Abstract
Forty-six Helicobacter cinaedi isolates from the same hospital were analyzed by multilocus sequence typing. Most H. cinaedi isolates exhibited clonal complex 9 and were mainly isolated from immunocompromised patients in the same ward. Three Helicobacter fennelliae isolates were obtained from the same ward and exhibited the same pulsed-field gel electrophoresis patterns. All isolates were resistant to clarithromycin and ciprofloxacin. H. cinaedi and H. fennelliae must be carefully monitored to prevent nosocomial infection.
TEXT
Helicobacter cinaedi and H. fennelliae are enterohepatic Helicobacter species that inhabit the colons of humans and animals. Isolation of H. cinaedi and H. fennelliae has been reported sporadically (1–10), although reports of H. cinaedi are increasing in frequency. The transmission route of H. cinaedi is unclear. Since it is prevalent in animals such as dogs and hamsters, these domestic pets may be a natural reservoir for H. cinaedi infection in humans (11). However, an association between animals and H. cinaedi infection has not been demonstrated. H. cinaedi infection is frequent in bacteremia in immunocompromised hosts; therefore, the pathogen may be prevalent in the human colon and invade the blood when the host's immune system is weakened.
In 2008, we reported nosocomial infection with H. cinaedi in our hospital (12). We developed a multilocus sequence typing (MLST) method for H. cinaedi strains by comparing the results of MLST and pulsed-field gel electrophoresis (PFGE) and confirmed that most H. cinaedi isolates from this outbreak were identical (13). After the outbreak, we isolated Helicobacter spp. from patients in this hospital to follow the path of H. cinaedi infection. In this study, we obtained H. cinaedi and H. fennelliae isolates from the same hospital for 5 years: the isolates were typed, and their antimicrobial susceptibilities were determined.
Helicobacter isolates were collected at Sapporo City General Hospital in Sapporo, Japan, during 2008 to 2012. Helicobacter spp. were isolated from 3 to 8 ml blood in Bactec Plus aerobic/F culture vials (Becton, Dickinson and Company, Franklin Lakes, NJ) after incubation for 3 to 8 days. The isolates were recovered by culture on sheep blood (T) (with Trypticase soy, catalog no. 251148; (Becton, Dickinson and Company, Japan) under microaerobic conditions (AnaeroPack; Mitsubishi Gas Chemical Company, Inc., Tokyo, Japan) for 7 days at 35°C. Isolates from feces were obtained from rectal swabs using Transystem (Copan Diagnostics, Inc., Murrieta, CA) by culture on modified Skirrow's agar plates (Nissui Pharmaceutical Co., Ltd., Tokyo, Japan) under microaerobic conditions (AnaeroPack) for 7 days. The isolates were identified by morphological analysis and DNA sequencing of the 16S rRNA and the 23S rRNA genes. All Helicobacter isolates were subcultured in brucella agar (Becton, Dickinson) with 5% horse blood under microaerobic conditions with hydrogen obtained by the gas replacement method using an anaerobic gas mixture (H2, 10%; CO2, 10%; and N2, 80%). During 2008 to 2012, 46 H. cinaedi isolates were obtained from different inpatients. Twelve H. cinaedi isolates were obtained in 2008; these were described previously (13). Since then, 12, 16, and 5 isolates were obtained in 2009, 2010, and 2011, respectively, and 1 H. cinaedi isolate was obtained in 2012; these are described here. Of 46 H. cinaedi isolates, 26 were obtained from females and 20 from males. The mean age of the H. cinaedi-infected patients was 60.9 years (range, 32 to 79 years). H. cinaedi-infected patients had diseases such as malignant lymphoma (n = 22), acute lymphoid leukemia (n = 2), acute myelogenous leukemia (n = 3), autoimmune disease (n = 5), myeloma (n = 2), adult T-cell leukemia (n = 1), nonhematological malignancy (n = 9), chronic renal failure (n = 1), and ischemic enteritis (n = 1). Eight of the H. cinaedi isolates were from feces; all others were from blood. Five (10.9%) patients had cellulitis. Of the 46 H. cinaedi isolates, 35 were obtained from patients in ward B on the 5th floor and another 6 were from patients in ward C on the same floor. Ward B is mainly reserved for immunocompromised patients; therefore, a weakened immune system is a certain risk factor for H. cinaedi infection.
MLST was performed as described previously (13). All sequences were registered in the H. cinaedi MLST Database (http://pubmlst.org/hcinaedi/); the allele number, sequence type (ST), and clonal complex (CC) were also defined by the MLST database and the eBURST software program (http://eburst.mlst.net/). As shown in Table 1, MLST analysis divided the 46 H. cinaedi isolates into 8 STs and 5 CC categories. Since CC9 (ST10 and ST11) was the most prevalent isolate from this hospital, the isolates were divided into 2 groups: those that belonged to CC9, and those that did not (non-CC9). The wards were compared between groups (Table 2). Of 35 CC9 isolates, 31 (88.6%) were isolated in ward B. Non-CC9 isolates were mainly from wards B and C and were isolated sporadically from other wards. The time courses of isolation of CC9 H. cinaedi strains during 2008 to 2012 are shown in Fig. 1. Once CC9 was identified in ward B, it was consistently found in other patients. Among these isolates, ST11 was isolated only in 2008 (n = 8) and 2009 (n = 1). ST10 was isolated every year from 2008 to 2012. Isolates belonging to other CCs were sporadically isolated from different wards during 2008 to 2012. We previously demonstrated that the CC9 strains possessed the same PFGE pattern (13). Therefore, the CC9 isolates have likely been continuously transmitted from patient to patient. To understand why CC9 was prevalent in this hospital, the prevalence of CC9 in each patient's characteristics was compared by Fisher's exact test using GraphPad Prism software (GraphPad Software, Inc., La Jolla, CA); however, no significant differences were observed: 78% (21/27) were found in females versus 74% (14/19) in males (P = 1.000); 79% (30/38) of were from blood versus 63% (5/8) from feces (P = 0.153); 76% (13/17) were found in patients aged >65 years versus 76% (22/29) in patients aged <65 years (P = 1.000); and 80% (4/5) were found in cellulitis-positive patients versus 76% (31/41) in cellulitis-negative patients (P = 0.420). Therefore, H. cinaedi CC9 isolates may possess a specific factor for transmission or pathogenicity. H. cinaedi expresses cytolethal distending toxin (CDT), but other pathogenetic factors have not been identified. Further investigation of these pathogenetic factors may explain why H. cinaedi CC9 is prevalent in this hospital.
Table 1.
Sequence types and allelic profiles of H. cinaedi isolates
| Sequence type | Clonal complex | No. (%) of isolates | Allelic profile (allele no.) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| 23S rRNA | ppa | aspA | aroE | atpA | tkt | cdtB | |||
| ST2 | CC1 | 1 (2.2) | 1 | 1 | 1 | 1 | 1 | 4 | 2 |
| ST3 | CC1 | 4 (8.7) | 1 | 1 | 1 | 1 | 1 | 4 | 1 |
| ST4 | CC4 | 1 (2.2) | 3 | 3 | 3 | 1 | 2 | 2 | 1 |
| ST10 | CC9 | 26 (56.5) | 4 | 2 | 2 | 2 | 2 | 1 | 2 |
| ST11 | CC9 | 9 (19.6) | 2 | 2 | 2 | 2 | 2 | 1 | 2 |
| ST15 | CC7 | 2 (4.3) | 4 | 3 | 5 | 4 | 3 | 4 | 1 |
| ST16 | CC16 | 1 (2.2) | 4 | 3 | 2 | 2 | 6 | 4 | 3 |
| ST17 | CC7 | 2 (4.3) | 4 | 1 | 5 | 4 | 3 | 4 | 1 |
Table 2.
Relationship between the clonal complex of H. cinaedi isolates and the wards from which they were isolated
| Floor | Ward | No. (%) of H. cinaedi isolates that belonged to: |
|
|---|---|---|---|
| CC9 (n = 35) | Non-CC9 (n = 11) | ||
| 4th | A | 1 (2.9) | 0 (0.0) |
| 5th | B | 31 (88.6) | 4 (36.4) |
| 5th | C | 2 (5.7) | 4 (36.4) |
| 6th | D | 1 (2.9) | 1 (9.1) |
| 10th | E | 0 (0.0) | 1 (9.1) |
| 2nd | Outpatient | 0 (0.0) | 1 (9.1) |
Fig 1.

Time course of CC9 H. cinaedi isolation.
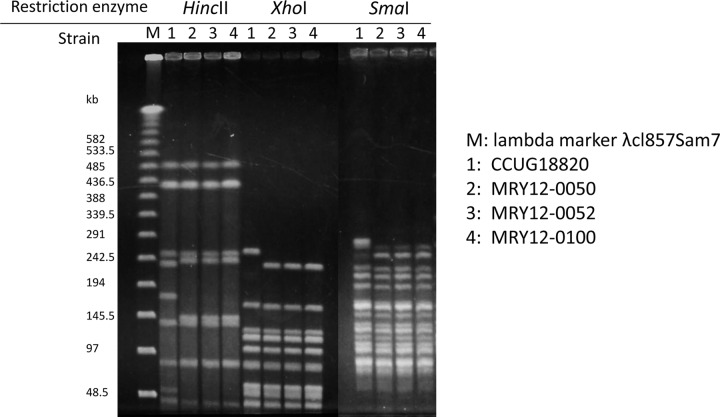
H. fennelliae (n = 3) isolates obtained from different patients at the same hospital in July 2011, August 2011, and February 2012 were also analyzed. Two isolates were from the blood of patients with malignant lymphoma (n = 2), and 1 isolate was from the blood of a patient with autoimmune disease (n = 1). Three isolates shared strong sequence identity in the 16S (97%) and 23S (96%) RNA sequences of H. fennelliae CCUG18820 and low sequence identity in the 16S and 23S rRNA sequences of H. cinaedi CCUG18818. These isolates were also tested by API Campy (bioMérieux) and verified as H. fennelliae. To our knowledge, H. fennelliae isolation from humans in Japan has not been reported previously. All isolates were obtained from patients in ward B. PFGE for H. fennelliae was performed with 3 restriction enzymes (HincII, XhoI, and SmaI) according to the method developed for H. cinaedi (13); the restriction patterns were identical between all isolates from the same hospital, while the patterns were different from H. fennelliae CCUG18820 (Fig. 2). We also amplified 11 housekeeping genes and confirmed that these sequences were the same in each isolate (data not shown). Therefore, H. fennelliae is most likely transferred from patient to patient, as is H. cinaedi. All H. fennelliae isolates were obtained from ward B; therefore, a weakened immune system could be a risk factor for H. fennelliae infection. H. fennelliae lacks the CDT that drives virulence in H. cinaedi (14). However, the clinical manifestations and backgrounds of H. fennelliae-infected and H. cinaedi-infected patients were similar. Since the number of H. fennelliae-infected patients in this study is small, further investigation is needed to identify the differences in H. cinaedi and H. fennelliae pathogenicities.
Fig 2.
PFGE patterns of 3 H. fennelliae isolates from different patients in the same hospital.
The antimicrobial susceptibilities of H. cinaedi and H. fennelliae isolates were measured by the agar dilution method (13) and are shown in Table 3. All isolates had high MICs for ciprofloxacin and clarithromycin, consistent with prior findings (13). The MIC level of clarithromycin was lower in H. fennelliae (8 mg/liter) than that in H. cinaedi (MIC90, >128 mg/liter). To determine the mechanism of ciprofloxacin resistance, GyrA and GyrB were sequenced in all H. cinaedi and H. fennelliae isolates. Compared to H. cinaedi CCUG18818, which is susceptible to ciprofloxacin, the H. cinaedi isolates possessed mutations in GyrA and GyrB. All mutation patterns and MICs of ciprofloxacin are shown in Table 4. H. cinaedi strains with mutations at position 84 in GyrA and position 423 or 442 in GyrB or 2 mutations at positions 84 and 88 in GyrA tended to exhibit high MICs for ciprofloxacin in comparison to isolates with a mutation at GyrA position 84. Since mutations in GyrB confer high-level resistance to fluoroquinolones in H. pylori and other species (15), the mutation in GyrB likely confers high-level resistance to ciprofloxacin in H. cinaedi. In the H. fennelliae isolates, the residue at position 86 of GyrA that corresponded to Thr84 in H. cinaedi was lysine. It has been previously shown that ciprofloxacin-susceptible H. fennelliae strains possess a threonine at position 84 (16). Therefore, the mutation at position 86 could be the major factor that decreases susceptibility to ciprofloxacin in H. fennelliae. The 23S rRNA gene sequences were analyzed in all isolates to identify clarithromycin resistance. All H. cinaedi isolates possessed an adenine-to-guanine mutation at position 2018, corresponding to position 2143 of the 23S rRNA gene in Helicobacter pylori, which confers clarithromycin resistance in that species. Three H. fennelliae isolates had no mutation at position 2327, which corresponds to position 2018 in H. cinaedi and position 2143 in H. pylori. However, the H. fennelliae isolates carried an adenine-to-guanine mutation at position 2879, corresponding to position 2694 of the 23S rRNA gene in H. pylori, conferring low-level resistance to clarithromycin (17). This mutation was not observed in H. fennelliae CCUG18820, which was susceptible to clarithromycin (0.125 mg/liter). Thus, clarithromycin resistance in the H. fennelliae isolates is likely due to mutation at position 2879 of 23S rRNA.
Table 3.
MICs of H. cinaedi and H. fennelliae isolates from a hospital in Japan
| Isolate and parametera | MIC (mg/liter) of: |
|||||
|---|---|---|---|---|---|---|
| Amoxicillin | Imipenem | Clarithromycin | Ciprofloxacin | Minocycline | Gentamicin | |
| H. cinaedi | ||||||
| Clinical isolates (n = 46) | ||||||
| Range | 0.5–32 | 0.031–0.25 | 2–>128 | 16–128 | 0.016–0.25 | 0.25–1 |
| 50% | 4 | 0.063 | 64 | 64 | 0.063 | 0.5 |
| 90% | 8 | 0.125 | >128 | 128 | 0.125 | 0.5 |
| CCUG18818 | 8 | 0.063 | 0.008 | 0.25 | 0.25 | 0.25 |
| H. fennelliae | ||||||
| Clinical isolates (n = 3), range | 1–2 | 0.031 | 8 | 64–>128 | 0.063–0.125 | 0.25–0.5 |
| CCUG18820 | 1 | 0.031 | 0.125 | 0.5 | 0.125 | 0.5 |
50% and 90%, MIC50 and MIC90, respectively.
Table 4.
Mutations in gyrases A and B and MICs of ciprofloxacin for H. cinaedi isolates
| No. of isolates | Mutation(s) in: |
Ciprofloxacin MIC (mg/liter) |
||
|---|---|---|---|---|
| GyrA | GyrB | Range | 50%a | |
| 16 | T84I | 16–64 | 16 | |
| 25 | T84I | D423N | 64–128 | 64 |
| 1 | T84I | P442S | 64 | |
| 2 | T84I, D88G | 64 | ||
| 1 | T84I, D88H | |||
| 1 | T84I, D88N | 64 | ||
50%, MIC50.
In conclusion, we analyzed 46 H. cinaedi and 3 H. fennelliae isolates from the same hospital in Japan during 2008 to 2012. Most H. cinaedi isolates belonged to the same clonal complex and were isolated from the same ward, which mainly hosts immunocompromised patients. H. fennelliae strains were isolated from the same ward and possessed the same PFGE patterns, indicating these isolates had undergone human-to-human transmission. Antimicrobial susceptibilities were similar between species; however, the mutations that conferred resistance to clarithromycin differed from those in H. cinaedi.
ACKNOWLEDGMENTS
This study was supported by a Grant-in-Aid for Scientific Research (C) from the Japan Society for Promotion of Science (no. 22590410), a Grant-in-Aid for Young Scientists (B) from the Japan Society for Promotion of Science (no. 25860105), and a grant from the Ministry of Health, Labor and Welfare of Japan (H24-Shinkou-Ippan-010).
Footnotes
Published ahead of print 8 May 2013
REFERENCES
- 1. Burman WJ, Cohn DL, Reves RR, Wilson ML. 1995. Multifocal cellulitis and monoarticular arthritis as manifestations of Helicobacter cinaedi bacteremia. Clin. Infect. Dis. 20:564–570 [DOI] [PubMed] [Google Scholar]
- 2. Burnens AP, Stanley J, Schaad UB, Nicolet J. 1993. Novel Campylobacter-like organism resembling Helicobacter fennelliae isolated from a boy with gastroenteritis and from dogs. J. Clin. Microbiol. 31:1916–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hsueh PR, Teng LJ, Hung CC, Chen YC, Yang PC, Ho SW, Luh KT. 1999. Septic shock due to Helicobacter fennelliae in a non-human immunodeficiency virus-infected heterosexual patient. J. Clin. Microbiol. 37:2084–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kiehlbauch JA, Brenner DJ, Cameron DN, Steigerwalt AG, Makowski JM, Baker CN, Patton CM, Wachsmuth IK. 1995. Genotypic and phenotypic characterization of Helicobacter cinaedi and Helicobacter fennelliae strains isolated from humans and animals. J. Clin. Microbiol. 33:2940–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lasry S, Simon J, Marais A, Pouchot J, Vinceneux P, Boussougant Y. 2000. Helicobacter cinaedi septic arthritis and bacteremia in an immunocompetent patient. Clin. Infect. Dis. 31:201–202 [DOI] [PubMed] [Google Scholar]
- 6. Mammen MP, Jr, Aronson NE, Edenfield WJ, Endy TP. 1995. Recurrent Helicobacter cinaedi bacteremia in a patient infected with human immunodeficiency virus: case report. Clin. Infect. Dis. 21:1055–1056 [DOI] [PubMed] [Google Scholar]
- 7. Murakami H, Goto M, Ono E, Sawabe E, Iwata M, Okuzumi K, Yamaguchi K, Takahashi T. 2003. Isolation of Helicobacter cinaedi from blood of an immunocompromised patient in Japan. J. Infect. Chemother. 9:344–347 [DOI] [PubMed] [Google Scholar]
- 8. Smuts HE, Lastovica AJ. 2011. Molecular characterization of the 16S rRNA gene of Helicobacter fennelliae isolated from stools and blood cultures from paediatric patients in South Africa. J. Pathog. 2011:217376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tee W, Street AC, Spelman D, Munckhof W, Mijch A. 1996. Helicobacter cinaedi bacteraemia: varied clinical manifestations in three homosexual males. Scand. J. Infect. Dis. 28:199–203 [DOI] [PubMed] [Google Scholar]
- 10. Totten PA, Fennell CL, Tenover FC, Wezenberg JM, Perine PL, Stamm WE, Holmes KK. 1985. Campylobacter cinaedi (sp. nov.) and Campylobacter fennelliae (sp. nov.): two new Campylobacter species associated with enteric disease in homosexual men. J. Infect. Dis. 151:131–139 [DOI] [PubMed] [Google Scholar]
- 11. Gebhart CJ, Fennell CL, Murtaugh MP, Stamm WE. 1989. Campylobacter cinaedi is normal intestinal flora in hamsters. J. Clin. Microbiol. 27:1692–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Minauchi K, Takahashi S, Sakai T, Kondo M, Shibayama K, Arakawa Y, Mukai M. 2010. The nosocomial transmission of Helicobacter cinaedi infections in immunocompromised patients. Intern. Med. 49:1733–1739 [DOI] [PubMed] [Google Scholar]
- 13. Rimbara E, Mori S, Matsui M, Suzuki S, Wachino J, Kawamura Y, Shen Z, Fox JG, Shibayama K. 2012. Molecular epidemiologic analysis and antimicrobial resistance of Helicobacter cinaedi isolated from seven hospitals in Japan. J. Clin. Microbiol. 50:2553–2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Young VB, Knox KA, Schauer DB. 2000. Cytolethal distending toxin sequence and activity in the enterohepatic pathogen Helicobacter hepaticus. Infect. Immun. 68:184–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rimbara E, Noguchi N, Kawai T, Sasatsu M. 2012. Fluoroquinolone resistance in Helicobacter pylori: role of mutations at position 87 and 91 of GyrA on the level of resistance and identification of a resistance conferring mutation in GyrB. Helicobacter 17:36–42 [DOI] [PubMed] [Google Scholar]
- 16. Husmann M, Feddersen A, Steitz A, Freytag C, Bhakdi S. 1997. Simultaneous identification of campylobacters and prediction of quinolone resistance by comparative sequence analysis. J. Clin. Microbiol. 35:2398–2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rimbara E, Noguchi N, Kawai T, Sasatsu M. 2008. Novel mutation in 23S rRNA that confers low-level resistance to clarithromycin in Helicobacter pylori. Antimicrob. Agents Chemother. 52:3465–3466 [DOI] [PMC free article] [PubMed] [Google Scholar]