Abstract
Infective endocarditis (IE) is a life-threatening infection of the heart endothelium and valves. Staphylococcus aureus is a predominant cause of severe IE and is frequently associated with infections in health care settings and device-related infections. Multilocus sequence typing (MLST), spa typing, and virulence gene microarrays are frequently used to classify S. aureus clinical isolates. This study examined the utility of these typing tools to investigate S. aureus epidemiology associated with IE. Ninety-seven S. aureus isolates were collected from patients diagnosed with (i) IE, (ii) bloodstream infection related to medical devices, (iii) bloodstream infection not related to medical devices, and (iv) skin or soft-tissue infections. The MLST clonal complex (CC) for each isolate was determined and compared to the CCs of members of the S. aureus population by eBURST analysis. The spa type of all isolates was also determined. A null model was used to determine correlations of IE with CC and spa type. DNA microarray analysis was performed, and a permutational analysis of multivariate variance (PERMANOVA) and principal coordinates analysis were conducted to identify genotypic differences between IE and non-IE strains. CC12, CC20, and spa type t160 were significantly associated with IE S. aureus. A subset of virulence-associated genes and alleles, including genes encoding staphylococcal superantigen-like proteins, fibrinogen-binding protein, and a leukocidin subunit, also significantly correlated with IE isolates. MLST, spa typing, and microarray analysis are promising tools for monitoring S. aureus epidemiology associated with IE. Further research to determine a role for the S. aureus IE-associated virulence genes identified in this study is warranted.
INTRODUCTION
Infective endocarditis (IE) is a life-threatening infection of the endocardial surfaces of the heart and heart valves (1, 2). Antibiotic therapy is often difficult due to causation by multidrug-resistant bacteria and the capacity of infecting bacteria to form biofilms on the heart endothelium or implanted medical devices (3). Staphylococcus aureus is the predominant cause of the most severe cases of IE and is the most common pathogen isolated from cases related to intravenous drug use, health care environments, and medical devices such as prosthetic heart valves and intravenous catheters (4–8). The potential of S. aureus to colonize the heart endothelium and cause IE is thought to be closely associated with the genotype of infecting strains (9–11).
Genotyping methods, such as multilocus sequence typing (MLST) and spa typing, have been developed to classify and monitor the S. aureus population. MLST involves assigning strains a sequence type (ST) based on allelic variations in seven genes in the core genome (arc, aroE, glpF, gmk, pta, tpi, and yqi). Strains that diverge at no more than one of the seven MLST loci are considered to belong to the same clonal complex (CC). Double-locus variants (dlvs) are included in the CC if the linking single-locus variant (slv) is present in the MLST database (http://eburst.mlst.net). S. aureus isolated from patients with IE have generally been classified into CC1, CC5, CC8, CC15, CC30, and CC45 (10–12), with the majority belonging to CC30 (10). It has indeed been suggested that CC30 is more closely associated with IE than with other types of S. aureus infections (10). However, other studies have found no association between CC and invasive disease (12–14).
spa typing involves amplification and sequencing of the hypervariable region of the spa gene, which encodes the surface-exposed staphylococcal protein A, a major S. aureus virulence factor (15). A spa type is assigned to a S. aureus strain according to the number and combination of variable short repeats (24 bp) which are located in the C terminus of the spa gene and are flanked by well-conserved regions. Associations between spa type and IE have not been reported.
Bacterial pathogenicity and the development of IE are underpinned by the presence of virulence genes that encode factors such as adhesins, toxins, and exoproteins. Fibronectin-binding protein A and B (FnBPA/FnBPB) and Clumping Factor A (ClfA) are well-characterized Microbial Surface Components Recognizing Adhesive Matrix Molecules (MSCRAMMs) that mediate attachment and colonization of S. aureus to the heart endothelium and medical devices (16–19). The capsular polysaccharide is another important virulence factor expressed by most clinical strains of S. aureus. The capsule prevents phagocytosis of the bacterium by host immune cells, thus facilitating the persistence of S. aureus within the host and development of disease (20, 21).
A recent analysis of individual S. aureus virulence genes and S. aureus IE strains found no correlation between S. aureus genotype and IE (12). However, complex bacterial phenotypes such as the capacity to cause IE are likely to be underpinned by multiple genetic determinants. This study therefore examined associations of CC, spa type, and carriage of multiple virulence-associated genes with S. aureus IE. We identify CCs and spa types that are significantly associated with IE isolates and define a set of virulence-associated genes and allelic variants that, when combined, are significantly correlated with S. aureus IE isolates compared to non-IE isolates.
MATERIALS AND METHODS
Bacterial isolates.
Ninety-seven S. aureus clinical isolates were collected between 2003 and 2007 from patients admitted to metropolitan hospitals in Brisbane, Australia. These hospitals included the Royal Brisbane and Women's Hospital, Ipswich Hospital, and Princess Alexandra Hospital. Isolates were categorized according to the clinical outcome associated with the respective patients: (i) IE (n = 24), (ii) bloodstream infection related to a medical device (BSI-D; n = 17), (iii) bloodstream infection not related to a medical device (BSI-ND; n = 32), and (iv) skin or soft-tissue infection (SSTI; n = 24). S. aureus endocarditis was defined according to the modified Duke Criteria (22).
Preparation of genomic DNA.
Bacteria were grown overnight on Columbia blood agar. To prepare genomic DNA, six to eight colonies were suspended in 100 μl of lysis solution, which contained 0.05 mg lysostaphin (Sigma, Steinheim, Germany), 2 mg lysozyme (Sigma), 2 mg RNase A (Sigma), 2 μl 20 mM Tris-HCl (pH 8.0), 2 μl 2 mM EDTA, and 1 μl Triton X-100. The suspension was incubated at 37°C for 45 min with aeration (300 rpm). Proteinase K (10 μl) and buffer AL (100 μl) (DNeasy kit; QIAgen, Hilden, Germany) were added, and the suspension was incubated at 56°C for 45 min. The DNA was then processed using a QIAgen EZ1 device according to the manufacturer's tissue lysis protocol.
Multilocus sequence typing and eBURST analysis.
MLST was performed as previously described (23). Briefly, primers were designed to PCR amplify seven defined S. aureus genes (arc, aroE, glpF, gmk, pta, tpi, and yqi). The PCR products were purified using a PCR purification kit (QIAgen), and both strands were sequenced using a CEQ 8000 genetic analysis system (Beckman Coulter, High Wycombe, United Kingdom). The sequences were aligned and analyzed using BioNumerics software (Applied Maths, St-Martens-Latem, Belgium). Allelic profiles and STs were assigned, and an MLST tree was constructed using the MLST database (http://www.mlst.net). STs were analyzed and assigned into CCs by the use of eBURST software (http://eburst.mlst.net). Strains that diverged at no more than one of the seven MLST loci were considered to belong to the same CC. Double-locus variants were included in the CC if the linking single-locus variant was present in the MLST database. The STs for strains SA4, SA6, SA7, and SA18 were assigned based on >99% sequence conservation for yqi, tpi, aroE, and tpi alleles, respectively. The isolates were compared with each other as well as with the entire S. aureus population available in the MLST database at the time of the study.
spa typing.
The hypervariable region of the spa gene was amplified using Taq DNA polymerase (QIAgen) according to the manufacturer's protocol and using primers designed as previously described (24): 1095F (5′-AGACGATCCTTCGGTGAGC) and 1517R (5′-GCTTTTGCAATGTCATTTACTG). PCR cycling conditions were as follows: an initial 3 min at 94°C; 30 cycles of 30 s at 94°C, 30 s at 55°C, and 1 min at 72°C; and a final 10 min at 72°C. The PCR product was purified using a Wizard SV Gel and PCR Clean Up system (Promega, Madison, WI). The forward and reverse strands of the purified PCR product were sequenced using BigDye Terminator v3.1 according to the manufacturer's protocol (Applied Biosystems, Foster City, CA). The sequences were analyzed and spa types assigned using the spaType Finder (http://fortinbras.us/cgi-bin/spaTyper/spaTyper.pl) and the Ridom SpaServer database (http://spa.ridom.de).
DNA microarray analysis.
The DNA microarray and reagents were obtained from Alere Technologies (Jena, Germany). The principle of the assay and related procedures have been described previously (25, 26). Briefly, the assay employs a linear PCR-like approach to amplify and label target DNA. In this approach, the array is stained by horseradish peroxidase-based tetramethylbenzidine precipitation for 10 to 30 min, after which the resulting pattern is recorded and analyzed using an ATR01 reader (CLONDIAG) and IconoClust software (CLONDIAG). The raw data were interpreted as previously described (25, 26).
The DNA array targets, primers, and probes used in the study have been previously published (27). The array covered 352 target sequences consisting of a variety of species markers, markers for accessory gene regulator (agr) alleles and capsule types, virulence factors, resistance genes, staphylococcal superantigen-like and exotoxin-like genes (set and ssl), and genes encoding adhesins. Depending upon the nomenclature used, the target sequences correspond to 185 distinct genes and their allelic variants. All probes were specific to their target gene, were not self-hybridizing, and were similar with respect to GC content, length, and melting temperature. The probes were analyzed against the GenBank database (http://www.ncbi.nlm.nih.gov/BLAST/) to eliminate probes that might cause false-positive or false-negative reactions.
Statistical analysis of CC, spa type, and microarray data.
To determine the correlation between the presence of each CC, spa type, and IE isolates, a null model with 1,000 permutations was used. In each permutation, each CC/spa type was randomly assigned to a strain. The difference between the percentage of IE strains with that CC/spa type and the percentage likely to be obtained by chance was calculated. The percentages obtained in the 1,000 permutations were then used to calculate the likelihood of the observed difference occurring by chance. A correlation matrix was built to explore the correlation between the presence of each CC and each spa type.
Permutational analysis of multivariate variance (PERMANOVA) was used to determine if there were genotypic differences between IE and non-IE strains. A principal coordinates (PCO) analysis was used to represent the genotype in a two-dimensional graphical space. To identify groups of genes that were present in IE isolates, the correlation coefficient values corresponding to the presence or absence of each gene and the first two axes of the PCO were calculated. Groups of genes or alleles with correlations that corresponded to regions associated with IE isolates in the multivariate space were then identified.
RESULTS
Patients and isolates.
Ninety-seven S. aureus clinical isolates were collected from patients representing four disease categories: IE, BSI-D, BSI-ND, and SSTI. A summary of the characteristics of patients for each type of infection is presented in Table 1. There was no significant difference in the median ages of patients and the clinical outcomes. More males were diagnosed with IE than females (7:1). Only five of the 97 S. aureus isolates were methicillin-resistant S. aureus (MRSA) isolates. All five MRSA isolates were from bloodstream infections not associated with IE.
Table 1.
Characteristics of 97 patients diagnosed with S. aureus IE, BSI-D, BSI-ND, and SSTIa
| Characteristic | Values |
|||
|---|---|---|---|---|
| IE (n = 24) | BSI-D (n = 17) | BSI-ND (n = 32) | SSTI (n = 24) | |
| Median age in yr (range) | 55 (28–83) | 58 (0–88) | 67 (0–92) | 34 (4–86) |
| No. (%) of female patients | 3 (13) | 9 (53) | 10 (31) | 15 (63) |
| No. (%) of patients with MRSA infection | 0 (0) | 2 (12) | 3 (9) | 0 (0) |
BSI-D, bloodstream infection related to a medical device; BSI-ND, bloodstream infection not related to a medical device; SSTI, skin or soft-tissue infection; MRSA, methicillin-resistant S. aureus.
Analysis of CCs associated with IE S. aureus isolates.
Previous studies have suggested an association between S. aureus CC and the type and severity of infection (9–11). The ST of all 97 S. aureus isolates was therefore determined, and 27 different STs were identified. Sixteen different STs were identified among the 24 IE isolates (Table 2).
Table 2.
Characteristics of MSSA clinical isolates associated with IEa
| Isolate | Sex of patient | Age of patient (yr) | ST | Capsule |
|---|---|---|---|---|
| 1 | M | 66 | 12 | 8 |
| 2 | M | 59 | 15 | 8 |
| 3 | M | 35 | 72 | 5 |
| 4 | M | 83 | 12 | 8 |
| 5 | M | 74 | 1 | 8 |
| 6 | M | 35 | 8 | 5 |
| 7 | F | 73 | 78 | 8 |
| 8 | M | 77 | 20 | 5 |
| 9 | M | 62 | 641 | 5 |
| 10 | M | 27 | 5 | 5 |
| 11 | M | 31 | 20 | 5 |
| 12 | F | 33 | 20 | 5 |
| 13 | M | 21 | 97 | 5 |
| 14 | M | 69 | 1273 | 8 |
| 15 | M | 70 | 30 | 8 |
| 16 | M | 49 | 78 | 8 |
| 17 | M | 76 | 101 | 8 |
| 18 | M | 56 | 188 | 8 |
| 19 | M | 53 | 795 | 8 |
| 20 | M | 39 | 30 | 8 |
| 21 | M | 45 | 5 | 5 |
| 22 | M | 28 | 8 | 5 |
| 23 | M | 30 | 291 | 5 |
| 24 | F | 65 | 8 | 5 |
F, female; M, male; ST, sequence type; Capsule, capsular serotype.
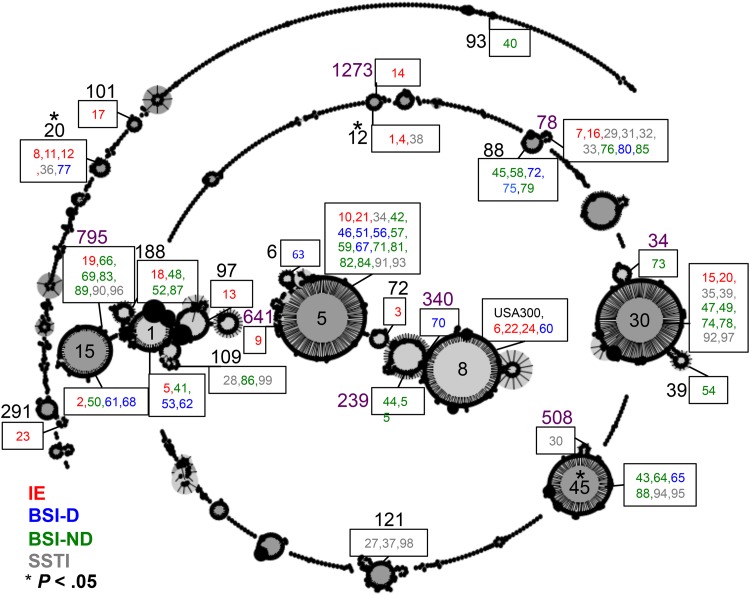
An eBURST analysis was conducted to group STs into CCs and to determine if particular CCs were associated with IE. Strains that diverged at no more than one of the seven MLST loci were considered to belong to the same CC. The clustering of STs into CCs allowed visualization of how the isolates were distributed among the members of the global S. aureus population (Fig. 1). The IE isolates were distributed across several CCs, and isolates with different clinical outcomes were assigned the same CC. We found that CC12 and CC20 were significantly positively associated with IE isolates and that CC45 was negatively associated with IE isolates in this study (Table 3).
Fig 1.
eBURST analysis showing phylogenetic relatedness of the 97 clinical S. aureus isolates with the S. aureus population. The number of individual isolates is written in a box next to the respective ST. The isolates are color coded according to clinical outcome: red for IE, blue for BSI-D, green for BSI-ND, and gray for SSTI. The ST number is provided beside the respective box or within the founder group. The CC number is shown in black, whereas a ST belonging to the CC is shown in purple. CCs were defined as all isolates within the group having at least six of the seven alleles identical with those of at least one other isolate in the group (http://eburst.mlst.net). CC12 and CC20 are significantly positively associated with IE, and CC45 is significantly negatively associated with IE (P < 0.05).
Table 3.
Relationship between CC and spa type with IE strainsa
| CC | % difference | P | spa type | % difference | P |
|---|---|---|---|---|---|
| CC12 | 11.13 | 0.007 | t160 | 6.96 | 0.003 |
| CC20 | 9.76 | 0.012 | t021 | 4.167 | 0.08 |
| CC8 | 7.02 | 0.06 | t024 | 4.167 | 0.08 |
| CC101 | 4.167 | 0.27 | t164 | 4.167 | 0.08 |
| CC291 | 4.167 | 0.27 | t267 | 4.167 | 0.08 |
| CC72 | 4.167 | 0.27 | t537 | 4.167 | 0.08 |
| CC97 | 4.167 | 0.27 | t888 | 4.167 | 0.08 |
| CC1 | 0.057 | 0.49 | t937 | 4.167 | 0.08 |
| CC188 | 0.057 | 0.49 | t1333 | 4.167 | 0.08 |
| CC39 | −1.37 | 0.43 | t1987 | 4.167 | 0.08 |
| CC6 | −1.37 | 0.43 | t11474 | 4.167 | 0.08 |
| CC93 | −1.37 | 0.43 | t11475 | 4.167 | 0.08 |
| CC15 | −4 | 0.241 | t002 | 2.91 | 0.238 |
| CC30 | −4 | 0.241 | t037 | −4.11 | 0.14 |
| CC109 | −4.11 | 0.24 | t371 | −4.11 | 0.14 |
| CC121 | −4.11 | 0.24 | t186 | −5.48 | 0.06 |
| CC5 | −6.68 | 0.18 | |||
| CC88 | −8.11 | 0.07 | |||
| CC45 | −9.59 | 0.04 |
% difference values represent the difference between the percentage of IE strains with that CC/spa type and the percentage likely to be obtained by chance. P values represent the probability that the difference between the percentages of IE and non-IE strains associated with each type was obtained by chance. Negative correlations are indicated with boldface characters.
Analysis of spa types associated with IE S. aureus isolates.
Two IE isolates contained novel spa sequences and were therefore designated new spa types from the Ridom SpaServer: t11474 for isolate 22 and t11475 for isolate 24. The prevalence of IE and non-IE isolates within each spa type was analyzed, and a significant association was found between spa type t160 and IE isolates (P < 0.05). A significant correlation was also found between CC12 and spa type t160 (P < 0.05). However, most CCs in this study were associated with a number of spa types (Fig. 2).
Fig 2.
A phylogenetic tree based on the ST of the 97 clinical S. aureus isolates and USA300. The spa type and isolate number are shown next to the respective ST. Isolates are color coded according to clinical outcome: red for IE, blue for BSI-D, green for BSI-ND, and gray for SSTI. The tree was constructed using the MLST database (http://www.mlst.net). CC12 and t160 were significantly associated with IE (P < 0.05).
Analysis of virulence genes associated with IE S. aureus isolates.
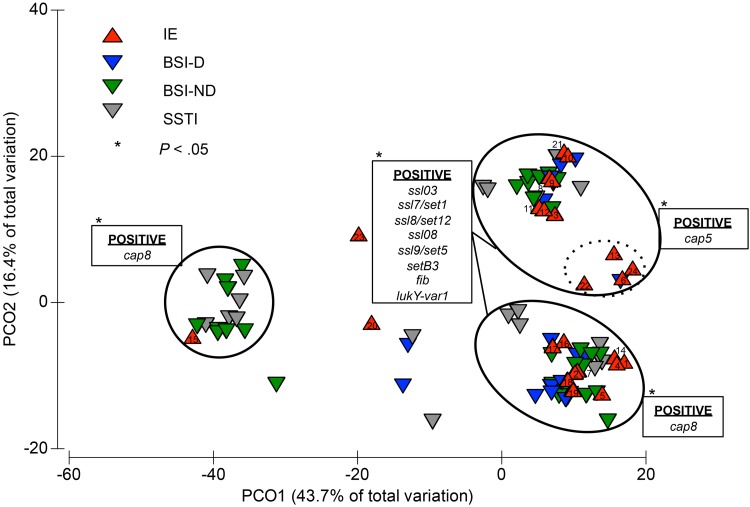
A number of genes, including the protease sspA, sspB, and sspP genes, the biofilm-associated icaA, icaC, and icaD genes, the microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) staphylococcal protein A (spa), clumping factor A (clfA), clumping factor B (clfB), enolase (eno), and fibronectin-binding protein A (fnbA) genes, and immune evasion transferrin-binding protein A (isdA) and type I site-specific DNase subunit (hsdSx) genes, were identified in all 97 isolates. The genes that showed variation between the isolates were analyzed to determine if any gene or gene set was more closely associated with the IE isolates than with the non-IE isolates. PERMANOVA results suggest that there is a marked difference between IE and non-IE strains (Sheudo-F = 3.0241; P = 0.01) The PCO shows the ordination of the different genes and alleles in a compressed two-dimensional space (Fig. 3). Isolates that clustered shared similar genotypes, whereas isolates that were spatially separated had greater genotypic variation.
Fig 3.
Ordination of the 97 clinical S. aureus isolates based on the first two axes of a PCO corresponding to the presence or absence of 352 target sequences. The presence of the genes and allelic variants was determined using a DNA microarray, and the 265 targets showing variation among the isolates were analyzed by a PERMANOVA and distributed in a PCO. Isolates are color coded according to clinical outcome: red for IE, blue for BSI-D, green for BSI-ND, and gray for SSTI. Genes are listed in the boxes next to the cluster containing isolates positive for the respective genes. The genes in the set consisting of ssl03, ssl7-set1, ssl8-set12, ssl08, ssl9-set5, setB3, fib, and lukY-var1 are significantly associated with the IE isolates (P < 0.05). The isolates within the dotted cluster differ from other cap5-positive IE isolates by subspecies allelic variations in several genes (lukS, sspA, ssl1-set6, ssl01, ssl11-set2, bbp, clfA, clfB, fnbA, fnbB, sdrC, sasG, hsdS3, Q2FXCO, and vwb).
The majority of the IE isolates (n = 21; 88%) clustered on the right of the plot and contained the ssl03, ssl7-set1, ssl8-set12, ssl08, ssl9-set5, and setB3 genes, the fibrinogen binding protein (fib) gene, and the leukocidin/hemolysin toxin family protein lukY-var1 gene (Fig. 3). In contrast, a largely non-IE isolate cluster on the left of the plot (which included IE isolate 15) did not contain these genes or contained allelic variants of these genes. A different set of genes and alleles, including ssl9-set5 (MRSA252), setB3 (MRSA252), lukY-var2, type I site-specific DNase subunit, 2nd locus (hsdS2) (MRSA252), ssl4-set9 (MRSA252, SAR0425), ssl5-set3 (MRSA252), and setB2, was present in this largely non-IE isolate cluster. The gene set ssl03, ssl7-set1, ssl8-set12, ssl08, ssl9-set5, setB3, fib, and lukY-var1 was significantly associated with IE isolates compared with the non-IE isolates (P < 0.05).
Variation was also observed in the genes that encode serotype-specific capsular polysaccharides. The S. aureus isolates in this study contained either cap5 genes (n = 32; 33%) or cap8 genes (n = 65; 67%). Among the 24 IE isolates, the cap5 and cap8 genes were equally distributed. Four of the cap5 IE isolates clustered into a tight subgroup (Fig. 3; dotted oval), the majority of whose members were IE isolates (n = 4; 80%). The other isolate within this subgroup was from a patient with a bloodstream infection related to a hemodialysis intravascular device. The differentiation of this subgroup from the other IE cap5 isolates is attributable to subspecies allelic variations between several genes, including those encoding glutamyl endopeptidase (sspA) and bone sialoprotein-binding protein (bbp), clfA, clfB, fnbA, fnbB, and the gene encoding von Willebrand factor-binding protein (vwb).
DISCUSSION
In this study, we determined the CC, spa type, and virulence-associated gene content of 24 S. aureus isolates associated with IE and compared these profiles to those of a set of S. aureus strains isolated from patients with different clinical syndromes from the same geographic location. Two CCs, one spa type, and a discrete set of virulence-associated genes and allelic variants displayed significant correlations with IE.
Although some studies have found no significant associations between CC and invasive disease (12, 13), it has also been suggested that IE S. aureus isolates generally belong to CC1, CC5, CC8, CC15, CC30, and CC45 (9, 10, 12). In this study, a total of 19 CCs were identified from the 97 clinical S. aureus isolates; these included 13 CCs containing IE isolates. Only 11 of the 24 IE isolates belonged to CC1, CC5, CC8, CC15, CC30, or CC45. Two IE isolates (8%) compared with nine non-IE isolates (12%) belonged to CC30; thus, there was no significant association between CC30 and IE in our study set. In contrast, we found that CC12 and CC20 were significantly positively associated with IE isolates and that CC45 was negatively associated with IE. The difference between this study and previous studies is most likely due to the different strain sets analyzed; for example, genetic variation exists between strains from different geographic locations (10).
As no previous study has reported a correlation between spa type and IE, spa typing was performed in this work to determine if particular spa types were associated with IE. spa type t160 was significantly associated with IE. A significant correlation was also found between CC12 and spa type t160. Most CCs in this study were associated with a number of spa types.
A DNA microarray and PERMANOVA were also performed to determine if known S. aureus virulence genes were associated with IE isolates. Analysis of the DNA microarray data clustered isolates into three main groups. Isolates between clusters differed in the presence of the genes and allelic variants ssl03, ssl7-set1, ssl8-set12, ssl08, ssl9-set5, setB3, fib, and lukY-var1 and the presence of cap5 or cap8 genes. The set of genes and allelic variants was significantly associated with IE isolates compared with non-IE isolates (P < 0.05). This result differs from those of a recent study by Tristan et al. in which no difference between IE and non-IE S. aureus isolates in the prevalences of individual virulence-associated genes was found. A two-tailed Fisher's exact test was used in this recent study, which analyzed the results obtained with each gene independently (12). However, the PERMANOVA used in the current work was able to identify correlations between gene sets and clinical outcome. As multiple genes are likely to contribute to complex phenotypes such as the capacity to cause bacterial IE, the PERMANOVA is better suited to predict a gene repertoire that correlates with disease.
The identified S. aureus-IE gene set contains a number of genes that encode superantigen-like proteins (Ssls), previously known as staphylococcal exotoxin-like proteins (Sets). All known ssl-set genes are located on mobile genetic elements, with set6–15 located in a 19-kb region on staphylococcal pathogenicity island 2 (28, 29). Ssl/Set proteins are structurally homologous to staphylococcal superantigens but lack superantigenic properties such as mitogenicity, pyrogenicity, and inducement of endotoxic shock (28, 30). Although defined functions have not yet been assigned to every Ssl/Set protein, the members of this family of proteins are collectively known to mediate immune evasion by inhibiting complement activation and binding to IgA and IgG and preventing the recruitment and function of neutrophils (31).
The fib gene encodes an extracellular fibrinogen binding protein (Efb), which is secreted by S. aureus during infection (32). Efb plays a number of important roles during pathogenesis. The protein binds to host fibrinogen (Fg) and platelets and therefore blocks the binding of Fg to neutrophils and inhibits platelet aggregation (33–35). Efb also binds to C3b, which minimizes B and T cell responses, and inhibits complement-mediated opsonization and phagocytosis (36, 37).
The lukY gene encodes a leukocidin subunit which is not associated with the well-characterized Panton-Valentine leukocidin (PVL) lukF-PV and lukS-PV genes. It is important to highlight that, according to our analysis (as partially supported by previous results [12]), the gene sets identified as positively or negatively related to IE appear to have a synergistic effect. It could therefore be misleading to infer that each independent member of each gene subset has a direct independent effect on IE.
The PCO analysis revealed that some isolates from different CCs clustered, such as isolate 12 (CC97) clustering with isolates 6, 22, and 24 (CC8). This observation is not surprising, as virulence-associated genes are readily passed horizontally between unrelated strains via homologous recombination or acquisition of mobile elements (38). In contrast, genes in the core genome, such as the seven MLST genes, are vertically acquired from a common ancestor and evolve more slowly. The CC or spa type alone does not indicate the potential of an isolate to cause disease (13).
It is likely that there are other S. aureus virulence determinants associated with IE that were not identified due to their absence from the DNA microarray. In addition, all IE strains analyzed in this study were methicillin-susceptible S. aureus (MSSA), and it is possible that there are differences between MSSA and MRSA strains in IE-associated genes. For example, MSSA and MRSA utilize different mechanisms for biofilm growth (39). MRSA strains typically utilize protein adhesins such as FnBP to form biofilms, whereas biofilm formation by MSSA strains is generally dependent on production of the polysaccharide adhesin polymeric N-acetylglucosamine. In addition, a combination of host factors (e.g., age, gender, and immune status) may contribute to the capacity of S. aureus to cause IE in an individual.
In conclusion, we have determined that CC12, CC20, and t160 are significantly associated with the IE-related S. aureus isolates used in this study. These two CCs and this spa type could be used to differentiate S. aureus strains which have the potential to cause IE from less virulent S. aureus strains. We have also identified S. aureus genes and allelic variants, namely, ssl03, ssl7-set1, ssl8-set12, ssl08, ssl9-set5, setB3, fib, and lukY-var1, which are significantly associated with IE. These IE-associated genes may provide a framework for further study to investigate the virulence of S. aureus strains associated with IE.
ACKNOWLEDGMENTS
Cara Nethercott is the recipient of an Australian National Health and Medical Research (http://www.nhmrc.gov.au/) Ph.D. scholarship.
This work was supported by grants from the PA Research Foundation and the Australian National Health and Medical Research Council.
Footnotes
Published ahead of print 24 April 2013
REFERENCES
- 1. Fowler VG, Jr, Bayer AS. 2012. Infective endocarditis, p 464–473 In Goldman L, Schafer AI. (ed), Goldman's Cecil medicine, 24th ed, vol 1 Elsevier Inc., New York, NY [Google Scholar]
- 2. Que Y-A, Moreillon P. 2011. Infective endocarditis. Nat. Rev. Cardiol. 8:322–336 [DOI] [PubMed] [Google Scholar]
- 3. Sousa C, Botelho C, Rodrigues D, Azeredo J, Oliveira R. 2012. Infective endocarditis in intravenous drug abusers: an update. Eur. J. Clin. Microbiol. Infect. Dis. 31:2905–2910 [DOI] [PubMed] [Google Scholar]
- 4. Hill EE, Herijgers P, Claus P, Vanderschueren S, Herregods M-C, Peetermans WE. 2007. Infective endocarditis: changing epidemiology and predictors of 6-month mortality: a prospective cohort study. Eur. Heart J. 28:196–203 [DOI] [PubMed] [Google Scholar]
- 5. Murdoch DR, Corey G, Hoen B, Miro JM, Fowler VG, Jr, Bayer AS, Karchmer AW. 2009. Clinical presentation, etiology, and outcome of infective endocarditis in the 21st century: the international collaboration on endocarditis—prospective cohort study. Arch. Intern. Med. 169:463–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Roig I, Darouiche R, Musher D, Trautner B. 2012. Device-related infective endocarditis, with special consideration of implanted intravascular and cardiac devices in a predominantly male population. Scand. J. Infect. Dis. 44:753–760 [DOI] [PubMed] [Google Scholar]
- 7. Fowler VG, Miro JM, Hoen B, Cabell CH, Abrutyn E, Rubinstein E, Corey R. 2005. Staphylococcus aureus endocarditis: a consequence of medical progress. JAMA 293:3012–3021 [DOI] [PubMed] [Google Scholar]
- 8. Wang A, Athan E, Pappas P, Fowler V, Olaison L, Pare C, Almirante B. 2007. Contemporary clinical profile and outcome of prosthetic valve endocarditis. JAMA 297:1354–1361 [DOI] [PubMed] [Google Scholar]
- 9. Fowler VG, Nelson CL, McIntyre LM, Kreiswirth BN, Monk A, Archer GL, Federspiel J, Naidich S, Remortel B, Rude T, Brown P, Reller LB, Corey GR, Gill SR. 2007. Potential associations between hematogenous complications and bacterial genotype in Staphylococcus aureus infection. J. Infect. Dis. 196:738–747 [DOI] [PubMed] [Google Scholar]
- 10. Nienaber JJC, Sharma Kuinkel BK, Clarke-Pearson M, Lamlertthon S, Park L, Rude TH, Barriere S, Woods CW, Chu VH, Marín M, Bukovski S, Garcia P, Corey GR, Korman T, Doco-Lecompte T, Murdoch DR, Reller LB, Fowler VG, for the International Collaboration on Endocarditis-Microbiology I 2011. Methicillin-susceptible Staphylococcus aureus endocarditis isolates are associated with clonal complex 30 genotype and a distinct repertoire of enterotoxins and adhesins. J. Infect. Dis. 204:704–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gill SR, McIntyre LM, Nelson CL, Remortel B, Rude T, Reller LB, Fowler VG., Jr 2011. Potential associations between severity of infection and the presence of virulence-associated genes in clinical strains of Staphylococcus aureus. PLoS One 6:e18673. 10.1371/journal.pone.0018673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tristan A, Rasigade J-P, Ruizendaal E, Laurent F, Bes M, Meugnier H, Lina G, Etienne J, Celard M, Tattevin P, Monecke S, Le Moing V, Vandenesch F, the French AsGoIE 2012. Rise of CC398 lineage of Staphylococcus aureus among infective endocarditis isolates revealed by two consecutive population-based studies in France. PLoS One 7:e51172. 10.1371/journal.pone.0051172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Feil EJ, Cooper JE, Grundmann H, Robinson DA, Enright MC, Berendt T, Peacock SJ, Smith JM, Murphy M, Spratt BG, Moore CE, Day NPJ. 2003. How clonal is Staphylococcus aureus? J. Bacteriol. 185:3307–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chi C-Y, Wang S-M, Lin C-C, Liu C-C. 2010. Microbiological characteristics of community-associated Staphylococcus aureus causing uncomplicated bacteremia and infective endocarditis. J. Clin. Microbiol. 48:292–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hallin M, Friedrich AW, Struelens MJ. 2009. Spa typing for epidemiological surveillance of Staphylococcus aureus. Methods Mol. Biol. 551:189–202 [DOI] [PubMed] [Google Scholar]
- 16. Buck AW, Fowler VG, Yongsunthon R, Liu J, DiBartola AC, Que Y-A, Moreillon P, Lower SK. 2010. Bonds between fibronectin and fibronectin-binding proteins on Staphylococcus aureus and Lactococcus lactis. Langmuir 26:10764–10770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Que Y-A, Haefliger J-A, Piroth L, François P, Widmer E, Entenza JM, Sinha B, Herrmann M, Francioli P, Vaudaux P, Moreillon P. 2005. Fibrinogen and fibronectin binding cooperate for valve infection and invasion in Staphylococcus aureus experimental endocarditis. J. Exp. Med. 201:1627–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Widmer E, Que Y-A, Entenza JM, Moreillon P. 2006. New concepts in the pathophysiology of infective endocarditis. Curr. Infect. Dis. Rep. 8:271–279 [DOI] [PubMed] [Google Scholar]
- 19. Piroth L, Que Y-A, Widmer E, Panchaud A, Piu S, Entenza JM, Moreillon P. 2008. The fibrinogen- and fibronectin-binding domains of Staphylococcus aureus fibronectin-binding protein A synergistically promote endothelial invasion and experimental endocarditis. Infect. Immun. 76:3824–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thakker M, Park J-S, Carey V, Lee JC. 1998. Staphylococcus aureus serotype 5 capsular polysaccharide is antiphagocytic and enhances bacterial virulence in a murine bacteremia model. Infect. Immun. 66:5183–5189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Karakawa WW, Sutton A, Schneerson R, Karpas A, Vann WF. 1988. Capsular antibodies induce type-specific phagocytosis of capsulated Staphylococcus aureus by human polymorphonuclear leukocytes. Infect. Immun. 56:1090–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li JS, Sexton DJ, Mick N, Nettles R, Fowler VG, Ryan T, Bashore T, Corey GR. 2000. Proposed modifications to the Duke criteria for the diagnosis of infective endocarditis. Clin. Infect. Dis. 30:633–638 [DOI] [PubMed] [Google Scholar]
- 23. Enright MC, Day NPJ, Davies CE, Peacock SJ, Spratt BG. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Harmsen D, Claus H, Witte W, Rothgänger J, Claus H, Turnwald D, Vogel U. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442–5448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Monecke S, Jatzwauk L, Weber S, Slickers P, Ehricht R. 2008. DNA microarray-based genotyping of methicillin-resistant Staphylococcus aureus strains from eastern Saxony. Clin. Microbiol. Infect. 14:534–545 [DOI] [PubMed] [Google Scholar]
- 26. Monecke S, Slickers P, Ehricht R. 2008. Assignment of Staphylococcus aureus isolates to clonal complexes based on microarray analysis and pattern recognition. FEMS Immunol. Med. Microbiol. 53:237–251 [DOI] [PubMed] [Google Scholar]
- 27. Coombs G, Monecke S, Pearson J, Tan H-L, Chew Y-K, Wilson L, Ehricht R, O'Brien F, Christiansen K. 2011. Evolution and diversity of community-associated methicillin-resistant Staphylococcus aureus in a geographical region. BMC Microbiol. 11:215. 10.1186/1471-2180-11-215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wilson GJ, Seo KS, Cartwright RA, Connelley T, Chuang-Smith ON, Merriman JA, Guinane CM, Park JY, Bohach GA, Schlievert PM, Morrison WI, Fitzgerald JR. 2011. A novel core genome-encoded superantigen contributes to lethality of community-associated MRSA necrotizing pneumonia. PLoS Pathog. 7:e1002271. 10.1371/journal.ppat.1002271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, Lian J, Ito T, Kanamori M, Matsumaru H, Maruyama A, Murakami H, Hosoyama A, Mizutani-Ui Y, Takahashi NK, Sawano T, Inoue R, Kaito C, Sekimizu K, Hirakawa H, Kuhara S, Goto S, Yabuzaki J, Kanehisa M, Yamashita A, Oshima K, Furuya K, Yoshino C, Shiba T, Hattori M, Ogasawara N, Hayashi H, Hiramatsu K. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225–1240 [DOI] [PubMed] [Google Scholar]
- 30. Arcus VL, Langley R, Proft T, Fraser JD, Baker EN. 2002. The three-dimensional structure of a superantigen-like protein, SET3, from a pathogenicity island of the Staphylococcus aureus genome. J. Biol. Chem. 277:32274–32281 [DOI] [PubMed] [Google Scholar]
- 31. Fraser JD, Proft T. 2008. The bacterial superantigen and superantigen-like proteins. Immunol. Rev. 225:226–243 [DOI] [PubMed] [Google Scholar]
- 32. Colque-Navarro P, Palma M, Söderquist B, Flock J-I, Möllby R. 2000. Antibody responses in patients with staphylococcal septicemia against two Staphylococcus aureus fibrinogen binding proteins: clumping factor and an extracellular fibrinogen binding protein. Clin. Diagn. Lab. Immunol. 7:14–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Palma M, Shannon O, Quezada HC, Berg A, Flock J-I. 2001. Extracellular fibrinogen-binding protein, Efb, from Staphylococcus aureus blocks platelet aggregation due to its binding to the α-chain. J. Biol. Chem. 276:31691–31697 [DOI] [PubMed] [Google Scholar]
- 34. Shannon O, Flock J-I. 2004. Extracellular fibrinogen binding protein, Efb, from Staphylococcus aureus binds to platelets and inhibitis platelet aggregation. Thromb Haemost. 91:779–789 [DOI] [PubMed] [Google Scholar]
- 35. Ko Y-P, Liang X, Smith CW, Degen JL, Höök M. 2011. Binding of Efb from Staphylococcus aureus to fibrinogen blocks neutrophil adherence. J. Biol. Chem. 286:9865–9874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee LYL, Liang X, Höök M, Brown EL. 2004. Identification and characterization of the C3 binding domain of the Staphylococcus aureus extracellular fibrinogen-binding protein (Efb). J. Biol. Chem. 279:50710–50716 [DOI] [PubMed] [Google Scholar]
- 37. Ricklin D, Ricklin-Lichtsteiner SK, Markiewski MM, Geisbrecht BV, Lambris JD. 2008. Cutting edge: members of the Staphylococcus aureus extracellular fibrinogen-binding protein family inhibit the interaction of C3d with complement receptor 2. J. Immunol. 181:7463–7467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Turner KME, Feil EJ. 2007. The secret life of the multilocus sequence type. Int. J. Antimicrob. Agents 29:129–135 [DOI] [PubMed] [Google Scholar]
- 39. Pozzi C, Waters EM, Rudkin JK, Schaeffer CR, Lohan AJ, Tong P, Loftus BJ, Pier GB, Fey PD, Massey RC, O'Gara JP. 2012. Methicillin resistance alters the biofilm phenotype and attenuates virulence in Staphylococcus aureus device-associated infections. PLoS Pathog. 8:e1002626. 10.1371/journal.ppat.1002626 [DOI] [PMC free article] [PubMed] [Google Scholar]