Abstract
Rapid antibiotic susceptibility testing is in high demand in health care fields as antimicrobial-resistant bacterial strains emerge and spread. Here, we describe an optical screening system (oCelloScope) which, based on time-lapse imaging of 96 bacteria-antibiotic combinations at a time, introduces real-time detection of bacterial growth and antimicrobial susceptibility with imaging material to support the automatically generated graphs. Automated antibiotic susceptibility tests of a monoculture showed statistically significant antibiotic effects within 6 min and within 30 min in complex samples from pigs suffering from catheter-associated urinary tract infections. The oCelloScope system provides a fast high-throughput screening method for detecting bacterial susceptibility that might entail an earlier diagnosis and introduction of appropriate targeted therapy and thus combat the threat from multidrug-resistant pathogenic bacteria. The oCelloScope system can be employed for a broad range of applications within bacteriology and might present new vistas as a point-of-care instrument in clinical and veterinary settings.
INTRODUCTION
The emergence and spread of antimicrobial-resistant bacteria and increasingly rapid transmission across international borders are serious global health threats (1, 2). These developments are associated with the extensive and increasing use of antimicrobial agents, and because of this combined with the paucity of development of new antimicrobial agents, the treatment options for resistant pathogens have become drastically limited (3–6). Infections with resistant pathogens are associated with higher mortality and morbidity rates and higher health care costs, and early targeted antibiotic treatment is an important prognostic factor, especially for seriously ill patients (7, 8). There is evidence that the avoidance of inappropriate broad-spectrum antimicrobial therapy can prevent antimicrobial resistance (9). There is also a demand for rapid high-throughput methods that can determine antibiotic susceptibility and enable targeted therapy as early in treatment as possible. Conventional culture-based methods, such as the disc diffusion test, the Etest (bioMérieux), and broth or agar dilution, used to determine the MICs of antibiotics, are time-consuming endpoint methods (10). Commercial instruments such as the Phoenix 100 (BD Biosciences) and the Vitek 2 (bioMérieux) allow automation and reduce hands-on and incubation times (11). Both instruments operate with colorimetric or fluorimetric indicators for bacterial identification and estimation of growth rates (12). The efficacy of the two systems in rapid and accurate bacterial identification and antibiotic susceptibility tests (ASTs) has been demonstrated (11–14). The average times for identification are 4.3 h for the Phoenix and 3 to 5.7 h for the Vitek 2, depending on the bacterial type, while the mean times for ASTs are 12.1 h and 9.8 h, respectively (11, 15, 16). Translating these times to results into clinical practice implies that a switch from broad-spectrum antibiotic therapy to narrow-spectrum targeted therapy will only be accomplished the following working day.
In order to improve sensitivity and speed, several biosensors based on either chip calorimetry (17), electrical conductivity (18, 19), millifluidic droplet analysis (20), or surface plasmon resonance (21) have been developed. Presently, these techniques are limited by single-sample analysis and the requirement for specialized technical personnel. Molecular methods, such as real-time PCR (22), mass spectrometry (23), microarrays (24), and flow cytometry (25), have been developed for rapid bacterial identification and ASTs because of their high sensitivity and promptness. A wide range of bacteria can be identified within minutes using mass spectrometry (26), and flow cytometry predicts antimicrobial susceptibility within 90 to 120 min (25). These techniques require expensive equipment, special probes, and/or skilled personnel.
The oCelloScope is a small portable platform developed to provide (i) fast and reliable susceptibility testing, (ii) high sensitivity to allow the use of low bacterial concentrations, and (iii) internal verification of results. This system allows real-time analysis of bacterial processes, including antimicrobial efficacy screening using an automated optical detection system. Here, we demonstrate proof of concept and show that results for automated ASTs can be obtained within 6 min in monoculture experiments and within 30 min in complex samples such as urine specimens from catheter-associated urinary tract infections (CAUTIs). The oCelloScope might present new vistas as a point-of-care instrument in clinical and veterinary settings.
MATERIALS AND METHODS
Bacterial strains were selected to represent families which differ in morphology and metabolism and to represent the most frequently isolated bacteria in clinical and veterinary infections. The Gram-negative facultative aerobe Escherichia coli 9910045-1:O149 was isolated from the intestinal contents of a pig with weaning diarrhea (27). The human facultative aerobe Staphylococcus aureus CCUG 4151 (Culture Collection, University of Gothenburg, Sweden) was included as a Gram-positive representative. The Gram-negative facultative aerobe Salmonella enterica serovar Typhimurium 3389-1 DT12 was isolated from a clinical case of salmonellosis in pigs and was kindly donated by D. L. Baggesen at the Technical University of Denmark, National Veterinary Institute (28). The common Gram-positive pig gut bacterium Streptococcus alactolyticus (type strain DSM 20728; DSMZ, Braunschweig, Germany) was included as an anaerobe representative. The E. coli and S. Typhimurium strains were grown aerobically at 37°C in Luria-Bertani (LB) medium (Merck, Darmstadt, Germany). S. aureus was grown aerobically at 37°C in brain heart infusion (BHI) broth (Merck), whereas S. alactolyticus was grown anaerobically at 37°C in broth prepared as described by Holdeman et al. (29), except that rumen extract was replaced by porcine colon extract (CGCM broth).
In all cases, 0.1 ml of an overnight culture was inoculated in 8 ml of medium for 2 h (37°C) to reach the exponential phase. These cell suspensions were standardized by adjusting the concentration of inoculates to approximately 2.3 × 107 organisms/ml, determined by measurements of the optical density at 600 nm (OD600) (performed on a UV-3100 PC spectrophotometer; VWR, Herlev, Denmark), and subsequently diluting to a final bacterial cell suspension of 7.8 × 105 bacteria ml−1. Beads were added to bacterial cell suspensions (2 × 104 6-μm beads/ml, microsphere standard, B-7277; Invitrogen, Naerum, Denmark) and loaded onto an F-base microtiter plate (50 μl/well) (TPP; Sigma-Aldrich, Brondby, Denmark) either untreated (control samples) or treated with antibiotics (E. coli, 2 μg/ml polymyxin B, P1004; S. aureus, 50 μg/ml penicillin G50, P3032; S. Typhimurium, 250 μg/ml spectinomycin, S2647; and S. alactolyticus, 0.5 μg/ml rifampin, R3501; Sigma-Aldrich, Brondby, Denmark). All experiments were done in triplicates consisting of eight iterations.
To validate the oCelloScope instrument, identical experimental setups measuring bacterial growth were analyzed (i) manually using a UV-3100 PC spectrophotometer (VWR, Herlev, Denmark) at OD600 with 1.5-ml disposable cuvettes, (ii) automatically using a PowerWaveX microplate reader (BioTek Instruments, Holm & Halby, Brondby, Denmark) at OD655, and (iii) automatically by the oCelloScope instrument. F-base microtiter plates (50 μl/well) (TPP; Sigma-Aldrich, Brondby, Denmark) were used for the spectrophotometer and the oCelloScope.
For antibiotic susceptibility tests (ASTs) of catheter-associated urinary tract infections (CAUTIs), three pigs (4 months old, crossbred [(Danish Landrace × Yorkshire) × Duroc], 40 to 45 kg) were catheterized. The animals did not show any clinical signs of infections or diarrhea. All experiments followed ethical guidelines approved by the Danish National Authority (2008/561-1493). On day 7, urine samples were centrifuged at 2,400 × g for 5 min at room temperature to discard any porcine leukocytes or epithelial cells. The supernatant was mixed 50% (vol/vol) with Müller-Hinton broth with N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES) (MH broth) (T3462, Sensititre; Thermo Scientific, Slangerup, Denmark) and incubated for 2 h at 37°C. According to the manufacturer's instructions, the cell concentration was adjusted to a 0.5 McFarland standard; 0.1-ml cell suspensions were transferred into 11 ml MH broth and mixed before 50 μl/well was loaded onto an NF Trek plate (Sensititre) and subsequently transferred onto an F-base microtiter plate (50 μl/well) (TPP; Sigma-Aldrich, Brondby, Denmark).
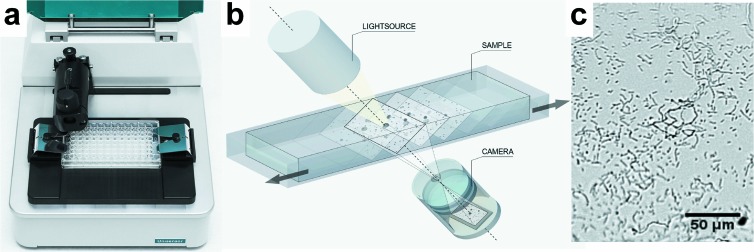
The basic principle behind the oCelloScope detection system (Unisensor Ltd.) is digital time-lapse microscopy scanning through a fluid sample, generating series of images. The instrument is shown in Fig. 1a together with a conceptual drawing showing the principles of the method (Fig. 1b) and a picture from a sample containing S. alactolyticus (Fig. 1c). The imaging system consists of an illumination unit, a lens, and a digital camera unit (shown in Fig. 1b). The optical axis of the imaging system is tilted 6.25° relative to the horizontal plane of the stage to enable scanning of volumes and extraction of phase information. The lens and digital camera unit contain a proprietary lens system and a 5-megapixel complementary metal oxide semiconductor (CMOS) camera chip (with length-by-height dimensions of 5.6 mm by 4 mm) designed to have a focus depth of approximately 10 μm, an absolute magnification factor of ×4, and an optical resolution of 1.3 μm, which is comparable to ×200 magnification in a standard light microscope. The images recorded by the imaging system form an image stack, which, due to the tilted imaging plane, constitutes a parallelepiped that covers a detection volume in the sample of Vsample = Lsample × Hsample × Wsample × sin(6.25). Lsample and Hsample are the camera chip dimensions downscaled with the magnification factor (i.e., 5.6 mm/4 and 4 mm/4), and Wsample is the scan length, n × step_length, where n is the total number of longitudinal steps and step_length is the minimum incremental step length, which is 7.5 μm for the instrument used. In the present study, an image stack consisting of 2 images separated by 6 steps (n = 6) was used. The perpendicular displacement between the object planes of the images thereby becomes zplane = sin(6.25) × 6 × 7.5 μm = 4.9 μm, and the covered volume is Vsample = 1.4 mm × 1 mm × 6 × 7.5 μm × sin(6.25) = 6.9 nl. Due to the higher refractive index of the solution, the object plane is displaced deeper into the medium than in an air sample. This has a direct impact on the scanning depth of the sample viewed at an oblique angle (6.25°) and effectively results in a total viewing depth that equals the free space viewing depth multiplied by the refractive index ratio between water and air. The true observed volume, therefore, is Vtrue sample × nwater = 9.1 nl, where nwater is the refractive index of water (which is nwater = 1.33 at 37°C for light at a wavelength of 505 nm). The higher refractive index of the medium also affects the perpendicular distance between the object planes, which then becomes ztrue plane = sin(6.25) × 6 × 7.5 μm × 1.33 = 6.5 μm. Each well was scanned repeatedly every 15 min, if not stated otherwise.
Fig 1.
The oCelloScope detection system. (a) Picture of the detection system with a standard 96-well plate inserted. (b) Simplified engineering drawing of the detection principle. A volume of 50 μl of a growing bacterial culture is scanned, resulting in a stack of images. Each image can be transformed to a two-dimensional (2D) picture. (c) 2D picture of S. alactolyticus from the scanning process. The magnitude is comparable to an ×200 magnification in a standard light microscope.
Time-lapse experiments, digital analysis, and image processing were conducted by a custom automation script in MATLAB version 8.0.0.783 (R2012b; The MathWorks, Inc., Natick, MA). Two algorithms, based on either pixel histogram summation (PHS) or contrast segmentation and extraction of surface area (SESA), were developed to determine bacterial growth kinetics as a result of image stack processing. The oCelloScope was placed within an Innova 44 incubator (New Brunswick Scientific), which allowed precise regulation of the temperature.
All data are expressed as mean values ± standard deviations (SDs). Statistical differences were analyzed using multiple t tests with the false-discovery rate set to 1%. GraphPad Prism version 6.00 for Windows (GraphPad Software, San Diego, CA) was used for statistical analysis.
RESULTS
Method evaluation.
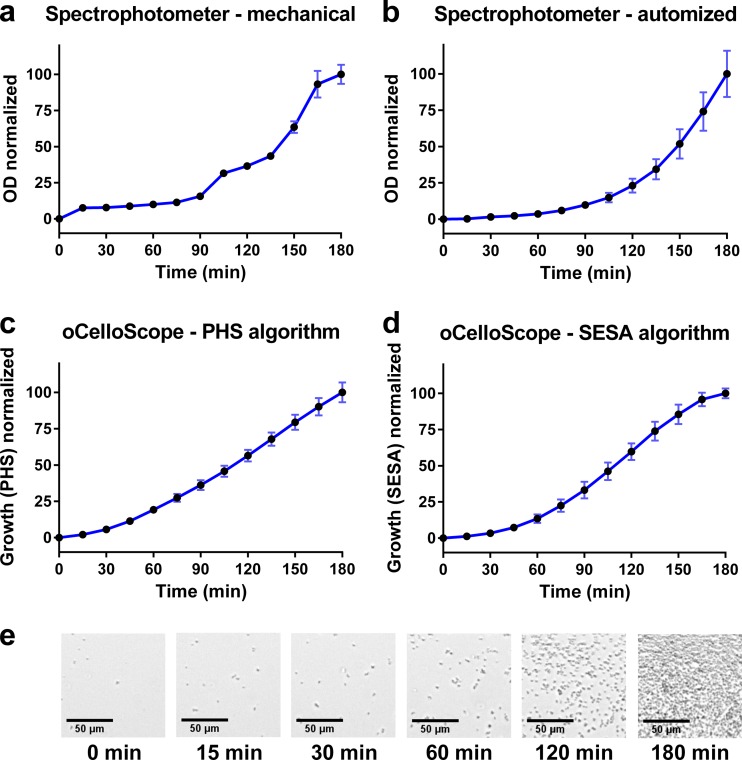
The oCelloScope detection system was used to detect bacterial growth and perform ASTs. A comparison of the oCelloScope to standard optical density methods was carried out (Fig. 2). These data demonstrated that the oCelloScope was able to record real-time bacterial growth of S. aureus in a 3-h experiment with the same accuracy as that for conventional optical density (OD) measurements but showed improved performance in the lower range of bacterial numbers. OD measurements by a standard spectrophotometer showed irregular results in the lower scale of bacterial concentrations (approximately <2 × 107 bacteria ml−1) (Fig. 2a), whereas more reliable results were obtained by an automated plate reader system (Fig. 2b). It should be noted that the initial bacterial concentrations were at the detection limits for these techniques. For the measurement of bacterial growth by the oCelloScope, two algorithms were designed based on pixel histogram summation (PHS) (Fig. 2c) and segmentation and extraction of surface area (SESA) (Fig. 2d). The PHS algorithm was the most efficient in detecting exponential growth (0 to 60 min) but displayed a more linear detection after 60 min, which was not in agreement with the image material. The overall detection level was enhanced by the SESA algorithm compared to the image material (Fig. 2d). This algorithm uses segmentation to summarize the number of bacteria, resulting in a refined detection limit in the lower bacterial concentrations compared to those for traditional OD measurements and a continued exponential growing pattern compared to that for PHS measurements performed by the oCelloScope.
Fig 2.
Bacterial growth of S. aureus assessed by the oCelloScope and traditional OD measurements. (a) Growth was measured by optical density in a standard laboratory spectrophotometer with one cuvette. The absorbance was measured at 600 nm. (b) Growth was measured by optical density (absorbance, 655 nm) using a standard laboratory plate reader with a 96-well plate. (c) Growth was measured by optical density using the oCelloScope pixel histogram summation (PHS) algorithm. (d) Growth was measured by the oCelloScope segmentation and extraction of surface area (SESA) algorithm. (e) Pictures taken by the oCelloScope showing bacterial growth to different time points. All experiments were done as eight replicates, and standard derivations are shown as error bars on the curves. Scale bar, 50 μm.
Determination of bacterial growth.
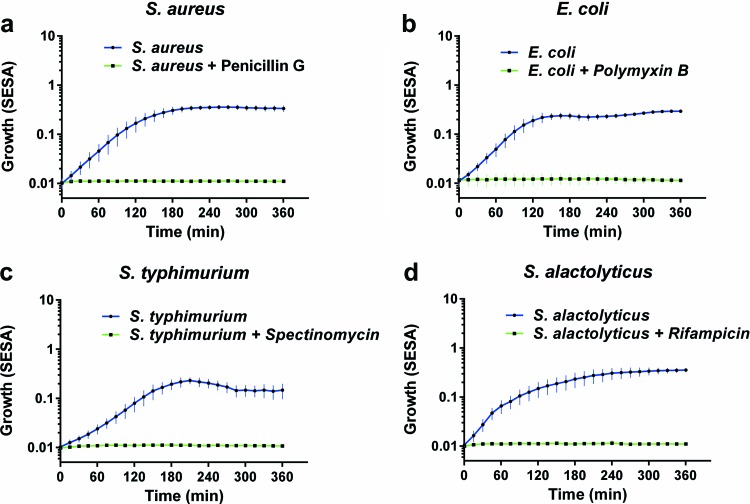
The SESA algorithm was based on the previous finding used to detect real-time bacterial growth of two Gram-positive and two Gram-negative bacteria (Fig. 3). All four bacteria, the facultative aerobe S. aureus, E. coli, S. Typhimurium, and the anaerobe S. alactolyticus, showed exponential growth patterns from the start of the experiment. Each of the four bacteria was incubated with an antibiotic with known inhibitory effects. A movie of the real-time detection is presented in Video S1 in the supplemental material.
Fig 3.
Bacterial growth of aerobic and anaerobic bacteria with (green line) or without (blue line) an antibiotic measured by the oCelloScope using the SESA algorithm. (a) S. aureus grown for 6 h with or without penicillin G; (b) E. coli grown for 6 h with or without polymyxin B; (c) S. Typhimurium grown for 6 h with or without spectinomycin; (d) S. alactolyticus grown for 6 h with or without rifampin. All experiments were done in triplicate, each consisting of eight iterations. Standard derivations are shown as error bars on the curves.
Rapid determination of antibiotic susceptibility in bacterial monocultures.
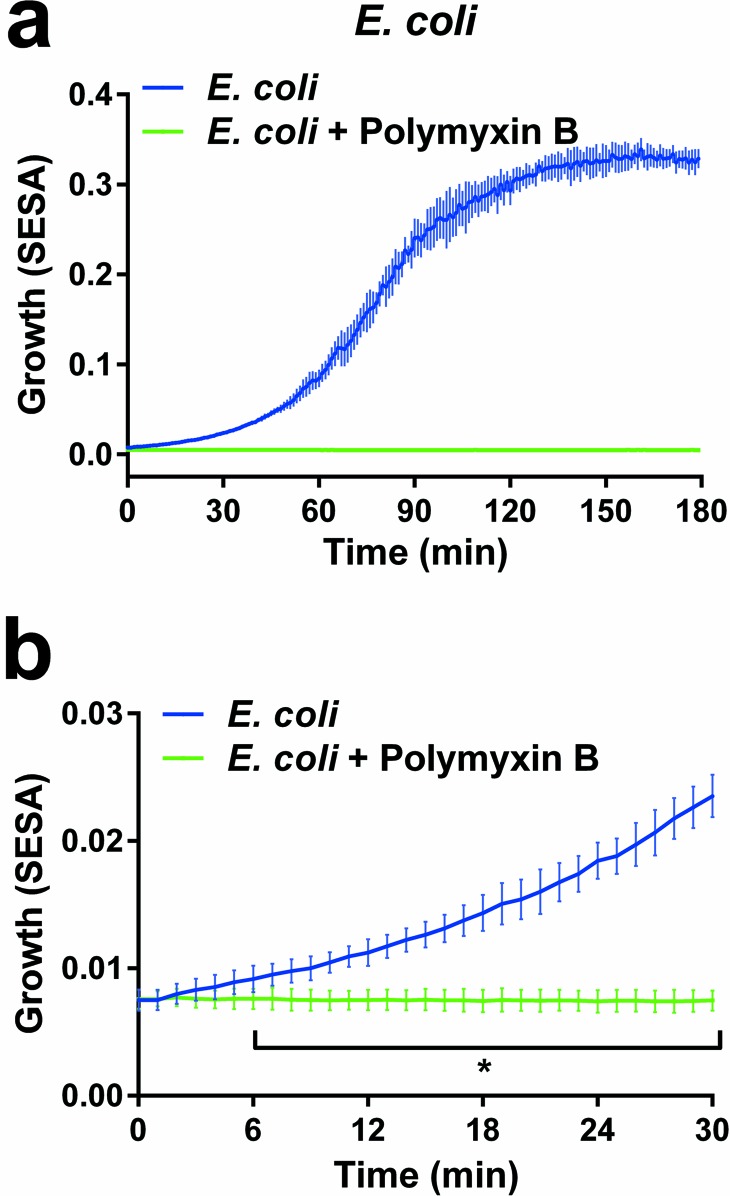
To test how fast samples incubated with or without antibiotics can be detected, the repetition time was changed to scan every minute in an experiment where E. coli was treated with or without polymyxin B (Fig. 4). The results showed that a statistically significant difference can be measured after 6 min. Real-time detection where the antibiotic is added following 65 min of growth is presented in Video S2 in the supplemental material. The technique was further evaluated by the application to veterinary samples.
Fig 4.
Bacterial growth measured by the oCelloScope in short time intervals. (a) Growth of E. coli with (green line) or without (blue line) polymyxin B. Bacteria were grown for 3 h and scanned every minute. (b) The first 30 min from panel a are shown close up. Results are expressed as mean values (±SD) from triplicate experiments, each representing 8 iterations. Statistical differences were analyzed using multiple t tests with the false-discovery rate set to 1%. The asterisk indicates that values are significantly (P < 0.01) different from those for bacteria incubated with polymyxin B.
Determination of antibiotic susceptibility in urine samples.
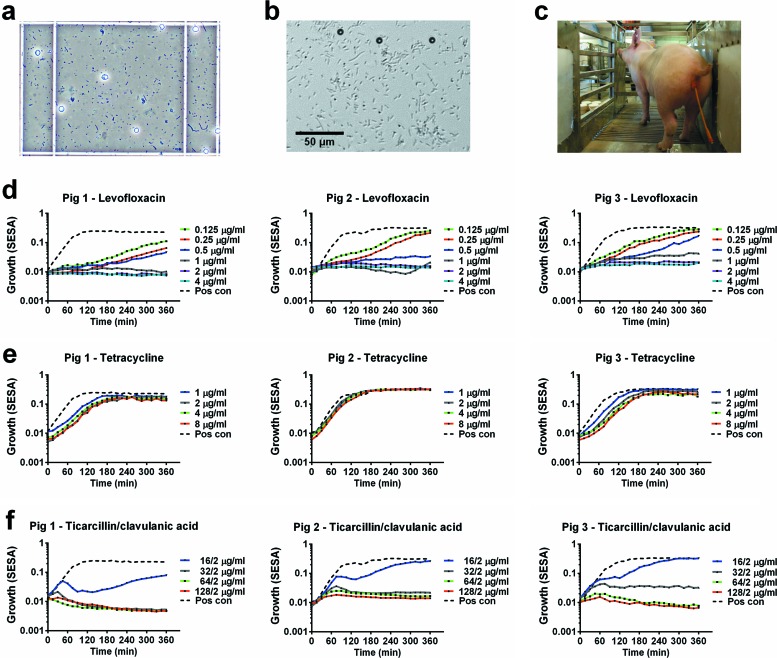
Urine samples from catheterized pigs suffering from CAUTIs (Fig. 5c) with bacterial infections as detected in a light microscope (Fig. 5a) were used with standard Trek Sensititre plates to evaluate the technology. Urine samples were preincubated for 2 h in MH broth in order to obtain exponential growth before measurement on the oCelloScope. Trek Sensititre NF plates with predosed and dried antibiotic panels were used to obtain antibiotic susceptibilities for 23 different antibiotics. The growth was followed for 6 h and reached the stationary phase after approximately 2 h. Figure 5d and e show the growth curves obtained for three different antibiotics, levofloxacin (0.125 to 4 μg/ml), tetracycline (1 to 8 μg/ml), and a combination of ticarcillin and clavulanic acid (16/2 to 128/2 μg/ml). Bacterial growth was inhibited by levofloxacin in all tested concentrations compared with controls, and the susceptibility values were estimated to be 1 μg/ml for pig 1 and 2 μg/ml for pigs 2 and 3. The bacteria from all three pigs were almost unaffected by tetracycline in the tested concentrations. The susceptibility values for the combination of ticarcillin and clavulanic acid were 32/2 μg/ml. The result was less clear when 16/2 μg/ml ticarcillin-clavulanic acid was used. As shown in Video S3 in the supplemental material, the image material reveals initial growth of bacteria with long rod-shaped morphology followed by the growth of bacteria with shortened rod-shaped morphology. Tentative identification of the microbial community of the initial urine sample using terminal restriction fragment length polymorphism analysis showed that the majority of the bacteria were Escherichia spp. with a minor representation of Enterococcus spp. (data not shown). For further identification of the specific bacteria grown under the influence of ticarcillin and clavulanic acid, additional experiments are necessary.
Fig 5.
Bacterial growth in urine samples from pigs suffering from catheter-associated urinary tract infections (CAUTIs) measured by the oCelloScope and analyzed by the SESA algorithm. Urine samples were collected from three pigs with CAUTIs, and bacterial growth was analyzed in samples incubated with or without antibiotics. (a) Picture of infected urine samples taken by a light microscope. The total magnification is ×100. Erythrocytes and bacteria are visible in the counting chamber. (b) Picture of a urine sample from a pig taken by the oCelloScope. (c) Picture of a pig with a catheter for urine collection. (d) Analysis of urine samples, from three different pigs, diluted with Müller-Hinton (MH) broth and incubated with levofloxacin. (e) Analysis of urine samples, from three different pigs, diluted with MH broth and incubated with tetracycline. (f) Analysis of urine samples, from three different pigs, diluted with MH broth and incubated with a combination of ticarcillin and clavulanic acid. Pos con, positive control. (A movie of the bacteria from pig 1 is available as Video S3 in the supplemental material.)
DISCUSSION
Here, we demonstrate how a small portable instrument using optical detection can revolutionize the speed and efficiency of ASTs of bacterial cultures. It records and determines bacterial growth and antibiotic susceptibilities with supporting imaging material. The tested bacterial strains represent three bacterial families frequently isolated in clinical or veterinary infections. Four different media were used to test the ability of the technology to process differences in background color.
The performance of the oCelloScope was compared to that of standard optical density methods. Turbidity measurements are commonly used to determine bacterial concentrations. Spectrophotometric determination has a lower limit of 0.01 OD600, which for E. coli corresponds to approximately 8.0 × 106 bacterial cells ml−1. To allow determinations at concentrations lower than 8.0 × 106 bacterial cells ml−1, a new algorithm was developed. The combination of the detection capability of the oCelloScope and the SESA algorithm greatly improved low-range bacterial measurements compared to those of standard spectrometric methods. The SESA algorithm also allowed us to detect a statistically significant antibiotic effect on E. coli within 6 min. The concentration-time relationship shown in Fig. S1 in the supplemental material indicates the lower limits of the instrument with regard to bacterial concentration. Preparation of the inoculum is critical for obtaining reliable results for all automated susceptibility methods, as they are sensitive to the number and the purity of the starting bacterial population. The oCelloScope generates pictures rapidly, and it is possible to visualize the starting material. Dormant bacterial cells, host cells, complex cultures, and contaminants are difficult to control in all automated susceptibility systems, and they affect the results. The oCelloScope detection system can exclude contaminants and host cells by optimized algorithms. Further developments will focus on parameters such as, e.g., bacterial cell swelling, changes in bacterial membrane morphology, and the refraction index to detect bacterial transformations in dormant bacteria. The detection of low bacterial cell concentrations can be further improved by increasing the number of scans, thereby increasing the analyzed sample volume.
Using the SESA algorithm, we detected a statistically significant antibiotic effect on E. coli within 6 min using a starting inoculum of 7.8 × 105 bacteria ml−1 and a two-image scan. The starting inoculum is slightly higher than the recommended starting inoculum for broth dilution of 5 × 105 CFU ml−1 (30). The concentration-time relationship displayed in Fig. S1 in the supplemental material indicates that a reduction in the concentration of the starting inoculum will entail an increase in the time to results. However, a simultaneous increase in the number of images per scan leads to a decrease in the time to results; thus, a reduction in the starting inoculum concentration can be improved further. Flow cytometry for bacterial cell count monitoring was recently shown to determine antimicrobial susceptibility in 90 to 120 min, depending on the bacterium (25). The advantage of this method is the possibility of measuring bacteria in the range of 1 to 6 × 104 cell counts; however, compared to the oCelloScope system, an AST can be performed for only one antibiotic at a time, and no image material that can reveal any erroneous results is available. The principles of several other methods of performing rapid ASTs have been presented (17–22, 31); a common limitation of these techniques is their inability to perform multiple-sample analysis.
A final proof of the efficiency of the technology was demonstrated by analyses of veterinary catheter urine samples. Porcine CAUTI samples with complex bacterial compositions were tested with standard NF Trek Sensititre plates preincubated with a panel of 23 antibiotics for Gram-negative bacteria. This technology can show if the antibiotics have elicited an effect within 30 min and can determine the inhibitory concentrations within 3 h. The nature of the CAUTI samples does not allow sample replicates, but the homogeneity of the results derived from the three pigs shows promise for the use of this model. Interpretation of the susceptibility data is complicated by the sample complexity. Consequently, the SESA algorithm will be further developed to determine the morphology of a pathogen. The imaging material substantiates the results by providing visual evidence for graphical findings (see Video S3 in the supplemental material). The short analysis time compared with the 18 h needed for the broth susceptibility test illustrates the potential of the instrument with regard to ASTs (10). A biosensor platform for rapid ASTs can provide susceptibility information from a clinical urine sample within 3.5 h using a similar experimental setup; however, this method is more laborious, as samples must be taken regularly at 15-min intervals for biosensor analysis (32).
The primary advantages of the oCelloScope over existing methods are that it is significantly faster, is portable, and requires low sample volumes. Another advantage is the ability to analyze up to 96 combinations of samples and antibiotics at once. The compact size of the instrument and the Windows-based software facilitate incorporation into existing fully automatic high-throughput systems. Based on its usability and size and the possibility of further development, the system has the potential for tailoring antibiotic treatment to individual patients in a point-of-care system, thereby reducing the inappropriate use of broad-spectrum antibiotics and improving survival rates for patients suffering from postoperative infections.
In conclusion, the oCelloScope system is suited for the imaging of fluid samples, such as bacterial cultures and urine samples. It is user-friendly and can accommodate a broad range of applications within bacteriology. Real-time detection of bacterial growth and antimicrobial susceptibility is linked to supporting imaging material. The technology can improve ASTs by reducing hands-on and incubation times and give rapid results that can lead to early targeted antibiotic therapy. A targeted narrow-spectrum antibiotic with adequate coverage will reduce bacterial antimicrobial resistance and reduce costs. The oCelloScope provides a way to carry out high-throughput bacterial susceptibility testing, which is essential for counteracting the rapid increase in antimicrobial-resistant bacteria.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Danish National Advanced Technology Foundation (grant 137–2012) and the Danish Agency for Science (grant 10-100105).
We thank Trine Poulsen for excellent technical assistance, Leif Tind for porcine urine samples, and Henriette Giese for valuable comments on the manuscript.
M.F. is employed in a joint research project between Aarhus University, Aalborg University, the Danish National Advanced Technology Foundation, and Unisensor Ltd. K.R.A. is employed at and owns warrants in Unisensor Ltd. E.J. is employed at Unisensor Ltd. T.O. is director of and owns stocks in Unisensor Ltd.
M.F. planned and carried out the experiments, analyzed data, and wrote the manuscript. K.R.A. and T.O. developed the oCelloScope. E.J. developed and implemented the algorithms. A.D. contributed to initial experimental work. B.B.J. contributed to the experimental design. F.S.R. contributed to the experimental design. T.E.S. was the project leader, analyzed data, and wrote the manuscript.
Footnotes
Published ahead of print 17 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00440-13.
REFERENCES
- 1. Bush K, Courvalin P, Dantas G, Davies J, Eisenstein B, Huovinen P, Jacoby GA, Kishony R, Kreiswirth BN, Kutter E, Lerner SA, Levy S, Lewis K, Lomovskaya O, Miller JH, Mobashery S, Piddock LJV, Projan S, Thomas CM, Tomasz A, Tulkens PM, Walsh TR, Watson JD, Witkowski J, Witte W, Wright G, Yeh P, Zgurskaya HI. 2011. Tackling antibiotic resistance. Nat. Rev. Microbiol. 9:894–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Okeke IN, Edelman R. 2001. Dissemination of antibiotic-resistant bacteria across geographic borders. Clin. Infect. Dis. 33:364–369 [DOI] [PubMed] [Google Scholar]
- 3. Butler MS, Cooper MA. 2011. Antibiotics in the clinical pipeline in 2011. J. Antibiot. 64:413–425 [DOI] [PubMed] [Google Scholar]
- 4. Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48:1–12 [DOI] [PubMed] [Google Scholar]
- 5. Wernli D, Haustein T, Conly J, Carmeli Y, Kickbusch I, Harbarth S. 2011. A call for action: the application of the international health regulations to the global threat of antimicrobial resistance. PLoS Med. 8:e1001022. 10.1371/journal.pmed.1001022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dellit TH, Owens RC, McGowan JE, Gerding DN, Weinstein RA, Burke JP, Huskins WC, Paterson DL, Fishman NO, Carpenter CF. 2007. Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America guidelines for developing an institutional program to enhance antimicrobial stewardship. Clin. Infect. Dis. 44:159–177 [DOI] [PubMed] [Google Scholar]
- 7. Cosgrove SE. 2006. The relationship between antimicrobial resistance and patient outcomes: mortality, length of hospital stay, and health care costs. Clin. Infect. Dis. 42:S82–S89 [DOI] [PubMed] [Google Scholar]
- 8. Kumar A, Roberts D, Wood KE, Light B, Parrillo JE, Sharma S, Suppes R, Feinstein D, Zanotti S, Taiberg L, Gurka D, Kumar A, Cheang M. 2006. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit. Care Med. 34:1589–1596 [DOI] [PubMed] [Google Scholar]
- 9. Zillich AJ, Sutherland JM, Wilson SJ, Diekema DJ, Ernst EJ, Vaugh TE, Doebeling BN. 2006. Antimicrobial use control measures to prevent and control antimicrobial resistance in US hospitals. Infect. Control Hosp. Epidemiol. 27:1088–1095 [DOI] [PubMed] [Google Scholar]
- 10. Jorgensen JH, Ferraro MJ. 2009. Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin. Infect. Dis. 49:1749–1755 [DOI] [PubMed] [Google Scholar]
- 11. Mittman SA, Huard RC, Della-Latta P, Whittier S. 2009. Comparison of BD Phoenix to Vitek 2, MicroScan MICroSTREP, and Etest for antimicrobial susceptibility testing of Streptococcus pneumoniae. J. Clin. Microbiol. 47:3557–3561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chatzigeorgiou KS, Sergentanis TN, Tsiodras S, Hamodrakas SJ, Bagos PG. 2011. Phoenix 100 versus Vitek 2 in the identification of Gram-positive and Gram-negative bacteria: a comprehensive meta-analysis. J. Clin. Microbiol. 49:3284–3291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gherardi G, Angeletti S, Panitti M, Pompilio A, Di Bonaventura G, Crea F, Avola A, Fico L, Palazzo C, Sapia GF. 2012. Comparative evaluation of the Vitek-2 compact and Phoenix systems for rapid identification and antibiotic susceptibility testing directly from blood cultures of Gram-negative and Gram-positive isolates. Diagn. Microbiol. Infect. Dis. 72:20–31 [DOI] [PubMed] [Google Scholar]
- 14. Duggal S, Gaind R, Tandon N, Deb M, Chugh TD. 2012. Comparison of an automated system with conventional identification and antimicrobial susceptibility testing. ISRN Microbiol. 2012:107203. 10.5402/2012/107203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kuper KM, Boles DM, Mohr JF, Wanger A. 2009. Antimicrobial susceptibility testing: a primer for clinicians. Pharmacotherapy 29:1326–1343 [DOI] [PubMed] [Google Scholar]
- 16. Sellenriek P, Holmes J, Ferrett R, Drury R, Storch GA. 2005. Comparison of MicroScan Walk-Away, Phoenix and Vitek-Two microbiology systems used in the identification and susceptibility testing of bacteria, abstr LR900. Abstr. 105th Gen. Meet. Am. Soc. Microbiol American Society for Microbiology, Washington, DC [Google Scholar]
- 17. Lerchner J, Mueller-Hagen D, Roehr H, Wolf A, Mertens F, Mueller R, Witte W, Klare I. 2011. Chip-calorimetric evaluation of the efficacy of antibiotics and bacteriophages against bacteria on a minute-timescale. J. Therm. Anal. Calorim. 104:31–36 [Google Scholar]
- 18. Huang Y, Sudibya HG, Chen P. 2011. Detecting metabolic activities of bacteria using a simple carbon nanotube device for high-throughput screening of anti-bacterial drugs. Biosens. Bioelectron. 26:4257–4261 [DOI] [PubMed] [Google Scholar]
- 19. Spiller E, Schöll A, Alexy R, Kümmerer K, Urban GA. 2006. A sensitive microsystem as biosensor for cell growth monitoring and antibiotic testing. Sensors Actuators A Phys. 130–131:312–321 [Google Scholar]
- 20. Baraban L, Bertholle F, Salverda MLM, Bremond N, Panizza P, Baudry J, de Visser JA, Bibette J. 2011. Millifluidic droplet analyser for microbiology. Lab Chip 11:4057–4062 [DOI] [PubMed] [Google Scholar]
- 21. Chiang YL, Lin CH, Yen MY, Su YD, Chen SJ, Chen H. 2009. Innovative antimicrobial susceptibility testing method using surface plasmon resonance. Biosens. Bioelectron. 24:1905–1910 [DOI] [PubMed] [Google Scholar]
- 22. Beuving J, Verbon A, Gronthoud FA, Stobberingh EE, Wolffs PFG. 2011. Antibiotic susceptibility testing of grown blood cultures by combining culture and real-time polymerase chain reaction is rapid and effective. PLoS One 6:e27689. 10.1371/journal.pone.0027689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sauer S, Kliem M. 2010. Mass spectrometry tools for the classification and identification of bacteria. Nat. Rev. Microbiol. 8:74–82 [DOI] [PubMed] [Google Scholar]
- 24. Cleven BEE, Palka-Santini M, Gielen J, Meembor S, Krönke M, Krut O. 2006. Identification and characterization of bacterial pathogens causing bloodstream infections by DNA microarray. J. Clin. Microbiol. 44:2389–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Broeren MAC, Maas Y, Retera E, Arents NLA. 2013. Antimicrobial susceptibility testing in 90 min by bacterial cell count monitoring. Clin. Microbiol. Infect. 19:286–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mellmann A, Cloud J, Maier T, Keckevoet U, Ramminger I, Iwen P, Dunn J, Hall G, Wilson D, LaSala P, Kostrzewa M, Harmsen D. 2008. Evaluation of matrix-assisted laser desorption ionization-time-of-flight mass spectrometry in comparison to 16S rRNA gene sequencing for species identification of nonfermenting bacteria. J. Clin. Microbiol. 46:1946–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Frydendahl K, Imberechts H, Lehmann S. 2001. Automated 5′ nuclease assay for detection of virulence factors in porcine Escherichia coli. Mol. Cell. Probes 15:151–160 [DOI] [PubMed] [Google Scholar]
- 28. Grøndahl ML, Jensen GM, Nielsen CG, Skadhauge E, Olsen JE, Hansen MB. 1998. Secretory pathways in Salmonella Typhimurium-induced fluid accumulation in the porcine small intestine. J. Med. Microbiol. 47:151–157 [DOI] [PubMed] [Google Scholar]
- 29. Holdeman LV, Cato EP, Moore EC. 1977. Anaerobe laboratory manual. Virginia Polytechnic Institute and State University, Blacksburg, VA [Google Scholar]
- 30. Wiegand I, Hilpert K, Hancock REW. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3:163–175 [DOI] [PubMed] [Google Scholar]
- 31. Tsou PH, Sreenivasappa H, Hong S, Yasuike M, Miyamoto H, Nakano K, Misawa T, Kameoka J. 2010. Rapid antibiotic efficacy screening with aluminum oxide nanoporous membrane filter-chip and optical detection system. Biosens. Bioelectron. 26:289–294 [DOI] [PubMed] [Google Scholar]
- 32. Mach KE, Mohan R, Baron EJ, Shih MC, Gau V, Wong PK, Liao JC. 2011. A biosensor platform for rapid antimicrobial susceptibility testing directly from clinical samples. J. Urol. 185:148–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.