Abstract
A clinical outbreak of bovine piroplasmosis was reported in Italy. The etiological agent was characterized as Babesia occultans, a parasite regarded as apathogenic and never detected before in continental Europe. This report paves the way for further studies to assess the occurrence of this tick-transmitted protozoan in other European regions.
TEXT
Bovine babesiosis is a tick-borne disease of cattle caused by apicomplexan protozoa of the genus Babesia which may induce clinical conditions characterized by hemolytic anemia and fever, with occasional hemoglobinuria and even death of animals. Babesia parasites are responsible for severe economic losses in cattle industry, with large social and epidemiological impacts (1). Among the main Babesia species infecting cattle, Babesia bovis and Babesia bigemina cause a severe and often fatal disease in untreated cattle of tropical and subtropical regions whereas Babesia divergens is associated with bovine and human infection in Europe (2). In addition, Babesia major (3) and Babesia occultans are less pathogenic. The latter species was detected more than 30 years ago in southern Africa, being transmitted by a Hyalomma marginatum tick (4). Babesia occultans has never been associated with clinical signs in animals, even after experimental infection of splenectomized individuals (4). For a long time, the distribution of B. occultans was believed to be confined to sub-Saharan Africa (4, 5), and this protozoan was detected only recently in H. marginatum in Tunisia (3) and in cattle in the Balearic Islands (6). However, even in this case, clinical disease was not associated with B. occultans infection. This study reports on a clinical outbreak of bovine piroplasmosis by B. occultans in continental Europe.
In May and June 2012, an outbreak of clinical piroplasmosis occurred in a cattle herd from the Apulia region of southern Italy following a severe tick infestation. At that time, the herd consisted of 48 brown Swiss cows kept in loose housing and fed with forages and purchased feeding stuffs. In early May of each year, all cows were subjected to deltamethrin treatments against ticks and facilities were treated with azamethiphos against flies. However, despite the antiparasitic treatments, tick infestation was repeatedly reported in the farm. Clinical signs were observed in 26 lactating cows (mainly primiparous animals) and consisted of marked pallor of the oral and genital mucosae, fever (up to 40.8°C), and a drop in milk production. Neither gastroenteric nor respiratory signs were reported. Two days after the onset of fever, the animals received oxytetracycline hydrochloride orally (p.o.) at a dose of 11 mg/kg of body weight every 12 h (q12h) for 7 days and two administrations, 36 h apart, of imidocarb dipropionate intramuscularly (i.m.) at a dose of 1.7 mg/kg. Deltamethrin treatments were also repeated in all animals at the farms. The therapy resulted in complete recovery of animals within about 10 days, although further clinical cases were sporadically reported in July and a single case occurred in August.
EDTA-blood samples collected from 8 febrile animals were used for hematological investigations, revealing mean hematological parameters generally below the reference ranges (red blood cells [RBC], 3.75 × 1012 ± 1.24 × 1012 liter−1; lower reference limit, 5.0 × 1012 liter−1; packed cell volume (PCV), 19.95% ± 4.78%; lower reference limit, 24%; hemoglobin (Hg), 66.1 ± 1.61 g liter−1; lower reference limit, 80 g liter−1). Blood samples were also submitted to a PCR assay for the detection of bovine piroplasms using the generic primers RLB-F2 (5′-GACACAGGGAGGTAGTGACAAG-3′) (7) and 18STBR (5′-GATCCTTCYGCAGGTTCACC-3′) as described elsewhere (8). Reactions were performed using the LA PCR kit version 2.1 (TaKaRa Bio Inc., Shiga, Japan) in 25-μl volumes containing 1 μmol liter−1 of primers, 1× LA PCR buffer (Mg2+ plus), 4 μl of a deoxynucleoside triphosphate (dNTP) mixture (corresponding to 400 μmol liter−1 of each dNTP), 1.25 units of TaKaRa LA Taq, and 5 μl of template DNA. The cycling protocol consisted of preheating at 94°C for 3 min followed by 40 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min, with a final extension at 72°C for 10 min. To rule out other pathogens responsible for anemia and/or fever in cattle, molecular analyses were carried out to detect Anaplasma marginale (9), Anaplasma centrale (10), bovine pestiviruses (11), bovine coronavirus (12), Schmallenberg virus (13), and bluetongue virus (14). Other eubacteria were searched for through amplification of 16S ribosomal DNA (15).
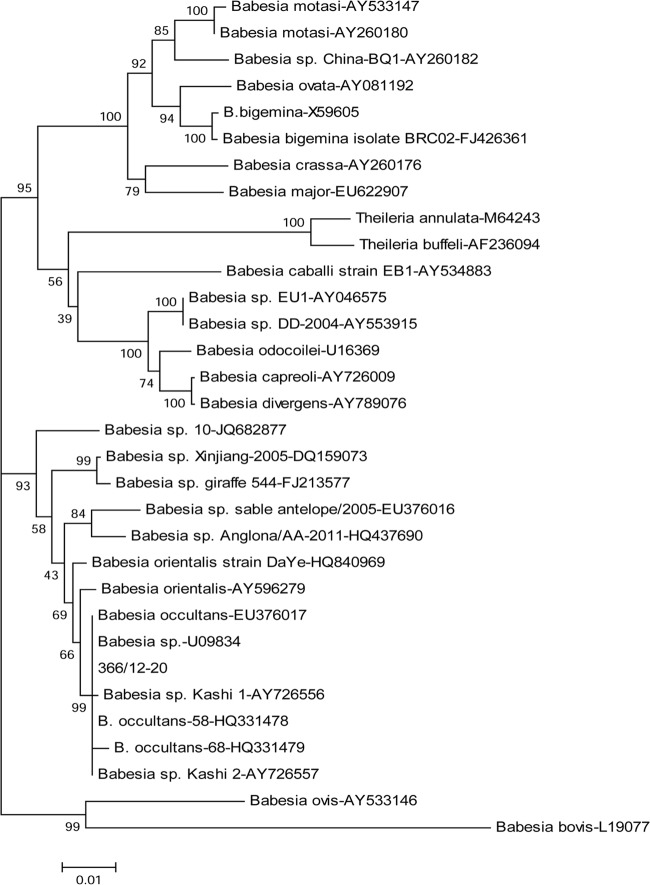
All samples tested positive for Babesia spp./Theileria spp. 18S rRNA genes without any evidence of coinfection by other pathogens of cattle. PCR products were purified using Montage PCR filter units (Millipore, Milan, Italy) and sequenced by BigDye 3.1 Ready reaction mix (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. Sequences were imported and assembled with the BioNumerics 5.0 software (Applied Maths, Saint-Martens-Latem, Belgium) and analyzed using the BioEdit software package (http://www.mbio.ncsu.edu/bioedit/bioedit.html) and NCBI's (http://www.ncbi.nlm.nih.gov) and EMBL's (http://www.ebi.ac.uk) analysis tools. The 18S rRNA gene sequences obtained from the diseased cows were 100% identical to each other and matched with the best nucleotide identity (99.7 to 100%) to analogous sequences of B. occultans retrieved from the GenBank database but not those of other protozoa within the genus Babesia. Phylogenetic analysis was carried out on a 1,050-nucleotide sequence generated from sample 366/12-20 (GenBank accession number KC157568), which was considered the prototype strain (Italy-366/12-20). The neighbor-joining tree obtained with the Mega4.1 software (http://www.megasoftware.net/mega4/mega41.html) over 1,000 replicates showed that strain Italy-366/12-20 clustered with the B. occultans clade, which also comprises Babesia orientalis, Babesia sp. Kashi 1, and Babesia sp. Kashi 2, but not with other piroplasms (Fig. 1). Babesia sp. Sable antelope and Babesia sp. Anglona, which was recently isolated from pigs in Italy (16), were also strictly related to the B. occultans cluster. This pattern of segregation was confirmed by the maximum-parsimony method (data not shown).
Fig 1.
Neighbor-joining tree based on partial 18S rRNA gene sequences of ruminant piroplasms. Statistical support was provided by bootstrapping more than 1,000 replicates. The scale bar indicates the estimated numbers of nucleotide substitutions per position.
In this article, we report a clinical outbreak of bovine piroplasmosis caused by B. occultans. Although there was no evidence of coinfections with other pathogens, less-common infectious agents of bovine anemia were not able to be ruled out definitively. Babesia occultans is considered a completely apathogenic or low-pathogenic parasite of cattle with a geographical distribution restricted to sub-Saharan Africa (4, 5). Although this piroplasm has been recently detected in the Balearic Islands (3, 6), no reports from continental Europe were available. Nonetheless, H. marginatum is diffused in southern Italy, where the infection by B. occultans might be more spread out than currently acknowledged (17). Babesia occultans was molecularly detected in cows that displayed fever, anemia, and severe alteration in the hematological parameters. Circumstantial evidence for the condition caused by this protozoan is also represented by the efficacy of imidocarb dipropionate in the animals' recovery. Phylogeny showed that other Babesia strains that have not been classified so far may likely belong to the species B. occultans. Importantly, the detection of this protozoan in continental Europe suggests that thorough surveillance programs should be undertaken for this tick-borne disease in order to implement effective control measures in cattle populations in which the proper tick species and the vectored pathogen occur.
Footnotes
Published ahead of print 8 May 2013
REFERENCES
- 1. Bock R, Jackson L, de Vos A, Jorgensen W. 2004. Babesiosis of cattle. Parasitology 129(Suppl):S247–S269 [DOI] [PubMed] [Google Scholar]
- 2. Zintl A, Mulcahy G, Skerrett HE, Taylor SM, Gray JS. 2003. Babesia divergens, a bovine blood parasite of veterinary and zoonotic importance. Clin. Microbiol. Rev. 16:622–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ros-García A, M'Ghirbi Y, Bouattour A, Hurtado A. 2011. First detection of Babesia occultans in Hyalomma ticks from Tunisia. Parasitology 138:578–582 [DOI] [PubMed] [Google Scholar]
- 4. Gray JS, De Vos AJ. 1981. Studies on a bovine Babesia transmitted by Hyalomma marginatum rufipes Koch, 1844. Onderstepoort J. Vet. Res. 48:215–223 [PubMed] [Google Scholar]
- 5. Dipeolu OO, Amoo A. 1984. The presence of kinetes of a Babesia species in the haemolymph smears of engorged Hyalomma ticks in Nigeria. Vet. Parasitol. 17:41–46 [DOI] [PubMed] [Google Scholar]
- 6. Ros-García A, García-Pérez AL, Verdera J, Juste RA, Hurtado A. 2012. Monitoring piroplasms infection in three cattle farms in Minorca (Balearic Islands, Spain) with previous history of clinical piroplamosis. Vet. Parasitol. 190:318–325 [DOI] [PubMed] [Google Scholar]
- 7. Georges K, Loria GR, Riili S, Greco A, Caracappa S, Jongejan F, Sparagano O. 2001. Detection of haemoparasites in cattle by reverse line blot hybridisation with a note on the distribution of ticks in Sicily. Vet. Parasitol. 99:273–286 [DOI] [PubMed] [Google Scholar]
- 8. Nagore D, García-Sanmartín J, García-Pérez AL, Juste RA, Hurtado A. 2004. Detection and identification of equine Theileria and Babesia species by reverse line blotting: epidemiological survey and phylogenetic analysis. Vet. Parasitol. 123:41–54 [DOI] [PubMed] [Google Scholar]
- 9. Carelli G, Decaro N, Lorusso A, Elia G, Lorusso E, Mari V, Ceci L, Buonavoglia C. 2007. Detection and quantification of Anaplasma marginale DNA in blood samples of cattle by real-time PCR. Vet. Microbiol. 124:107–114 [DOI] [PubMed] [Google Scholar]
- 10. Decaro N, Carelli G, Lorusso E, Lucente MS, Greco G, Lorusso A, Radogna A, Ceci L, Buonavoglia C. 2008. Duplex real-time polymerase chain reaction for simultaneous detection and quantification of Anaplasma marginale and Anaplasma centrale. J. Vet. Diagn. Invest. 20:606–611 [DOI] [PubMed] [Google Scholar]
- 11. Decaro N, Sciarretta R, Lucente MS, Mari V, Amorisco F, Colaianni ML, Cordioli P, Parisi A, Lelli R, Buonavoglia C. 2012. A nested PCR approach for unambiguous typing of pestiviruses infecting cattle. Mol. Cell. Probes 26:42–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Decaro N, Elia G, Campolo M, Desario C, Mari V, Radogna A, Colaianni ML, Cirone F, Tempesta M, Buonavoglia C. 2008. Detection of bovine coronavirus using a TaqMan-based real-time RT-PCR assay. J. Virol. Methods 151:167–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoffmann B, Scheuch M, Höper D, Jungblut R, Holsteg M, Schirrmeier H, Eschbaumer M, Goller KV, Wernike K, Fischer M, Breithaupt A, Mettenleiter TC, Beer M. 2012. Novel orthobunyavirus in Cattle, Europe, 2011. Emerg. Infect. Dis. 18:469–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Toussaint JF, Sailleau C, Breard E, Zientara S, De Clercq K. 2007. Bluetongue virus detection by two real-time RT-qPCRs targeting two different genomic segments. J. Virol. Methods 140:115–123 [DOI] [PubMed] [Google Scholar]
- 15. Wilson KH, Blitchington RB, Greene CE. 1990. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J. Clin. Microbiol. 28:1942–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zobba R, Parpaglia ML, Spezzigu A, Pittau M, Alberti A. 2011. First molecular identification and phylogeny of a Babesia sp. from a symptomatic sow (Sus scrofa Linnaeus 1758). J. Clin. Microbiol. 49:2321–2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dantas-Torres F, Otranto D. 2013. Species diversity and abundance of ticks in three habitats in southern Italy. Ticks Tick Borne Dis. 4:251–255 [DOI] [PubMed] [Google Scholar]