Abstract
From September 2010 to December 2011, 26 KPC-3-producing Enterobacter cloacae isolates were identified from 16 patients at a single hospital. Analyses revealed the blaKPC gene to be localized on multiple plasmids in a diverse nonclonal E. cloacae genetic background. These findings highlight the potential complexity of a KPC outbreak at a single hospital.
TEXT
Enterobacter cloacae ranks as the tenth most common organism isolated in Canadian hospitals (1). As with other Enterobacteriaceae, carbapenem-hydrolyzing activity has predominantly been associated with the Klebsiella pneumoniae carbapenemases (KPCs) encoded within the Tn4401 transposon carrying one of many variants of the blaKPC gene (2, 3). Starting in September 2010, the Jewish General Hospital (JGH), in Montréal, Canada, a tertiary-care hospital with 637 acute-care beds, began screening for the presence of carbapenem-resistant Enterobacteriaceae (CRE) from both clinical and screening specimens. This report discusses the subset of KPC-producing E. cloacae isolated at the JGH.
Screening was performed using rectal swabs transported in Amies media (Becton Dickinson, Franklin Lakes, NJ) that were then directly plated onto a CRE screening plate (4). Resistance to ertapenem in isolates from the screen plates were confirmed by Etest (AB Biodisk, Solna, Sweden), with interpretations based on iterations of CLSI document M100 (5). Isolates with an ertapenem MIC of ≥1 μg/ml were sent for confirmation of carbapenem resistance to the Laboratoire de Santé Publique du Québec (LSPQ). The LSPQ confirmed species identification using API 20E strips (bioMérieux, Lyon, France). Extended biochemical panel testing (6) with or without rpoB sequencing was used to resolve discrepancies in species identification. The presence of blaKPC was confirmed by PCR technique at the LSPQ (7, 8). The blaKPC subtypes were obtained by sequencing the amplicon using two additional internal primers (2). Further characterization of the genetic environment of blaKPC at five distinct intergenic regions within the Tn4401 transposon was performed by PCR and sequencing, as previously described (9). Pulsed-field gel electrophoresis (PFGE) using the XbaI and BioNumerics software (version 6.5; Applied Maths, Belgium) generated clustering of strains. A cutoff of ≥85% similarity was used for cluster determination. Plasmids harboring blaKPC were transferred using electroporation to Escherichia coli DH10B and fingerprinted using EcoRI as previously described (10). Plasmids were subsequently typed by a PCR-based replicon typing method as previously described (11). Antimicrobial susceptibility was evaluated among clinical isolates by various established methods (Etest, broth microdilution, and disk diffusion).
Between September 2010 and December 2011, 26 isolates of KPC-3-producing E. cloacae were identified from 16 patients: 12 men and 4 women (Fig. 1). Of the 26 isolates, 7 were recovered from clinical specimens (six patients): 2 from the blood, 3 from urine, and 2 from pus. In addition, 19 isolates from 11 patients were recovered from rectal screens. All of the patients except one were hospitalized at the time of their positive culture determination. The exception (strain G) was detected in the emergency room from a patient who had been recently hospitalized at the JGH. The other patients were located on six different wards at the time of the positive culture (Fig. 1). Macrorestriction analysis using PFGE revealed four clusters of three or more isolates (Fig. 1). The seven clinical isolates identified were found in four distinct PFGE types.
Fig 1.
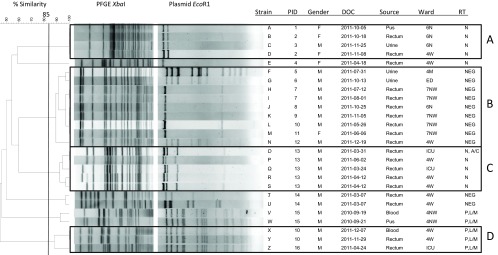
Dendrogram of all 26 strains of E. cloacae isolated at the JGH between September 2010 and December 2011. PID, patient identifier; DOC, date of collection; N, north; NW, northwest; W, west; M, main; ED, emergency department; ICU, intensive care unit; RT, replicon type; NEG, negative.
Transfer of the blaKPC plasmids to E. coli DH10B was successful for all isolates. Plasmid size ranged from 28 to 103 kb, with a median size of 58 kb. Subsequent molecular analysis revealed numerous incompatibility types, including IncN (n = 9), IncP,L/M (n = 5), and IncN,A/C (n = 1), and 11 with an unknown incompatibility type, labeled NEG (Fig. 1). Plasmids harboring IncN were identified in three different E. cloacae fingerprint groups. The five IncP,L/M plasmids were identified in two different E. cloacae fingerprint clusters, suggesting that this plasmid had transferred between strains during the course of the outbreak.
All isolates had a similar Tn4401 transposon structure surrounding blaKPC, suggesting that the same transposon was present among all isolates and plasmids. No deletion was observed in the polymorphic region between the istB of the ISKpn7 and the blaKPC gene (3).
Using MICs of ≤2 and ≥4 mg/liter to express, respectively, sensitivity and resistance to colistin, 6 of the 26 strains were found to be resistant to this antibiotic. For tigecycline, using cutoff MICs of ≤2 and ≥8 mg/liter to establish sensitivity and resistance, only one strain was found to be resistant, with six in the intermediate range with an MIC of 4 mg/liter. Only half of the strains underwent susceptibility testing to chloramphenicol, and all of these were sensitive to this antibiotic except for two in the intermediate range (5). These and other antimicrobial susceptibility profiles to non-beta-lactam antibiotics are detailed in Table 1.
Table 1.
Antimicrobial testing results using non-beta-lactam antibiotics for all KPC-positive E. cloacae strainsa
| Strain information |
Etest MIC (mg/liter) |
Broth microdilution MIC (mg/liter) |
Disk diffusion (mm) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain ID | Patient ID | AZT | TGC | CST | GEN | TOB | AMI | CIP | LEV | TMP/SMX | CHL |
| A | 1 | 256 | 4 | 4 | 64 | >64 | 2 | 16 | 8 | 6 | 16 |
| B | 2 | 256 | 4 | 4 | 64 | >64 | 4 | 16 | 6 | 6 | 15 |
| C | 3 | 128 | 4 | 4 | 64 | >64 | 4 | 8 | 8 | 6 | 20 |
| D | 2 | ≥256 | 4 | 2 | 64 | >64 | 8 | 8 | 9 | 6 | 21 |
| E | 4 | 256 | 2 | 4 | 32 | 16 | 2 | 0.5 | 23 | 6 | NA |
| F | 5 | 64 | 0.5 | 2 | 1 | 1 | 2 | 1 | 23 | 30 | 25 |
| G | 6 | 64 | 0.5 | 2 | 0.5 | 0.5 | 1 | 1 | 19 | 26 | 18 |
| H | 7 | ≥256 | 1 | 2 | 0.25 | 0.5 | 1 | 2 | 21 | 27 | 22 |
| I | 7 | 128 | 0.5 | 2 | 0.5 | 1 | 2 | 1 | 24 | 34 | 25 |
| J | 8 | 64 | 0.5 | 2 | 0.5 | 1 | 2 | 1 | 23 | 29 | 25 |
| K | 9 | 256 | 0.5 | 2 | 0.5 | 0.5 | 2 | 0.5 | 21 | 28 | 24 |
| L | 10 | 256 | 1 | 2 | 4 | 4 | 2 | 1 | 22 | 29 | NA |
| M | 11 | 64 | 0.5 | 2 | 0.5 | 0.5 | 1 | 1 | 22 | 15 | NA |
| N | 12 | 64 | 0.5 | 2 | 0.5 | 1 | 2 | 1 | 23 | 30 | 26 |
| O | 13 | ≥256 | 2 | 2 | 32 | 32 | 2 | 64 | 6 | 6 | NA |
| P | 13 | ≥256 | 2 | 2 | 32 | 32 | 2 | >64 | 6 | 6 | NA |
| Q | 13 | ≥256 | 4 | 2 | 32 | 32 | 2 | 4 | 15 | 6 | NA |
| R | 13 | ≥256 | 2 | 2 | 32 | 32 | 1 | 64 | 6 | 6 | NA |
| S | 13 | ≥256 | 2 | 2 | 32 | 32 | 2 | 64 | 6 | 6 | NA |
| T | 14 | ≥256 | 0.5 | 1 | 0.5 | 0.5 | 1 | 1 | 22 | 27 | NA |
| U | 14 | ≥256 | 0.5 | 2 | 0.5 | 0.5 | 1 | 1 | 22 | 27 | NA |
| V | 15 | ≥256 | 8 | 2 | 16 | 8 | 1 | 4 | 15 | 6 | NA |
| W | 15 | ≥256 | 4 | 4 | 8 | 16 | 0.5 | 2 | 14 | 6 | NA |
| X | 10 | ≥256 | 2 | 4 | 16 | 16 | 1 | 1 | 20 | 6 | 21 |
| Y | 10 | ≥256 | 2 | 2 | 16 | 16 | 1 | 0.5 | 25 | 6 | 22 |
| Z | 16 | 256 | 1 | 2 | 16 | 16 | 2 | 0.25 | 25 | 6 | NA |
AZT, aztreonam; TGC, tigecycline; CST, colistin; GEN, gentamicin; TOB, tobramycin; AMI, amikacin; CIP, ciprofloxacin; LEV, levofloxacin; TMP/SMX, trimethoprim-sulfamethoxazole; CHL, chloramphenicol; NA, not available.
To our knowledge, this study is unique with regard to the breadth and depth in the reporting of an outbreak of KPC-producing E. cloacae. The various analyses performed on this cluster of E. cloacae strains revealed a highly conserved Tn4401 transposon expressing the blaKPC-3 gene. As opposed to traditional outbreak investigations that reveal one or a few strains spreading among individuals, our findings in this outbreak instead suggest that multiple modes of spread were responsible for the dissemination of these strains. In some cases, a highly promiscuous plasmid carrying the Tn4401 transposon disseminated among polyclonal E. cloacae strains (Inc type P,L/M among strains V to Z). However, the multiple plasmid Inc types observed harboring KPC-3 also indicate a high mobility of Tn4401. Indeed, in some cases a single patient could harbor at the same time different E. cloacae strains with different plasmids (strains T and U of patient 14); in another instance, a single patient was found to have distinct E. cloacae strains with different plasmid Inc types over a period of a few months (strains L, X, and Y of patient 10). The variability of strains and plasmids observed could also be explained by multiple-point source outbreaks occurring at the same hospital during the time frame under study. A more detailed epidemiologic investigation and the use of whole-genome sequencing may provide answers to these questions.
Outbreaks of KPC-producing organisms are being increasingly reported and represent a challenge to infection control practitioners as well as to clinicians. The findings from our study suggest that sophisticated methods are increasingly required to develop a thorough understanding of the spread of KPC-producing organisms. Whereas traditional tools such as PFGE initially pointed to a group of polyclonal strains of phenotypically similar bacteria expressing carbapenem resistance, additional molecular methods have revealed that the plasmid or the transposon was the entity spreading among patients and not the bacteria themselves. Rather than investigating suspected outbreaks of KPC-producing organisms by focusing on clonal strains, infection control practitioners, clinicians, and medical microbiologists should supplement the traditional techniques of phenotypic typing and PFGE with the necessary molecular tools to focus on the genetic structure of the KPC-bearing transposon and plasmid in order to better identify and follow the spread of these resistant organisms.
Footnotes
Published ahead of print 1 May 2013
REFERENCES
- 1. Zhanel GG, Adam HJ, Low DE, Blondeau J, DeCorby M, Karlowsky JA, Weshnoweski B, Vashisht R, Wierzbowski A, Hoban DJ. 2011. Antimicrobial susceptibility of 15,644 pathogens from Canadian hospitals: results of the CANWARD 2007-2009 study. Diagn. Microbiol. Infect. Dis. 69:291–306 [DOI] [PubMed] [Google Scholar]
- 2. Bratu S, Landman D, Alam M, Tolentino E, Quale J. 2005. Detection of KPC carbapenem-hydrolyzing enzymes in Enterobacter spp. from Brooklyn, New York. Antimicrob. Agents Chemother. 49:776–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Naas T, Cuzon G, Villegas MV, Lartigue MF, Quinn JP, Nordmann P. 2008. Genetic structures at the origin of acquisition of the β-lactamase blaKPC gene. Antimicrob. Agents Chemother. 52:1257–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Leung V, Lévesque S, Domingo MC, Bourgault AM, Miller M. 2011. First Canadian Hospital Outbreak of KPC-producing Klebsiella pneumoniae, abstr K-824. Abstr. 51st Intersci. Conf. Antimicrob. Agents Chemother American Society for Microbiology, Washington, DC [Google Scholar]
- 5. Clinical and Laboratory Standards Institute 2012. Performance standards for antimicrobial susceptibility testing; 22nd informational supplement. Document M100-S22. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 6. Farmer JJ, III, Boatwright KD, Janda JM. Enterobacteriaceae: introduction and identification, p 649–669 In Murray PR, Baron EJ, Jorgensen JH, Landry ML, Pfaller MA. (ed), Manual of clinical microbiology, 9th ed American Society for Microbiology, Washington, DC [Google Scholar]
- 7. Queenan AM, Bush K. 2007. Carbapenemases: the versatile β-lactamases. Clin. Microbiol. Rev. 20:440–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Leung V, Loo VG, Frenette C, Domingo Bourgault M-C, Mulvey A-MMR, Robson HR. 2012. First Canadian outbreak of Enterobacteriaceae-expressing Klebsiella pneumoniae carbapenemase type 3. Can. J. Infect. Dis. Med. Microbiol. 23:117–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mathers AJ, Cox HL, Kitchel B, Bonatti H, Brassinga AKC, Carroll J, Scheld WM, Hazen KC, Sifri CD. 2011. Molecular dissection of an outbreak of carbapenem-resistant Enterobacteriaceae reveals intergenus KPC carbapenemase transmission through a promiscuous plasmid. mBio 2:e00204-11. 10.1128/mBio.00204-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mataseje LF, Boyd DA, Willey BM, Prayitno N, Kreiswith N, Gelosia A, Poutanen SM, Low DE, Jenkins SG, Katz K, Mulvey MR. 2011. Plasmid comparison and molecular analysis of Klebsiella pneumoniae harbouring blaKPC from New York City and Toronto. J. Antimicrob. Chemother. 66:1273–1277 [DOI] [PubMed] [Google Scholar]
- 11. Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219–228 [DOI] [PubMed] [Google Scholar]


