Abstract
During the past 5 years, matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (MS) has become a powerful tool for routine identification in many clinical laboratories. We analyzed our 11-year experience in routine identification of clinical isolates (40 months using MALDI-TOF MS and 91 months using conventional phenotypic identification [CPI]). Among the 286,842 clonal isolates, 284,899 isolates of 459 species were identified. The remaining 1,951 isolates were misidentified and required confirmation using a second phenotypic identification for 670 isolates and using a molecular technique for 1,273 isolates of 339 species. MALDI-TOF MS annually identified 112 species, i.e., 36 species/10,000 isolates, compared to 44 species, i.e., 19 species/10,000 isolates, for CPI. Only 50 isolates required second phenotypic identifications during the MALDI-TOF MS period (i.e., 4.5 reidentifications/10,000 isolates) compared with 620 isolates during the CPI period (i.e., 35.2/10,000 isolates). We identified 128 bacterial species rarely reported as human pathogens, including 48 using phenotypic techniques (22 using CPI and 37 using MALDI-TOF MS). Another 75 rare species were identified using molecular methods. MALDI-TOF MS reduced the time required for identification by 55-fold and 169-fold and the cost by 5-fold and 96-fold compared with CPI and gene sequencing, respectively. MALDI-TOF MS was a powerful tool not only for routine bacterial identification but also for identification of rare bacterial species implicated in human infectious diseases. The ability to rapidly identify bacterial species rarely described as pathogens in specific clinical specimens will help us to study the clinical burden resulting from the emergence of these species as human pathogens, and MALDI-TOF MS may be considered an alternative to molecular methods in clinical laboratories.
INTRODUCTION
Early and accurate microbial identification is a critical requisite for early, adequate antibiotic treatment. The number of newly described bacteria has risen impressively during the past few decades (1, 2). Notably, the identification of new pathogens in clinical microbiology has been spectacularly improved during previous decades by the use of molecular identification, especially 16S rRNA gene sequencing (3–8). Molecular identification is one of the most useful techniques but remains expensive and requires a workload that is not adapted for routine use. Moreover, clinical definitions of some species do not match those used for 16S rRNA identification, such as the mismatched definitions used for streptococci (9–11).
Bacterial identification directly from colonies and samples using matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (MS) has been described as a revolutionary tool perfectly adapted to the clinical microbiology laboratory (12, 13). MALDI-TOF MS has been used to identify bacterial species and subspecies (14, 15), and in some outbreaks, MALDI-TOF MS has been reported to be able to identify the lineages of strains (16–18). Recently, MALDI-TOF MS has also been used to detect clinical pathogens previously misidentified or ambiguously identified (19–24). Detection of antimicrobial resistance using MALDI-TOF MS has been reported for Staphylococcus aureus (25–32), Acinetobacter baumannii (26), Escherichia coli, and other members of the family Enterobacteriaceae (33–35). Several new bacterial species emerging as human pathogens have been identified using MALDI-TOF MS (36–45).
In the present study, we examined data from a large collection of clinical isolates routinely identified during the last 11 years in our laboratory to evaluate the performance of MALDI-TOF MS for routine bacterial identification compared with conventional phenotypic identification (CPI). Particularly, we evaluated the capacity of MALDI-TOF MS to identify bacterial species that were rarely reported as human pathogens compared with conventional phenotypic and molecular identifications.
MATERIALS AND METHODS
Specimen collection.
Clinical isolates were recovered from blood samples, cerebrospinal fluid samples, wounds, exudate samples, abscesses, respiratory tract samples, genitourinary samples, bone-joint infection samples, digestive samples, stools, and other clinical samples from 1 January 2002 through 31 December 2012, excluding December 2002 (data not available). In September 2008, an anaerobic laboratory with anaerobic chamber, preincubation of agar plates in strictly anaerobic condition, and a team of dedicated technicians was created with the opening of another laboratory at the North University Hospital, Marseille, France (600 beds) in our 4,000-bed university hospital.
Bacterial identification.
All isolates were identified after aerobic, microaerophilic, and anaerobic incubation of clinical specimens on 5% sheep blood, chocolate, Mueller-Hinton, Trypticase soy, and MacConkey agar plates (bioMérieux).
(i) Conventional phenotypic identification period.
In CPI, we used semiautomated Gram staining (Aerospray Wiescor; Elitech), determined catalase and oxidase activities, and used the Vitek 2 system (bioMérieux), with 330 microorganism strains as references or the API 20A identification strip for anaerobes (bioMérieux) to identify bacterial species from 1 January 2002 to 30 August 2009. Correct identification of an isolate using the Vitek 2 system was confirmed when the T index was ≥0.25; identification using the API system was confirmed when the percentage of identification was ≥90%, and the T index was ≥ 0.25 (46). We reidentified organisms by Gram staining rather than by using the Vitek 2 system. API identification strips included API 20A, API Coryne, API Campy, API 20E, API 20NE, API Strep, API Staph, API NH, and API Listeria strips (bioMérieux) as the second phenotypic identification in the CPI period to identify uncertainly identified isolates at the species level.
(ii) MALDI-TOF MS identification period. (a) MALDI-TOF MS analysis.
We used MALDI-TOF MS as a routine bacterial identification tool to categorize bacterial species from direct colonies, and the procedure was performed as previously described (12). We used a MALDI-TOF MS AutoFlex II system (Brüker Daltonik) for the first part of the MALDI-TOF MS identification period, from 1 September 2009 to 30 November 2010 and a MicroFlex LT mass spectrometer (Brüker Daltonik) for the second part of the MALDI-TOF MS identification period, from 1 December 2010 to 31 December 2012.
(b) MALDI-TOF mass spectrum database.
The Brüker database updated with a laboratory collection of spectra from clinical isolates identified by 16S rRNA gene sequencing was used from 1 September 2009 to 31 December 2012. For each organism updated, a consensus spectrum was obtained by using the Biotyper MSP (mean spectrum projection) creation standard method from a total of 12 spots made for each isolate, and the manipulation was repeated in two independent runs. The Fisher exact test was used to evaluate the reproducibility. We determined the sensitivity of MALDI-TOF MS by identification of 10 colonies of the same bacterial species in another independent run. Our MALDI-TOF mass spectrum database has 6,213 reference microorganism strain spectra, and we updated the primary Brüker database containing 3,993 microorganism spectra (3,670 of bacteria, 7 of Archaea, and 316 of Eukaryota) with laboratory bacterial spectra including spectra from well-typed bacterial strains and other human-pathogenic bacteria identified by using a molecular technique.
(c) MALDI-TOF MS identification.
Bacterial species were directly identified from one bacterial colony; each colony was covered with 2 ml of matrix solution (saturated α-cyano-4-hydroxycinnamic acid in 50% acetonitrile and 2.5% trifluoroacetic acid) without other supplements and extracted as previously described (12). We used MALDI Biotyper 3.0 software to compare the first 100 peaks of each spectrum to our MALDI-TOF mass spectrum database previously updated as described below. An isolate was considered correctly identified at the species level by using MALDI-TOF MS if 2 spectra had scores of ≥1.9. Uncertainly identified isolates at the species level (scores of <1.9) were identified with certainty by MALDI-TOF MS analysis of 2 additional spectra. A second run of MALDI-TOF MS identification with 4 spectra was done for unsatisfied species identification in the MALDI-TOF MS period.
(iii) Molecular identification.
Isolates misidentified by the second CPI or MALDI-TOF MS analyses were identified with certainty using molecular identification using 16S rRNA or rpoB gene sequencing as described elsewhere (4, 12, 47, 48). An isolate was correctly identified when (i) its 16S rRNA gene sequence yielded ≥98.7% identity with the sequence of the most closely related bacterial species in GenBank (49) or (ii) when its rpoB gene sequence yielded ≥97% identity with the sequence of the most closely related bacterial species in GenBank or a local database (12, 48).
Database analysis.
Our database included bacterial identification results and their associated clinical information; 500,174 identifications of clinical isolates were performed during the study period. All results were extracted into Microsoft Excel files for further analysis. Duplicate analyses were eliminated by retaining only a single bacterial identification per sample. We also excluded all samples for which there were phenotypic or molecular identifications of fungi, environmental isolates, Mycobacterium, and other intra- and extralaboratory strains that were not of human origin.
Meaning of rare species.
Rare species were defined as bacterial species with ≤10 reports designating them as human pathogens retrieved from the PubMed database (http://www.ncbi.nlm.nih.gov/pubmed/). The possibility of inaccurate classifications as rare species due to taxonomy changes was checked using the National Center for Biotechnology Information (NCBI) taxonomy database (http://www.ncbi.nlm.nih.gov/guide/taxonomy/).
Time, cost, and training requirement evaluation of a MALDI-TOF MS identification technique.
We evaluated the time required for the MALDI-TOF mass spectrometry identification as the period between the deposit of a bacterial colony on the MALDI-TOF MS plate by a technician and the completion of the informatics interpretation of the resulting spectra (i.e., identification ready to be transmitted to a clinician). The costs of identification were evaluated by adding the costs of matrix reagents, plates, positive controls, and technician salary, with provisions for 5-year depreciations of the apparatuses used (Gram staining apparatus, microscope, identification apparatus, and mass spectrometer) on the basis of ≈67,000 isolates analyzed per year (the number of samples analyzed in 2012 in our laboratory).
Statistical analysis.
Data analyses were performed using IBM SPSS Statistics software version 20.0. Proportions were compared using the chi-squared or Fisher's exact two-tailed tests. A P value of <0.05 was considered statistically significant.
RESULTS
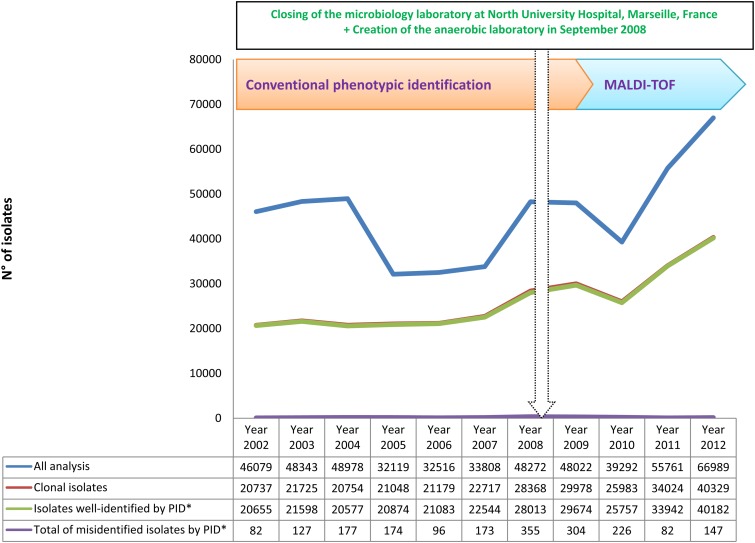
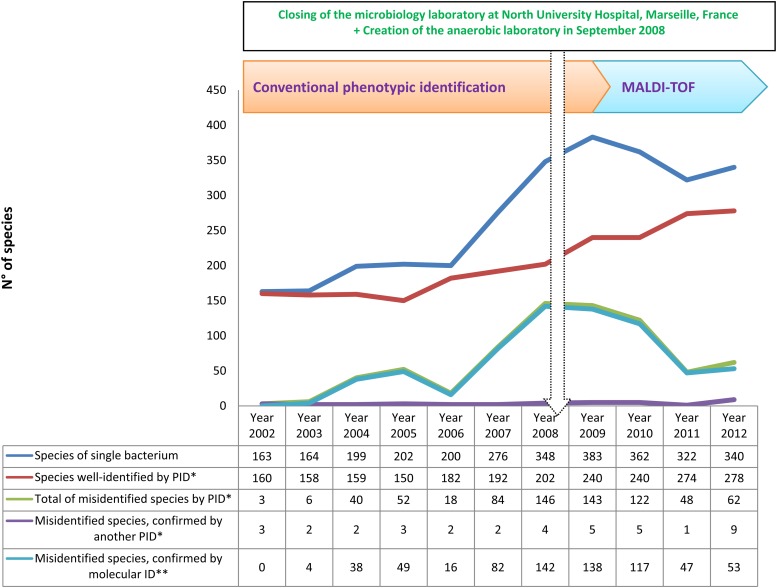
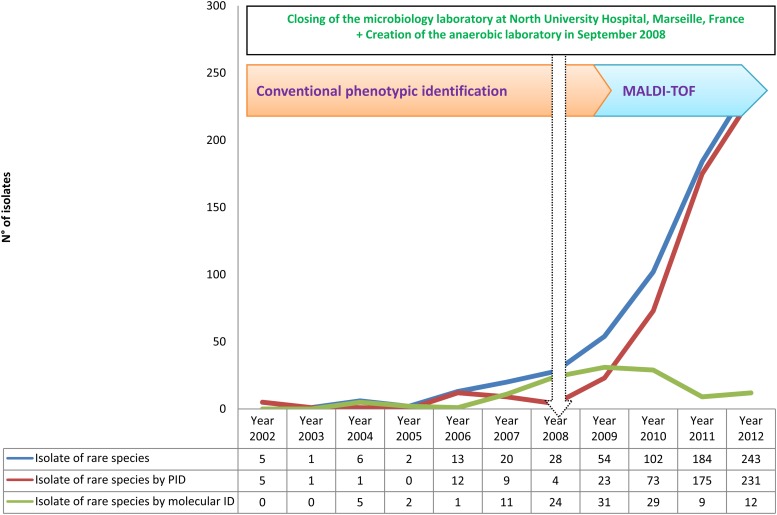
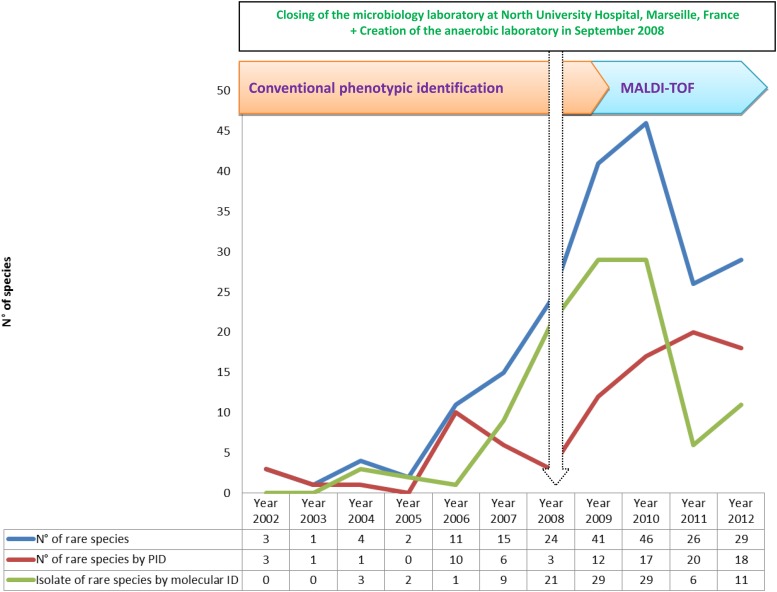
Over 11 years, we performed 500,179 bacterial identifications in our laboratory (Table 1). We grew our capacity for identification between 2002 and 2012, increasing the number of analyses from 46,079 per year to 66,989 per year, by creating an anaerobic laboratory and joining with another microbiology laboratory located at North University Hospital, Marseille, France, in September 2008 (Fig. 1). The implementation of a new tool for identification (MALDI-TOF MS) has spectacularly improved our capacity to identify more clinical isolates and more human-pathogenic bacteria. We identified 160 bacterial species during 2002 and 278 species during 2012 (Fig. 2).
Table 1.
Summary of 11 years of bacterial identification in our laboratorya
| Identification technique (study period [day-mo-yr]) | Study period (no. of months) | Total no. of analyses | No. of clonal isolates | No. of isolates identified by 1st PID | No. of species identified by 1st PID | No. of bacterial species identified/year | No. of isolates confirmed by 2nd PID | No. of isolates identified by molecular identification | No. of isolates misidentified by 1st PID | % misidentified |
|---|---|---|---|---|---|---|---|---|---|---|
| CPI period (1-Jan-02 to 30-Aug-09) | 91 | 322,291 | 175,999 | 174,636 | 336 | 44 | 620 | 743 | 1,363 | 0.77 |
| MALDI-TOF MS period (1-Sep-09 to 30-Dec-12) | 40 | 177,888 | 110,843 | 110,263 | 382 | 112 | 50 | 530 | 580 | 0.52 |
| AutoFlex II (1-Sep-09 to 30-Nov-10) | 15 | 52,695 | 34,839 | 34,497 | 264 | 211 | 32 | 310 | 342 | 0.98 |
| MicroFlex (1-Dec-10 to 31-Dec-12) | 25 | 125,193 | 76,004 | 75,766 | 340 | 163 | 18 | 220 | 238 | 0.31 |
| Total | 131 | 500,179 | 286,842 | 284,899 | 459 | 42 | 670 | 1,273 | 1,951 | 0.68 |
We identified 459 bacterial species among 284,899 clinical isolates during nearly 11 years. We identified 112 species per year using MALDI-TOF MS compared with 44 identified using conventional phenotypic identification (CPI) (Gram staining, API, Vitek 2 system identification). PID, phenotypic identification.
Fig 1.
Time course of the total numbers of isolates analyzed, clonal isolates analyzed, and clonal isolates identified and misidentified using phenotypic identification (PID*) during 11 years of routine identification in our clinical laboratory.
Fig 2.
Time course of the numbers of species of clonal bacteria identified, species identified using an initial phenotypic identification (PID*), total species misidentified, species confirmed by another PID*, and species confirmed by molecular identification (molecular ID**) over 11 years of routine identification in our clinical laboratory.
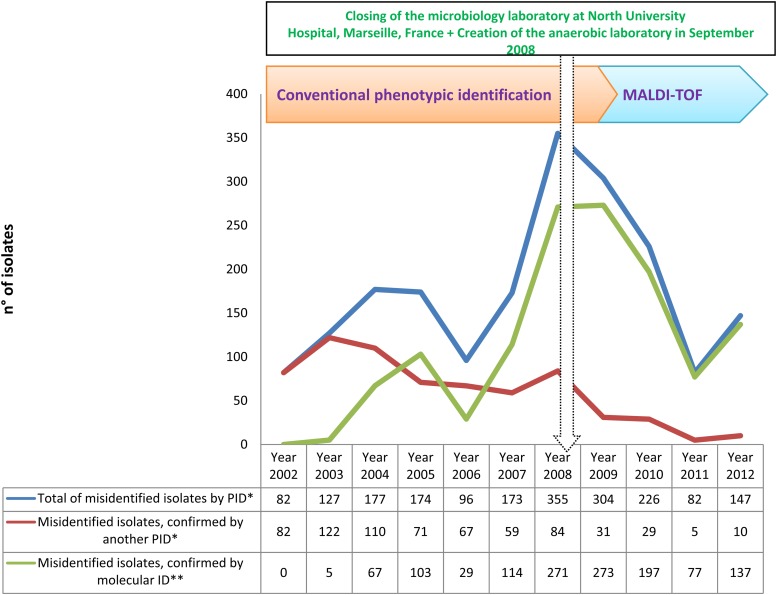
Among 286,842 clonal isolates identified, phenotypic identification methods (CPI or MALDI-TOF MS) correctly identified 284,899 isolates including 459 species of 134 genera and 6 phyla. Another 1,951 isolates were misidentified and required identification by another phenotypic or molecular method (Table 1 and Fig. 3).
Fig 3.
Time course of the numbers of total isolates misidentified using phenotypic identification (PID*), isolates confirmed by a second PID* and isolates confirmed by molecular identification (ID**) over 11 years of routine identification in our clinical laboratory.
CPI identified 174,636 isolates, including 336 species of 120 genera and 6 phyla, over the 91 months from 1 January 2002 through 30 August 2009, whereas MALDI-TOF MS identified 110,263 isolates classified in 382 species of 114 genera and 6 phyla over the 40 months from 1 September 2009 through 31 December 2012. Thus, MALDI-TOF MS yearly identified 32,430 isolates of 112 species, i.e., 36 species/10,000 isolates, compared with 22,692 isolates of 44 species, i.e., 19 species per 10,000 isolates, for CIP (P < 0.0001) (Table 1 and Fig. 4).
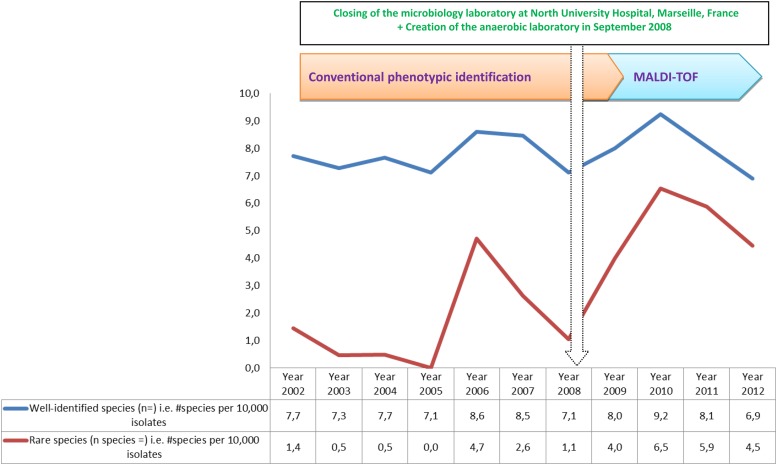
Fig 4.
Biodiversity of rare species identified in the routine identification of all clinical isolates tested (identified plus misidentified) during the last 11 years.
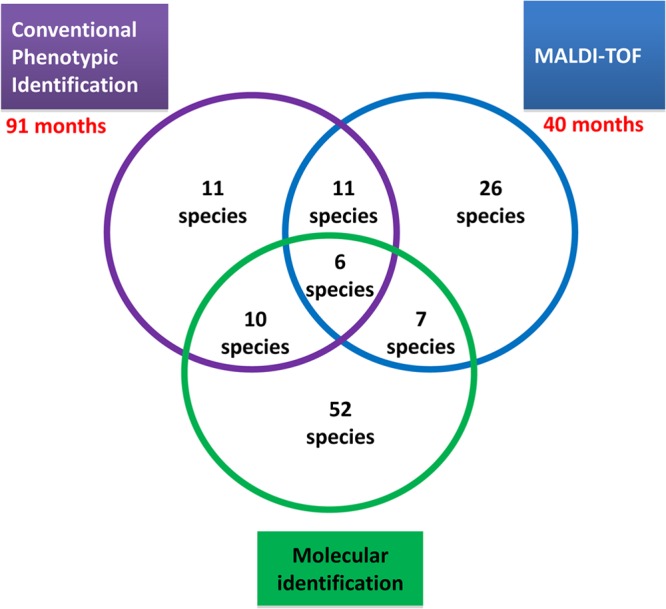
Among the 459 bacterial species identified during 2002 to 2012, 76 species (17%) were identified using only CPI over a 91-month period, 124 species (27%) were identified using only MALDI-TOF MS during a 40-month period (see Table S1 and Table S2 in the supplemental material), and 258 species (56%) were identified using both methods.
In the group of bacterial species identified only by CPI, 15 (20%) of the 76 isolates were absent from our MALDI-TOF mass spectrum database. In the phylum Actinobacteria, 16 species of 11 genera were identified using only CPI, and 3 species were absent from our MALDI-TOF MS database. In the phylum Bacteroidetes, 5 species of 3 genera were identified using CPI exclusively, and 1 species was absent from the MALDI-TOF MS database. In the phylum Firmicutes, 19 species of 10 genera were identified using only CPI, and 3 were missing from the MALDI-TOF MS database. In the phylum Fusobacteria, 3 species of 2 genera were identified using only CPI, and 1 was missing from the MALDI-TOF MS database. In the phylum Proteobacteria, 33 species of 22 genera were identified using CPI exclusively, and 7 were missing from the MALDI-TOF MS database (see Table S1 in the supplemental material).
In the group of bacterial species identified only by MALDI-TOF MS, 21 (17%) of the 124 isolates were present in the Vitek 2 database, whereas 103 (83%) were not (see Table S2 in the supplemental material). In the phylum Actinobacteria, 21 species of 12 genera were identified using only MALDI-TOF MS and were lacking in the Vitek 2 database. In the phylum Bacteroidetes, 10 species of 7 genera were identified by using MALDI-TOF MS exclusively, and 9 species were absent from the Vitek 2 database. In the phylum Firmicutes, 54 species of 18 genera were identified using only MALDI-TOF MS, and 41 were missing from the Vitek 2 database. In the phylum Fusobacteria, Fusobacterium periodonticum was identified using only MALDI-TOF MS and was missing from the Vitek 2 database. In the phylum Proteobacteria, 38 species of 20 genera were identified using MALDI-TOF MS exclusively, and 31 were missing from the Vitek 2 database. No species in the phylum Tenericutes was identified by using MALDI-TOF MS exclusively (see Table S2 in the supplemental material).
During the study period, 1,951 isolates were misidentified and required confirmation by another round of phenotypic identification for 670 isolates of 21 species (see Table S3 in the supplemental material) and by molecular identification for 1,273 isolates of 339 species (see Table S4 in the supplemental material). Among 339 species that required confirmation by molecular identification, 63 species were absent from the initial Brüker database, which contained 3,993 bacterial spectra, and only 24 were missing from our updated MALDI-TOF mass spectrum database (6,213 bacterial spectra). Among 24 bacterial species of 46 isolates missed from our MALDI-TOF MS database, 16 species of 32 isolates were identified by a molecular method in the CPI period, and 11 species of 14 isolates were identified by a molecular method in the MALDI-TOF MS period. Despite their presence in our MALDI-TOF database, 315 other species had to be examined by molecular identification; this included 228 species of 711 isolates and 196 species of 516 isolates in the CPI period and the MALDI-TOF MS period, respectively.
We identified 40 species of 1,506 anaerobic organisms before MALDI-TOF MS by using the API 20A system (bioMérieux), and we identified 103 species of 1,564 anaerobic organisms at the species level using MALDI-TOF MS identification.
During the CPI period, 1,363 isolates (0.77%) were misidentified; the 1,363 isolates included 620 isolates reidentified using a second CPI as described below (i.e., 35.2 per 10,000 isolates) and 743 confirmed using a molecular technique (i.e., 42 per 10,000 isolates). During the MALDI-TOF MS period, 580 isolates (0.52%) were misidentified; the 580 isolates included 50 isolates reidentified using a second run of identification by MALDI-TOF MS, i.e., 4.5 species per 10,000 isolates, and 530 isolates confirmed using a molecular technique, i.e., 47 species per 10,000 isolates (Table 1 and Fig. 3).
The molecular identification requirements were similar during the CPI and MALDI-TOF MS periods at 42 and 47 molecular identifications/10,000 isolates, respectively. However, a decreasing trend was observed during the final 2 years, with 47 and 53 during 2011 and 2012, respectively, compared with 142 molecular identifications in 2008 (Fig. 2 and Fig. 3).
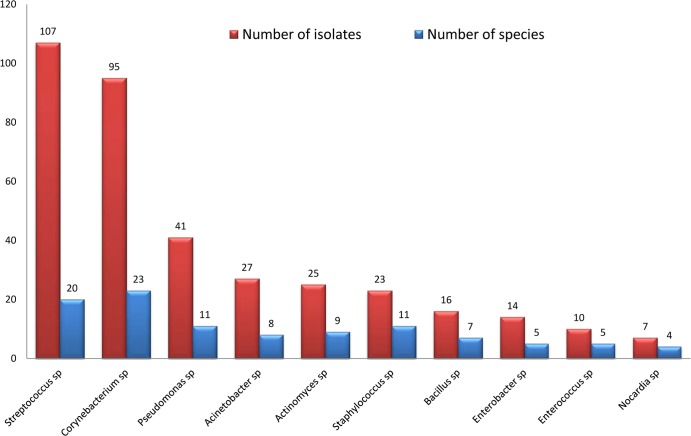
During 11 years of routine identification, we identified 123 rare species of bacteria that were reported to be human pathogens fewer than or equal to 10 times in the literature (PubMed database). Among these species, 48 were identified by phenotypic identification. Another 75 species were confirmed by molecular identification. In addition, CPI identified only 22 rare species during 91 months, and MALDI-TOF MS identified 37 such rare species during 40 months (Fig. 5, Fig. 6, and Fig. 7). Among 196 species of 516 isolates that were not satisfactorily identified in the MALDI-TOF MS period, 365 (71%) isolates represented 10 genera, including Streptococcus, Corynebacterium, Pseudomonas, Acinetobacter, Actinomyces, Staphylococcus, Bacillus, Enterobacter, Enterococcus, and Nocardia, that frequently required molecular identification (Fig. 8).
Fig 5.
Time course of the numbers of isolates of 128 rare species, 48 of which were identified using phenotypic identification (PID), and 75 of which were identified using molecular identification (ID).
Fig 6.
Time course for the numbers of species identified among 128 rare species, 48 of which were identified using phenotypic identification (PID) and 75 of which were identified using molecular identification (ID).
Fig 7.
Of 48 rare species identified using phenotypic techniques, MALDI-TOF MS identified 37 rare species and conventional phenotypic identification identified 22 rare species in 40 and 91 months of study, respectively. Seventy-five rare species were identified using molecular techniques.
Fig 8.
Ten genera of 365 (71%) isolates that frequently required molecular identification among 196 species of 516 isolates identified unsatisfactorily in the MALDI-TOF period.
Identification of 11 of the 48 rare species identified using phenotypic methods was performed using only CPI, and 26 other rare species were identified using only MALDI-TOF MS (Table 2). In the phylum Actinobacteria, 18 rare species were identified, including 9 exclusively identified using MALDI-TOF MS, 5 using CPI, and 4 species using both techniques. In the phylum Bacteroidetes, 6 rare species were identified; the 6 species included 2 exclusively identified using MALDI-TOF MS, 1 using CPI, and 3 using both techniques. In the phylum Firmicutes, 12 rare species were identified, including 7 exclusively identified using MALDI-TOF MS, 2 using CPI, and 3 using both techniques. In the phylum Fusobacteria, 2 rare species were totally identified using CPI. In the phylum Proteobacteria, 10 rare species were identified, including 8 exclusively identified using MALDI-TOF MS, 1 using CPI, and 1 using both techniques (Table 2).
Table 2.
Species of clinical isolates that were identified by phenotypic identification as species that had been rarely reported as human pathogensa
| Phylum | Genus | Bacterial rare species identified by PID | No. of isolates | Identification method(s) | No. of isolates identified by CPI | No. of isolates identified by MALDI-TOF MS | No. of reports in PubMed |
|---|---|---|---|---|---|---|---|
| Actinobacteria | Actinobaculum | Actinobaculum massiliense | 1 | MALDI-TOF MS | 0 | 1 | 4 |
| Actinomadura | Actinomadura cremea | 1 | CPI | 1 | 0 | 6 | |
| Actinomyces | Actinomyces europaeus | 12 | CPI and MALDI-TOF MS | 3 | 9 | 9 | |
| Actinomyces radicidentis | 3 | MALDI-TOF MS | 0 | 3 | 4 | ||
| Actinomyces radingae | 20 | CPI and MALDI-TOF MS | 5 | 15 | 10 | ||
| Arthrobacter | Arthrobacter cumminsii | 5 | CPI and MALDI-TOF MS | 3 | 2 | 4 | |
| Brevibacterium | Brevibacterium luteolum | 1 | CPI | 1 | 0 | 4 | |
| Brevibacterium massiliense | 1 | MALDI-TOF MS | 0 | 1 | 2 | ||
| Brevibacterium paucivorans | 1 | MALDI-TOF MS | 0 | 1 | 3 | ||
| Brevibacterium ravenspurgense | 1 | MALDI-TOF MS | 0 | 1 | 0 | ||
| Corynebacterium | Corynebacterium auriscanis | 3 | CPI | 3 | 0 | 5 | |
| Corynebacterium coyleae | 7 | CPI and MALDI-TOF MS | 2 | 5 | 7 | ||
| Corynebacterium fastidiosum | 2 | MALDI-TOF MS | 0 | 2 | 0 | ||
| Corynebacterium imitans | 2 | MALDI-TOF MS | 0 | 2 | 2 | ||
| Corynebacterium mucifaciens | 5 | MALDI-TOF MS | 0 | 5 | 6 | ||
| Microbacterium | Microbacterium schleiferi | 1 | MALDI-TOF MS | 0 | 1 | 6 | |
| Pseudoclavibacter | Pseudoclavibacter bifida | 1 | CPI | 1 | 0 | 1 | |
| Varibaculum | Varibaculum cambriense | 2 | CPI | 3 | 9 | 2 | |
| Bacteroidetes | Alistipes | Alistipes finegoldii | 3 | CPI and MALDI-TOF MS | 0 | 3 | 4 |
| Bacteroides | Bacteroides cellulosilyticus | 4 | MALDI-TOF MS | 5 | 15 | 2 | |
| Butyricimonas | Butyricimonas virosa | 1 | MALDI-TOF MS | 3 | 2 | 1 | |
| Porphyromonas | Porphyromonas somerae | 9 | CPI and MALDI-TOF MS | 1 | 0 | 1 | |
| Prevotella | “Candidatus Prevotella conceptionensis” | 3 | CPI and MALDI-TOF MS | 0 | 1 | 1 | |
| Prevotella massiliensis | 1 | CPI | 0 | 1 | 2 | ||
| Firmicutes | Acidaminococcus | Acidaminococcus intestini | 2 | CPI and MALDI-TOF MS | 0 | 1 | 2 |
| Anaerococcus | Anaerococcus lactolyticus | 3 | MALDI-TOF MS | 3 | 40 | 9 | |
| Anaerococcus octavius | 7 | MALDI-TOF MS | 2 | 5 | 3 | ||
| Eubacterium | Eubacterium tenue | 2 | MALDI-TOF MS | 0 | 2 | 6 | |
| Eubacterium yurii | 1 | MALDI-TOF MS | 0 | 2 | 10 | ||
| Facklamia | Facklamia languida | 1 | CPI | 0 | 5 | 2 | |
| Peptoniphilus | Peptoniphilus harei | 95 | CPI and MALDI-TOF MS | 0 | 1 | 7 | |
| Robinsoniella | Robinsoniella peoriensis | 3 | MALDI-TOF MS | 1 | 0 | 8 | |
| Sporosarcina | Sporosarcina ginsengisoli | 1 | CPI | 2 | 0 | 1 | |
| Streptococcus | Streptococcus massiliensis | 4 | MALDI-TOF MS | 1 | 2 | 1 | |
| Turicibacter | Turicibacter sanguinis | 3 | CPI and MALDI-TOF MS | 0 | 4 | 3 | |
| Veillonella | Veillonella montpellierensis | 1 | MALDI-TOF MS | 0 | 1 | 3 | |
| Fusobacteria | Leptotrichia | Leptotrichia goodfellowii | 1 | CPI | 1 | 8 | 5 |
| Leptotrichia trevisanii | 3 | CPI | 1 | 2 | 3 | ||
| Proteobacteria | Acinetobacter | Acinetobacter parvus | 2 | MALDI-TOF MS | 1 | 0 | 8 |
| Comamonas | Comamonas kerstersii | 2 | MALDI-TOF MS | 1 | 1 | 3 | |
| Enterobacter | Enterobacter cowanii | 3 | MALDI-TOF MS | 0 | 3 | 9 | |
| Enterobacter kobei | 272 | MALDI-TOF MS | 0 | 7 | 10 | ||
| Ochrobactrum | Ochrobactrum grignonense | 1 | MALDI-TOF MS | 0 | 2 | 8 | |
| Pandoraea | Pandoraea pulmonicola | 31 | MALDI-TOF MS | 0 | 1 | 7 | |
| Paracoccus | Paracoccus yeeii | 2 | CPI and MALDI-TOF MS | 1 | 0 | 1 | |
| Pseudomonas | Pseudomonas hibiscicola | 2 | MALDI-TOF MS | 11 | 84 | 4 | |
| Roseomonas | Roseomonas ludipueritiae | 1 | CPI | 0 | 3 | 4 | |
| Serratia | Serratia ureilytica | 1 | MALDI-TOF MS | 1 | 0 | 6 |
List of 48 species of 534 clinical isolates that were identified by phenotypic identification as species that had been rarely reported as human pathogens, with ≤10 reports in PubMed. PID, phenotypic identification; CPI, conventional phenotypic identification (Gram staining, API, Vitek 2 system identification).
Looking in detail at the group of 48 rare species identified using phenotypic methods, 4 of these were identified more than 10 times in our laboratory during the last 11 years, including 12 isolates of Actinomyces europaeus, 20 isolates of Actinomyces radingae, 31 isolates of Pandoraea pulmonicola, 95 isolates of Peptoniphilus harei, and 272 isolates of Enterobacter kobei (Table 2).
The rare species identified using phenotypic methods were mostly recovered from bloodstream and urinary traction infections (see Table S5 in the supplemental material). Enterobacter kobei was the most frequently identified among the 48 rare species (see Table S5 in the supplemental material). In the following analysis, using MALDI-TOF MS, we identified two bacterial species, Brevibacterium ravenspurgense and Corynebacterium fastidiosum, that had never been reported as human pathogens in PubMed (Table 2).
Moreover, molecular techniques identified 75 rare species among 124 isolates including 23 that were identified as rare species using phenotypic identification methods (Table 3). In all, 57 of the 75 rare species identified using molecular techniques were absent from the Brüker database and 18 were absent from our MALDI-TOF database. Among 57 bacterial rare species identified by molecular methods which spectrum present in our MALDI-TOF database, 39 species were recently created during the study. Fourteen of 18 rare species exclusively identified in the CPI period were recently created. Twenty-five of 39 rare species identified in the MALDI-TOF MS period were recently created in our database. Other 14 rare species that were present in the database but that needed molecular identification in the MALDI-TOF MS period were Actinomyces europaeus (2 isolates), Corynebacterium argentoratense (2 isolates), Corynebacterium confusum (1), Corynebacterium coyleae (4 isolates), Corynebacterium imitans (1 isolate), Corynebacterium kroppenstedtii (1 isolate), Corynebacterium mucifaciens (3 isolates), Corynebacterium riegelii (1 isolate), Corynebacterium ureicelerivorans (1 isolate), Microbacterium aurum (1 isolate), Streptococcus criceti (3 isolates), Streptococcus peroris (1 isolate), Enterobacter kobei (3 isolates), and Pandoraea pulmonicola (3 isolates).
Table 3.
Rare bacterial species identified using molecular identificationa
| Phylum | Genus | Bacterial species confirmed by molecular identification | No. of isolates | No. of isolates identified in the CPI period | No. of isolates identified in the MALDI-TOF MS period | No. of reports in PubMed | 48 rare species by PID | Presence/absence of species in our MALDI-TOF MS database | Presence/absence of species in MALDI-TOF MS database (Brüker) |
|---|---|---|---|---|---|---|---|---|---|
| Actinobacteria | Actinomyces | Actinomyces europaeus | 3 | 1 | 2 | 9 | Yes | Present | Present |
| Actinomyces lingnae | 1 | 0 | 1 | 1 | No | Absent | Absent | ||
| Actinomyces radingae | 5 | 3 | 2 | 10 | Yes | Present | Absent | ||
| Actinomyces urogenitalis | 2 | 0 | 2 | 4 | No | Present | Absent | ||
| Arthrobacter | Arthrobacter cumminsii | 5 | 4 | 1 | 4 | Yes | Present | Absent | |
| Bifidobacterium | Bifidobacterium scardovii | 1 | 1 | 0 | 5 | No | Present | Absent | |
| Brachybacterium | Brachybacterium muris | 1 | 0 | 1 | 3 | No | Present | Absent | |
| Brachybacterium sacelli | 1 | 0 | 1 | 3 | No | Absent | Absent | ||
| Brevibacterium | Brevibacterium massiliense | 1 | 1 | 0 | 2 | Yes | Absent | Absent | |
| Brevibacterium otitidis | 1 | 1 | 0 | 9 | No | Absent | Absent | ||
| Brevibacterium paucivorans | 2 | 1 | 1 | 3 | Yes | Present | Absent | ||
| Brevibacterium ravenspurgense | 1 | 1 | 0 | 0 | Yes | Present | Absent | ||
| Brevibacterium sanguinis | 1 | 1 | 0 | 2 | No | Present | Absent | ||
| Brevibacterium stationis | 1 | 0 | 1 | 10 | No | Present | Absent | ||
| Corynebacterium | Corynebacterium argentoratense | 2 | 0 | 2 | 3 | No | Present | Present | |
| Corynebacterium auriscanis | 3 | 3 | 0 | 5 | Yes | Present | Present | ||
| Corynebacterium confusum | 1 | 0 | 1 | 2 | No | Present | Present | ||
| Corynebacterium coyleae | 4 | 0 | 4 | 7 | Yes | Present | Present | ||
| Corynebacterium durum | 1 | 1 | 0 | 3 | No | Present | Absent | ||
| Corynebacterium fastidiosum | 1 | 0 | 1 | 0 | Yes | Absent | Absent | ||
| Corynebacterium imitans | 1 | 0 | 1 | 2 | Yes | Present | Present | ||
| Corynebacterium kroppenstedtii | 1 | 0 | 1 | 9 | No | Present | Present | ||
| Corynebacterium mucifaciens | 3 | 0 | 3 | 6 | Yes | Present | Present | ||
| Corynebacterium riegelii | 1 | 0 | 1 | 6 | No | Present | Present | ||
| Corynebacterium ureicelerivorans | 1 | 0 | 1 | 3 | No | Present | Present | ||
| Dietzia | Dietzia cinnamea | 1 | 1 | 0 | 10 | No | Present | Absent | |
| Janibacter | Janibacter hoylei | 1 | 0 | 1 | 2 | No | Present | Absent | |
| Microbacterium | Microbacterium aurum | 2 | 1 | 1 | 5 | No | Present | Present | |
| Microbacterium chocolatum | 1 | 1 | 0 | 1 | No | Absent | Absent | ||
| Microbacterium flavum | 1 | 0 | 1 | 5 | No | Present | Absent | ||
| Nesterenkonia | Nesterenkonia lacusekhoensis | 1 | 0 | 1 | 4 | No | Present | Absent | |
| Propionimicrobium | Propionimicrobium lymphophilum | 2 | 1 | 1 | 3 | No | Present | Absent | |
| Trueperella | Trueperella abortisuis | 1 | 1 | 0 | 5 | No | Present | Absent | |
| Zimmermannella | Zimmermannella bifida | 1 | 1 | 0 | 1 | Yes | Absent | Absent | |
| Bacteroidetes | Alistipes | Alistipes finegoldii | 1 | 1 | 0 | 4 | Yes | Present | Absent |
| Bacteroides | Bacteroides dorei | 1 | 1 | 0 | 8 | No | Absent | Absent | |
| Butyricimonas | Butyricimonas virosa | 2 | 0 | 2 | 1 | Yes | Present | Absent | |
| Chryseobacterium | Chryseobacterium hominis | 1 | 0 | 1 | 4 | No | Present | Absent | |
| Chryseobacterium vrystaatense | 1 | 0 | 1 | 3 | No | Absent | Absent | ||
| Peptoniphilus | Candidatus Peptoniphilus massiliensis | 1 | 0 | 1 | 0 | No | Absent | Absent | |
| Porphyromonas | Porphyromonas uenonis | 4 | 4 | 0 | 2 | No | Present | Absent | |
| Prevotella | “Candidatus Prevotella conceptionensis” | 1 | 1 | 0 | 1 | Yes | Present | Absent | |
| Wautersiella | Wautersiella falsenii | 2 | 1 | 1 | 4 | No | Present | Absent | |
| Firmicutes | Aerosphaera | Aerosphaera taetra | 1 | 1 | 0 | 0 | No | Present | Absent |
| Anaerococcus | Anaerococcus octavius | 2 | 2 | 0 | 3 | Yes | Present | Absent | |
| Anaerotruncus | Anaerotruncus colihominis | 2 | 1 | 1 | 2 | No | Present | Absent | |
| Lysinibacillus | Lysinibacillus massiliensis | 1 | 0 | 1 | 8 | No | Absent | Absent | |
| Catabacter | Catabacter hongkongensis | 1 | 1 | 0 | 6 | No | Absent | Absent | |
| Clostridium | Clostridium aldenense | 1 | 0 | 1 | 3 | No | Present | Absent | |
| Dialister | Dialister micraerophilus | 1 | 0 | 1 | 3 | No | Present | Absent | |
| Granulicatella | Granulicatella para-adiacens | 1 | 0 | 1 | 2 | No | Present | Absent | |
| Peptoniphilus | Peptoniphilus harei | 3 | 2 | 1 | 7 | Yes | Present | Absent | |
| Streptococcus | Streptococcus criceti | 3 | 0 | 3 | 10 | No | Present | Present | |
| Streptococcus massiliensis | 2 | 2 | 0 | 1 | Yes | Present | Present | ||
| Streptococcus peroris | 1 | 0 | 1 | 6 | No | Present | Present | ||
| Turicibacter | Turicibacter sanguinis | 1 | 1 | 0 | 3 | Yes | Present | Absent | |
| Fusobacteria | Leptotrichia | Leptotrichia trevisanii | 5 | 4 | 1 | 3 | Yes | Present | Absent |
| Proteobacteria | Acetobacter | Acetobacter indonesiensis | 2 | 2 | 0 | 9 | No | Absent | Absent |
| Acinetobacter | Acinetobacter parvus | 1 | 1 | 0 | 8 | Yes | Present | Present | |
| Acinetobacter septicus | 5 | 4 | 1 | 3 | No | Present | Absent | ||
| Aurantimonas | Aurantimonas altamirensis | 1 | 0 | 1 | 9 | No | Present | Absent | |
| Blastomonas | Blastomonas ursincola | 1 | 1 | 0 | 5 | No | Present | Present | |
| Desulfovibrio | Desulfovibrio intestinalis | 1 | 1 | 0 | 5 | No | Absent | Absent | |
| Enterobacter | Enterobacter kobei | 3 | 0 | 3 | 10 | Yes | Present | Present | |
| Hematobacter | Hematobacter massiliensis | 3 | 1 | 2 | 2 | No | Absent | Absent | |
| Pandoraea | Pandoraea pulmonicola | 3 | 0 | 3 | 7 | Yes | Present | Present | |
| Pantoea | Pantoea brenneri | 1 | 0 | 1 | 1 | No | Absent | Absent | |
| Pantoea eucrina | 1 | 0 | 1 | 2 | No | Present | Absent | ||
| Pseudochrobactrum | Pseudochrobactrum asaccharolyticum | 1 | 0 | 1 | 2 | No | Present | Absent | |
| Pseudomonas | Pseudomonas lurida | 1 | 0 | 1 | 3 | No | Present | Absent | |
| Ralstonia | Ralstonia insidiosa | 1 | 0 | 1 | 5 | No | Present | Absent | |
| Roseomonas | Roseomonas genomospecies 5 | 1 | 1 | 0 | 6 | No | Absent | Absent | |
| Rothia | Rothia aeria | 1 | 1 | 0 | 8 | No | Present | Absent | |
| Serratia | Serratia nematodiphila | 1 | 0 | 1 | 3 | No | Absent | Absent | |
| Sphingomonas | Sphingomonas mucosissima | 1 | 1 | 0 | 2 | No | Present | Absent |
List of 75 rare bacterial species identified using molecular identification; 18 of these species were absent from our MALDI-TOF database, and 57 species from the Brüker database. PID, phenotypic identification; CPI, conventional phenotypic identification (Gram staining, API, Vitek 2 system identification).
The time required for identification of one clinical isolate using MALDI-TOF MS was 6 to 8 min 30 s for the AutoFlex II system (Brüker Daltonik) and 1 min 46 s for the MicroFlex LT mass spectrometer (Brüker Daltonik). The cost of identification of one clinical isolate using MALDI-TOF MS was 1.43 euros for the AutoFlex II system (Brüker Daltonik) and 1.35 euros for the MicroFlex LT mass spectrometer (Brüker Daltonik) (Table 4). In comparison, the time required for identification for one clinical isolate using 16S rRNA or rpoB sequencing was 24 h. In addition, the cost of bacterial isolate identification using gene sequencing was 137.70 euros.
Table 4.
Comparison of time, cost, and level of training required for routine identification of one isolate using the different techniques in our clinical laboratory
| Identification technique | Time required for identification of one isolate | Cost (euros) | Level of training |
|---|---|---|---|
| Gram staining | 6 min | 0.6 | Medium to high |
| API system identification (bioMérieux) | 18–48 h | 4.6–6 | Medium |
| Vitek 2 system identification (bioMérieux) | 5–8 h | 5.9–8.23 | Medium |
| Molecular identification by 16S rRNA or rpoB sequencing | 24 h | 137.7 | Medium to high |
| MALDI-TOF MS by AutoFlex II system (Brüker Daltonik) | 6–8 min 30 s | 1.43 | Low to medium |
| MALDI-TOF MS by MicroFlex LT mass spectrometer (Brüker Daltonik) | 1 min 46 s | 1.35 | Low to medium |
DISCUSSION
During the last 11 years, our clinical laboratory has seen an increased ability to analyze bacteriological samples due to several reasons: first, the establishment of another laboratory at the North University Hospital, Marseille, France, and second, the creation of an anaerobic laboratory in September 2008. By optimizing the new tool of MALDI-TOF mass spectrometry for routine identification, we were able to increase our yearly analysis capacity from 46,079 analyses in 2002 to 66,989 in 2012.
In 2008, we evaluated the performance of MALDI-TOF MS to identify 1,660 clinical isolates in a 16-week period by comparing it with routine phenotypic identification methods, such as semiautomated Gram staining (Aerospray Wescor; Elitech), catalase and oxidase assays and automated identifications using the Vitek 2 and API 20A systems (bioMérieux). Since then, more than 300 scientific publications have confirmed that MALDI-TOF MS can be adapted to achieve performances similar to the routine identification methods used in clinical laboratories (14, 50–53). Many clinical laboratories have, like us, adopted bacterial identification using MALDI-TOF MS for biotyping microbes to replace all of the traditional phenotypic methods used for routine diagnoses directly from colony or clinical samples (13, 45, 54–58).
Recently, MALDI-TOF MS was used in culturomics studies to identify 32 new bacterial species and another 177 bacterial species that had never been reported to occur in the human gut microbiota that may explain the involvement of microorganisms in human diseases such as obesity (59, 60). MALDI-TOF MS has been used to identify 233 of 349 bacterial species from 4 stool samples by direct identification from 36,500 colonies. MALDI-TOF MS has also identified 116 unknown bacterial species with the score < 1.9 that was needed to identify by 16S rRNA gene sequencing. Seventy-one of 116 (61%) bacterial species were previously absent in our MALDI-TOF database. Among 45 (39%) species present in our MALDI-TOF database, 24 (20%) have only 1 reference spectrum, and only one serovar of 18 serovars of Acinetobacter pittii has more than 10 spectra in the database (59–61). We used an incremental database with each spectra identified by 16S rRNA gene sequencing from the first three stool samples that allowed us to use the culturomics study of Dubourg et al. (61) for the fourth stool sample; in the study of Dubourg et al., only 4 of 4,000 bacterial colonies needed molecular identification (61).
The capacity of MALDI-TOF MS to identify an unknown bacterial species before molecular identification has been previously observed by Bizzini et al. (62) and confirmed after updating the MALDI-TOF database. Among 410 bacterial strains that were not satisfactorily identified by the Vitek 2 and API systems (bioMérieux), 62% of them were concordantly identified by MALDI-TOF MS and 16S rRNA gene sequencing. Failure to identify 85 other bacterial species was due to the absence of spectra of 78 species in the MALDI-TOF database (62).
The 196 species (516 isolates) that were not identified included 57 rare bacterial species present in the MALDI-TOF database that needed molecular identification in the MALDI-TOF period can be attributed to two causes. The first cause is the absence of reference spectrum. The second cause was the presence of a low number of spectra in the database that does not allow MALDI-TOF to identify the bacteria in the groups with biodiversity within species. As an example, 10 genera that frequently needed molecular identification in the MALDI-TOF MS period in spite of the presence of some reference spectra were Streptococcus, Corynebacterium, Pseudomonas, Acinetobacter, Actinomyces, Staphylococcus, Bacillus, Enterobacter, Enterococcus, and Nocardia.
In addition to the capacity to analyze more isolates as shown in the present study, MALDI-TOF MS has annually identified 2.5 times more species than CPI, identifying 112 species (i.e., 36 species/10,000 isolates) compared with 44 species (i.e., 19 species/10,000 isolates), respectively. This performance of MALDI-TOF MS in annually identifying more species per isolate tested can be explained first by the increasing numbers of colonies analyzed from each clinical sample and a tendency to identify systematically all isolates from a polymicrobial clinical specimen. Second, the MALDI-TOF database is now 10 times larger than the Vitek 2 database (bioMérieux, Durham, NC), with 6,213 reference strains compared with 330 reference strains, respectively.
Another benefit of MALDI-TOF MS in routine identification revealed in this study is the reduced need for secondary phenotypic identification, which significantly decreased the cost and time required to provide results to clinicians. Only 50 secondary phenotypic identifications of 110,263 clonal-bacterial isolates tested (i.e., 4.5 reidentifications/10,000 isolates) were required during the MALDI-TOF MS period compared with 620 of 175,999 isolates during the CPI period (i.e., 35.2 reidentifications/10,000 isolates).
Over 3 years of experience in routine identification using MALDI-TOF MS, we observed a rise in the numbers of isolates and species that were identified using MALDI-TOF MS. The ability to expand the database by incorporation of laboratory spectra for bacteria that had been identified previously by molecular techniques has improved the performance of MALDI-TOF MS in identifying human-pathogenic bacteria.
Interestingly, MALDI-TOF MS identified more bacterial species that had been rarely reported as human pathogens than CPI did. A total of 37 of 48 rare species (77%) identified by phenotypic techniques were identified using MALDI-TOF MS. A systematic identification of all colonies derived from clinical samples will increase the capacity to identify more rare species in the future.
We also evaluated the time and cost-effectiveness of MALDI-TOF MS, which reduced by 55-fold and 169-fold the time required for identification and reduced by 5- and 96-fold the cost compared with CPI and gene sequencing, respectively (12). The time required for identification has been newly improved to 1 min 46 s using the MicroFlex LT mass spectrometer (Brüker Daltonik) compared with the AutoFlex II system, which took 6 to 8 min 30 s for identification of one isolate. The cost was evaluated at 1.35 euros for the MicroFlex LT mass spectrometer and 1.43 euros for the AutoFlex II system.
Conclusion.
We have shown the effectiveness and performance of MALDI-TOF MS in the identification of clinical isolates and bacterial species in routine bacterial identification in a clinical laboratory over 11 years of study.
The ability of MALDI-TOF MS to identify a large number of bacterial species well is leading many clinical laboratories to abandon traditional phenotypic identification. We have shown that MALDI-TOF MS is not only a powerful tool for routine bacterial identification in the clinical laboratory but also a powerful tool to identify rare bacterial species implicated in human infectious diseases.
This capacity to identify rare species as human pathogens using MALDI-TOF MS could be an alternative to molecular methods in the clinical laboratory. The rapid identification of bacterial species that were rarely or never previously described as pathogens in specific clinical specimens will help us to study the clinical burden due to the emergence of these species as human pathogens and to implement their real-time surveillance.
Supplementary Material
ACKNOWLEDGMENT
We thank Véronique Filosa for expert technical assistance in data extraction.
Footnotes
Published ahead of print 1 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00492-13.
REFERENCES
- 1. Woo PC, Lau SK, Teng JL, Tse H, Yuen KY. 2008. Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin. Microbiol. Infect. 14:908–934 [DOI] [PubMed] [Google Scholar]
- 2. Drancourt M, Raoult D. 2005. Sequence-based identification of new bacteria: a proposition for creation of an orphan bacterium repository. J. Clin. Microbiol. 43:4311–4315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Drancourt M, Bollet C, Carlioz A, Martelin R, Gayral JP, Raoult D. 2000. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J. Clin. Microbiol. 38:3623–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Drancourt M, Berger P, Raoult D. 2004. Systematic 16S rRNA gene sequencing of atypical clinical isolates identified 27 new bacterial species associated with humans. J. Clin. Microbiol. 42:2197–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fenollar F, Roux V, Stein A, Drancourt M, Raoult D. 2006. Analysis of 525 samples to determine the usefulness of PCR amplification and sequencing of the 16S rRNA gene for diagnosis of bone and joint infections. J. Clin. Microbiol. 44:1018–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Janda JM, Abbott SL. 2007. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J. Clin. Microbiol. 45:2761–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Al Masalma M, Armougom F, Scheld WM, Dufour H, Roche PH, Drancourt M, Raoult D. 2009. The expansion of the microbiological spectrum of brain abscesses with use of multiple 16S ribosomal DNA sequencing. Clin. Infect. Dis. 48:1169–1178 [DOI] [PubMed] [Google Scholar]
- 8. Schlaberg R, Simmon KE, Fisher MA. 2012. A systematic approach for discovering novel, clinically relevant bacteria. Emerg. Infect. Dis. 18:422–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Teles C, Smith A, Ramage G, Lang S. 2011. Identification of clinically relevant viridans group streptococci by phenotypic and genotypic analysis. Eur. J. Clin. Microbiol. Infect. Dis. 30:243–250 [DOI] [PubMed] [Google Scholar]
- 10. Ikryannikova LN, Lapin KN, Malakhova MV, Filimonova AV, Ilina EN, Dubovickaya VA, Sidorenko SV, Govorun VM. 2011. Misidentification of alpha-hemolytic streptococci by routine tests in clinical practice. Infect. Genet. Evol. 11:1709–1715 [DOI] [PubMed] [Google Scholar]
- 11. Maeda Y, Goldsmith CE, Coulter WA, Mason C, Dooley JS, Lowery CJ, Millar BC, Moore JE. 2011. Comparison of five gene loci (rnpB, 16S rRNA, 16S-23S rRNA, sodA and dnaJ) to aid the molecular identification of viridans-group streptococci and pneumococci. Br. J. Biomed. Sci. 68:190–196 [DOI] [PubMed] [Google Scholar]
- 12. Seng P, Drancourt M, Gouriet F, La Scola B, Fournier PE, Rolain JM, Raoult D. 2009. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 49:543–551 [DOI] [PubMed] [Google Scholar]
- 13. La Scola B, Raoult D. 2009. Direct identification of bacteria in positive blood culture bottles by matrix-assisted laser desorption ionisation time-of-flight mass spectrometry. PLoS One 4:e8041. 10.1371/journal.pone.0008041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Seng P, Rolain JM, Fournier PE, La Scola B, Drancourt M, Raoult D. 2010. MALDI-TOF-mass spectrometry applications in clinical microbiology. Future Microbiol. 5:1733–1754 [DOI] [PubMed] [Google Scholar]
- 15. Drancourt M. 2010. Detection of microorganisms in blood specimens using matrix-assisted laser desorption ionization time-of-flight mass spectrometry: a review. Clin. Microbiol. Infect. 16:1620–1625 [DOI] [PubMed] [Google Scholar]
- 16. Bittar F, Cassagne C, Bosdure E, Stremler N, Dubus JC, Sarles J, Reynaud-Gaubert M, Raoult D, Rolain JM. 2010. Outbreak of Corynebacterium pseudodiphtheriticum infection in cystic fibrosis patients, France. Emerg. Infect. Dis. 16:1231–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Williamson YM, Moura H, Woolfitt AR, Pirkle JL, Barr JR, Carvalho MDAG, Ades EP, Carlone GM, Sampson JS. 2008. Differentiation of Streptococcus pneumoniae conjunctivitis outbreak isolates by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 74:5891–5897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Griffin PM, Price GR, Schooneveldt JM, Schlebusch S, Tilse MH, Urbanski T, Hamilton B, Venter D. 2012. Use of matrix-assisted laser desorption ionization-time of flight mass spectrometry to identify vancomycin-resistant enterococci and investigate the epidemiology of an outbreak. J. Clin. Microbiol. 50:2918–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Werno AM, Christner M, Anderson TP, Murdoch DR. 2012. Differentiation of Streptococcus pneumoniae from nonpneumococcal streptococci of the Streptococcus mitis group by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 50:2863–2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hinic V, Lang C, Weisser M, Straub C, Frei R, Goldenberger D. 2012. Corynebacterium tuberculostearicum: a potentially misidentified and multiresistant Corynebacterium species isolated from clinical specimens. J. Clin. Microbiol. 50:2561–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Djelouadji Z, Roux V, Raoult D, Kodjo A, Drancourt M. 2012. Rapid MALDI-TOF mass spectrometry identification of Leptospira organisms. Vet. Microbiol. 158:142–146 [DOI] [PubMed] [Google Scholar]
- 22. Alvarez-Buylla A, Culebras E, Picazo JJ. 2012. Identification of Acinetobacter species: is Bruker biotyper MALDI-TOF mass spectrometry a good alternative to molecular techniques? Infect. Genet. Evol. 12:345–349 [DOI] [PubMed] [Google Scholar]
- 23. Lista F, Reubsaet FA, De Santis R, Parchen RR, de Jong AL, Kieboom J, van der Laaken AL, Voskamp-Visser IA, Fillo S, Jansen HJ, Van der Plas J, Paauw A. 2011. Reliable identification at the species level of Brucella isolates with MALDI-TOF-MS. BMC Microbiol. 11:267. 10.1186/1471-2180-11-267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fournier PE, Couderc C, Buffet S, Flaudrops C, Raoult D. 2009. Rapid and cost-effective identification of Bartonella species using mass spectrometry. J. Med. Microbiol. 58:1154–1159 [DOI] [PubMed] [Google Scholar]
- 25. Hrabak J, Walkova R, Studentova V, Chudackova E, Bergerova T. 2011. Carbapenemase activity detection by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 49:3222–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kempf M, Bakour S, Flaudrops C, Berrazeg M, Brunel JM, Drissi M, Mesli E, Touati A, Rolain JM. 2012. Rapid detection of carbapenem resistance in Acinetobacter baumannii using matrix-assisted laser desorption ionization-time of flight mass spectrometry. PLoS One 7:e31676. 10.1371/journal.pone.0031676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Edwards-Jones V, Claydon MA, Evason DJ, Walker J, Fox AJ, Gordon DB. 2000. Rapid discrimination between methicillin-sensitive and methicillin-resistant Staphylococcus aureus by intact cell mass spectrometry. J. Med. Microbiol. 49:295–300 [DOI] [PubMed] [Google Scholar]
- 28. Walker J, Fox AJ, Edwards-Jones V, Gordon DB. 2002. Intact cell mass spectrometry (ICMS) used to type methicillin-resistant Staphylococcus aureus: media effects and inter-laboratory reproducibility. J. Microbiol. Methods 48:117–126 [DOI] [PubMed] [Google Scholar]
- 29. Jackson KA, Edwards-Jones V, Sutton CW, Fox AJ. 2005. Optimisation of intact cell MALDI method for fingerprinting of methicillin-resistant Staphylococcus aureus. J. Microbiol. Methods 62:273–284 [DOI] [PubMed] [Google Scholar]
- 30. Du Z, Yang R, Guo Z, Song Y, Wang J. 2002. Identification of Staphylococcus aureus and determination of its methicillin resistance by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Chem. 74:5487–5491 [DOI] [PubMed] [Google Scholar]
- 31. Rajakaruna L, Hallas G, Molenaar L, Dare D, Sutton H, Encheva V, Culak R, Innes I, Ball G, Sefton AM, Eydmann M, Kearns AM, Shah HN. 2009. High throughput identification of clinical isolates of Staphylococcus aureus using MALDI-TOF-MS of intact cells. Infect. Genet. Evol. 9:507–513 [DOI] [PubMed] [Google Scholar]
- 32. Majcherczyk PA, McKenna T, Moreillon P, Vaudaux P. 2006. The discriminatory power of MALDI-TOF mass spectrometry to differentiate between isogenic teicoplanin-susceptible and teicoplanin-resistant strains of methicillin-resistant Staphylococcus aureus. FEMS Microbiol. Lett. 255:233–239 [DOI] [PubMed] [Google Scholar]
- 33. Camara JE, Hays FA. 2007. Discrimination between wild-type and ampicillin-resistant Escherichia coli by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Bioanal. Chem. 389:1633–1638 [DOI] [PubMed] [Google Scholar]
- 34. Russell SC, Edwards N, Fenselau C. 2007. Detection of plasmid insertion in Escherichia coli by MALDI-TOF mass spectrometry. Anal. Chem. 79:5399–5406 [DOI] [PubMed] [Google Scholar]
- 35. Sparbier K, Schubert S, Weller U, Boogen C, Kostrzewa M. 2012. Matrix-assisted laser desorption ionization-time of flight mass spectrometry-based functional assay for rapid detection of resistance against beta-lactam antibiotics. J. Clin. Microbiol. 50:927–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zbinden A, Mueller NJ, Tarr PE, Eich G, Schulthess B, Bahlmann AS, Keller PM, Bloemberg GV. 2012. Streptococcus tigurinus, a novel member of the Streptococcus mitis group, causes invasive infections. J. Clin. Microbiol. 50:2969–2973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tani A, Sahin N, Matsuyama Y, Enomoto T, Nishimura N, Yokota A, Kimbara K. 2012. High-throughput identification and screening of novel Methylobacterium species using whole-cell MALDI-TOF/MS analysis. PLoS One 7:e40784. 10.1371/journal.pone.0040784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chan JF, Lau SK, Curreem SO, To KK, Leung SS, Cheng VC, Yuen KY, Woo PC. 2012. First report of spontaneous intrapartum Atopobium vaginae bacteremia. J. Clin. Microbiol. 50:2525–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gouriet F, Million M, Henri M, Fournier PE, Raoult D. 2012. Lactobacillus rhamnosus bacteremia: an emerging clinical entity. Eur. J. Clin. Microbiol. Infect. Dis. 31:2469–2480 [DOI] [PubMed] [Google Scholar]
- 40. Angelakis E, Million M, Henry M, Raoult D. 2011. Rapid and accurate bacterial identification in probiotics and yoghurts by MALDI-TOF mass spectrometry. J. Food Sci. 76:M568–M572 [DOI] [PubMed] [Google Scholar]
- 41. Dridi B, Raoult D, Drancourt M. 2012. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry identification of Archaea: towards the universal identification of living organisms. APMIS 120:85–91 [DOI] [PubMed] [Google Scholar]
- 42. Fernandez-Olmos A, Morosini MI, Lamas A, Garcia-Castillo M, Garcia-Garcia L, Canton R, Maiz L. 2012. Clinical and microbiological features of a cystic fibrosis patient chronically colonized with Pandoraea sputorum identified by combining 16S rRNA sequencing and matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 50:1096–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ng LS, Sim JH, Eng LC, Menon S, Tan TY. 2012. Comparison of phenotypic methods and matrix-assisted laser desorption ionisation time-of-flight mass spectrometry for the identification of aero-tolerant Actinomyces spp. isolated from soft-tissue infections. Eur. J. Clin. Microbiol. Infect. Dis. 31:1749–1752 [DOI] [PubMed] [Google Scholar]
- 44. Huber H, Ziegler D, Pfluger V, Vogel G, Zweifel C, Stephan R. 2011. Prevalence and characteristics of methicillin-resistant coagulase-negative staphylococci from livestock, chicken carcasses, bulk tank milk, minced meat, and contact persons. BMC Vet. Res. 7:6. 10.1186/1746-6148-7-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. La Scola B, Fournier PE, Raoult D. 2011. Burden of emerging anaerobes in the MALDI-TOF and 16S rRNA gene sequencing era. Anaerobe 17:106–112 [DOI] [PubMed] [Google Scholar]
- 46. Carroll KC, Weinstein MP. 2007. Manual and automated systems for detection and identification of microorganisms, p 192–217 In Murray PR, Baron EJ, Jorgensen JH, Landry ML, Pfaller MA. (ed), Manual of clinical microbiology, 9th ed American Society for Microbiology, Washington, DC [Google Scholar]
- 47. Grosse-Herrenthey A, Maier T, Gessler F, Schaumann R, Bohnel H, Kostrzewa M, Kruger M. 2008. Challenging the problem of clostridial identification with matrix-assisted laser desorption and ionization-time-of-flight mass spectrometry (MALDI-TOF MS). Anaerobe 14:242–249 [DOI] [PubMed] [Google Scholar]
- 48. Khamis A, Raoult D, La Scola B. 2005. Comparison between rpoB and 16S rRNA gene sequencing for molecular identification of 168 clinical isolates of Corynebacterium. J. Clin. Microbiol. 43:1934–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Adekambi T, Drancourt M, Raoult D. 2009. The rpoB gene as a tool for clinical microbiologists. Trends Microbiol. 17:37–45 [DOI] [PubMed] [Google Scholar]
- 50. van Veen SQ, Claas EC, Kuijper EJ. 2010. High-throughput identification of bacteria and yeast by matrix-assisted laser desorption ionization-time of flight mass spectrometry in conventional medical microbiology laboratories. J. Clin. Microbiol. 48:900–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bizzini A, Durussel C, Bille J, Greub G, Prod'hom G. 2010. Performance of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J. Clin. Microbiol. 48:1549–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cherkaoui A, Hibbs J, Emonet S, Tangomo M, Girard M, Francois P, Schrenzel J. 2010. Comparison of two matrix-assisted laser desorption ionization–time of flight mass spectrometry methods with conventional phenotypic identification for routine identification of bacteria to the species level. J. Clin. Microbiol. 48:1169–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Eigner U, Holfelder M, Oberdorfer K, Betz-Wild U, Bertsch D, Fahr AM. 2009. Performance of a matrix-assisted laser desorption ionization-time-of-flight mass spectrometry system for the identification of bacterial isolates in the clinical routine laboratory. Clin. Lab. 55:289–296 [PubMed] [Google Scholar]
- 54. Stevenson LG, Drake SK, Murray PR. 2010. Rapid identification of bacteria in positive blood culture broths by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 48:444–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Christner M, Rohde H, Wolters M, Sobottka I, Wegscheider K, Aepfelbacher M. 2010. Rapid identification of bacteria from positive blood culture bottles by use of matrix-assisted laser desorption-ionization time of flight mass spectrometry fingerprinting. J. Clin. Microbiol. 48:1584–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ferreira L, Sanchez-Juanes F, Gonzalez-Avila M, Cembrero-Fucinos D, Herrero-Hernandez A, Gonzalez-Buitrago JM, Munoz-Bellido JL. 2010. Direct identification of urinary tract pathogens from urine samples by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 48:2110–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Christensen JJ, Dargis R, Hammer M, Justesen US, Nielsen XC, Kemp M, Danish MALDI-TOF MS Study Group 2012. Matrix-assisted laser desorption ionization-time of flight mass spectrometry analysis of Gram-positive, catalase-negative cocci not belonging to the Streptococcus or Enterococcus genus and benefits of database extension. J. Clin. Microbiol. 50:1787–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yan Y, Meng S, Bian D, Quinn C, Li H, Stratton CW, Tang YW. 2011. Comparative evaluation of Bruker Biotyper and BD Phoenix systems for identification of bacterial pathogens associated with urinary tract infections. J. Clin. Microbiol. 49:3936–3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lagier JC, Armougom F, Million M, Hugon P, Pagnier I, Robert C, Bittar F, Fournous G, Gimenez G, Maraninchi M, Trape JF, Koonin EV, La Scola B, Raoult D. 2012. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin. Microbiol. Infect. 18:1185–1193 [DOI] [PubMed] [Google Scholar]
- 60. Lagier JC, Million M, Hugon P, Armougom F, Raoult D. 2012. Human gut microbiota: repertoire and variations. Front. Cell. Infect. Microbiol. 2:136. 10.3389/fcimb.2012.00136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dubourg G, Lagier JC, Armougom F, Robert C, Hamad I, Brouqui P, Raoult D. 2013. The gut microbiota of a patient with resistant tuberculosis is more comprehensively studied by culturomics than by metagenomics. Eur. J. Clin. Microbiol. Infect. Dis. 32:637–645 [DOI] [PubMed] [Google Scholar]
- 62. Bizzini A, Jaton K, Romo D, Bille J, Prod'hom G, Greub G. 2011. Matrix-assisted laser desorption ionization-time of flight mass spectrometry as an alternative to 16S rRNA gene sequencing for identification of difficult-to-identify bacterial strains. J. Clin. Microbiol. 49:693–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.