture sensitivity (3). The PCR panel was less affected than culture by prior antibiotics. In some subjects, PCR was positive after more than 1 month of antibiotic treatment, demonstrating persistence of bacterial DNA in bone and joint samples following antimicrobial therapy (26, 27).
Our panel includes anaerobic bacteria (Propionibacterium and Actinomyces species in one assay and Gram-positive anaerobic cocci and B. fragilis group in two others). At least one anaerobic assay was positive in 14 PJI subjects (9.7%), emphasizing the need to detect anaerobes (1).
A multiplex approach appears more useful than broad-range PCR for rapid microbial PJI diagnosis. Compared with the Achermann and Portillo studies (2, 16), our panel includes common PJI bacteria, including Propionibacterium and Corynebacterium species, and anaerobic Gram-positive cocci. In the case of negativity of cultures and the PCR panel, fungal culture(s)/PCR, 16S rRNA PCR, and molecular/serologic testing for esoteric bacteria might be useful to detect unusual organisms not detected by the panel.
Rapid detection of polymicrobial PJI is an advantage of the panel, with 11 cases detected. Such cases would not be easily diagnosed with broad-range PCR. Results with PCR were concordant with tissue and/or sonicate fluid cultures for six subjects; for five, the panel detected organisms not cultured.
The tissue culture sensitivity reported herein (70.1%) was higher than in our prior study (60.8%), approaching that of sonicate fluid culture in this and our prior study (72.9 and 78.5%, respectively) (3). We extended the anaerobic tissue culture incubation duration shortly before the study end, although we do not believe this impacted results of this study (28). The number of cultured tissues per PJI subject was similar. A possible explanation is that in our prior study 61% of patients with PJI had received antibiotics within 4 weeks prior to surgery, compared to 43% in this study. The sensitivity of sonicate culture (modified by adding a concentration step) was slightly lower than previously described.
Study limitations included lack of a gold standard definition of PJI and assay multiplexing. Culture is required for detection of unusual microorganisms and antimicrobial susceptibility testing, although conceivably, resistance genes (such as mecA) could be incorporated into a PCR panel.
The results of this study highlight the need to culture multiple tissues and for ≥2 to be positive with the same organism to signify PJI. The specificity of a single positive tissue culture was lower than that of ≥2 positive tissues (84.5 and 97.9%, respectively; P < 0.001).
Despite the new technique, there remained PCR-negative cases of apparent PJI, even in subjects who had not received prior antimicrobial agents. For most, tissue and sonicate fluid cultures were also negative. Our PCR panel was not designed to detect rare causes of PJI (29). Case misclassification may account for some cases; two subjects with negative cultures and PCR results (cases 52 and 53) were considered to not have PJI after medical record review.
Failure of the Staphylococcus PCR assay to detect some cases of staphylococcal PJI was further investigated; poor lysis of staphylococci appeared to explain this finding (PCR inhibitors were not detected). We measured the limit of detection of the Staphylococcus PCR assay using staphylococcal isolates from three subjects with Staphylococcus PCR-negative sonicate fluids and, surprisingly, the limit was ≥10,000 CFU/ml of sonicate fluid for all three isolates, which is markedly higher than the limit of detection of the control isolates used for assay development, one of which (IRDL-7371) was a PJI isolate (Table 1). Adding a lysis step with 10 mg/ml lysostaphin (Sigma-Aldrich, St. Louis, MO) to the DNA extraction resulted in 100- to 1,000-fold improvement in the limit of detection when testing the isolates from cases 259, 342, and 428. Some staphylococci associated with PJI may, therefore, be challenging to lyse.
It has been suggested that some aseptic failures are missed PJI cases (10). Despite improved sensitivity, PCR did not identify a substantial number of infected cases among aseptic failures, suggesting that typical PJI bacteria may not be involved in aseptic failure pathogenesis (3, 7).
In summary, we have described here a real-time PCR panel performed on implant sonicate fluid which is more sensitive than tissue culture, and more rapid than culture, for the diagnosis of PJI.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under award number R01 AR056647 and the Collège des Universitaires des Maladies Infectieuses et Tropicales.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We thank James R. Uhl, Terry M. Therneau, Elie F. Berbari, and the outstanding staff of the Mayo Clinic Clinical Microbiology Laboratory for technical assistance and Robert T. Trousdale, David G. Lewallen, Mark W. Pagnano, Miguel E. Cabanela, Robert H. Cofield, Daniel J. Berry, Joaquin Sanchez-Sotelo, Michael J. Yaszemski, Michael J. Stuart, Franklin H. Sim, Bernard F. Morrey, Rafael J. Sierra, Tad M. Mabry, Diane L. Dahm, Stephen A. Sems, Joseph R. Cass, Bruce A. Levy, Michael E. Torchia, and Michael J. Taunton for submitting explanted prostheses for this study.
Published ahead of print 8 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00335-13.
REFERENCES
- 1. Del Pozo JL, Patel R. 2009. Clinical practice. Infection associated with prosthetic joints. N. Engl. J. Med. 361:787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Achermann Y, Vogt M, Leunig M, Wust J, Trampuz A. 2010. Improved diagnosis of periprosthetic joint infection by multiplex PCR of sonication fluid from removed implants. J. Clin. Microbiol. 48:1208–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Trampuz A, Piper KE, Jacobson MJ, Hanssen AD, Unni KK, Osmon DR, Mandrekar JN, Cockerill FR, Steckelberg JM, Greenleaf JF, Patel R. 2007. Sonication of removed hip and knee prostheses for diagnosis of infection. N. Engl. J. Med. 357:654–663 [DOI] [PubMed] [Google Scholar]
- 4. Piper KE, Jacobson MJ, Cofield RH, Sperling JW, Sanchez-Sotelo J, Osmon DR, McDowell A, Patrick S, Steckelberg JM, Mandrekar JN, Fernandez Sampedro M, Patel R. 2009. Microbiologic diagnosis of prosthetic shoulder infection by use of implant sonication. J. Clin. Microbiol. 47:1878–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vergidis P, Greenwood-Quaintance KE, Sanchez-Sotelo J, Morrey BF, Steinmann SP, Karau MJ, Osmon DR, Mandrekar JN, Steckelberg JM, Patel R. 2011. Implant sonication for the diagnosis of prosthetic elbow infection. J. Shoulder Elbow Surg. 20:1275–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bergin PF, Doppelt JD, Hamilton WG, Mirick GE, Jones AE, Sritulanondha S, Helm JM, Tuan RS. 2010. Detection of periprosthetic infections with use of ribosomal RNA-based polymerase chain reaction. J. Bone Joint Surg. Am. 92:654–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dempsey KE, Riggio MP, Lennon A, Hannah VE, Ramage G, Allan D, Bagg J. 2007. Identification of bacteria on the surface of clinically infected and non-infected prosthetic hip joints removed during revision arthroplasties by 16S rRNA gene sequencing and by microbiological culture. Arthritis Res. Ther. 9:R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dora C, Altwegg M, Gerber C, Bottger EC, Zbinden R. 2008. Evaluation of conventional microbiological procedures and molecular genetic techniques for diagnosis of infections in patients with implanted orthopedic devices. J. Clin. Microbiol. 46:824–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Panousis K, Grigoris P, Butcher I, Rana B, Reilly JH, Hamblen DL. 2005. Poor predictive value of broad-range PCR for the detection of arthroplasty infection in 92 cases. Acta Orthop. 76:341–346 [PubMed] [Google Scholar]
- 10. Tunney MM, Patrick S, Curran MD, Ramage G, Anderson N, Davis RI, Gorman SP, Nixon JR. 1999. Detection of prosthetic joint biofilm infection using immunological and molecular techniques. Methods Enzymol. 310:566–576 [DOI] [PubMed] [Google Scholar]
- 11. De Man FH, Graber P, Luem M, Zimmerli W, Ochsner PE, Sendi P. 2009. Broad-range PCR in selected episodes of prosthetic joint infection. Infection 37:292–294 [DOI] [PubMed] [Google Scholar]
- 12. Fihman V, Hannouche D, Bousson V, Bardin T, Liote F, Raskine L, Riahi J, Sanson-Le Pors MJ, Bercot B. 2007. Improved diagnosis specificity in bone and joint infections using molecular techniques. J. Infect. 55:510–517 [DOI] [PubMed] [Google Scholar]
- 13. Vandercam B, Jeumont S, Cornu O, Yombi JC, Lecouvet F, Lefevre P, Irenge LM, Gala JL. 2008. Amplification-based DNA analysis in the diagnosis of prosthetic joint infection. J. Mol. Diagn. 10:537–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gomez E, Cazanave C, Cunningham SA, Greenwood-Quaintance KE, Steckelberg JM, Uhl JR, Hanssen AD, Karau MJ, Schmidt SM, Osmon DR, Berbari EF, Mandrekar J, Patel R. 2012. Prosthetic joint infection diagnosis using broad-range PCR of biofilms dislodged from knee and hip arthroplasty surfaces using sonication. J. Clin. Microbiol. 50:3501–3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Trampuz A, Piper KE, Hanssen AD, Osmon DR, Cockerill FR, Steckelberg JM, Patel R. 2006. Sonication of explanted prosthetic components in bags for diagnosis of prosthetic joint infection is associated with risk of contamination. J. Clin. Microbiol. 44:628–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Portillo ME, Salvado M, Sorli L, Alier A, Martinez S, Trampuz A, Gomez J, Puig L, Horcajada JP. 2012. Multiplex PCR of sonication fluid accurately differentiates between prosthetic joint infection and aseptic failure. J. Infect. 65:541–548 [DOI] [PubMed] [Google Scholar]
- 17. Osmon DR, Berbari EF, Berendt AR, Lew D, Zimmerli W, Steckelberg JM, Rao N, Hanssen A, Wilson WR. 2013. Diagnosis and management of prosthetic joint infection: clinical practice guidelines by the Infectious Diseases Society of America. Clin. Infect. Dis. 56:e1–e25 [DOI] [PubMed] [Google Scholar]
- 18. Hughes JG, Vetter EA, Patel R, Schleck C, Harmsen S, Turgeant LT, Cockerill FR., III 2001. Culture with BACTEC Peds Plus/F bottle compared with conventional methods for detection of bacteria in synovial fluid. J. Clin. Microbiol. 39:4468–4471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Piper KE, Jacobson MJ, Steckelberg JM, Patel R. 2008. Microbiologic diagnosis of hip and knee prosthetic joint infection using explanted prostheses sonication followed by concentration of sonicate fluid, abstr D-1103. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother., 46th Infect. Dis. Soc. Am American Society for Microbiology and Infectious Diseases Society of America, Washington, DC [Google Scholar]
- 20. Cazanave C, Greenwood-Quaintance KE, Hanssen AD, Patel R. 2012. Corynebacterium prosthetic joint infection. J. Clin. Microbiol. 50:1518–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Qin X, Emerson J, Stapp J, Stapp L, Abe P, Burns JL. 2003. Use of real-time PCR with multiple targets to identify Pseudomonas aeruginosa and other nonfermenting gram-negative bacilli from patients with cystic fibrosis. J. Clin. Microbiol. 41:4312–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sakai H, Procop GW, Kobayashi N, Togawa D, Wilson DA, Borden L, Krebs V, Bauer TW. 2004. Simultaneous detection of Staphylococcus aureus and coagulase-negative staphylococci in positive blood cultures by real-time PCR with two fluorescence resonance energy transfer probe sets. J. Clin. Microbiol. 42:5739–5744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bernard L, Lubbeke A, Stern R, Bru JP, Feron JM, Peyramond D, Denormandie P, Arvieux C, Chirouze C, Perronne C, Hoffmeyer P. 2004. Value of preoperative investigations in diagnosing prosthetic joint infection: retrospective cohort study and literature review. Scand. J. Infect. Dis. 36:410–416 [DOI] [PubMed] [Google Scholar]
- 24. Trampuz A, Hanssen AD, Osmon DR, Mandrekar J, Steckelberg JM, Patel R. 2004. Synovial fluid leukocyte count and differential for the diagnosis of prosthetic knee infection. Am. J. Med. 117:556–562 [DOI] [PubMed] [Google Scholar]
- 25. Parvizi J, Zmistowski B, Berbari EF, Bauer TW, Springer BD, Della Valle CJ, Garvin KL, Mont MA, Wongworawat MD, Zalavras CG. 2011. New definition for periprosthetic joint infection: from the workgroup of the Musculoskeletal Infection Society. Clin. Orthop. Relat. Res. 469:2992–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van der Heijden IM, Wilbrink B, Vije AE, Schouls LM, Breedveld FC, Tak PP. 1999. Detection of bacterial DNA in serial synovial samples obtained during antibiotic treatment from patients with septic arthritis. Arthritis Rheum. 42:2198–2203 [DOI] [PubMed] [Google Scholar]
- 27. Canvin JM, Goutcher SC, Hagig M, Gemmell CG, Sturrock RD. 1997. Persistence of Staphylococcus aureus as detected by polymerase chain reaction in the synovial fluid of a patient with septic arthritis. Br. J. Rheumatol. 36:203–206 [DOI] [PubMed] [Google Scholar]
- 28. Shannon SK, Mandrekar J, Gustafson DR, Rucinski SL, Dailey AL, Segner RE, Burman MK, Boelman KJ, Lynch DT, Rosenblatt JE, Patel R. 2013. Anaerobic thioglycolate broth culture for recovery of Propionibacterium acnes from shoulder tissue and fluid specimens J. Clin. Microbiol. 51:731–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marculescu CE, Berbari EF, Cockerill FR, III, Osmon DR. 2006. Fungi, mycobacteria, zoonotic and other organisms in prosthetic joint infection. Clin. Orthop. Relat. Res. 451:64–72 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.