Abstract
A number of diagnostic tests are available for dengue virus (DENV) detection, including a variety of nucleic acid amplification tests (NAATs). However, reports describing a direct comparison of different NAATs have been limited. In this study, we report the design of an internally controlled real-time reverse transcriptase PCR (rRT-PCR) that detects all four DENV serotypes but does not distinguish between them (the pan-DENV assay). Two hundred clinical samples were then tested using four different DENV RT-PCR assays: the pan-DENV assay, a commercially produced, internally controlled DENV rRT-PCR (the Altona assay), a widely used heminested RT-PCR, and a serotype-specific multiplex rRT-PCR assay. The pan-DENV assay had a linear range extending from 1.0 to 7.0 log10 cDNA equivalents/μl and a lower limit of 95% detection ranging from 1.7 to 7.6 cDNA equivalents/μl, depending on the serotype. When measured against a composite reference standard, the pan-DENV assay proved to be more clinically sensitive than either the Altona or heminested assays, with a sensitivity of 98.0% compared to 72.3% and 78.8%, respectively (P ≤ 0.0001 for both comparisons). The pan-DENV assay detected DENV in significantly more samples collected on or after day 5 of illness and in a subgroup of patients with detectable anti-DENV IgM at presentation. No significant difference in sensitivity was observed between the pan-DENV assay and the multiplex rRT-PCR, despite the presence of an internal control in the former. The detection of DENV RNA late in the course of clinical illness should serve to lengthen the period during which a confirmed molecular diagnosis of DENV infection can be provided.
INTRODUCTION
Dengue virus (DENV) is the most common vector-borne human viral pathogen worldwide (1). Infection with one or more of the four closely related virus serotypes (designated DENV-1 to -4) results in a range of clinical manifestations spanning asymptomatic infection, dengue fever (DF), and severe dengue, a category that includes entities previously classified as dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS) (1, 2). Infection with one serotype (primary infection) results in immunity to that serotype, but infection can still occur with any of the remaining serotypes (secondary infection). Secondary DENV infection has been shown to be a significant risk factor for the development of DHF or DSS (3, 4). DENV transmission largely occurs in the tropical and subtropical regions of the world, though the number of countries where DENV is endemic has been increasing (1, 5). Recent reports estimate that 230 million DENV infections occur annually worldwide, including 2 million cases of severe disease and 21,000 deaths (6). Over 3.6 billion people live in regions where this pathogen is endemic and are at risk for infection (6). DF is also one of the most common causes of a systemic febrile illness in travelers returning from countries where the virus is endemic and remains a major concern for military personnel stationed in these areas (7–9).
A wide array of diagnostic tests for DENV have been developed and remain in use throughout the world. These include laboratory-based testing for anti-DENV IgM, anti-DENV IgG, and the DENV nonstructural protein 1 (NS1). Point-of-care tests for these analytes are also available. In addition, molecular methods are performed, including heminested RT-PCR and real-time reverse transcriptase RT-PCR (rRT-PCR) assays (1, 10). Currently, the WHO supports the use of a number of these tests depending on individual laboratory capabilities and the goals of testing, as no gold standard for DENV diagnosis has been established (1).
Consistent with the state of DENV diagnostics as a whole, a variety of nucleic acid amplification tests (NAATs) have been reported in the literature. A heminested RT-PCR for the detection and serotyping of DENV, a version of which remains in wide use, was first reported in 1992 (11, 12). Though numerous DENV species-specific and serotype-specific assays have since been developed, direct comparisons between these assays are notably rare (11, 13–17). Rather, studies evaluating new DENV molecular diagnostic tests typically use samples collected within the first 5 days of fever from patients who had positive testing by viral isolation, seroconversion, or both (10, 18–25). This practice ensures that only the highest viral loads are evaluated and it likely does not reflect clinical reality, where patients may present at any time during their illness, including five or more days after fever onset.
The majority of DENV NAATs have been designed with the dual goals of sensitive DENV detection and the ability to provide a serotype-specific diagnosis (11, 13, 14, 23, 24). While serotyping capability is useful for epidemiologic surveillance, the relative contribution of serotype-specific information to the care of individual patients remains unclear (1, 10, 26). For laboratories outside regions where the disease is endemic that primarily test returning travelers, serotype-specific DENV diagnosis may not be required (10, 20, 26). Pan-DENV assays may still be able to provide information that has been associated with the development of severe disease, including the detection of DENV RNA at defervescence (27–30).
Despite the large number of reported DENV NAATs, few of these assays have been designed to include an internal control (IC), either as an extrinsic molecule spiked into each sample before or after extraction or as a heterologous intrinsic target that is coextracted with DENV RNA (14, 17, 21, 25, 31). It has been advocated that ICs be used in settings where PCR inhibitors present a significant source of false-negative results, which may be particularly important in the performance of NAATs in countries where dengue is endemic (26, 32).
In this study, we report the development of a species-specific internally controlled rRT-PCR utilizing hydrolysis probes for DENV detection (here referred to as the pan-DENV assay). We also report an independent evaluation of the Altona RealStar dengue RT-PCR kit, which is a commercially produced internally controlled rRT-PCR kit for DENV detection. Furthermore, using 200 clinical samples, we directly compared four molecular tests for DENV: the pan-DENV and Altona assays, a version of the heminested RT-PCR (11), and the serotype-specific DENV multiplex rRT-PCR (16).
MATERIALS AND METHODS
Pan-DENV assay design.
The pan-DENV assay utilizes the same primers as the multiplex rRT-PCR assay, though the primer concentrations differ. The primer design has been described previously, and these primers are listed in Table 1 (16). Separate hydrolysis probes were designed for the pan-DENV assay based on the 95% consensus sequence of the 5′ untranslated region (UTR) and capsid gene. RealTimeDesign software (Biosearch Technologies, Novato, CA) was used to create BHQplus probes, which utilize a duplex stabilizing technology to increase melting temperature and allow for reduced probe length. Four probes were designed to account for sequence diversity; these probe sequences are listed in Table 2 and were given letter designations A to D. The probes were tested in silico using BLASTn to query the NCBI nucleotide database. Searches excluding the DENV group (identification no. 11052) were also performed to identify the best non-DENV sequence matches in the database, as well as matches in the Flaviviridae family.
Table 1.
Primer sequences for the pan-DENV assay
| Primer name | Primer sequence (5′ → 3′) | Concn (nM)a | DENV genomic locationb |
|---|---|---|---|
| DENV-1, -2, -3 forward | CAGATCTCTGATGAACAACCAACG | 350 | 86 |
| DENV-2 forward C→T | CAGATCTCTGATGAATAACCAACG | 350 | 87 |
| DENV-3 forward C→T | CAGATTTCTGATGAACAACCAACG | 300 | 85 |
| DENV-4 forward | GATCTCTGGAAAAATGAAC | 450 | 81 |
| DENV-1, -3 reverse | TTTGAGAATCTCTTCGCCAAC | 300 | 199 (DENV-1), 198 (DENV-3) |
| DENV-2 reverse | AGTTGACACGCGGTTTCTCT | 350 | 171 |
| DENV-2 reverse A→G | AGTCGACACGCGGTTTCTCT | 350 | 171 |
| DENV-4 reverse | AGAATCTCTTCACCAACC | 450 | 190 |
| RNase P forward | AGATTTGGACCTGCGAGCG | 100 | NA |
| RNase P reverse | GAGCGGCTGTCTCCACAAGT | 100 | NA |
The concentration of each primer in the final reaction mixture. Predicted amplicon sizes are as follows: DENV-1, 114 bp; DENV-2, 85 bp; DENV-3, 114 bp; DENV-4, 110 bp.
Genomic locations are provided for the 5′ base complementary to each primer using the following reference virus sequences: DENV-1 US/Hawaii/1944 (GenBank accession no. EU848545.1), DENV-2 New Guinea C strain (GenBank accession no. AF038403.1), DENV-3 strain H87 (GenBank accession no. M93130.1), and DENV-4 strain H241 (GenBank accession no. AY947539.1). NA, not applicable.
Table 2.
DENV and RNase P probe sequences for the pan-DENV assay
| Probe | 5′ Fluor | Probe sequence (5′ → 3′) | 3′ Quencher | Concn (nM)b |
|---|---|---|---|---|
| DENV probe A | FAMa | CTCGCGCGTTTCAGCATAT | BHQplus | 200 |
| DENV probe B | FAM | CTCTCGCGTTTCAGCATAT | BHQplus | 200 |
| DENV probe C | FAM | CTCTCACGTTTCAGCATATTG | BHQplus | 200 |
| DENV probe D | FAM | CTCACGCGTTTCAGCATAT | BHQplus | 200 |
| RNase P | Cal Fluor Red 610 | TTCTGACCTGAAGGCTCTGCGCG | BHQ-2 | 50 |
FAM, 6-carboxyfluorescein.
The concentration for each probe in the final reaction mixture.
A previously described assay for the detection of RNase P was modified for use as the IC in this assay (33). The primer and probe sequences were maintained as originally published (see Tables 1 and 2); however, the RNase P probe was labeled with Cal Fluor Red 610 to facilitate multiplexing. The IC primer and probe concentrations were selected based on titration experiments, such that samples that were negative for DENV routinely had crossing threshold (CT) values between 25 and 30.
RT-PCR assays.
The pan-DENV assay was performed using the SuperScript III Platinum one-step qRT-PCR kit (Invitrogen, Carlsbad, CA). Reaction mixtures were scaled down from the manufacturer-recommended 50-μl volume to 25 μl per reaction mixture, and 5 μl of RNA template was added to each reaction mixture. The primer and probe concentrations in the final reaction mixtures are listed in Tables 1 and 2, respectively. During the analytical evaluation, 5 μl of pooled RNA extracts from domestic patient plasma samples was spiked into the reaction mixture to mimic the IC in the clinical samples. RT-PCRs were performed using the Rotor-Gene Q instrument (Qiagen, Valencia, CA). Cycling conditions differed from those of the multiplex rRT-PCR and were as follows: 52°C for 15 min (RT step), 94°C for 2 min, 45 cycles of 94°C for 15 s, 55°C for 20 s, and 68°C for 20 s (run time, 110 min). Detection was performed in the green and orange channels at 55°C, and the gain was set at 4.67 for green and at 10 for orange. During analysis, the first five cycles were cropped from the green channel to improve baseline normalization; slope correction was performed for both channels. The threshold was set at 0.025 for the green channel, and any curve crossing this threshold prior to cycle 40 was considered to be positive for DENV. All results after cycle 40 were evaluated individually, and any sample generating an exponential curve that crossed the threshold after cycle 40 was also considered positive. The threshold was set at 0.1 for the orange RNase P channel. In samples that were negative for DENV, RNase P was considered to be detected if the curve crossed this threshold at any cycle. Samples in which DENV and RNase P RNA were not amplified were considered to have failed extraction or to contain an inhibitory substance.
The Altona assay was performed using the RealStar dengue PCR kit 1.0 (Altona Diagnostics, Hamburg, Germany). Reaction mixtures were scaled down from the recommended 50-μl volume to 25 μl, with each reaction mixture containing 2.5 μl master mix A, 10 μl master mix B, 1.25 μl of internal control solution, and 6.25 μl of water. Five microliters of RNA template was added to each reaction mixture, which was decreased from the manufacturer-recommended 25 μl. An IC, included with the kit, was added to the reaction mixture following the setup of the negative-control sample on each run. RT-PCRs were again performed using the Rotor-Gene Q. Cycling conditions were: 50°C for 10 min (RT step), 95°C for 2 min, 45 cycles of 95°C for 15 s, 55°C for 45 s, and 72°C for 15 s (run time, 120 min). Detection was performed in the green and yellow channels at 55°C, and the gain was set at 10 for both channels. During analysis, slope correction was performed for both channels. The threshold was set at 0.1 for the green channel, and curves were interpreted with the same algorithm used for the pan-DENV assay. The threshold was set at 0.05 for the yellow IC channel. In samples that were negative for DENV, the Altona IC was required to be detected within 45 cycles; samples in which the Altona IC was not amplified were considered inhibited.
The heminested RT-PCR and the serotype-specific multiplex rRT-PCR were performed as previously described (16). However, for the heminested assay, 5 μl of RNA template was added in the RT-PCR step, and 5 μl of RT-PCR product was added to the heminested reaction mixture. For all clinical samples, the pan-DENV, multiplex rRT-PCR, and heminested assays were performed concurrently. Samples underwent one further freeze-thaw cycle prior to use in the Altona assay.
Reference virus RNA.
Genomic RNA from reference strains of the four DENV serotypes, DENV-1 Hawaii 1944, DENV-2 New Guinea C strain, DENV-3 strain H87, and DENV-4 strain H241 were obtained from Vircell (Granada, Spain). Genomic RNA of three strains of West Nile virus (WNV) (NY 1999, a clinical isolate previously reported as NAL strain, and B956) and a single strain each of Japanese encephalitis virus (JEV) and tick-borne encephalitis virus (TBEV) was obtained from the St. Orsola-Malpighi Hospital, Regional Reference Centre for Microbiological Emergencies, in Bologna, Italy (34).
Plasmid generation, quantitation, and sequencing.
The generation and sequencing of plasmids has been described previously (16). Plasmid sequences were identical to the expected sequences for each DENV reference strain. The concentration of plasmid DNA was quantified using the Quant-iT PicoGreen double-stranded DNA (dsDNA) reagent and kit (Invitrogen, Carlsbad, CA). Twenty-five- and 50-fold dilutions were tested in triplicate. A standard curve was generated, and the concentration of plasmid in the initial eluate was calculated.
Linearity, lower limit of detection, and precision.
The analytical evaluation of the pan-DENV assay was performed according to previously published recommendations (32). For each serotype, linearity studies were performed on serial 10-fold dilutions of both quantified plasmid DNA and reference virus RNA. For the plasmid DNA, dilutions from 7.0 log10 copies/μl to 1 copy/μl were tested in triplicate on a single run. Reference virus RNA concentrations were originally quantified by the manufacturer in ng/μl of total RNA. Ten-fold dilutions from 1 ng/μl to 0.01 pg/μl RNA were tested in triplicate on a single run. Using the standard curve generated with dilutions of plasmid DNA, the concentration in DENV cDNA equivalents/μl was calculated for the highest concentration of RNA (1 ng/μl) for each serotype. The linear range was established by fitting a best-fit line to the data by regression analysis and included the range where the R2 value for this line was ≥0.99.
To establish the lower limit of 95% detection (95% LLOD), the lowest concentrations of RNA at which all replicates were detectable during the linear range study were used as the starting point. Ten replicates of four 2-fold dilutions from this concentration were tested on a single run. The 95% LLOD was then calculated using probit analysis.
The precision of the pan-DENV assay was determined using three dilutions of RNA controls (high positive, low positive, and limit of quantitation). These were performed five times on three separate days. Fresh dilutions were made on the day of each run from aliquots of high-concentration stocks (1 ng/μl, high positive). Intra- and interrun variability were calculated from the log10 concentration of the samples.
To establish the linear range and estimate the intrarun precision for the Altona assay, serial 10-fold dilutions from 1 ng/μl to 0.01 pg/μl of reference virus RNA for each DENV serotype were tested in triplicate on a single run. Testing to establish the 95% LLOD was performed as described for the pan-DENV assay.
Specificity.
Specificity was evaluated by testing genomic RNA from the WNV, JEV, and TBEV isolates, as well as from the yellow fever 17D (YF-17D) vaccine strain. The JEV, TBEV, and WNV strains are described above. For YF-17D, genomic RNA (Vircell, Granada, Spain) was tested at concentrations of ≥12,500 copies/μl and 250 copies/μl, as quantitated by the manufacturer. An additional 60 domestic samples with detectable levels of hepatitis C virus (HCV) were extracted and tested. All specificity experiments were performed for the pan-DENV and Altona assays.
Clinical samples.
A total of 201 clinical samples from patients presenting with a systemic febrile illness were tested, including 161 archived deidentified samples from Nicaragua and 40 deidentified prospectively collected samples from Sri Lanka. The Nicaraguan samples were collected between 23 September 2008 and 23 December 2011 as part of the Nicaraguan Pediatric Dengue Cohort study, as well as a hospital-based study to assess the risk factors for severe dengue in inpatients of the infectious diseases ward of the Hospital Infantil Manuel de Jesús Rivera (Managua, Nicaragua). The study design and methods for both of these studies have been described previously (35–37). Of these patients, 121 were suspected dengue cases and had been tested with an earlier version of the heminested RT-PCR assay on site (12). For 40 patients, another etiology was felt to be more likely than DENV (referred to as C cases). C cases have been described previously; these samples had not been tested for DENV (36). Results from the C cases were included in the overall sensitivity analysis for each assay but were separated for analysis based on the day of illness on which the sample was obtained. For all samples, RNA was extracted from aliquots of plasma that had been stored at −80°C.
Samples were also collected from 40 children presenting to the Lady Ridgeway Hospital (Colombo, Sri Lanka) with an acute febrile illness that was clinically suspected to be dengue. These samples have been described elsewhere (16). Briefly, samples were collected from 18 March to 28 May 2012, and patients were tested with the Hexagon GmbH dengue assay (HUMAN Diagnostics, Wiesbaden, Germany), which is a rapid assay detecting anti-DENV IgM and IgG antibodies.
For the clinical samples, a composite reference was utilized as the reference standard. Samples that tested positive by two or more RT-PCR assays were considered to be positive by the composite reference. Those that tested positive by only one RT-PCR assay or that tested negative by all assays were considered negative by the composite reference. The clinical sensitivity and specificity of each assay were then calculated using this composite reference standard. Throughout the paper, sensitivity refers to clinical sensitivity. The phrase analytical sensitivity is used when this characteristic is being explicitly discussed.
Nucleic acid extraction from clinical specimens.
Nucleic acid extraction was performed using the QIAamp viral RNA mini kit (Qiagen, Valencia, CA). All extractions were carried out according to the manufacturer's recommendations. Extractions were performed using 140 μl of patient plasma and eluted into 60 μl of buffer AVE.
A second centrifugation step that is recommended following the addition of buffer AW2 to the columns was performed for all clinical samples. In order to assess the causes of IC failures in the Altona assay, a subset of HCV-positive plasma samples were reextracted, and this centrifugation step was lengthened to 5 min. HCV-positive plasma samples that displayed evidence of inhibition were also heat treated at 65°C for 10 min, and two 10-fold dilutions were performed to eliminate potential inhibitors.
Ethics.
The protocols for the Nicaraguan Pediatric Dengue Cohort study and the Pediatric Hospital-Based Dengue study were reviewed and approved by the institutional review boards (IRB) of the University of California—Berkeley and the Nicaraguan Ministry of Health. The parents or legal guardians of all subjects provided written informed consent, and subjects aged ≥6 years provided assent. The IRB at Stanford University waived the review of this study, as all samples were precollected and deidentified.
Statistics.
Basic statistical analysis, including the calculation of means and standard deviations was performed using Excel software (Microsoft, Bellevue, WA). Two-tailed Fisher's exact tests and paired t tests were performed using GraphPad software (GraphPad, La Jolla, CA). Probit analysis was performed using SPSS (IBM, Armonk, NY).
RESULTS
Pan-DENV analytical evaluation.
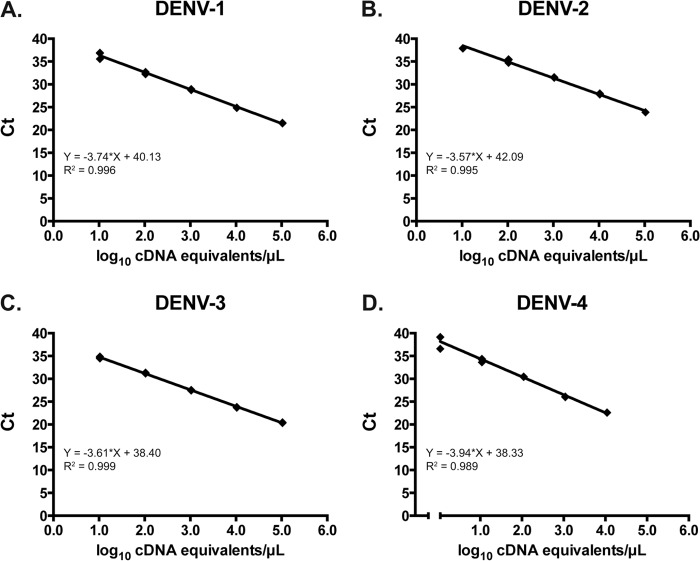
The primers and probes used in the pan-DENV assay are listed in Tables 1 and 2, respectively. Using serial dilutions of plasmid DNA, the linear range for each serotype extended from 1.0 to 7.0 log10 cDNA equivalents/μl (Fig. 1). Based on the standard curves generated using these plasmids, the concentration of DENV in cDNA equivalents/μl for the highest-concentration reference virus RNA was calculated. Using serial dilutions of reference virus RNA, the linear range of the pan-DENV assay for each serotype, expressed in cDNA equivalents/μl, was 0.16 to 5.16 log10 for DENV-1, 0.83 to 4.83 log10 for DENV-2, 0.03 to 5.03 log10 for DENV-3, and 1.11 to 4.11 log10 for DENV-4 (data not shown).
Fig 1.
Linearity of the pan-DENV assay using serial 10-fold dilutions of plasmid containing the amplicons for DENV-1 (A), DENV-2 (B), DENV-3 (C), and DENV-4 (D). Ct, crossing threshold.
The 95% LLOD was determined for each serotype in the pan-DENV assay by probit analysis using reference virus RNA dilutions. The 95% LLOD was calculated to be 1.7 for DENV-1, 1.7 for DENV-2, 2.2 for DENV-3, and 7.6 cDNA equivalents/μl for DENV-4. Both intra- and interrun precision measures were calculated for each serotype at three concentrations (high positive, low positive, and limit of quantitation) using reference virus RNA. Geometric mean concentrations in log10 cDNA equivalents/μl, as well as intra- and interrun precision measures, are shown in Table 3.
Table 3.
Precision of the pan-DENV assay
| DENV serotype | Dilution | Interrun precision |
Intrarun precision |
||||
|---|---|---|---|---|---|---|---|
| Concn (log10 cDNA equivalents/μl) | SD | % CoVa | Concn (Range, log10 cDNA equivalents/μl) | SD range | % CoV rangea | ||
| DENV-1 | High positive | 5.05 | 0.17 | 3.4 | 4.95–5.16 | 0.06–0.18 | 1.2–3.7 |
| Low positive | 3.25 | 0.16 | 4.8 | 3.05–3.38 | 0.04–0.07 | 1.3–2.3 | |
| Limit of quantitation | 1.20 | 0.15 | 12.2 | 1.03–1.32 | 0.05–0.10 | 4.0–9.5 | |
| DENV-2 | High positive | 4.61 | 0.19 | 4.1 | 4.43–4.83 | 0.02–0.10 | 0.1–2.0 |
| Low positive | 2.62 | 0.08 | 3.2 | 2.57–2.74 | 0.051–0.054 | 1.9–2.1 | |
| Limit of quantitation | 0.57 | 0.28 | 48.1 | 0.39–0.76 | 0.18–0.29 | 23.9–56.0 | |
| DENV-3 | High positive | 5.02 | 0.18 | 3.7 | 4.89–5.03 | 0.07–0.21 | 1.3–4.2 |
| Low positive | 2.35 | 0.13 | 5.5 | 2.18–2.44 | 2.8–7.8 | 1.2–3.2 | |
| Limit of quantitation | 0.32 | 0.24 | 7.6 | 0.20–0.45 | 0.27–0.17 | 38.0–127 | |
| DENV-4 | High positive | 4.04 | 0.14 | 3.6 | 3.96–4.16 | 0.06–0.18 | 1.4–4.6 |
| Low positive | 2.32 | 0.12 | 5.1 | 2.20–2.39 | 0.07–0.09 | 3.1–3.8 | |
| Limit of quantitation | 1.43 | 0.18 | 12.6 | 1.27–1.60 | 0.08–0.16 | 4.9–12.8 | |
% CoV, percent coefficient of variation.
Altona assay analytical evaluation.
The linear range of the Altona assay was established using serial dilutions of reference virus RNA for each DENV serotype. The linear range for this assay, expressed in cDNA equivalents/μl, extended from 1.04 to 5.04 to log10 for DENV-1, 1.61 to 4.61 log10 for DENV-2, 1.02 to 5.02 log10 for DENV-3, and 0.04 to 4.04 log10 for DENV-4 (Fig. 2). The 95% LLOD was determined for each serotype in the Altona assay by probit analysis using reference virus RNA dilutions. The 95% LLOD was calculated to be 7.6 for DENV-1, 21.7 for DENV-2, 1.2 for DENV-3, and 0.97 cDNA equivalents/μl for DENV-4.
Fig 2.
Linearity of the Altona dengue assay using serial 10-fold dilutions of reference virus RNA for DENV-1 (A), DENV-2 (B), DENV-3 (C), and DENV-4 (D). Ct, crossing threshold.
Intrarun precision was determined for the Altona assay using DENV reference virus RNA. The standard deviation (percent coefficient of variation) at the high and low ends of the linear range, respectively, for each serotype was 0.02 (0.37%) and 0.20 (20.17%) for DENV-1, 0.005 (0.1%) and 0.11 (6.72%) for DENV-2, 0.02 (0.39%) and 0.06 (5.38%) for DENV-3, and 0.01 (0.13%) and 0.10 (9.54%) for DENV-4. These results for the Altona assay were comparable to the results obtained with the pan-DENV assay.
Specificity.
The pan-DENV primers and probes were evaluated in silico by querying the NCBI nucleotide database for related sequences. As described previously, the primers demonstrated limited homologies to other members of the Flaviviridae family, as well as to non-DENV sequences (16). No organism represented the best match for both the forward and reverse primers for any of the primer pairs. The best non-DENV matches for probes A, B, C, and D were Brassica rapa, Schistosoma mansoni, Drosophila ananassae, and Desulfatibacillum alkenivorans AK-01, respectively (query coverage, 89 to 94%; E value, 0.28 to 4.4). The best matches within the Flaviviridae family were strains of JEV (query coverage, 71 to 78%; E value, 0.44 to 0.52), though these sequences would not be predicted to amplify using the primers in this assay.
No amplification was observed in the pan-DENV or Altona assays when RNA from other closely related flaviviruses (three strains of WNV, one strain each of JEV and TBEV, and the YF-17D strain tested at two concentrations) was used. Sixty HCV-positive clinical samples were also tested by both assays; these samples had a median HCV viral load of 6.05 log10 IU/ml of patient plasma (range, <1.63 to 7.66). No amplification was observed in either assay for any of these samples.
IC performance.
The performance of the ICs for the pan-DENV and Altona assays was evaluated using 201 clinical samples from Nicaragua and Sri Lanka. A single sample resulted in an IC failure on the pan-DENV assay, indicating a failed extraction or the presence of inhibitors. This sample was not included in further analysis for any assay. All samples that were negative for DENV RNA by the pan-DENV assay (n = 45) had similar amounts of RNase P RNA (mean CT, 29.69; standard deviation, 2.76). Samples with CT values of <20 in the green channel (DENV) did not have a detectable RNase P signal, whereas samples with a CT of >25 in the green channel yielded consistently detectable signals for RNase P (data not shown).
The Altona assay experienced a high number of IC failures during testing with the clinical samples. Overall, IC failure occurred in 49 samples (24.5%). These samples were not interpretable by the Altona assay and were excluded from further analysis for this test. Of these samples, DENV RNA was detected in 25 (51.0%) samples by the pan-DENV assay, 24 (49.0%) by the multiplex rRT-PCR, and 12 (48.0%) by the heminested assay. The percentages of IC failures were similar in both groups of clinical samples (Nicaragua, 39/160 [24.4%], and Sri Lanka, 10/40 [25%]). Three IC failures also occurred in the HCV-positive plasma samples described above (3/60 [5%]). These failures were repeated and confirmed on separate runs (data not shown). After 10- and 100-fold dilution of these samples, IC results were similar to the IC-only control. Prolonged centrifugation during the extraction or heating of the eluate did not improve results (data not shown).
Samples from areas of endemicity.
All clinical samples were tested by the pan-DENV, Altona, heminested, and DENV multiplex rRT-PCR assays. Each assay was compared against a composite reference standard, defined as the detection of DENV RNA in two or more RT-PCR assays (Table 4). The composite reference comprised 200 samples for the pan-DENV, multiplex rRT-PCR, and heminested assays. The sensitivity and specificity of each assay were then calculated using this reference. The pan-DENV and multiplex rRT-PCR had similar sensitivities (98.0% and 100%, respectively; P = 0.248). Both assays proved to be more sensitive than the heminested assay (sensitivity, 78.8%; P < 0.0001 for both comparisons). The clinical specificities of the pan-DENV (85.7%), multiplex rRT-PCR (87.8%), and heminested assays (93.9%) were not significantly different from one another (P > 0.05 for all comparisons).
Table 4.
Comparison of the pan-DENV, Altona, multiplex rRT-PCR, and heminested assays with a composite reference
| Assay | Assay result | Composite reference results (n) |
||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| Pan-DENV | Positive | 148 | 7 | 155 |
| Negative | 3 | 42 | 45 | |
| Total | 151 | 49 | 200 | |
| Altonab | Positive | 94 | 0 | 94 |
| Negative | 36 | 21 | 57 | |
| Total | 130 | 21 | 151 | |
| Multiplex rRT-PCR | Positive | 151 | 6 | 157 |
| Negative | 0 | 43 | 43 | |
| Total | 151 | 49 | 200 | |
| Heminested | Positive | 119 | 3 | 122 |
| Negative | 32 | 46 | 78 | |
| Total | 151 | 49 | 200 | |
A single sample failed RNase P amplification and was excluded from all comparisons.
Forty-nine IC failures were excluded from the Altona analysis.
The sensitivity and specificity for the Altona assay were calculated using a composite reference comprising 151 samples. Test characteristics for the other assays were also calculated using this subset for comparison. The Altona assay had the lowest sensitivity of any assay tested (72.3%; P < 0.0001 for comparison with the pan-DENV or multiplex rRT-PCR assays, P = 0.035 for comparison with the heminested assay). All samples detected by the Altona assay were also detected using the pan-DENV and multiplex rRT-PCR assays. The clinical specificity of the Altona assay was 100% compared against the composite reference, but this was not significantly different from the specificities of the other three assays (P > 0.05 for all comparisons). A total of 16 samples had DENV RNA detected by only a single assay (Table 5). The mean CT values for these samples, when applicable, are also displayed in Table 5.
Table 5.
Results for 16 clinical samples with DENV RNA detected using a single assay
| Assay | n | Mean CT (SD)a | Serotype (no.) |
|---|---|---|---|
| Pan-DENV | 7 | 38.88 (1.34) | NAb |
| Multiplex rRT-PCR | 6 | 41.19 (1.66) | DENV-1 (5), DENV-3 (1) |
| Heminested | 3 | NAb | DENV-3 (3) |
CT, crossing threshold.
NA, not applicable.
To understand the clinical performance of these tests in more detail, we specifically analyzed the results for the C cases and IgM-positive samples, and we evaluated test performance based on the day of illness of sample collection.
C cases.
Of the 40 samples collected from C cases, which were febrile patients thought to have a nondengue disease, a total of 19 (47.5%) samples had DENV RNA detected by at least one assay. This included 12 (30%) positive samples as detected by either the pan-DENV or multiplex rRT-PCR and 10 (25%) positive samples as detected by the heminested assay (data not shown). Seven samples (all DENV-3) tested positive by the pan-DENV, heminested, and multiplex rRT-PCR assays, which include the five samples that tested positive using the Altona assay. Five samples were detected only by the pan-DENV assay, and five different samples were detected only using the multiplex rRT-PCR assay. The average CT for these 10 samples was 40.2 (standard deviation, 1.8) compared to 20.8 for the seven samples that tested positive by both assays (standard deviation, 9.3; P < 0.0001). Two samples (both DENV-3) were detected only using the heminested assay. The alternative diagnoses for these patients were not available for this study.
IgM-positive Sri Lankan samples.
Given the high analytical and clinical sensitivity of the pan-DENV assay, we next compared assay performance measures in the subset of specimens that were positive for anti-DENV IgM antibodies. The 40 samples from Sri Lanka were collected relatively late in the clinical course (mean day of illness, 5.9; standard deviation, 1.4), and 36 patients already had detectable anti-DENV IgM at presentation. Complete IgM data were not available for the Nicaraguan samples.
In this IgM-positive group, the pan-DENV assay detected DENV RNA in 35 samples (97.2%), whereas the heminested assay detected DENV RNA in 18 samples (50.0%; P < 0.0001). The pan-DENV assay was also more sensitive than the Altona assay when only those samples with interpretable results by the latter were considered (26/26 [100%] versus 13/26 [50.0%]; P < 0.0001). The multiplex rRT-PCR assay detected DENV RNA in the same samples detected by the pan-DENV assay; these results have been reported previously (16).
Day-of-illness analysis.
Test results for 158 samples with available data were stratified according to the day of illness of sample collection (40 C cases plus 2 additional samples did not have the day of illness recorded). The difference in sensitivity between the pan-DENV or multiplex rRT-PCR assays and either the Altona or the heminested RT-PCR varied based on the day of illness on which the sample was obtained (Table 6). In samples collected on ≤4 days of illness, DENV RNA was detected in 61/67 (91.0%) samples by the pan-DENV assay compared to 59/67 (88.1%) samples by the heminested assay. This difference was not significant (P > 0.05). There was also no significant difference between the assays when only the subset of samples with interpretable Altona results was considered (P > 0.05, data not shown). When the sample was obtained on ≥5 days of illness (range, 5 to 9 days), DENV RNA was detected in 81/91 (89.0%) samples by the pan-DENV assay compared to 52/91 (57.1%) samples by the heminested RT-PCR (P < 0.0001). For samples with interpretable Altona results, the pan-DENV was more sensitive than the Altona assay with samples obtained on ≥5 days of illness (64/67 versus 35/67, respectively; P < 0.0001). The difference between the Altona and heminested assays (44/67 detected) did not reach statistical significance (P = 0.16). There was no significant difference between the pan-DENV and multiplex rRT-PCR assays based on the day of illness (data not shown).
Table 6.
Clinical samples with detectable DENV RNA stratified by the day of illness of sample collectiona
| Day of illness | Pan-DENV | Altonab | Multiplex rRT-PCR | Heminested |
|---|---|---|---|---|
| 2 | 27/27 (100) | 27/27 (100) | 27/27 (100) | 27/27 (100) |
| 3 | 14/16 (87.5) | 12/14 (85.7) | 14/16 (87.5) | 13/16 (81.3) |
| 4 | 20/24 (83.3) | 15/20 (75.0) | 21/24 (87.5) | 19/24 (79.2) |
| 5 | 35/42 (83.3) | 17/31 (54.8) | 36/42 (85.7) | 24/42 (57.1) |
| >5 | 46/49 (93.9) | 18/36 (50.0) | 45/49 (91.8) | 28/49 (57.1) |
| Total | 142/158 (89.9) | 89/128 (69.5) | 143/158 (89.9) | 111/158 (70.2) |
Results are reported as number detected/total number (%). Data were not available for the 40 C cases and two additional samples.
IC failures were removed from the Altona assay analysis.
DISCUSSION
This study presents the development and evaluation of the pan-DENV assay, which is a species-specific internally controlled rRT-PCR for pan-DENV detection. A second commercially produced internally controlled rRT-PCR assay, the Altona assay, was also independently evaluated. Both assays were then included in a large clinical comparison of four molecular assays for the detection of DENV, including a heminested RT-PCR that remains in wide use throughout the world and a serotype-specific multiplex rRT-PCR previously developed in our laboratory (1, 16).
Comparisons of different DENV molecular tests, such as those performed in the current study, are scarce, despite the large number of diagnostic assays that have been reported for DENV (11, 13–15, 17, 38). Two earlier studies, published in 1998 and 2002, compared conventional RT-PCR assays to the original version of the heminested RT-PCR. In both of these studies, the heminested assay was equivalent to or more sensitive than the newer assays (17, 38). Four other groups have reported the comparison of DENV NAATs (three rRT-PCRs and one transcription-mediated amplification assay [TMA]) to a second molecular test (11, 13–15). Ito et al. (15) described the development of a hydrolysis probe-based rRT-PCR for DENV and compared its performance to that of two conventional RT-PCR assays (39). The new assay proved more sensitive when tested using 35 clinical specimens from returning travelers to Japan (15). Two groups have reported the comparison of serotype-specific multiplex rRT-PCR assays to a version of the heminested assay used in this study (11, 13). In both of these studies, however, the heminested assay remained more sensitive than either rRT-PCR assay (11, 13). A commercial TMA assay (14) was compared with one of the rRT-PCRs and proved to be more sensitive, though it was not compared to the heminested RT-PCR.
The current comparison offers two DENV rRT-PCRs that proved to be more sensitive than the heminested and Altona assays. The pan-DENV and multiplex assays were more sensitive when testing clinical samples obtained on ≥5 days of illness, whereas all four assays performed comparably when testing samples collected before that time. Results reported in DENV NAAT studies are often obtained using samples collected on ≤5 days of illness, when, based on our findings, it may be difficult to distinguish assays with relatively poor sensitivity (14, 23, 40). In studies where samples are tested on ≥5 days of illness, test sensitivity decreases markedly (41, 42). Both the pan-DENV and multiplex rRT-PCR assays also remained sensitive (97.2%) when samples obtained from acutely ill patients with detectable anti-DENV IgM levels were tested. It has generally been reported that as anti-DENV IgM levels become detectable, DENV viral load declines below detectable levels (1). As a result, patients with a positive IgM are infrequently tested in DENV NAAT studies. In cases where such patients are tested, the sensitivity of DENV detection has generally been poor, ranging from 0 to 69% (20, 22, 43–45). The improved sensitivities of the pan-DENV and multiplex rRT-PCR assays may result from the use of a highly conserved target region in the 5′ UTR and capsid gene, compared to previous assays that utilized targets in other regions of the genome, including the C-prM, E, NS3, NS5, and 3′ UTR (11, 13, 15, 20, 24).
Despite the improved sensitivities of the pan-DENV and multiplex rRT-PCR assays, the specificities of these two assays did not differ significantly from those of the heminested or Altona assays. Furthermore, no amplification was observed in the pan-DENV assay using samples from 60 patients with HCV, as well as with reference virus RNA from other closely related flaviviruses. Similar results for the multiplex rRT-PCR have been published previously (16). The samples that were detected by only one of these two assays and counted as false positives had very low concentrations of DENV RNA, making it difficult to confirm or refute the results obtained by other means, such as sequencing. While the Altona assay had a specificity of 100%, this came at the expense of 36 false-negative results and poor assay sensitivity in the samples collected on ≥5 days of illness.
The pan-DENV assay provides species-specific DENV detection but cannot be used for serotype determination in patient samples. For the management of acutely ill patients, however, the provision of a serotype-specific diagnosis may not be necessary and currently does not affect clinical care (1, 10, 20). Also, outside areas where dengue is endemic, the arguments for needing serotyping capability are less pertinent (10, 26). Given the differences in quantitation between the different serotypes, quantitative results from the pan-DENV assay would only be accurate if serotype information was also available. If serial samples are tested over time, however, relative quantitation can be performed, as well as sensitive detection on the day of defervescence, a finding that has been associated with the development of severe disease (30). With further testing of the pan-DENV assay, thresholds may be established for these parameters that identify patients who are at increased risk for the development of severe dengue. The somewhat-lower cost of the pan-DENV compared to the multiplex rRT-PCR reagents ($2.00 versus $2.60 per reaction mixture) may also allow for its use as a screening test or for serial monitoring, as mentioned above.
A further benefit of the pan-DENV assay is the multiplex detection of RNase P, which serves as a heterologous intrinsic IC. Of the method comparisons mentioned previously, only the conventional RT-PCR developed in 1998 and the TMA assay contained an IC (14, 17). IC results were also not reported as a part of an external quality-control evaluation of laboratories that were performing molecular DENV testing (26). The presence of IC data may have allowed the examination of extraction efficiency and reaction inhibition as potential contributors to the poor test sensitivity that is observed in a number of participating laboratories. The utility of an IC is further highlighted in this study by the large number of failures for the Altona assay, which uses an extrinsic molecule spiked into the reaction mixture. The fact that controls diluted in water (rather than extracted RNA eluted in buffer AVE) yielded consistently positive IC results suggests that the Altona reaction mixture may have been inhibited by eluates from the QIAamp viral RNA minikit. Ethanol contamination is one specific concern, though we decreased this risk by including an optional centrifugation step during RNA extraction. However, prolonged centrifugation did not improve the results. Eluate dilution did eliminate the observed inhibition, though this would not be a practical step for use with clinical samples. While the high IC failure rate did not fully explain the decreased sensitivity of the Altona assay, it remains a point of concern regarding the use of the current version of this test.
A number of samples (7/40 [17.5%]) from acutely ill patients who were initially suspected to have an illness other than dengue (C cases) tested positive for DENV by at least three of the RT-PCRs used in this study. The viral load was high among these samples, as indicated by the CT values, consistent with true-positive results. Twelve additional samples (30%) were positive by only one assay, albeit at very low viral loads. Because the samples were obtained during high DENV activity in Nicaragua, we favor the interpretation that these are also true positive. A large number of inapparent DENV infections occur, estimated at 50% to >90% of infections, with large variation from year to year and between studies (depending on the case capture procedures used) (46–49); thus, it is likely that some proportion of C cases were experiencing an inapparent DENV infection.
Despite the inclusion of 200 clinical samples from two areas where DENV is endemic, there were no clinical samples with detectable levels of the DENV-4 serotype. While the analytical sensitivity for DENV-4 has been documented in our laboratory for all of the assays included in this study, the performance of these tests needs to be confirmed with DENV-4-positive clinical specimens. In addition, we cannot rule out the possibility that very high levels of IC may inhibit DENV detection, though the analytical and clinical sensitivities of the pan-DENV assay suggest that IC competition is unlikely to be a significant problem in plasma samples. Nevertheless, the pan-DENV assay may be even more sensitive if performed without IC primers and probes. Further limitations of this study include concerns that are common to all NAATs regarding the emergence of divergent viral strains that do not match the primer or probe designs of a given assay. A specific limitation of the pan-DENV assay in this regard is the use of BHQplus moieties to allow for the design of shorter DENV probes with higher melting temperatures. While this improves the specificity of the assay, the binding of BHQplus probes may be more susceptible to single base-pair changes than longer hydrolysis probes without this modification (50).
In conclusion, we report here the design and evaluation of the pan-DENV assay, which is a species-specific internally controlled rRT-PCR for pan-DENV detection. In a comparison of four molecular diagnostic assays for DENV, the pan-DENV assay was shown to be equally sensitive to a previously described multiplex rRT-PCR, as well as more sensitive than a commercially produced rRT-PCR and a widely used heminested assay. This was despite the addition of an IC that should serve to improve quality assurance, particularly when sample preparation and assay setup are performed under suboptimal conditions. The improved sensitivities of the pan-DENV and multiplex rRT-PCR assays resulted from the ability to detect DENV RNA in samples collected on ≥5 days of illness and samples with detectable anti-DENV IgM, thereby lengthening the period of time during which a confirmed molecular diagnosis of DENV infection can be obtained.
ACKNOWLEDGMENTS
This research was supported by the National Institutes of Health grant RC4 TW008781-01. The studies in Nicaragua were supported by R01AI099631 (to A.B.), U54AI65359 (to A.B.), and HHSN2722001000026C (to E.H. and A.B.) from the National Institutes of Health and VE-1 (to E.H.) from the Pediatric Dengue Vaccine Initiative. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We thank the Stanford SPARK program for their support over the course of this project. We also thank Vittorio Sambri, Anna Pierro, and Paolo Gaibani for providing genomic flavivirus RNA for the specificity experiments. Finally, we thank Altona Diagnostics for providing the RealStar dengue RT-PCR kits for evaluation.
Footnotes
Published ahead of print 1 May 2013
REFERENCES
- 1. World Health Organization 2009. Dengue: guidelines for diagnosis, treatment, prevention and control. WHO Press, Geneva, Switzerland: [PubMed] [Google Scholar]
- 2. World Health Organization 1997. Dengue haemorrhagic fever: diagnosis, treatment, prevention and control, 2nd ed World Health Organization, Geneva, Switzerland [Google Scholar]
- 3. Halstead SB, Nimmannitya S, Cohen SN. 1970. Observations related to pathogenesis of dengue hemorrhagic fever. IV. Relation of disease severity to antibody response and virus recovered. Yale J. Biol. Med. 42:311–328 [PMC free article] [PubMed] [Google Scholar]
- 4. Martina BEE, Koraka P, Osterhaus AD. 2009. Dengue virus pathogenesis: an integrated view. Clin. Microbiol. Rev. 22:564–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gubler DJ. 2012. The economic burden of dengue. Am. J. Trop. Med. Hyg. 86:743–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beatty ME, Letson GW, Margolis HS. 2008. Estimating the global burden of dengue. In Abstract book: dengue 2008. Proceedings of the 2nd International Conference on Dengue and Dengue Haemorrhagic Fever Phuket, Thailand [Google Scholar]
- 7. Freedman DO, Weld LH, Kozarsky PE, Fisk T, Robins R, von Sonnenburg F, Keystone JS, Pandey P, Cetron MS, GeoSentinel Surveillance Network 2006. Spectrum of disease and relation to place of exposure among ill returned travelers. N. Engl. J. Med. 354:119–130 [DOI] [PubMed] [Google Scholar]
- 8. Burnette WN, Hoke CH, Jr, Scovill J, Clark K, Abrams J, Kitchen LW, Hanson K, Palys TJ, Vaughn DW. 2008. Infectious disease investment decision evaluation algorithm: a quantitative algorithm for prioritization of naturally occurring infectious disease threats to the U.S. military. Mil. Med. 173:174–181 [DOI] [PubMed] [Google Scholar]
- 9. Flores-Figueroa J, Okhuysen PC, von Sonnenburg F, DuPont HL, Libman MD, Keystone JS, Hale DC, Burchard G, Han PV, Wilder-Smith A, Freedman DO, GeoSentinel Surveillance Network 2011. Patterns of illness in travelers visiting Mexico and Central America: the GeoSentinel experience. Clin. Infect. Dis. 53:523–531 [DOI] [PubMed] [Google Scholar]
- 10. Peeling RW, Artsob H, Pelegrino JL, Buchy P, Cardosa MJ, Devi S, Enria DA, Farrar J, Gubler DJ, Guzman MG, Halstead SB, Hunsperger E, Kliks S, Margolis HS, Nathanson CM, Nguyen VC, Rizzo N, Vázquez S, Yoksan S. 2010. Evaluation of diagnostic tests: dengue. Nat. Rev. Microbiol. 8(12 Suppl):S30–S38. 10.1038/nrmicro2459 [DOI] [PubMed] [Google Scholar]
- 11. Chien LJ, Liao TL, Shu PY, Huang JH, Gubler DJ, Chang GJJ. 2006. Development of real-time reverse transcriptase PCR assays to detect and serotype dengue viruses. J. Clin. Microbiol. 44:1295–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV. 1992. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J. Clin. Microbiol. 30:545–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Johnson BW, Russell BJ, Lanciotti RS. 2005. Serotype-specific detection of dengue viruses in a fourplex real-time reverse transcriptase PCR assay. J. Clin. Microbiol. 43:4977–4983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Muñoz-Jordán J, Collins CS, Vergne E, Santiago GA, Peterson L, Sun W, Linnen JM. 2009. Highly sensitive detection of dengue virus nucleic acid in samples from clinically ill patients. J. Clin. Microbiol. 47:927–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ito M, Takasaki T, Yamada KI, Nerome R, Tajima S, Kurane I. 2004. Development and evaluation of fluorogenic TaqMan reverse transcriptase PCR assays for detection of dengue virus types 1 to 4. J. Clin. Microbiol. 42:5935–5937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Waggoner J, Abeynayake J, Sahoo MK, Gresh L, Tellez Y, Gonzalez K, Ballesteros G, Pierro AM, Gaibani P, Guo FP, Sambri V, Balmaseda A, Karunarante K, Harris E, Pinsky BA. 2013. Single-reaction, multiplex, real-time RT-PCR for the detection, quantitation, and serotyping of dengue viruses. PLoS Negl. Trop. Dis. 7:e2116. 10.1371/journal.pntd.0002116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harris E, Roberts TG, Smith L, Selle J, Kramer LD, Valle S, Sandoval E, Balmaseda A. 1998. Typing of dengue viruses in clinical specimens and mosquitoes by single-tube multiplex reverse transcriptase PCR. J. Clin. Microbiol. 36:2634–2639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Callahan JD, Wu SJ, Dion-Schultz A, Mangold BE, Peruski LF, Watts DM, Porter KR, Murphy GR, Suharyono W, King CC, Hayes CG, Temenak JJ. 2001. Development and evaluation of serotype- and group-specific fluorogenic reverse transcriptase PCR (TaqMan) assays for dengue virus. J. Clin. Microbiol. 39:4119–4124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Drosten C, Göttig S, Schilling S, Asper M, Panning M, Schmitz H, Günther S. 2002. Rapid detection and quantification of RNA of Ebola and Marburg viruses, Lassa virus, Crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, dengue virus, and yellow fever virus by real-time reverse transcription-PCR. J. Clin. Microbiol. 40:2323–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dumoulin A, Marti H, Panning M, Hatz C, Hirsch HH. 2008. Pan-dengue virus detection by PCR for travelers returning from the tropics. J. Clin. Microbiol. 46:3104–3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hue KD, Tuan TV, Thi HT, Bich CT, Anh HH, Wills BA, Simmons CP. 2011. Validation of an internally controlled one-step real-time multiplex RT-PCR assay for the detection and quantitation of dengue virus RNA in plasma. J. Virol. Methods 177:168–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kong YY, Thay CH, Tin TC, Devi S. 2006. Rapid detection, serotyping and quantitation of dengue viruses by TaqMan real-time one-step RT-PCR. J. Virol. Methods 138:123. [DOI] [PubMed] [Google Scholar]
- 23. Lai Y, Chung YK, Tan HC, Yap HF, Yap G, Ooi EE, Ng LC. 2007. Cost-effective real-time reverse transcriptase PCR (RT-PCR) to screen for Dengue virus followed by rapid single-tube multiplex RT-PCR for serotyping of the virus. J. Clin. Microbiol. 45:935–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leparc-Goffart I, Baragatti M, Temmam S, Tuiskunen A, Moureau G, Charrel R, de Lamballerie X. 2009. Development and validation of real-time one-step reverse transcription-PCR for the detection and typing of dengue viruses. J. Clin. Virol. 45:61–66 [DOI] [PubMed] [Google Scholar]
- 25. Naze F, Le Roux K, Schuffenecker I, Zeller H, Staikowsky F, Grivard P, Michault A, Laurent P. 2009. Simultaneous detection and quantitation of Chikungunya, Dengue and West Nile viruses by multiplex RT-PCR assays and Dengue virus typing using high resolution melting. J. Virol. Methods 162:1–7 [DOI] [PubMed] [Google Scholar]
- 26. Domingo C, Niedrig M, Teichmann A, Kaiser M, Rumer L, Jarman RG, Donoso-Mantke O. 2010. 2nd international external quality control assessment for the molecular diagnosis of Dengue infections. PLoS Negl. Trop. Dis. 4:e833. 10.1371/journal.pntd.0000833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Duyen HT, Ngoc TV, Ha do T, Hang VT, Kieu NT, Young PR, Farrar JJ, Simmons CP, Wolbers M, Wills BA. 2011. Kinetics of plasma viremia and soluble nonstructural protein 1 concentrations in dengue: differential effects according to serotype and immune status. J. Infect. Dis. 203:1292–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Libraty DH, Young PR, Pickering D, Endy TP, Kalayanarooj S, Green S, Vaughn DW, Nisalak A, Ennis FA, Rothman AL. 2002. High circulating levels of the dengue virus nonstructural protein NS1 early in dengue illness correlate with the development of dengue hemorrhagic fever. J. Infect. Dis. 186:1165–1168 [DOI] [PubMed] [Google Scholar]
- 29. Tricou V, Minh NN, Farrar J, Tran HT, Simmons CP. 2011. Kinetics of viremia and NS1 antigenemia are shaped by immune status and virus serotype in adults with dengue. PLoS Negl. Trop. Dis. 5:e1309. 10.1371/journal.pntd.0001309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vaughn DW, Green S, Kalayanarooj S, Innis BL, Nimmannitya S, Suntayakorn S, Endy TP, Raengsakulrach B, Rothman AL, Ennis FA, Nisalak A. 2000. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J. Infect. Dis. 181:2–9 [DOI] [PubMed] [Google Scholar]
- 31. Wang WK, Lee CN, Kao CL, Lin YL, King CC. 2000. Quantitative competitive reverse transcription-PCR for quantification of dengue virus RNA. J. Clin. Microbiol. 38:3306–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Burd EM. 2010. Validation of laboratory-developed molecular assays for infectious diseases. Clin. Microbiol. Rev. 23:550–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. WHO/CDC 2009. CDC protocol of realtime RTPCR for swine influenza A (H1N1). WHO Collaborating Centre for Influenza at CDC Atlanta, United States of America, Atlanta, GA [Google Scholar]
- 34. Rossini G, Carletti F, Bordi L, Cavrini F, Gaibani P, Landini MP, Pierro A, Capobianchi MR, Di Caro A, Sambri V. 2011. Phylogenetic analysis of West Nile virus isolates, Italy, 2008-2009. Emerg. Infect. Dis. 17:903–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gutierrez G, Standish K, Narvaez F, Perez MA, Saborio S, Elizondo D, Ortega O, Nuñez A, Kuan G, Balmaseda A, Harris E. 2011. Unusual dengue virus 3 epidemic in Nicaragua, 2009. PLoS Negl. Trop. Dis. 5:e1394. 10.1371/journal.pntd.0001394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kuan G, Gordon A, Avilés W, Ortega O, Hammond SN, Elizondo D, Nuñez A, Coloma J, Balmaseda A, Harris E. 2009. The Nicaraguan pediatric dengue cohort study: study design, methods, use of information technology, and extension to other infectious diseases. Am. J. Epidemiol. 170:120–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Narvaez F, Gutierrez G, Pérez MA, Elizondo D, Nuñez A, Balmaseda A, Harris E. 2011. Evaluation of the traditional and revised WHO classifications of Dengue disease severity. PLoS Negl. Trop. Dis. 5:e1397. 10.1371/journal.pntd.0001397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Raengsakulrach B, Nisalak A, Maneekarn N, Yenchitsomanus PT, Limsomwong C, Jairungsri A, Thirawuth V, Green S, Kalayanarooj S, Suntayakorn S, Sittisombut N, Malasit P, Vaughn D. 2002. Comparison of four reverse transcription-polymerase chain reaction procedures for the detection of dengue virus in clinical specimens. J. Virol. Methods 105:219–232 [DOI] [PubMed] [Google Scholar]
- 39. Yamada K, Takasaki T, Nawa M, Kurane I. 2002. Virus isolation as one of the diagnostic methods for dengue virus infection. J. Clin. Virol. 24:203–209 [DOI] [PubMed] [Google Scholar]
- 40. Dos Santos HW, Poloni TR, Souza KP, Muller VD, Tremeschin F, Nali LC, Fantinatti LR, Amarilla AA, Castro HL, Nunes MR, Casseb SM, Vasconcelos PF, Badra SJ, Figueiredo LT, Aquino VH. 2008. A simple one-step real-time RT-PCR for diagnosis of dengue virus infection. J. Med. Virol. 80:1426–1433 [DOI] [PubMed] [Google Scholar]
- 41. Chutinimitkul S, Payungporn S, Theamboonlers A, Poovorawan Y. 2005. Dengue typing assay based on real-time PCR using SYBR Green I. J. Virol. Methods 129:8–15 [DOI] [PubMed] [Google Scholar]
- 42. Gurukumar KR, Priyadarshini D, Patil JA, Bhagat A, Singh A, Shah PS, Cecilia D. 2009. Development of real time PCR for detection and quantitation of dengue viruses. Virol. J. 6:10. 10.1186/1743-422X-6-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hertz JT, Munishi OM, Ooi EE, Howe S, Lim WY, Chow A, Morrissey AB, Bartlett JA, Onyango JJ, Maro VP, Kinabo GD, Saganda W, Gubler DJ, Crump JA. 2012. Chikungunya and dengue fever among hospitalized febrile patients in northern Tanzania. Am. J. Trop. Med. Hyg. 86:171–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Laue T, Emmerich P, Schmitz H. 1999. Detection of dengue virus RNA in patients after primary or secondary dengue infection by using the TaqMan automated amplification system. J. Clin. Microbiol. 37:2543–2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shu PY, Chang SF, Kuo YC, Yueh YY, Chien LJ, Sue CL, Lin TH, Huang JH. 2003. Development of group- and serotype-specific one-step SYBR green I-based real-time reverse transcription-PCR assay for dengue virus. J. Clin. Microbiol. 41:2408–2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Balmaseda A, Standish K, Mercado JC, Matute JC, Tellez Y, Saborio S, Hammond SN, Nuñez A, Avilés W, Henn MR, Holmes EC, Gordon A, Coloma J, Kuan G, Harris E. 2010. Trends in patterns of dengue transmission over 4 years in a pediatric cohort study in Nicaragua. J. Infect. Dis. 201:5–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Endy TP, Anderson KB, Nisalak A, Yoon IK, Green S, Rothman AL, Thomas SJ, Jarman RG, Libraty DH, Gibbons RV. 2011. Determinants of inapparent and symptomatic dengue infection in a prospective study of primary school children in Kamphaeng Phet, Thailand. PLoS Negl. Trop. Dis. 5:e975. 10.1371/journal.pntd.0000975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Endy TP, Chunsuttiwat S, Nisalak A, Libraty DH, Green S, Rothman AL, Vaughn DW, Ennis FA. 2002. Epidemiology of inapparent and symptomatic acute dengue virus infection: a prospective study of primary school children in Kamphaeng Phet, Thailand. Am. J. Epidemiol. 156:40–51 [DOI] [PubMed] [Google Scholar]
- 49. Standish K, Kuan G, Avilés W, Balmaseda A, Harris E. 2010. High dengue case capture rate in four years of a cohort study in Nicaragua compared to national surveillance data. PLoS Negl. Trop. Dis. 4:e633. 10.1371/journal.pntd.0000633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kutyavin IV, Afonina IA, Mills A, Gorn VV, Lukhtanov EA, Belousov ES, Singer MJ, Walburger DK, Lokhov SG, Gall AA, Dempcy R, Reed MW, Meyer RB, Hedgpeth J. 2000. 3′-minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res. 28:655–661 [DOI] [PMC free article] [PubMed] [Google Scholar]