Abstract
Microbiological diagnosis is pivotal to the appropriate management and treatment of infective endocarditis. We evaluated PCR-electrospray ionization mass spectrometry (PCR/ESI-MS) for bacterial and candidal detection using 83 formalin-fixed paraffin-embedded heart valves from subjects with endocarditis who had positive valve and/or blood cultures, 63 of whom had positive valvular Gram stains. PCR/ESI-MS yielded 55% positivity with concordant microbiology at the genus/species or organism group level (e.g., viridans group streptococci), 11% positivity with discordant microbiology, and 34% with no detection. PCR/ESI-MS detected all antimicrobial resistance encoded by mecA or vanA/B and identified a case of Tropheryma whipplei endocarditis not previously recognized.
INTRODUCTION
Infective endocarditis (IE) is a serious infection that was historically a disease of children and young adults with underlying chronic rheumatic heart disease (1) but today is additionally associated with prosthetic heart valves, injection drug use, and intravascular catheters (2). Accurate and prompt detection of the infecting organism(s) is essential to ensure appropriate management. Diagnosis requires evidence of both bloodstream infection and cardiac valvular involvement, with blood cultures and echocardiography being essential components (3, 4). Determination of the infecting organism guides antibiotic treatment, but IE is not confirmed through culture in all cases (5). A challenge in the diagnosis of IE is the occurrence of negative blood and valve cultures. These may be caused by a number of factors, including the use of antibiotics prior to cultures being collected or the presence of fastidious organisms (6, 7). In such circumstances, serologic studies and/or pathogen detection by molecular methods might be helpful (8).
In some instances, cultures are not obtained due to a lack of clinical suspicion for IE. This can occur when a patient undergoes valve surgery for reasons other than suspected IE, and histopathologic analysis unexpectedly demonstrates microorganisms. Another challenge for management of IE is presented by cases of polymicrobial endocarditis because many of the commonly used diagnostics do not detect multiple organisms in individual specimens (9). Finally, the antimicrobial susceptibility of the implicated organism(s) is a key component of IE management. Isolating the infectious agent(s) in culture and determining antimicrobial susceptibility can delay appropriate treatment (10); unfortunately, phenotypic antimicrobial susceptibility testing is not possible in culture-negative cases.
In situations where cultures are negative, inconclusive, or unavailable, molecular testing of explanted heart valves for microbial DNA may be useful. Broad-range bacterial PCR targeting a conserved (e.g., 16S rRNA) gene is one option (11, 12), as is PCR targeting individual microorganisms (e.g., Bartonella species, Coxiella burnetii, and Tropheryma whipplei) (13). Recently, PCR has been combined with electrospray ionization mass spectrometry (ESI-MS) for detection and characterization of amplified DNA (14). Unlike mass spectrometry adapted for identification of bacteria and fungi isolated from cultures, PCR/ESI-MS, as a result of a PCR-amplification step, offers the possibility for direct detection of an organism(s) from tissue. The PCR/ESI-MS assay studied herein (the PLEX-ID BAC detection assay [Abbott Laboratories, Abbott Park, IL]) utilizes 18 primer pairs multiplexed into 16 wells of a microtiter well plate to detect bacterial and candidal organisms (14, 15). Identification of bacteria and Candida species is accomplished through broad-range and targeted PCR with subsequent base composition analysis of amplified product(s) via ESI-MS. The PCR/ESI-MS database studied represents over 3,400 bacterial species and over 40 Candida species. In addition, primers amplify genes associated with resistance to vancomycin (vanA and vanB), carbapenems (blaKPC), and most commonly used β-lactams (mecA). Unlike most broad-range PCR approaches, this technology readily detects polymicrobial infection (14).
In the clinical microbiology setting, PCR/ESI-MS has been used for genotyping of pneumococci (16), detection of pathogens in positive blood culture bottles (17, 18), identification of respiratory pathogens (19), and simultaneous detection of influenza A and B viruses (20), as well as general identification and genotyping of microbes (21, 22). Other applications include identification of plant pathogens (23) and of pathogenic Vibrio species in aquatic environments (24). The aforementioned studies were done using DNA extracted from bacteria cultured using solid or liquid medium. The technology has also been used to detect microorganisms in synovial fluid from patients with joint infections (25). PCR/ESI-MS detected not only organisms present in culture-negative synovial fluids but also those in polymicrobial infections, when a single organism had been initially suspected. To our knowledge, the study described herein is the first to use PCR/ESI-MS for direct detection of microbial DNA in formalin-fixed paraffin-embedded (FFPE) tissue. We have shown that PCR/ESI-MS may be used to detect bacteria and select resistance markers in FFPE native and prosthetic heart valve tissue from patients with IE.
MATERIALS AND METHODS
Case selection.
A convenience sample of subjects with IE identified through the Mayo Clinic Tissue Registry was selected based on availability of FFPE tissue blocks of heart valves and positive blood and/or valve cultures (or in one instance, positive Bartonella serology and PCR). Medical records were reviewed. Subgroup analyses were performed comparing those subjects with positive and negative valvular Gram stains. Tissue blocks had been stored in the Mayo Clinic Tissue Registry, an off-site storage facility which files and stores microscopic slides and paraffin blocks at controlled room temperature for the Division of Anatomic Pathology, Mayo Clinic. Fresh cuts and fresh Gram stains were performed to determine the best tissue samples to use.
Specimen processing.
A 40-μm-thick section of an FFPE tissue block was cut by hand using a microtome or by the Histology Laboratory at the Mayo Clinic. For all samples, an 8- to 10-μm section was first removed using a razor blade to minimize contamination of the block; a fresh blade was then used to cut a 40-μm section for testing. Microtome blades were changed between each specimen. Samples were placed in glass tubes, 500 μl xylene (Sigma-Aldrich, St. Louis, MO) was added, and the samples were incubated at room temperature for 5 min, vortexed, and centrifuged at 20,800 × g for 30 s. The xylene was removed and the process was repeated. Following the second xylene wash, 0.5 ml of 95% ethanol (Sigma-Aldrich, St. Louis, MO) was added, and the mixture was then incubated for 5 min, vortexed, and centrifuged for 3 min at 20,800 × g. The ethanol was removed. The tissue was resuspended in 1 ml of water and transferred to a 2-ml PLEX-ID bead tube (Abbott Laboratories). Then 25 μl of proteinase K (Abbott Laboratories) and 150 μl SDS-1 solution (Abbott Laboratories) were added, and the tube was vortexed and placed in a 55°C heat block (Thermomixer R; Eppendorf, Hauppauge, NY) overnight. The bead tube was processed using a MagNA Lyser (Roche, Indianapolis, IN) for 1 min at 7,000 rpm, following which the tubes were centrifuged for 3 min at 20,800 × g; 1 ml of the supernatant was transferred to a KingFisher (Thermo Scientific, Waltham, MA) 24-well extraction plate for DNA extraction.
DNA extraction.
DNA extraction was performed using a KingFisher DNA extraction instrument (Thermo Scientific) with 24 deep-well plates and reagents from Abbott Laboratories. Briefly, four wash plates were prepared, one with 2 ml wash buffer 1 (4.7 M guanidinium thiocyanate, 10% Tween 20, 100 mM Tris [pH 7.8], and 33.3% ethanol) and three with 2 ml wash buffer 2 (0.01 M Tris [pH 8.0], 0.05 mM EDTA, and 70% ethanol) in each well. An elution plate was prepared with 280 μl elution buffer (0.01 M Tris [pH 8.0] and 0.05 mM EDTA) in each well. The sample plate was prepared by adding 1 ml of wash buffer 1 and 160 μl of magnetic particles to each well, followed by 1 ml of the designated sample following tissue processing. The plates were loaded onto the KingFisher instrument and isolated under the specific extraction program.
Broad-range PCR.
PCR plates (BAC detection 2.0 assay plates, Abbott Laboratories) (14) were thawed and centrifuged at 1,800 × g (IEC Centra MP4R, Thermo Scientific) for 1 min. PCR plates were loaded from the extracted DNA plates using a CAS 1200 precision liquid-handling system (Corbett Research [Qiagen], Valencia, CA). Sixteen wells were filled with 10 μl of extracted sample from each specimen, allowing testing of 6 samples per 96-well plate. Following sample loading, the plates were sealed with Easy Pierce 20-μm heat-sealing tape (catalog no. AB-1720; Thermo Scientific) at 175°C for 1.5 s (ThermoSci ALPS 50V; Thermo Scientific), centrifuged at 1,800 × g for 1 min, and placed in an Eppendorf Mastercycler proS thermocycler. PCR was performed using the following thermocycling protocol: 95°C for 10 min followed by 8 cycles of 95°C for 30 s, 48°C for 30 s, and 72°C for 30 s, followed by 37 cycles of 95°C for 15 s, 56°C for 20 s, and 72°C for 20 s, and then by 72°C for 2 min and 99°C for 20 min.
Mass spectrometry.
Following PCR, the plate was placed in the desalting/ESI-MS instrument (PLEX-ID, Abbott Laboratories) and the PCR product was desalted and analyzed using ESI-MS to determine the weight of the amplified products. The assay was run using 1.3 RC5 software under version IVD.32.35.26, with analysis done under version GenX-V08R002. The protocol for reporting organisms and their cutoff levels was 4.0.0.8480. Four control valves from subjects who underwent valve excision for noninfectious causes were tested and all yielded negative results.
Data analysis.
Identification with concordant microbiology was defined as detection that agreed with microbiological culture results. Identification with discordant microbiology was defined as detection of an organism discordant with known microbiology. Samples that were undetected had negative results by PCR/ESI-MS (but previous microbiology indicating a specific infection).
Along with organism identification, the ESI-MS analysis includes a level and a Q score. The level indicates the amount of amplified DNA present in the sample. This is reported as genome equivalents/well, and calculated referent to the internal calibrant, as previously described (15). The Q score, a rating between 0 (low) and 1 (high), represents a relative measure of the strength of the data supporting identification; only Q scores of ≥0.90 were reported. Proportions of interest were compared using Fisher's exact test to assess statistical differences in detection.
RESULTS
Overview of cases selected.
We studied 83 cases of IE, which were differentiated on the basis of Gram stain and valve storage for more or less than a decade. Cases were selected from 1999 to 2012 with the majority (64/83) having a positive Gram stain. The average subject age was 57 years, with men predominating (n = 66), and native valves (n = 59) being more common than prosthetic valves (n = 24). Table 1 summarizes the results found with PCR/ESI-MS, separated into categories of detection with concordant microbiology, detection with discordant microbiology, and no detection. Overall there was 55% detection with concordant microbiology, 11% detection with discordant microbiology, and 34% no detection (Table 1).
Table 1.
PCR/ESI-MS assay results
| PCR/ESI-MS assay result | No. (%) of indicated valvular Gram stain results within the date range shown |
Total no. (%) of valves (n = 83) |
|||
|---|---|---|---|---|---|
| Positive (n = 64) |
Negative (n = 19) |
||||
| 1999–2002 (n = 33) | 2008–2010 (n = 30) | 2000–2002 (n = 13) | 2008–2012 (n = 7) | 1999–2012 | |
| Antibiotic resistance marker (if applicable) detection | 14 (100) | 1 (100) | 10 (100) | 1 (100) | 26 (100) |
| Detection | 22 (67) | 23 (77) | 6 (46) | 4 (57) | 55 (66) |
| Concordant microbiology | 19 (58) | 19 (63)a,b | 4 (30)c | 4 (57)b,d | 46 (55) |
| Discordant microbiology | 3 (9) | 4 (13) | 2 (15) | 0 (0) | 9 (11) |
| No detection | 11 (33) | 7 (23) | 7 (54) | 3 (43) | 28 (34) |
Detection of additional organisms in addition to the concordant microbiology (n = 5).
Candida tropicalis contaminant detected (n = 6).
Detection of an additional organism in addition to the concordant microbiology (n = 1).
Detection of additional organisms in addition to the concordant microbiology (n = 2).
Detection of a targeted antimicrobial resistance marker was correct in all cases where antimicrobial susceptibility was known (26/26, 100%) (Table 1). PCR/ESI-MS correctly identified the presence/absence of mecA in all 22 cases of staphylococcal endocarditis caused by staphylococci of known methicillin susceptibility (based on testing of blood/valve culture isolates) (Tables 2 and 3). PCR/ESI-MS correctly identified the presence/absence of vanA/B in all four enterococcal endocarditis cases of known vancomycin susceptibility (Table 2).
Table 2.
PCR/ESI-MS assay results for formalin-fixed paraffin-embedded heart valve tissues from subjects with positive valvular Gram stainsb
| Valve typea | Blood culture result | Valve culture result | Time from positive blood culture to surgery (days) | Age of FFPE block (yr) | PCR/ESI-MS result | Levelc | Q score |
|---|---|---|---|---|---|---|---|
| P | MSSA | MSSA | 6 | 12 | Staphylococcus aureuse | 67 | 1 |
| P | MSSA | Negative | 25 | 11 | S. aureuse | 4 | 1 |
| P | MSSA | Negative | 18 | 11 | S. aureuse | 349 | 1 |
| N | MSSA | MSSA | 8 | 11 | S. aureuse | 189 | 1 |
| N | MSSA | MSSA | 4 | 11 | S. aureuse | 126 | 0.99 |
| N | MSSA | Negative | 5 | 10 | S. aureuse | 161 | 1 |
| N | MRSA | MRSA | 4 | 10 | S. aureus | 61 | 1 |
| mecA positive | 27 | 1 | |||||
| N | MRSA | S. aureus | 2 | 11 | S. aureus | 174 | 1 |
| mecA positive | 3 | 1 | |||||
| P | Not done | S. aureus (oxacillin susceptibility unknown) | NAd | 12 | S. aureuse | 19 | 0.99 |
| P | MSSA | Negative | 14 | 4 | S. aureuse | 83 | 0.99 |
| N | MSSA | Negative | 28 | 4 | S. aureuse | 269 | 1 |
| N | MSSA | Negative | 31 | 3 | S. aureuse | 81 | 0.99 |
| N | MSSA | Negative | 53 | 4 | S. aureuse | 7 | 0.98 |
| N | MRSA | MRSA | 2 | 3 | S. aureus | 67 | 0.99 |
| mecA positive | 144 | 0.98 | |||||
| P | Staphylococcus epidermidis (oxacillin susceptible) | Negative | 20 | 12 | S. epidermidise | 148 | 1 |
| N | VGS | Negative | 10 | 13 | Streptococcus sp. | 6 | 0.97 |
| N | Negative | VGS | NA | 13 | Streptococcus oralis | 84 | 1 |
| N | No cultures obtained | VGS | NA | 12 | Streptococcus mutans | 46 | 0.98 |
| P | VGS | Negative | 8 | 4 | Streptococcus sanguinis | 233 | 0.99 |
| N | Streptococcus mitis | Negative | 2 | 3 | S. mitis group | 45 | 0.98 |
| N | VGS | Negative | 0 | 2 | S. mitis group | 5 | 0.98 |
| N | S. mutans | Negative | 25 | 3 | S. mutans | 77 | 0.98 |
| N | Streptococcus bovis group | Negative | 7 | 12 | S. equinus/bovis complex | 83 | 1 |
| P | Cardiobacterium hominis | Negative | 308 | 11 | C. hominis | 233 | 0.99 |
| N | Enterococcus faecalis (vancomycin susceptible) | E. faecalis | 6 | 10 | E. faecalisf | 174 | 1 |
| N | E. faecalis (vancomycin susceptible) | Negative | 5 | 11 | E. faecalisf | 231 | 1 |
| N | E. faecalis (vancomycin susceptible) | Negative | 11 | 11 | E. faecalisf | 62 | 0.99 |
| N | Enterococcus faecium (vancomycin resistant) | VGS | 2 | 13 | E. faecium | 83 | 1 |
| vanA positive | 39 | 1 | |||||
| N | E. faecalis | E. faecalis | 13 | 2 | E. faecalis | 158 | 0.99 |
| N | Staphylococcus lugdunensis (methicillin susceptible) | Not done | 5 | 3 | S. lugdunensise | 77 | 0.98 |
| N | CoNS (methicillin susceptible) | Negative | 5 | 4 | S. lugdunensise | 225 | 0.99 |
| N | CoNS (methicillin-resistant) | CoNS | 8 | 4 | S. epidermidis | 162 | 0.99 |
| mecA positive | 252 | 0.98 | |||||
| P | CoNS (methicillin-resistant) | Negative | 1 | 4 | S. epidermidis/Staphylococcus caprae | 116 | 1 |
| mecA positive | 176 | 0.98 | |||||
| P | CoNS (methicillin-resistant) | Negative | 10 | 4 | S. epidermidis | 211 | 0.99 |
| mecA positive | 181 | 0.98 | |||||
| P | Propionibacterium sp. | Negative | 66 | 4 | P. acnes | 264 | 0.99 |
| N | Haemophilus parainfluenzae | Negative | 21 | 4 | H. parainfluenzae | 424 | 0.99 |
| Candida tropicalis | 6 | 0.98 | |||||
| N | Aerococcus sp. | Negative | 8 | 4 | Granulicatella adiacens | 94 | 0.97 |
| Aerococcus urinae | 103 | 0.98 | |||||
| C. tropicalis | 46 | 0.98 | |||||
| N | VGS | Negative | 4 | 4 | Streptococcus pneumoniae | 164 | 0.99 |
| C. tropicalisg | 16 | 0.98 | |||||
| N | VGS | Negative | 16 | 11 | S. pneumoniaeg | 123 | 0.97 |
| N | S. mitis | Negative | 8 | 12 | S. pneumoniaeg | 14 | 0.94 |
| N | S. mitis | Negative | 3 | 11 | S. pneumoniaeg | 101 | 0.97 |
| P | Granulicatella elegans | Negative | 0 | 4 | S. epidermidise | 65 | 0.99 |
| C. tropicalisg | 42 | 0.96 | |||||
| N | E. faecalis | Negative | 59 | 4 | S. epidermidise | 49 | 0.99 |
| C. tropicalisg | 37 | 0.97 | |||||
| P | S. lugdunensis | Not done | 7 | 4 | Tropheryma whippleig | 502 | 0.99 |
| N | MSSA | Negative | 19 | 12 | No detection | ||
| P | S. epidermidis | Negative | 5 | 12 | No detection | ||
| N | VGS | Negative | 13 | 12 | No detection | ||
| N | VGS | Negative | 110 | 11 | No detection | ||
| N | VGS | Negative | 9 | 11 | No detection | ||
| N | VGS | Negative | 13 | 12 | No detection | ||
| N | S. mitis (VGS) | Negative | 22 | 12 | No detection | ||
| N | Streptococcus salivarius (VGS) | Negative | 66 | 11 | No detection | ||
| N | S. salivarius (VGS) | Negative | 135 | 12 | No detection | ||
| N | H. parainfluenzae | H. parainfluenzae | 8 | 12 | No detection | ||
| P | Streptomyces sp. | Negative | 3 | 11 | No detection | ||
| P | MSSA | Negative | 13 | 4 | No detection | ||
| P | CoNS | Negative | 60 | 4 | No detection | ||
| N | S. epidermidis | Negative | 6 | 3 | No detection | ||
| N | VGS | Negative | 3 | 2 | No detection | ||
| N | S. mitis | Negative | 2 | 1 | No detection | ||
| N | E. faecalis | Negative | 5 | 2 | No detection | ||
| N | E. faecalis | Negative | 94 | 2 | No detection |
Infection of a native (N) or prosthetic (P) valve.
MSSA, methicillin-susceptible S. aureus; MRSA, methicillin-resistant S. aureus; VGS, viridans group Streptococcus sp.
The amount of amplified DNA present in the sample, reported as genome equivalents/well.
NA, not applicable.
The sample was mecA negative.
The sample was vanA/vanB negative.
Discordant with previous microbiology.
Table 3.
PCR/ESI-MS results for formalin-fixed paraffin-embedded heart valve tissues from subjects with negative valvular Gram stainsb
| Valve typea | Blood culture result | Valve culture result | Time from positive blood culture to surgery (days) | Age of FFPE block (yr) | PCR/ESI-MS result | Levelc | Q score |
|---|---|---|---|---|---|---|---|
| P | MSSA | MSSA | 6 | 13 | Staphylococcus aureus (mecA negative) | 22 | 0.99 |
| N | MSSA | Negative | 52 | 11 | S. aureus (mecA negative) | 7 | 0.98 |
| N | MSSA | Negative | 26 | 2 | S. aureus (mecA negative) | 104 | 0.99 |
| N | VGS | Negative | 12 | 12 | Streptococcus gordonii | 79 | 1 |
| P | VGS | Negative | 84 | 11 | Streptococcus sp. | 3 | 0.97 |
| N | Peptostreptococcus sp. | Negative | 60 | 4 | Gemella morbillorumd | 178 | 0.98 |
| N | Granulicatella sp. | Negative | 11 | 4 | G. adiacens | 169 | 0.99 |
| Candida tropicalis | 43 | 0.98 | |||||
| N | Escherichia coli | Negative | 15 | 4 | E. coli | 256 | 0.99 |
| Staphylococcus epidermidis | 30 | 0.97 | |||||
| P | MSSA | Negative | 12 | 11 | Propionibacterium acnese | 22 | 0.98 |
| P | CoNS | Negative | 17 | 12 | P. acnese | 78 | 0.99 |
| P | MSSA | Negative | 32 | 10 | No detection | ||
| N | MSSA | Negative | 24 | 11 | No detection | ||
| N | S. epidermidis | CoNS | 5 | 13 | No detection | ||
| P | CoNS | Negative | 21 | 12 | No detection | ||
| N | VGS | Negative | 20 | 12 | No detection | ||
| P | Enterococcus faecalis | Negative | 21 | 12 | No detection | ||
| N | Cardiobacterium hominis | Negative | 22 | 12 | No detection | ||
| N | E. faecalis | Negative | 61 | 4 | No detection | ||
| N | E. faecalis | Negative | 32 | 4 | No detection | ||
| N | Positive Bartonella serology and PCR | Negative | 0 | 4 | No detection |
Infection of a native (N) or prosthetic (P) valve.
MSSA, methicillin-susceptible S. aureus; VGS, viridans group Streptococcus sp.; CoNS, coagulase-negative Staphylococcus sp.
The amount of amplified DNA present in the sample, reported as genome equivalents/well.
Synonym, “Peptostreptococcus morbillorum.”
Discordant with previous microbiology.
Positive valvular Gram stain.
Among 63 valves with positive Gram stains, 45 (71%) were detected, of which 38 (60%) had concordant microbiology (Tables 1 and 2). Detections with concordant microbiology included Staphylococcus aureus (n = 14), coagulase-negative staphylococci (CoNS) (n = 6), Enterococcus faecalis/faecium (n = 5), viridans group streptococci (VGS) (n = 6), Streptococcus species (n = 1), Streptococcus bovis species group (n = 1), Cardiobacterium hominis (n = 1), Propionibacterium acnes (n = 1), Mycobacterium fortuitum (n = 1), Haemophilus parainfluenzae (n = 1), and Aerococcus species (n = 1). Detection with discordant microbiology consisted of four VGS misidentified as Streptococcus pneumoniae, one Granulicatella elegans misidentified as Staphylococcus epidermidis, one E. faecalis misidentified as S. epidermidis, and Staphylococcus lugdunensis misidentified as T. whipplei. Since T. whipplei was detected at a high level (502 genome equivalents/well), this case was further investigated (see below). Undetected valves had known prior microbiology indicating VGS (n = 9), CoNS (n = 3), S. aureus (n = 2), E. faecalis (n = 2), H. parainfluenzae (n = 1), and Streptomyces species (n = 1) infections (Table 2).
T. whipplei case.
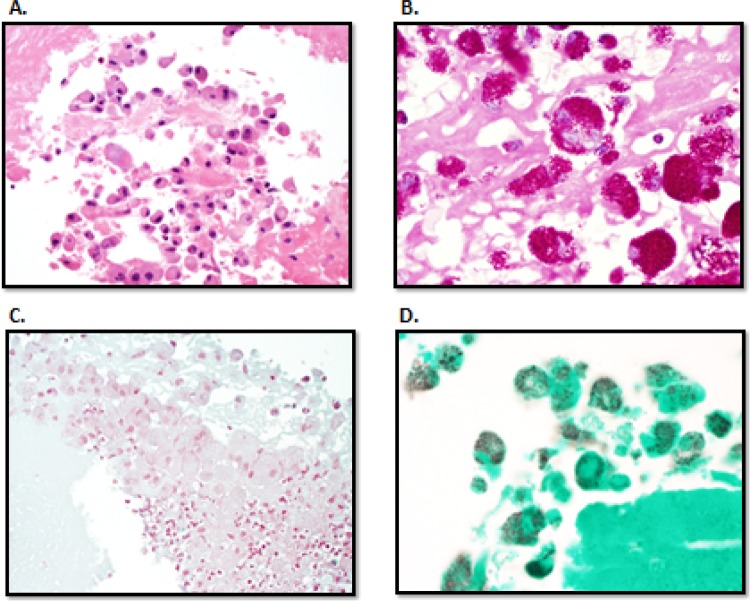
In one case, T. whipplei was detected despite S. lugdunensis being listed as the infectious agent of record (Table 2). A thorough review of the medical record indicated growth of S. lugdunensis in 1 of 10 blood cultures, a result that in retrospect was considered to be attributable to a probable contaminant. Valve cultures had not been performed. The patient had a negative blood PCR for T. whipplei. Periodic acid-Schiff (PAS) staining of heart valve tissue revealed classic histopathologic findings of Whipple disease (Fig. 1) (26). To confirm the presence of T. whipplei, a T. whipplei-specific real-time PCR assay (27) was performed on heart valve tissue and was positive.
Fig 1.
Histological identification of T. whipplei. (A) Routine hematoxylin and eosin staining revealed a vegetation with scattered macrophages and rare neutrophils (400× original magnification). (B) The macrophages contained periodic acid-Schiff-positive diastase-resistant rods consistent with Whipple bacilli (1,000× original magnification). The bacilli stained poorly with routine stains used for valve histopathology examination, including tissue Gram stain (C) (1,000× original magnification) and Gomori methenamine silver stain (D) (1,000× original magnification).
Negative valvular Gram stain.
Among 20 valves with negative Gram stains, 10 (50%) were detected, of which 8 (40%) had concordant microbiology (Tables 1 and 3). Detections with concordant microbiology included S. aureus (n = 3), VGS (n = 1), Gemella morbillorum (n = 1), Granulicatella species (n = 1), Escherichia coli (n = 1), and Streptococcus species (n = 1). Detections with discordant microbiology consisted of one each of S. aureus and CoNS misidentified as P. acnes. Samples that were undetected had previous microbiology indicating S. aureus (n = 2), CoNS (n = 2), VGS (n = 1), E. faecalis (n = 3), and C. hominis (n = 1) infections. The final undetected case was in a patient with positive Bartonella serology and PCR results (28).
There were no differences in the detection of common infectious agents with respect to whether the valves were more or less than a decade old (P = 0.42). Additionally, there were no differences in detection in samples with positive and negative Gram stains (P = 0.35). In cases with positive Gram stains compared to those from the same time periods with negative Gram stains, detection was not enhanced in valves more than a decade (P = 0.31) or those less than a decade old (P = 0.36) (Table 1).
DISCUSSION
Over the last 30 years, molecular diagnostics have played an increasingly important role in the diagnosis of IE (29). The data indicate that PCR/ESI-MS has potential for use as a diagnostic tool for IE. PCR/ESI-MS has the potential to decrease turnaround time to 12 to 16 h compared to 24 to 48 h for standard tissue processing and culture (providing it is performed on fresh tissue). Beyond organism detection and classification, select antimicrobial resistance is also defined.
A major benefit of this method over other molecular methods is the ability to detect more than one organism simultaneously in a sample. ESI-MS may be more sensitive than other molecular methods. Imrit et al. identified causative organisms of IE using FFPE heart valves coupled with 16S rRNA gene sequencing. They found that the length of time the heart valves were stored in the paraffin block affected the sensitivity of the assay, with 70% positivity for valves stored for less than 5 years versus 18% positivity for those stored longer (12). This is in contrast to the data presented herein, which showed no difference in detection between FFPE heart valves stored for more than and less than a decade (P = 0.42).
This is the first large-scale PCR/ESI-MS study to use FFPE samples of infected tissue. A case report has described the use of PCR/ESI-MS to detect Bartonella species in an abdominal aortic mycotic aneurysm (30), and a recent publication has described the use of this technology for the analysis of synovial fluid from subjects with prosthetic joint infections (25). Organisms detected in this study included common IE pathogens (S. aureus, S. epidermidis, and E. faecalis) and not-so-common pathogens (T. whipplei, Granulicatella adiacens, G. morbillorum, C. hominis, and M. fortuitum) (31, 32), which speaks to the potential applications of the assay studied.
In eight cases with concordant microbiology, an additional bacterium was detected (G. adiacens [n = 1], S. epidermidis [n = 1], and Candida tropicalis [n = 6]) using PCR/ESI-MS; these apparently false-positive results may relate to contamination of the tissue block during storage, sectioning, or processing for testing or PCR contamination. The specimens studied were inherently nonsterile and subject to contamination during cutting of the blocks (i.e., from technologists or razor blades). Further, PCR/ESI-MS is not a closed system, so it allows for possible amplified product contamination. All C. tropicalis detection occurred with a single assay plate, a finding that strongly supports contamination.
Not all IE cases were detected, possibly because the valves studied came from FFPE blocks. Fixation with formalin degrades DNA, lowering the yield of amplifiable DNA compared to that of fresh tissue (33). Although not evaluated herein, testing of fresh tissue is a possible clinical application of this assay. Finally, it is possible that the infectious agent was not equally distributed throughout the valves, in which case, due to sampling error, a 40-μm cut may have missed the microbial DNA.
Misidentification of VGS as S. pneumoniae, noted in four instances, was likely due to an error in the identification algorithms used. This can possibly be corrected by targeting a more specific gene for S. pneumoniae during the upfront PCR step or by reporting all S. pneumoniae detected as “VGS/S. pneumoniae.” Identification of T. whipplei IE from a case originally misdiagnosed through blood culture as S. lugdunensis IE illustrates that this system can identify and diagnose cases that might otherwise be missed using conventional methods.
There are several limitations of the technology evaluated and of the study design. DNA extracted from FFPE tissue was diluted to have enough volume to distribute to 16 PCR wells, potentially impacting assay sensitivity. As a consequence, some positive specimens may not have been identified due to low concentrations of target DNA. Formalin fixation may lead to DNA cross-linking, preventing PCR amplification. The PCR products generated with the assay studied herein are very short (∼80 to 120 bp), maximizing the likelihood of detection since cross-linking occurs at approximately every 100 bp. A major limitation in study design was that only culture-positive valves were studied; future studies should assess culture-negative valves. Finally, limited antimicrobial resistance markers were studied, although mecA, vanAB, and blaKPC do represent key early decision markers for the inclusion or exclusion of related antimicrobials in the treatment regimen.
In summary, PCR/ESI-MS might be a useful tool in the diagnosis of IE cases where a microbiologic etiology is not defined by current laboratory methods. If these findings are supported by those demonstrated in future investigations, this tool might have a meaningful impact on both early and subsequent antimicrobial selection, which might positively influence patient outcomes.
Footnotes
Published ahead of print 17 April 2013
REFERENCES
- 1. Normand J, Bozio A, Etienne J, Sassolas F, Le Bris H. 1995. Changing patterns and prognosis of infective endocarditis in childhood. Eur. Heart J. 16(Suppl B):28–31 [DOI] [PubMed] [Google Scholar]
- 2. Wilson LE, Thomas DL, Astemborski J, Freedman TL, Vlahov D. 2002. Prospective study of infective endocarditis among injection drug users. J. Infect. Dis. 185:1761–1766 [DOI] [PubMed] [Google Scholar]
- 3. Li JS, Sexton DJ, Mick N, Nettles R, Fowler VG, Jr, Ryan T, Bashore T, Corey GR. 2000. Proposed modifications to the Duke criteria for the diagnosis of infective endocarditis. Clin. Infect. Dis. 30:633–638 [DOI] [PubMed] [Google Scholar]
- 4. Habib G, Badano L, Tribouilloy C, Vilacosta I, Zamorano JL, Galderisi M, Voigt JU, Sicari R, Cosyns B, Fox K, Aakhus S. 2010. Recommendations for the practice of echocardiography in infective endocarditis. Eur. J. Echocardiogr. 11:202–219 [DOI] [PubMed] [Google Scholar]
- 5. Que YA, Moreillon P. 2011. Infective endocarditis. Nat. Rev. Cardiol. 8:322–336 [DOI] [PubMed] [Google Scholar]
- 6. Houpikian P, Raoult D. 2005. Blood culture-negative endocarditis in a reference center: etiologic diagnosis of 348 cases. Medicine (Baltimore) 84:162–173 [DOI] [PubMed] [Google Scholar]
- 7. Brouqui P, Raoult D. 2001. Endocarditis due to rare and fastidious bacteria. Clin. Microbiol. Rev. 14:177–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Houpikian P, Raoult D. 2002. Diagnostic methods: current best practices and guidelines for identification of difficult-to-culture pathogens in infective endocarditis. Infect. Dis. Clin. North Am. 16:377–392 [DOI] [PubMed] [Google Scholar]
- 9. Tran C, Cometta A, Letovanec I, Jaton K, Wenger A, Ruchat P, Jaussi A. 2007. Candida dubliniensis in recurrent polymicrobial tricuspid endocarditis. Echocardiography 24:756–759 [DOI] [PubMed] [Google Scholar]
- 10. Thuny F, Grisoli D, Collart F, Habib G, Raoult D. 2012. Management of infective endocarditis: challenges and perspectives. Lancet 379:965–975 [DOI] [PubMed] [Google Scholar]
- 11. Vondracek M, Sartipy U, Aufwerber E, Julander I, Lindblom D, Westling K. 2011. 16S rDNA sequencing of valve tissue improves microbiological diagnosis in surgically treated patients with infective endocarditis. J. Infect. 62:472–478 [DOI] [PubMed] [Google Scholar]
- 12. Imrit K, Goldfischer M, Wang J, Green J, Levine J, Lombardo J, Hong T. 2006. Identification of bacteria in formalin-fixed, paraffin-embedded heart valve tissue via 16S rRNA gene nucleotide sequencing. J. Clin. Microbiol. 44:2609–2611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tang YW. 2009. Duplex PCR assay simultaneously detecting and differentiating Bartonella quintana, B. henselae, and Coxiella burnetii in surgical heart valve specimens. J. Clin. Microbiol. 47:2647–2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kaleta EJ, Clark AE, Johnson DR, Gamage DC, Wysocki VH, Cherkaoui A, Schrenzel J, Wolk DM. 2011. Use of PCR coupled with electrospray ionization mass spectrometry for rapid identification of bacterial and yeast bloodstream pathogens from blood culture bottles. J. Clin. Microbiol. 49:345–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hofstadler SA, Sampath R, Blyn LB, Eshoo MW, Hall TA, Jiang Y, Drader JJ, Hannis JC, Sannes-Lowery KA, Cummins LL, Libby B, Walcott DJ, Schink A, Massire C, Ranken R, Gutierrez J, Manalili S, Ivy C, Melton R, Levene H, Barrett-Wilt G, Li F, Zapp V, White N, Samant V, McNeil JA, Knize D, Robbins D, Rudnick K, Desai A, Moradi E, Ecker DJ. 2005. TIGER: the universal biosensor. Int. J. Mass Spectrom. 242:23–41 [Google Scholar]
- 16. Massire C, Gertz RE, Jr, Svoboda P, Levert K, Reed MS, Pohl J, Kreft R, Li F, White N, Ranken R, Blyn LB, Ecker DJ, Sampath R, Beall B. 2012. Concurrent serotyping and genotyping of pneumococci by use of PCR and electrospray ionization mass spectrometry. J. Clin. Microbiol. 50:2018–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jeng K, Gaydos CA, Blyn LB, Yang S, Won H, Matthews H, Toleno D, Hsieh YH, Carroll KC, Hardick J, Masek B, Kecojevic A, Sampath R, Peterson S, Rothman RE. 2012. Comparative analysis of two broad-range PCR assays for pathogen detection in positive-blood-culture bottles: PCR-high-resolution melting analysis versus PCR-mass spectrometry. J. Clin. Microbiol. 50:3287–3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kaleta EJ, Clark AE, Cherkaoui A, Wysocki VH, Ingram EL, Schrenzel J, Wolk DM. 2011. Comparative analysis of PCR-electrospray ionization/mass spectrometry (MS) and MALDI-TOF/MS for the identification of bacteria and yeast from positive blood culture bottles. Clin. Chem. 57:1057–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ecker DJ, Sampath R, Blyn LB, Eshoo MW, Ivy C, Ecker JA, Libby B, Samant V, Sannes-Lowery KA, Melton RE, Russell K, Freed N, Barrozo C, Wu J, Rudnick K, Desai A, Moradi E, Knize DJ, Robbins DW, Hannis JC, Harrell PM, Massire C, Hall TA, Jiang Y, Ranken R, Drader JJ, White N, McNeil JA, Crooke ST, Hofstadler SA. 2005. Rapid identification and strain-typing of respiratory pathogens for epidemic surveillance. Proc. Natl. Acad. Sci. U. S. A. 102:8012–8017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tang YW, Lowery KS, Valsamakis A, Schaefer VC, Chappell JD, White-Abell J, Quinn CD, Li H, Washington CA, Cromwell J, Giamanco CM, Forman M, Holden J, Rothman RE, Parker ML, Ortenberg EV, Zhang L, Lin YL, Gaydos CA. 2013. Clinical accuracy of a PLEX-ID Flu device for simultaneous detection and identification of influenza viruses A and B. J. Clin. Microbiol. 51:40–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ecker DJ, Massire C, Blyn LB, Hofstadler SA, Hannis JC, Eshoo MW, Hall TA, Sampath R. 2009. Molecular genotyping of microbes by multilocus PCR and mass spectrometry: a new tool for hospital infection control and public health surveillance. Methods Mol. Biol. 551:71–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Baldwin CD, Howe GB, Sampath R, Blyn LB, Matthews H, Harpin V, Hall TA, Drader JJ, Hofstadler SA, Eshoo MW, Rudnick K, Studarus K, Moore D, Abbott S, Janda JM, Whitehouse CA. 2009. Usefulness of multilocus polymerase chain reaction followed by electrospray ionization mass spectrometry to identify a diverse panel of bacterial isolates. Diagn. Microbiol. Infect. Dis. 63:403–408 [DOI] [PubMed] [Google Scholar]
- 23. Postinikova E, Baldwin C, Whitehouse CA, Sechler A, Schaad NW, Sampath R, Harpin V, Li F, Melton R, Blyn L, Drader J, Hofstadler S, Schneider WL. 2008. Identification of bacterial plant pathogens using multilocus polymerase chain reaction/electrospray ionization-mass spectrometry. Phytopathology 98:1156–1164 [DOI] [PubMed] [Google Scholar]
- 24. Whitehouse CA, Baldwin C, Sampath R, Blyn LB, Melton R, Li F, Hall TA, Harpin V, Matthews H, Tediashvili M, Jaiani E, Kokashvili T, Janelidze N, Grim C, Colwell RR, Huq A. 2010. Identification of pathogenic Vibrio species by multilocus PCR-electrospray ionization mass spectrometry and its application to aquatic environments of the former Soviet Republic of Georgia. Appl. Environ. Microbiol. 76:1996–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jacovides CL, Kreft R, Adeli B, Hozack B, Ehrlich GD, Parvizi J. 2012. Successful identification of pathogens by polymerase chain reaction (PCR)-based electron spray ionization time-of-flight mass spectrometry (ESI-TOF-MS) in culture-negative periprosthetic joint infection. J. Bone Joint Surg. Am. 94:2247–2254 [DOI] [PubMed] [Google Scholar]
- 26. Bonhomme CJ, Renesto P, Desnues B, Ghigo E, Lepidi H, Fourquet P, Fenollar F, Henrissat B, Mege JL, Raoult D. 2009. Tropheryma whipplei glycosylation in the pathophysiologic profile of Whipple's disease. J. Infect. Dis. 199:1043–1052 [DOI] [PubMed] [Google Scholar]
- 27. Sloan LM, Rosenblatt JE, Cockerill FR., III 2005. Detection of Tropheryma whipplei DNA in clinical specimens by LightCycler real-time PCR. J. Clin. Microbiol. 43:3516–3518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lin EY, Tsigrelis C, Baddour LM, Lepidi H, Rolain JM, Patel R, Raoult D. 2010. Candidatus Bartonella mayotimonensis and endocarditis. Emerg. Infect. Dis. 16:500–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Millar BC, Moore JE. 2004. Current trends in the molecular diagnosis of infective endocarditis. Eur. J. Clin. Microbiol. Infect. Dis. 23:353–365 [DOI] [PubMed] [Google Scholar]
- 30. Koo M, Manalili S, Bankowski MJ, Sampath R, Hofstadler SA, Koo J. 2010. A “silent culture-negative” abdominal aortic mycotic aneurysm: rapid detection of Bartonella species using PCR and high-throughput mass spectrometry. Hawaii Med. J. 69:68–69 [PMC free article] [PubMed] [Google Scholar]
- 31. Akiyama K, Taniyasu N, Hirota J, Iba Y, Maisawa K. 2001. Recurrent aortic valve endocarditis caused by Gemella morbillorum—report of a case and review of the literature. Jpn. Circ. J. 65:997–1000 [DOI] [PubMed] [Google Scholar]
- 32. Al-Tawfiq JA, Kiwan G, Murrar H. 2007. Granulicatella elegans native valve infective endocarditis: case report and review. Diagn. Microbiol. Infect. Dis. 57:439–441 [DOI] [PubMed] [Google Scholar]
- 33. Srinivasan M, Sedmak D, Jewell S. 2002. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am. J. Pathol. 161:1961–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]