Abstract
The human cannabinoid 1 receptor (hCB1), a ubiquitous G protein-coupled receptor (GPCR), transmits cannabinergic signals that participate in diverse (patho)physiological processes. Pharmacotherapeutic hCB1 targeting is considered a tractable approach for treating such prevalent diseases as obesity, mood disorders, and drug addiction. The hydrophobic nature of the transmembrane helices of hCB1 presents a formidable difficulty to its direct structural analysis. Comprehensive experimental characterization of functional hCB1 by mass spectrometry (MS) is essential to the targeting of affinity probes that can be used to define directly hCB1 binding domains using a ligand-assisted experimental approach. Such information would greatly facilitate the rational design of hCB1-selective agonists/antagonists with therapeutic potential. We report the first high-coverage MS analysis of the primary sequence of the functional hCB1 receptor, one of the few such comprehensive MS-based analyses of any GPCR. Recombinant C-terminal hexa-histidine-tagged hCB1 (His6-hCB1) was expressed in cultured insect (Spodoptera frugiperda) cells, solubilized by a procedure devised to enhance receptor purity following metal-affinity chromatography, desalted by buffer exchange, and digested in solution with (chymo)-trypsin. “Bottom-up” nanoLC-MS/MS of the (chymo)tryptic digests afforded a degree of overall hCB1 coverage (>94%) thus far reported for only two other GPCRs. This MS-compatible procedure devised for His6-hCB1 sample preparation, incorporating in-solution (chymo)trypsin digestion in the presence of a low concentration of CYMAL-5 detergent, may be applicable to the MS-based proteomic characterization of other GPCRs. This work should help enable future ligand-assisted structural characterization of hCB1 binding motifs at the amino-acid level using rationally designed and targeted covalent cannabinergic probes.
Keywords: transmembrane protein, G protein-coupled receptor, protein expression, affinity purification, endocannabinoid signaling system, proteomic analysis, signal transduction, ligand-assisted protein analysis, ligand targeting, drug design, binding motifs, nanoLC-MS/MS
Introduction
As the largest family of membrane proteins, G protein-coupled receptors (GPCRs) bind diffusible signaling molecules presented at the cell surface and G-protein subunits at their intracellular, cytoplasmic aspect.1 Their pervasive role as signal transducers that mediate a wide variety of cellular responses and their status as the single largest class of drug targets have made GPCRs the focus of intense research and translational interest.1,2 Information on the biochemistry, structure, and binding domain of individual GPCRs is avidly sought to inform rational, targeted drug design.2 Traditional GPCRs share the hallmark features of an extracellular amino terminus, an intracellular carboxyl terminus, and an extensive membrane-spanning region containing seven transmembrane α-helices (TMHs) connected by intra- and extracellular loops.1,3 The nature of GPCRs as heptahelical, hydrophobic, integral-membrane proteins has made it challenging to express and purify a sufficient quantity of most any GPCR in intact, functionally active form for direct experimental definition of its structural properties and binding interactions.4
A component of the ubiquitous endogenous cannabinoid (endocannabinoid) signaling system, the cannabinoid 1 receptor (CB1) is a prominent class-A GPCR localized primarily with-in the vertebrate central nervous system (CNS) as the most abundant GPCR in mammalian brain.5 Central CB1 activation by the phytocannabinoid Δ9-tetrahydrocannabinol mediates most of the psychobehavioral effects associated with cannabis use.6 In the CNS, endocannabinoid signaling through CB1 helps control human motor, cognitive, emotional, and sensory functions and influences pain perception, hormonal activity, thermo-regulation, and cardiovascular status.7 In peripheral organs such as liver and pancreas, CB1 helps regulate energy homeostasis and lipid metabolism/deposition.8 CB1 involvement in diverse physiological functions and the limitations of the first-in-class CB1 antagonist/inverse agonist pharmaceutical (rimonabant) have made CB1 a prominent drug-discovery target.9 Rational design of agonist and antagonist ligands that safely and effectively modulate CB1 transmission for therapeutic benefit against over-weight/obesity and allied cardiometabolic risk factors, drug dependence, pain, and neurodegeneration would be greatly facilitated by detailed information on the ligand-binding domains of functional human CB1 (hCB1).5,7,9,10
An experimentally derived, three-dimensional structure of CB1 from any species is currently unavailable, in lieu of which provisional hCB1 computational models have been constructed based upon indirect data from mutational studies and bovine rhodopsin crystal structures.11–16 The applicability of rhodopsin X-ray structures to hCB1 homology modeling and ligand design remains equivocal due to the many biochemical and pharmacological differences betwen these two GPCRs, not the least of which is rhodopsin’s light- (vs ligand-) activated nature.16,17 Despite the attendant uncertainties, hCB1 homology models11–15 have served as surrogates for inferring putative small-molecule chemotypes and receptor amino acids critical to the binding of prospective hCB1-targeted drugs. Crystal structures of only a very few GPCRs (i.e., the human β2 adrenergic,18,19 A2A adenosine,20 and opsin21,22 receptors and the turkey β1 adrenergic receptor23) have been solved since the first X-ray diffraction structure of a class-A GPCR (bovine rhodopsin) was reported a decade ago.16 These advances notwithstanding, the overall similarity among class-A GPCRs is low; for example, there is 23, 26, and 27% identity between the TMHs of hCB1 and bovine rhodopsin, the human β2 adrenergic receptor, and the A2A adenosine receptor, respectively.3,24 Therefore, to reconcile homology-generated hCB1 computational models with the native organization of hCB1 ligand-binding site(s) and motifs, experimental hCB1 characterization is required. Such considerations invite new experimental approaches for direct interrogation of hCB1 ligand-binding domains at the molecular (i.e., amino-acid) level. This laboratory has formulated a ligand-assisted approach involving the design of novel high-affinity probes that bind covalently to amino acid residues at (or very near) their interaction site(s) within a functional protein target. Incorporation of chemically reactive, targeted cannabinoid ligands into this paradigm has allowed direct identification of amino acid residues critical to the interaction of small molecules with cannabinoid-system enzymes25 and GPCRs26,27 as potential drugs that modulate cannabinergic signal transduction for therapeutic gain. The resulting data contribute to the experimental characterization of small-molecule interaction domains within the target proteins.
To apply a ligand-assisted approach for interrogating directly the structure of hCB1 ligand-binding domains, sufficient amounts of functional, purified hCB1 are required. We previously expressed functional, C-terminal hexa-histidine (His6)-tagged hCB1 (His6-hCB1) and N-terminal FLAG-tagged/C-terminal His6-tagged hCB1 (FLAG-His6-hCB1) in insect (Spodoptera frugiperda) cells using a baculovirus system.28 This system circumvented some formidable technological road-blocks encountered in prior attempts at hCB1 expression17,29–31 and allowed satisfactory isolation of recombinant hCB1 proteins by immobilized metal-affinity chromatography (IMAC). However, matrix-assisted laser desorption/ionization time-off-light mass spectrometric (MALDI-TOF MS) analysis of in-gel digested His6-hCB1 samples afforded limited sequence coverage of only 46% of the receptor’s TMHs (most thoroughly, TMHs 3 and 7 with little coverage of the other five TMHs).28 This limited MS coverage represents a serious impediment to characterizing hCB1 binding domains, for critical hCB1 ligand-recognition sites and responses to both activating ligands and the surrounding membrane matrix have been associated with this receptor’s (and, more generally, GPCR) TMHs.10–12,32 Therefore, we have devised and validated an experimental protocol that enabled purification of recombinant hCB1 and generation of proteolytic peptides for “bottom-up” MS-based proteomic analysis. Virtually the entire hCB1 amino-acid sequence, including all seven of its TMHs, was observed directly with nanocapillary liquid chromatography-tandem mass spectrometry (nanoLC-MS/MS). The >94% sequence coverage attained in this first comprehensive hCB1 MS analysis elevates hCB1 into the select company of only five other GPCRs (out of the 5000 unique GPCRs estimated33 to be present in 13 species whose genomes are fully sequenced) with ≥80% overall reported MS sequence coverage.34–38 These data should help facilitate future characterization of hCB1 binding motifs and conformational features, inform the design of covalent affinity probes useful for identifying amino acid residues critical to CB1 ligand binding, and promote the synthesis of small molecules as hCB1-directed therapeutics.
Materials and Methods
Reagents and Materials
Unless otherwise specified, standard laboratory chemicals and buffers were purchased from Sigma Chemical Co. (St. Louis, MO) and Fisher (Pittsburgh, PA). BioSafe Coomassie Staining Protocol, SDS-PAGE gels, and related electrophoresis buffers and reagents were from Bio-Rad (Hercules, CA). n-Dodecyl-β-d-maltoside (DM) and 5-cyclohexyl-1-pentyl-β-d-maltoside (CYMAL-5) were purchased from Anatrace (Maumee, OH). Trypsin Gold, MS grade, (>15 000 Units/mg protein) was from Promega (Madison, WI), and bovine pancreatic α-chymotrypsin (C3142) (>40 Units/mg protein) was from Sigma.
Expression of Recombinant hCB1 Proteins and Membrane Isolation
Functional His6-hCB1 and FLAG-His6-hCB1 were expressed in suspension-cultured fallworm (Spodoptera frugiperda) Sf21 cells using the optimized baculovirus expression system detailed.28 Membrane pellets from Sf21 cells expressing His6-hCB1 or FLAG-His6-hCB1 were prepared from whole-cell lysates and either processed immediately for either saturation-binding assay or receptor isolation or were snap-frozen in liquid nitrogen and stored at −80 °C.28
Saturation-Binding Assay
Saturation-binding assays with [3H]CP-55,940 as radioligand were performed in a 96-well plate format and analyzed as previously detailed.28
His6-hCB1 Solubilization and Purification
A two-step procedure was devised that allows selective His6-hCB1 extraction from Sf21 cell membranes. The lysate-derived membrane pellet (above) was first resuspended in 50 mM sodium phosphate buffer, pH 8.0, containing 300 mM NaCl, 1% DM, and 0.5% Sigma protease inhibitor cocktail (20 mg membrane protein/1.0 mL buffer) with manual homogenization in a Dounce-type homogenizer. Membranes were solubilized by gently mixing the homogenate in a rotator for 2 h at 4 °C. Following centrifugation of the solubilized membrane preparation at 27 000× g for 30 min at 4 °C, the pellet was recovered and resuspended in denaturing buffer (50 mM sodium phos-phate buffer, pH 8.0, containing 300 mM NaCl, 0.5% DM, 0.5% Sigma protease inhibitor cocktail, and 8 M urea) (5 mg membrane protein/1.0 mL buffer) and manually homogenized as before. Protein extraction was completed by gently mixing the homogenate in a rotator for 1 h at room temperature. Insoluble material was precipitated by centrifugation at 27 000× g for 30 min at room temperature. The supernatant was collected for His6-hCB1 purification according to the procedure detailed28 using cobalt-based IMAC with BD Talon resin (Clontech Laboratories, Mountainview, CA). The IMAC column eluate fractions were collected and monitored by SDS/PAGE.
SDS-PAGE and Immunoblotting
Samples of IMAC column eluate were incubated in Laemmli buffer containing 5%β-mer-captoethanol at room temperature for 30 min and resolved on 10% Tris-HCl gels (running buffer: 25 mM Tris, pH 8.3, containing 250 mM glycine and 0.1% SDS). Protein bands were either visualized with Commassie blue or transferred to polyvinylidene difluoride membranes using semidry blotting at room temperature (10 V for 10 min followed by 15 V for 15 min) for Western analysis. The protein blots were visualized by enzyme immunodetection with a 1:10 000 dilution of anti-5His-horseradish peroxidase-antibody conjugate (Qiagen, Valencia, CA) following the procedures outlined in the ECL Western blotting analysis system (GE Healthcare, Piscataway, NJ). In all cases, membrane preparations from Sf21 cells infected with nonrecombinant wild-type virus were used as negative controls.
In-Solution Digestion
Pooled IMAC fractions containing purified His6-hCB1 were reduced with dithiothreitol (17 mM) at 56 °C for 1 h, alkylated with iodoacetamide (55 mM) at room temperature for 1 h in the dark, desalted on Micro Bio Spin P6 columns with exchange buffer (25 mM NH4HCO3, 0.05% CYMAL-5), and digested overnight with either trypsin (5 ng/µL sample) at 37 °C or α-chymotrypsin (10 ng/µL sample) at 30 °C.35
NanoLC-MS/MS
(Chymo)trypsin digests of purified His6-hCB1 were resolved and subjected to peptide identification and sequencing by nanoLC-MS/MS using a hybrid quadrupole-linear ion trap mass spectrometer (4000 Q-Trap; Applied Biosystems/MDS Sciex, Framingham, MA). Mobile phase A consisted of 98% water, 2% acetonitrile, and 0.1% formic acid, by volume. Mobile phase B consisted of 98% acetonitrile, 2% water, and 0.1% formic acid, by volume. For concentrating and desalting, 1.5-µL sample volumes at 10 µL/min were injected onto a trap column (150-µm × 4-mm manually packed with Oasis HLB, 15 µm) (Waters Corp. Milford, MA) over 5 min in mobile phase A using a Tempo nanomultidimentional-LC system (Applied Biosystems). Gradient separation was carried out on a 75-µm × 100-mm reverse-phase C4 nano-LC column (5-µm, 120-Å, manually packed) (YMC Europe GmbH, Dinslaken, Germany). A linear gradient from 20% to 70% mobile phase B over 50 min followed by a 10-min isocratic wash with 90% B was employed at a constant flow rate of 350 nL/min. Eluted peaks were introduced into the mass spectrometer via positive-mode nanospray ionization. Instrument conditions were: source temp, 180 °C; curtain gas, 15; nebulizer gas, 8; source voltage, 2200 V; declustering potential, 80 V. Peptides were analyzed using information-dependent acquisition, scanning over a mass range of 400 to 1600 m/z. Specifically, the charge states of the three most intense ions in an enhanced, multiply charged survey scan were determined. Those ions that met the selection criteria were fragmented and analyzed in MS/MS mode. Rolling collision energies were applied based upon the detected precursor’s m/z. Dynamic fill time of the linear trap was used to eliminate space-charging effects. Acquired spectra were searched with Protein Pilot software (Applied Biosystems) using a single-entry hCB1 database (accession P21554) to identify and sequence hCB1 peptides. The Protein Pilot Paragon search algorithm was set to perform a thorough identification, including over 90 biological modifications at a detected protein threshold of 1.3 (95%) for all samples.
Results and Discussion
His6-hCB1 Expression, Purification, and Sample Preparation for nanoLC-MS/MS
We have expressed hCB1 (as His6-hCB1 and FLAG-His6-hCB1) in cultured Sf21 cells at unprecedented yields using a baculovirus system.28 Moreover, Sf21 cell membranes containing His6-hCB1 or FLAG-His6-hCB1 evidence specific, high-affinity binding of the CB1 full agonist [3H]CP-55,940 and antagonist [3H]SR141716A, and forskolin-stimulated cyclic adenosine-3′,5′-monophosphate (cAMP) production is inhibited by CP-55,940 in Sf21 cells expressing His6-hCB1 (Table 1 and data not shown).28 Although these results demonstrate the functionality of the recombinant hCB1 proteins, several major obstacles arose during previous attempts at recombinant hCB1 “bottom-up” MS analysis.28 SDS/PAGE revealed that the His6-hCB1 expressed in insect cells using a baculovirus system was a mixture of monomers, dimers, and oligomers. The His6-hCB1 preparation also contained N- and C-terminal truncated receptors, necessitating an additional SDS/PAGE separation prior to (chymo)trypsin digestion. MALDI-TOF analysis of in-gel (chymo)trypsin digestions provided ~80% coverage of hCB1 hydrophilic regions, but very limited coverage (<50%) of its TMHs.28 After in-gel digestion, the highly hydrophobic TMHs-based peptides appear extremely resistant to extraction from the polyacrilamide gel matrix. Indeed, following similar sample preparation procedures, only the most hydrophilic TMHs (i.e., 3 and 7) were entirely identified by MALDI-TOF analysis of an in-gel digest of hCB1 expressed in insect28 or yeast31 cells. These technological problems surrounding in-gel digestion compromised the overall His6-hCB1 MS signal and obviated detection of low-abundance, very hydrophobic TMH peptides, thereby severely limiting the MALDI-TOF MS-based coverage of this physiologically and therapeutically important GPCR.28
Table 1.
Saturation-Binding Parameters of Expressed hCB1sa
| construct | Kd (nM) | Bmax (pmol/mg) |
|---|---|---|
| His6-hCB1 | 3.3 ± 0.3 | 5.7 ± 0.2 |
| FLAG-His6-hCB1 | 5.1 ± 0.6 | 0.6 ± 0.1 |
Binding affinities (as Kd) and receptor densities (as Bmax) were determined in saturation-binding assays with [3H]CP-55,940 and membrane preparations from Sf21 cells expressing a tagged hCB1, as detailed.28 Data are means ±range from the average of two independent determinations, each performed in triplicate.
Since extraction and purification of expressed hCB1 from other membrane proteins is a sine qua non for its MS-based proteomic characterization, the current work initially addressed aspects of sample preparation critical to obtaining recombinant hCB1 in suitable quantities for and under conditions compatible with MS analysis. Our success in purifying FLAG-His6-human cannabinoid 2 receptor (hCB2) from Sf21 cells using detergent solubilization and anti-FLAG M2 affinity chromatography28 prompted us to apply an analogous procedure to FLAG-His6-hCB1. However, three findings undermined this approach (data not shown): (a) The expression of recombinant FLAG-His6-hCB1 was consistently lower than that of His6-hCB1.28 (b) FLAG-His6-hCB1 proved extraordinarily resistant to solubilization by a variety of detergents, including those effective for FLAG-His6-hCB2.35 (c) Membrane treatment under denaturing conditions with urea followed by dilution in an attempt to make the final urea concentration compatible with subsequent analytical procedures generated FLAG-His6-hCB1 that failed to bind to anti-FLAG affinity resin. Plausible reasons for the binding incompetency include adverse effects of residual urea on the immobilized anti-FLAG antibody and of the denaturing conditions on FLAG-tag accessibility. Collectively, these data rendered FLAG-His6-hCB1 refractory to purification by anti-FLAG M2 affinity chromatography.
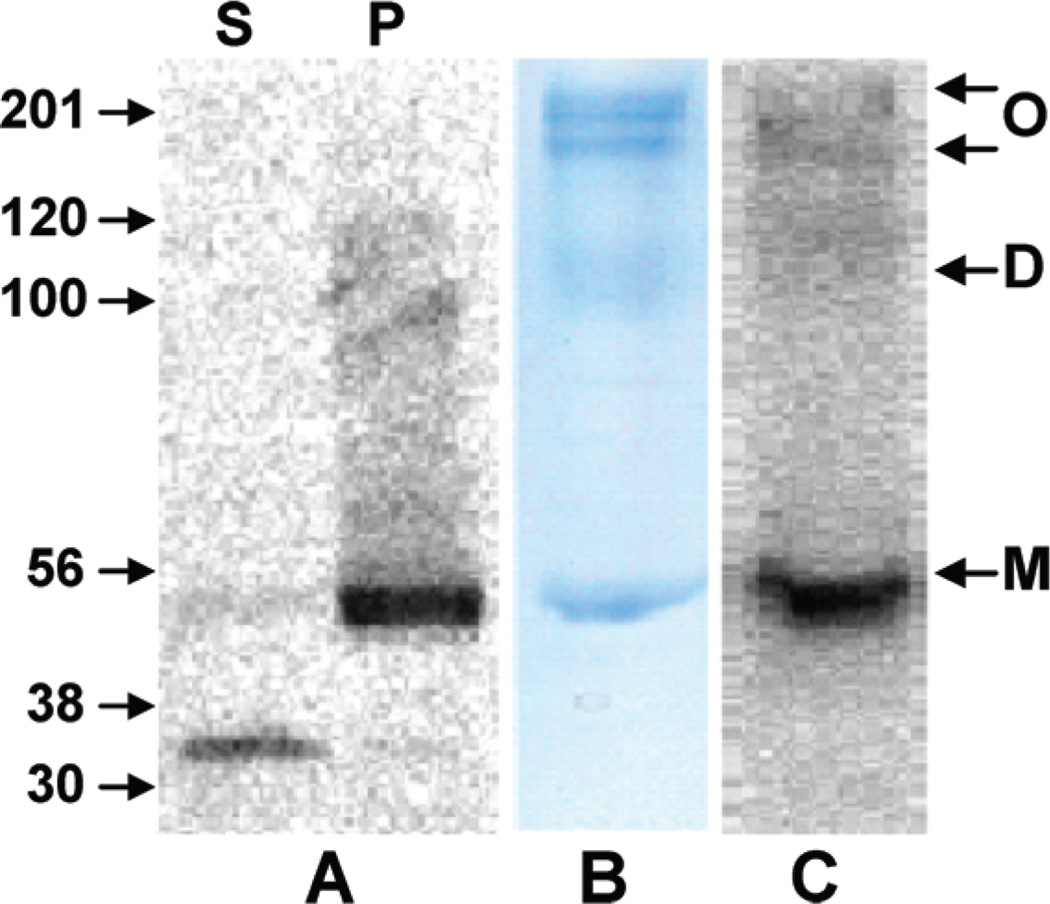
We therefore focused on developing an alternative sample-preparation protocol capable of yielding purified His6-hCB1 in sufficient quantity for subsequent proteolytic processing and high-coverage MS-based proteomic analysis. A two-step solubilization procedure was devised and validated (Materials and Methods) that incorporated an initial membrane extraction under native conditions with buffered DM. The extracted membrane pellet consisted predominantly of His6-hCB1 that proved amenable to solubilization under denaturing conditions with DM-urea and IMAC final purification (Figure 1). The His6-hCB1 protein was observed on SDS/PAGE as a ~54 KDa band (calculated MW, 53 681 Da).
Figure 1.
(A) Western-blot analysis of supernatant (S) and pellet (P) fractions obtained from native detergent solubilization of Sf21 cell membranes containing functional His6-hCB1. (B) Coomassiestained gel and (C) Western blot of IMAC-purified His6-hCB1. The arrows at the right indicate His6-hCB1 monomer (M), dimer (D), and oligomer (O) forms. Immunodetection was performed with an anti-5His-horseradish peroxidase-antibody conjugate.
Due to the high quality of the purified His6-hCB1 protein using the two-step solubilization procedure and IMAC, SDS-PAGE separation was not required. This result allowed us to obviate the difficulties associated with hCB1 hydrophobic peptide extraction after in-gel digestion28 by utilizing in-solution (chymo)trypsin digestion.35 Accordingly, the purified His6-hCB1 was sequentially reduced, alkylated, desalted in the presence of a low concentration of CYMAL-5, and digested in solution with (chymo)trypsin. The saccharide-based nonionic detergent, CYMAL-5, was selected because of its proven ability to solubilize membrane proteins at concentrations not detrimental to electrospray-ionization MS.39
MS Characterization of Purified His6-hCB1
NanoLC-MS/MS is a well-established, sensitive technique for peptide separation and analysis.40 His6-hCB1 (chymo)tryptic peptides were resolved using nanoLC and characterized using a 4000 Q-Trap hybrid triple quadrupole/linear ion trap mass spectrometer. Several advanced capabilities of this instrument facilitated peptide analysis. Specifically, the enhanced multiple-charge scan mode, in concert with an information-dependent acquisition workflow, allowed selective isolation of multiply charged peptides for tandem MS identification over singly charged chemical background ions, including any CYMAL-5 detergent aggregates that may have formed.
The amino acid sequence of hCB1 (as His6-hCB1) is presented in Figure 2. Approximately half of the receptor’s 478 amino acids are within its TMHs, and only TMH3 contains a discrete trypsin-cleavage site. Trypsin digestion of His6-hCB1 produced, on average, 33-mers for each of the seven TMH domains. The average tryptic peptide length for the extra-and intracellular loops and the N- and C-terminal chains was approximately 8 amino acids. MS analysis of large hydrophobic tryptic peptides such as those from the hCB1 TMHs can be complicated by their irreversible binding to chromatographic media and the formation of multiply charged ions outside the instrument’s mass range.41 To circumvent this difficulty, chymotrypsin was also used to prepare His6-hCB1digests for nanoLC-MS/MS in view of its distinct cleavage specificity for aromatic and hydrophobic amino acid residues relative to that of trypsin.
Figure 2.
Experimentally determined amino acid sequence of His6-hCB1. The hydrophobic, core peptides in each of the seven TMH domains are sequentially labeled and bolded.
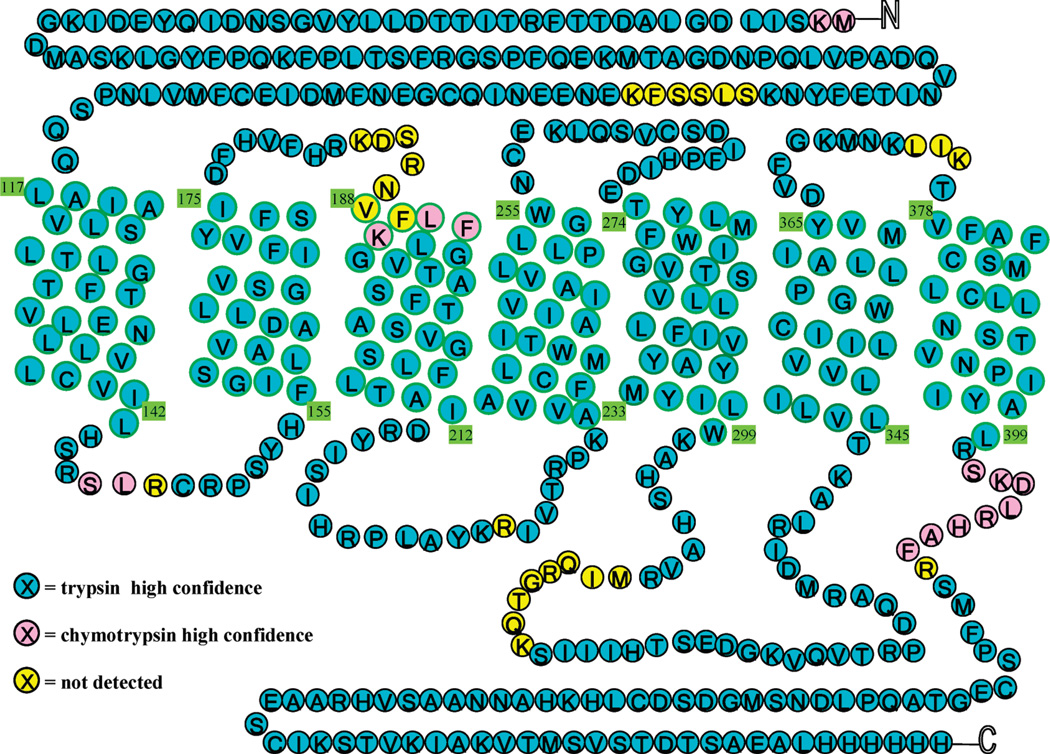
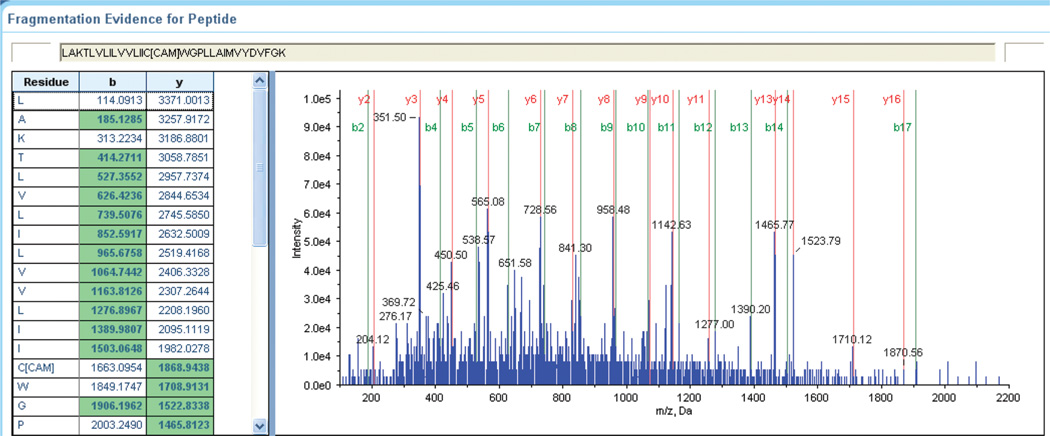
A two-dimensional His6-hCB1 serpentine diagram is presented in Figure 3. The vast majority (91.4%) of hCB1 could be identified from the trypsin digest alone, validating the adequacy of our sample-preparation and analytical conditions for His6-hCB1 detection and sequencing over all seven of its TMH domains. Exemplary tandem MS data used to sequence individual peptides are given in Figure 4. The standard y-and b-ion series shown represents the complete sequencing of the peptide containing hCB1 TMH6 (LAKTLVLILVVLIIC*WGPLLAIMVYDVFGK, where C* designates a cysteine that had been carbamidomethylated consequent to experimental iodoacetamide alkylation). These sequence data now allow definitive identification of this TMH6 cysteine residue as the amino acid provisionally implicated in ligand-induced hCB1 activation through functional analysis of hCB1 mutants.11 Figure 3 illustrates that virtually complete sequence coverage of the hCB1 hydrophobic TMH domains was achieved, a noteworthy finding given the structural and functional importance of GPCR (including CB1) TMH domains to receptor activation.1–3,10–12,32 Although there are three putative N-glycosylation sites within the N-terminal portion of hCB1, we did not observe glycosylation of the recombinant receptor. This finding is consonant with the limited capacity of (baculovirus-infected) insect cells for N-glycosylation (especially with respect to the more compositionally complex, terminally sialylated N-glycans of mammalian proteins) and the robust postglycosylation trimming pathways present in insect cells.42,43 The lack of detectable glycosylation of recombinant hCB1, however, did not affect the receptor’s ligand-binding or functionality (Table 1 and data not shown),28 likely because its candidate glycosylation sites are outsideofthefunctionallycriticalTMHandC-terminalregions.11–15,17
Figure 3.
(Chymo)trypsin His6-hCB1 MS coverage. Amino acids are color-coded to represent the enzyme used in their identification: Blue, trypsin; Pink, chymotrypsin. The 27 amino acids in yellow were not identified.
Figure 4.
Tandem MS sequence annotation of the His6-hCB1 TMH6 tryptic peptide LAKTLVLILVVLIIC*WGPLLAIMVYDVFGK (C*, carbamidomethylated cysteine).
Comparison of the theoretical hCB1 tryptic peptides with those identified experimentally by (chymo)trypsin digestion is presented in Table 2. As calculated using the sequenced peptides generated by both proteases, 94.4% of the His6-hCB1 sequence was determined. The remaining 5.6% is accounted for by unidentified small peptides (six amino acids or less). It is likely that these peptides were not detected due to their exclusion by the enhanced multiple-charge scan mode employed, for such small peptide ions typically do not support multiple charges and reside as singly charged species. Since these regions are considered not to be involved in ligand binding by class-A GPCRs such as hCB1,1–3,10–12,32 and in light of the exceptionally high33–38 total MS coverage (94.4%) we attained with our His6-hCB1 sample preparation, we did not pursue their identification. A single-entry database for hCB1 (Swiss-Prot # P21554) was used for simplicity and search efficiency; however, the coverage was identical when the entire Swiss-Prot database was used (data not shown). A high resolution “zoom” scan determined the charge state of selected ions throughout the entire data acquisition to assist in accurate database searching with Protein Pilot software. The charge states of the identified peptides Table 2 ranged from +2 to +6 (Table 2). A search of the entire Swiss-Prot database produced several incomplete matches to CB1 proteins from nonhuman species (data not shown).
Table 2.
Theoretical Tryptic Digestion of His6-hCB1 and Peptides Identified by MSa
| theoretical MW (Da) |
detected MW (Da) |
start | stop | sequence | charge states |
|---|---|---|---|---|---|
| 1410.723 | 1410.707 | 1 | 13 | MKSILDGLADTTF | 2 |
| 1307.672 | 1307.624 | 3 | 14 | SILDGLADTTFR | 3 |
| 2300.142 | 2300.068 | 15 | 34 | TITTDLLYVGSNDIQYEDIK | 3 |
| 2889.396 | 2889.366 | 15 | 40 | TITTDLLYVGSNDIQYEDIKGDMASK | 3 |
| 1699.909 | 1699.814 | 41 | 54 | LGYFPQKFPLTSFR | 2 |
| 2473.279 | 2473.052 | 41 | 61 | LGYFPQKFPLTSFRGSPFQEK | 3 |
| 2564.222 | 2564.12 | 62 | 84 | MTAGDNPQLVPADQVNITEFYNK | 3 |
| 668.361 | 85 | 90 | SLSSFK | ||
| 2258.856 | 2258.926 | 91 | 108 | ENEENIQCGENFMDIECF | 3 |
| 4075.294 | 4076.94 | 109 | 145 | MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSR | 4 |
| 2797.578 | 2797.57 | 123 | 147 | SLTLGTFTVLENLLVLCVILHSRSL | 3 |
| 175.119 | 148 | 148 | R | ||
| 3969.019 | 3969.608 | 149 | 182 | CRPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHR | 5 |
| 147.113 | 183 | 183 | K | ||
| 377.178 | 184 | 186 | DSR | ||
| 767.445 | 187 | 192 | NVFLFK | ||
| 1138.639 | 1138.534 | 190 | 200 | LFKLGGVTASF | 2 |
| 2182.163 | 2181.931 | 193 | 214 | LGGVTASFTASVGSLFLTAIDR | 3 |
| 3523.919 | 3523.529 | 193 | 225 | LGGVTASFTASVGSLFLTAIDRYISIHRPLAYK | 4 |
| 175.119 | 226 | 226 | R | ||
| 3782.085 | 3781.702 | 227 | 259 | IVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEK | 4 |
| 3086.652 | 3083.351 | 233 | 259 | AVVAFCLMWTIAIVIAVLPLLGWNCEK | 3 |
| 5745.927 | 5745.559 | 260 | 307 | LQSVCSDIFPHIDETYLMFWIGVTSVLLLFIVYAYMYILWKAHSHAVR | 5 |
| 547.302 | 308 | 311 | MIQR | ||
| 433.241 | 312 | 315 | GTQK | ||
| 1198.619 | 1198.607 | 316 | 326 | SIIIHTSEDGK | 2 |
| 2349.24 | 2349.193 | 316 | 336 | SIIIHTSEDGKVQVTRPDQAR | 3 |
| 1168.631 | 1168.549 | 327 | 336 | VQVTRPDQAR | 2 |
| 3828.225 | 3827.687 | 337 | 370 | MDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGK | 4 |
| 4259.409 | 4260.01 | 337 | 374 | MDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMNK | 6 |
| 3743.172 | 3742.843 | 341 | 373 | LAKTLVLILVVLIICWGPLLAIMVYDVFGKMNK | 4 |
| 3058.785 | 3058.411 | 344 | 370 | TLVLILVVLIICWGPLLAIMVYDVFGK | 3 |
| 3430.956 | 3431.542 | 344 | 373 | TLVLILVVLIICWGPLLAIMVYDVFGKMNK | 4 |
| 373.281 | 374 | 376 | LIK | ||
| 2802.427 | 2802.443 | 377 | 400 | TVFAFCSMLCLLNSTVNPIIYALR | 3 |
| 1312.736 | 1312.672 | 398 | 408 | ALRSKDLRHAF | 3 |
| 175.119 | 409 | 409 | R | ||
| 2783.13 | 2782.984 | 410 | 434 | SMFPSCEGTAQPLDNSMGDSDCLHK | 3 |
| 1075.527 | 1075.448 | 435 | 444 | HANNAASVHR | 2 |
| 3190.633 | 3192.19 | 444 | 473 | RAAESCIKSTVKIAKVTMSVSTDTSAEALH | 5 |
| 1992.086 | 1992.072 | 444 | 461 | RAAESCIKSTVKIAKVTM | 3 |
| 1216.557 | 1216.48 | 462 | 473 | SVSTDTSAEALH | 2 |
| 2233.008 | 2233.833 | 459 | 478 | VTMSVSTDTSAEALHHHHHH | 4 |
The amino acids or peptides listed in bold were not found. Peptides in italics are from the chymotrypsin digest. Total coverage with both digestion methods was 94.4%.
Conclusion
Notwithstanding the importance of GPCRs as signal-transduction elements and pharmacotherapeutic targets for various diseases, their hydrophobic, transmembrane, heptahelical character complicates direct GPCR analysis. Consequently, experimental data characterizing the structures of even prevalent GPCRs such as hCB1 are conspicuously lacking.2,3,13,14,24 Formidable challenges surround the isolation and direct structure analysis of functionally active GPCRs. Agents used to solubilize and extract recombinant GPCRs expressed at the plasma membrane, for instance, are often incompatible with modern MS-based proteomic analyses.34,39–41 The work presented here addresses and solves such problems for hCB1, the most abundant GPCR in the CNS,5–7 by devising a method for solubilizing and purifying functional recombinant His6-hCB1 in quantities sufficient for direct experimental characterization through “bottom-up” MS-based proteomics. Three key methodological features enabled us to obtain high sequence coverage of His6-hCB1: (a) membrane solubilization under native conditions that enriched the subsequent protein extract for His6-hCB1 analyte; (b) in-solution proteolytic digestion of purified and desalted His6-hCB1 that overcame the poor extraction efficiency of hydrophobic peptides generated from in-gel digestion; and (c) nanoLC separation with enhanced MS/MS detection of analyte ions. Consequently, we were able to achieve high (>94%) overall sequence coverage of His6-hCB1 primary structure and virtually complete coverage of all of its seven, functionally critical10–12 TMH domains. Most (chymo)-tryptic hCB1 peptide fragments were identified as multiply charged species, and their sequences were confirmed with high confidence. The experimental MS characterization of functional hCB1 detailed should help inform future work involving this prominent GPCR, for example, probing higher-order hCB1 architectural detail, defining conformational correlates between hCB1 binding interactions and signal transmission, and designing site-directed ligands as either affinity probes or potential therapeutics. The MS primary structures of merely ~0.1% of the GPCRs estimated33 to occur among the fully sequenced genomes of thirteen species have been determined experimentally at ≥80% sequence coverage, and only two (rhodopsin and hCB2) at virtually complete coverage.34–38 It is suggested, therefore, that at least some features of our experimental approach may be profitably applied to advance the MS-based proteomic characterization of other GPCRs.
Acknowledgment
This work has been supported by National Institutes of Health grants from the National Institute on Drug Abuse: DA09158, DA00493, DA03801, DA07215, and DA07312 (A.M.).
Abbreviations
- cAMP
cyclic adenosine 3′,5′-monophosphate
- CB1
cannabinoid 1
- CB2
cannabinoid 2
- hCB1
human cannabinoid 1 receptor
- hCB2
human cannabinoid 2 receptor
- CNS
central nervous system
- CYMAL-5
5-cyclohexyl-1-pentyl-β-d-maltoside
- DM
n-dodecyl-β-d-maltoside
- FLAG
DYKDDDK peptide tag
- FLAG-His6-hCB1
N-terminal FLAG/C-terminal hexa-histidine-tagged hCB1 receptor
- FLAG-His6-hCB2
N-terminal FLAG-tagged/C-terminal hexa-histidine-tagged hCB2 receptor
- GPCR
G protein-coupled receptor
- His6
hexa-histidine peptide tag
- His6-hCB1
C-terminal hexa-histidine-tagged hCB1 receptor
- IMAC
immobilized metal affinity chromatography
- LC
liquid chromatography
- MALDI-TOF MS
matrix-assisted laser desorption/ionization time-of-flight mass spectrometry
- MS
mass spectrometry
- m/z
mass to charge ratio
- TMH
transmembrane helix.
References
- 1.Rosenbaum DM, Rasmussen SGF, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Langerström MC, Schiöth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat. Rev. Drug Discovery. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 3.Mustafi D, Palczewski K. Topology of class A G protein-coupled receptors: insights gained from crystal structures of rhodopsins, adrenergic and adenosine receptors. Mol. Pharmacol. 2009;75:1–12. doi: 10.1124/mol.108.051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCusker EC, Bane SE, O’Malley MA, Robinson AS. Heterologous GPCR expression: a bottleneck to obtaining crystal structures. Biotechnol. Prog. 2007;23:540–547. doi: 10.1021/bp060349b. [DOI] [PubMed] [Google Scholar]
- 5.Mackie K. Cannabinoid receptors: where they are and what they do. J. Neuroendocrinol. 2008;20(suppl. 1):10–14. doi: 10.1111/j.1365-2826.2008.01671.x. [DOI] [PubMed] [Google Scholar]
- 6.Cooper ZD, Haney M. Actions of Δ9-tetrahydrocannabinol in cannabis: relation to use, abuse, dependence. Int. Rev. Psychiatry. 2009;21:104–112. doi: 10.1080/09540260902782752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Marzo V. The endocannabinoid system: its general strategy of action, tools for its pharmacological manipulation and potential therapeutic exploitation. Pharmacol. Res. 2009;60:77–84. doi: 10.1016/j.phrs.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 8.Vettor R, Pagano C. The role of the endocannabinoid system in lipogenesis and fatty acid metabolism. Best Pract. Res. Clin. Endocrinol. Metab. 2009;23:51–63. doi: 10.1016/j.beem.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 9.Janero DR, Makriyannis A. Cannabinoid receptor antagonists: pharmacological opportunities, clinical experience, and translational prognosis. Expert Opin. Emerging Drugs. 2009;14:43–65. doi: 10.1517/14728210902736568. [DOI] [PubMed] [Google Scholar]
- 10.Tiburu EK, Gulla SV, Tiburu M, Janero DR, Budil DE, Makriyannis A. Dynamic conformational responses of a human cannabinoid receptor-1 helix domain to its membrane environment. Biochemistry. 2009;48:4895–4904. doi: 10.1021/bi802235w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Picone RP, Khanolkar AD, Xu W, Ayotte LA, Thakur GA, Hurst DP, Abood ME, Reggio PH, Fournier DJ, Makriyannis A. (-)-7′-Isothiocyanato11-hydroxy1′ ,1′-dimethyl-heptylhexahydrocannabinol (AM841), a high-affinity electrophilic ligand, interacts covalently with a cysteine in helix six and activates the CB1 cannabinoid receptor. Mol. Pharmacol. 2005;68:1623–1635. doi: 10.1124/mol.105.014407. [DOI] [PubMed] [Google Scholar]
- 12.Kapur A, Samaniego P, Thakur GA, Makriyannis A, Abood ME. Mapping the structural requirements in the CB1 cannabinoid receptor transmembrane helix II for signal transduction. J. Pharmacol. Exp. Ther. 2008;325:341–348. doi: 10.1124/jpet.107.133256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bramblett RD, Panu AM, Ballesteros JA, Reggio PH. Construction of a 3D model of the cannabinoid CB1 receptor: determination of helix ends and helix orientation. Life Sci. 1995;56:1971–1982. doi: 10.1016/0024-3205(95)00178-9. [DOI] [PubMed] [Google Scholar]
- 14.Salo OM, Lahtela-Kakkonen M, Gynther J, Jarvinen T, Poso A. Development of a 3D model for the human cannabinoid CB1 receptor. J. Med. Chem. 2004;47:3048–3057. doi: 10.1021/jm031052c. [DOI] [PubMed] [Google Scholar]
- 15.Montero C, Campillo NE, Goya P, Paez JA. Homology models of the cannabinoid CB1 and CB2 receptors. A docking analysis study. Eur. J Med. Chem. 2005;40:75–83. doi: 10.1016/j.ejmech.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 17.Andersson H, D’Antona AM, Kendall DA, Von Heijne G, Chin CN. Membrane assembly of the cannabinoid receptor 1: impact of a long N-terminal tail. Mol. Pharmacol. 2003;64:570–577. doi: 10.1124/mol.64.3.570. [DOI] [PubMed] [Google Scholar]
- 18.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered humanβ2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, Schertler GF, Weis WI, Kobilka BK. Crystal structure of the humanβ2 adrenergic G-proteincoupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 20.Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EY, Lane JR, IJzerman AP, Stevens RC. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 22.Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, Choe HW, Hofmann KP, Ernst OP. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497– 502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 23.Serrano-Vega MJ, Magnani F, Shibata Y, Tate CG. Conformational thermostabilization of theβ1-adrenergic receptor in a detergent-resistant form. Proc. Natl. Acad. SciU.S A. 2008;105:877–882. doi: 10.1073/pnas.0711253105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mobarec JC, Filizola M. Advances in the development and application of computational methodologies for structural modeling of G-protein-coupled receptors. Exp. Opin. Drug Discovery. 2008;3:343–355. doi: 10.1517/17460441.3.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zvonok N, Pandarinathan L, Williams J, Johnston M, Karageorgos I, Janero DR, Krishnan S, Makriyannis A. Covalent inhibitors of human monoacylglycerol lipase: ligand-assisted characterization of the catalytic site by mass spectrometry and mutational analysis. Chem. Biol. 2008;15:854–862. doi: 10.1016/j.chembiol.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Picone RP, Fournier DJ, Makriyannis A. Ligand based structural studies of the CB1 cannabinoid receptor. J. Pept. Res. 2002;60:348–356. doi: 10.1034/j.1399-3011.2002.21069.x. [DOI] [PubMed] [Google Scholar]
- 27. Pei Y, Mercier R, Anday J, Thakur G, Zvonok A, Hurst D, Reggio P, Janero DR, Makriyannis A. Ligand-binding architecture of human CB2 cannabinoid receptor: evidence for receptor subtype-specific binding motif and modeling GPCR activation. Chem. Biol. 2008;15:1207–1219. doi: 10.1016/j.chembiol.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu W, Filppula SA, Mercier R, Yaddanapudi S, Pavlopoulos S, Cai J, Pierce WM, Makriyannis A. Purification and mass spectroscopic analysis of human CB1 cannabinoid receptor functionally expressed using the baculovirus system. J. Pept. Res. 2005;66:138–150. doi: 10.1111/j.1399-3011.2005.00283.x. [DOI] [PubMed] [Google Scholar]
- 29.Calandra B, Tucker J, Shire D, Grisshammer R. Expression in Escherichia coli and characterization of the human central CB1 and peripheral CB2 cannabinoid receptors. Biotechnol. Lett. 1997;19:425–428. [Google Scholar]
- 30.Farrens DL, Dunham TD, Fay JF, Dews IC, Caldwell J, Nauert B. Design, expression, and characterization of a synthetic human cannabinoid receptor and cannabinoid receptor/G-protein fusion protein. J. Pept. Res. 2002;60:336–347. doi: 10.1034/j.1399-3011.2002.21066.x. [DOI] [PubMed] [Google Scholar]
- 31.Kim TK, Zhang R, Feng W, Cai J, Pierce W, Song ZH. Expression and characterization of human CB1 cannabinoid receptor in methylotrophic yeast Pichia pastoris. Protein Expr. Purif. 2005;40:60–70. doi: 10.1016/j.pep.2004.10.026. [DOI] [PubMed] [Google Scholar]
- 32.Strander CD, Fong TM, Graziano MP, Tota MR. The family of G-protein-coupled receptors. FASEB J. 1995;9:745–754. [PubMed] [Google Scholar]
- 33.Fredriksson F, Schiöth HB. The repertoire of G-protein-coupled receptors in fully sequenced genomes. Mol. Pharmacol. 2005;67:1414–1425. doi: 10.1124/mol.104.009001. [DOI] [PubMed] [Google Scholar]
- 34.Ablonczy Z, Crouch RK, Knapp DR. Mass spectrometric analysis of integral membrane proteins at the subpicomolar level: application to rhodopsin. J. Chromatogr. 2005;825:169–175. doi: 10.1016/j.jchromb.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 35.Zvonok N, Yaddanapudi S, Williams J, Dai S, Dong K, Rejtar T, Karger BL, Makriyannis A. Comprehensive proteomic mass spectrometric characterization of human cannabinoid CB2 receptor. J. Proteome Res. 2007;6:2068–2079. doi: 10.1021/pr060671h. [DOI] [PubMed] [Google Scholar]
- 36.Alves ID, Sachon E, Bolbach G, Millstine L, Lavielle S, Sagan S. Analysis of an intact G-protein coupled receptor by MALDI-TOF mass spectrometry: molecular heterogeneity of the tachykinin NK-1 receptor. Anal. Chem. 2007;79:2189–2198. doi: 10.1021/ac062415u. [DOI] [PubMed] [Google Scholar]
- 37.Sansuk K, Balog CIA, van der does AM, Booth R, de Grip WJ, Deelder AM, Bakker RA, Leurs R, Hensbergen PJ. GPCR proteomics: mass spectrometric and functional analysis of histamine H1 receptor after baculovirus-driven and in vitro cell free expression. J. Proteome Res. 2008;7:621–629. doi: 10.1021/pr7005654. [DOI] [PubMed] [Google Scholar]
- 38.Ho JTC, White JF, Grisshammer R, Hess S. Analysis of a G-protein coupled receptor for neurotensin by liquid chromatography-electrospray ionization mass spectrometry. Anal. Biochem. 2008;376:13–24. doi: 10.1016/j.ab.2007.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katayama H, Tabata T, Ishihama Y, Sato T, Oda Y, Nagasu T. Efficient in-gel digestion procedure using 5-cyclohexyl-1-pentyl-β-D-maltoside as an additive for gel-based membrane proteomics Rapid Commun. Mass Spectrom. 2004;18:2388–2394. doi: 10.1002/rcm.1637. [DOI] [PubMed] [Google Scholar]
- 40.Ishihama Y. Proteomic LC-MS systems using nanoscale liquid chromatography with tandem mass spectrometry. J. Chromatogr. 2005;1067:73–83. doi: 10.1016/j.chroma.2004.10.107. [DOI] [PubMed] [Google Scholar]
- 41.Issaq HJ, Blonder J. Electrophoresis and liquid chromatography/tandem mass spectrometry in disease biomarker discovery. J. Chromatogr B. 2009;877:1222–1228. doi: 10.1016/j.jchromb.2008.12.028. [DOI] [PubMed] [Google Scholar]
- 42.Altmann F, Staudacher E, Wilson IBH, März L. Insect cells as hosts for the expression of recombinant glycoproteins. Glycoconj. J. 1999;16:109–123. doi: 10.1023/a:1026488408951. [DOI] [PubMed] [Google Scholar]
- 43.Hollister J, Grabenhorst E, Nimtz M, Conradt H, Jarvis DL. Engineering the protein N-glycosylation pathway in insect cells for production of biantennary, complex N-glycans. Biochemistry. 2002;41:15093–15104. doi: 10.1021/bi026455d. [DOI] [PMC free article] [PubMed] [Google Scholar]