ABSTRACT
In the human fungal pathogen Candida albicans, the CUG codon is translated 97% of the time as serine and 3% of the time as leucine, which potentially originates an array of proteins resulting from the translation of a single gene. Genes encoding cell surface proteins are enriched in CUG codons; thus, CUG mistranslation may influence the interactions of the organism with the host. To investigate this, we compared a C. albicans strain that misincorporates 28% of leucine at CUGs with a wild-type parental strain. The first strain displayed increased adherence to inert and host molecules. In addition, it was less susceptible to phagocytosis by murine macrophages, probably due to reduced exposure of cell surface β-glucans. To prove that these phenotypes occurred due to serine/leucine exchange, the C. albicans adhesin and invasin ALS3 was expressed in Saccharomyces cerevisiae in its two natural isoforms (Als3p-Leu and Als3p-Ser). The cells with heterologous expression of Als3p-Leu showed increased adherence to host substrates and flocculation. We propose that CUG mistranslation has been maintained during the evolution of C. albicans due to its potential to generate cell surface variability, which significantly alters fungus-host interactions.
IMPORTANCE
The translation of genetic information into proteins is a highly accurate cellular process. In the human fungal pathogen Candida albicans, a unique mistranslation event involving the CUG codon occurs. The CUG codon is mainly translated as serine but can also be translated as leucine. Leucine and serine are two biochemically distinct amino acids, hydrophobic and hydrophilic, respectively. The increased rate of leucine incorporation at CUG decoding triggers C. albicans virulence attributes, such as morphogenesis, phenotypic switching, and adhesion. Here, we show that CUG mistranslation masks the fungal cell wall molecule β-glucan that is normally recognized by the host immune system, delaying its response. Furthermore, we demonstrate that two different proteins of the adhesin Als3 generated by CUG mistranslation confer increased hydrophobicity and adhesion ability on yeast cells. Thus, CUG mistranslation functions as a mechanism to create protein diversity with differential activities, constituting an advantage for a mainly asexual microorganism. This could explain its preservation during evolution.
Introduction
Many pathogenic microorganisms switch surface antigens through constant variation of cell surface molecules. This process enables microbes to occupy diverse niches within the host and reduces microbe recognition by the immune system. Various mechanisms of antigenic switching have been described in both commensal and pathogenic organisms, including Plasmodium falciparum, Trypanosoma brucei, Pneumocystis carinii, and Candida glabrata (1–9).
Antigenic switching can also influence microbial adherence, an important virulence attribute that prevents a microorganism from being removed from a favorable location, thus providing the ability to infect specific niches within the host (10). The C. albicans ALS (agglutinin-like sequence) gene family encodes adhesins that bind to various substrates. Some of these Als proteins also mediate biofilm formation, host cell invasion, and iron acquisition (11–13). Some ALS genes are differentially expressed by yeast versus hyphae. However, unlike the Saccharomyces cerevisiae FLO and C. glabrata EPA genes, which also specify adhesins, C. albicans ALS genes are not located near the telomeres and are not subject to subtelomeric silencing. Interestingly, the coding sequences of ALS genes contain a high number of CUG codons (14, 15). This codon is translated as leucine in most organisms. However, in C. albicans and other Candida species, the CUG codon is dually translated as serine (95 to 97% of the time) and leucine (3 to 5% of the time) (15). This atypical CUG translation is mediated by a Ser-tRNACAG that, in contrast to other tRNAs, is recognized by two aminoacyl-tRNA synthetases, seryl- and leucyl-tRNA synthetases (16). Because serine is hydrophilic, whereas leucine is hydrophobic, the variable incorporation of these two amino acids into a protein has the potential to generate a family of proteins with altered structure and function (17). The first successful manipulation of genetic code ambiguity in C. albicans resulted in the increase of virulence factor expression, in particular phenotypic switching, morphogenesis, and adhesion (18). However, the adhesion phenotype was not quantitatively measured, and it was not clear whether adhesion resulted from the high rates of spontaneous transitions of white to opaque and yeast to hyphal forms.
We hypothesized that Ser/Leu incorporation variability in cell surface proteins of C. albicans may influence its adherence to various substrates as well as its interactions with host immune effector cells. To investigate this hypothesis, we used a highly CUG-mistranslating strain of C. albicans that has increased incorporation of leucine at CUG codons (18). We found that CUG mistranslation increased adherence to host substrates, by changing cell surface hydrophobicity. Furthermore, the diversity in the cell wall proteome created by CUG mistranslation altered the cell surface exposure of 1,3-β-glucan and reduced C. albicans phagocytosis by macrophages. These results suggest that protein diversity caused by variable translation of the CUG codon has important functional consequences for the interactions of C. albicans with the host.
RESULTS
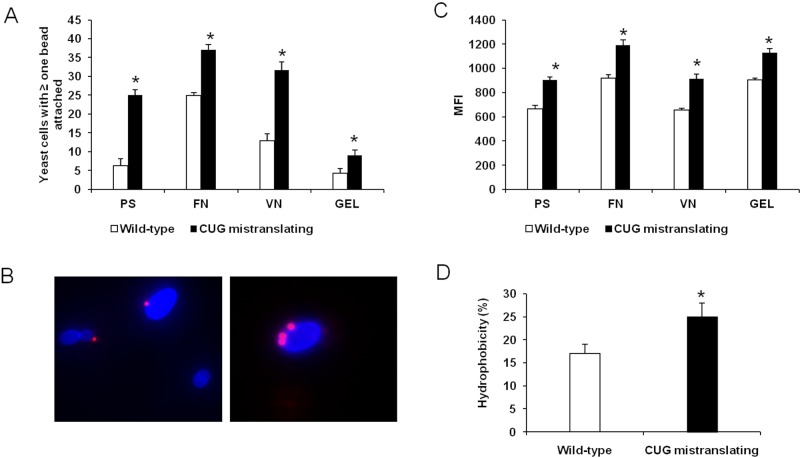
Forced CUG mistranslation increases adhesion and cell surface hydrophobicity.
To test whether variable serine/leucine incorporation at CUG sites potentiates adhesion and hydrophobicity, we compared yeast forms of a wild-type C. albicans strain (pUA12) with those of a mutant strain that has a high level of CUG mistranslation (pUA15), displaying 28% leucine incorporation rather than the usual 3% leucine incorporation in the wild-type C. albicans (15, 18). We used a quantitative flow cytometry assay to characterize the adherence of the two C. albicans strains to polystyrene and the representative host ligands fibronectin, vitronectin, and gelatin (collagen) (19). In this assay, fluorescent polystyrene beads uncoated or coated with various substrates were incubated with nonfluorescent C. albicans yeasts (Fig. 1B). The number of organisms with at least one bead attached (Fig. 1A) and the mean fluorescence intensity (MFI) emitted by each yeast-bead were quantified by flow cytometry (Fig. 1C; see also Fig. S1 in the supplemental material). MFI is directly proportional to the number of beads attached to each organism (Fig. 1C; see also Fig. S1). The majority of wild-type organisms did not bind to any beads (Fig. 1A and B; see also Fig. S1). A small percentage of these cells were attached to a single bead, and even fewer cells were attached to two or more beads (Fig. S1). In contrast, a high percentage of cells of the highly CUG-mistranslating strain bound to at least one bead, and many of them bound to multiple beads (Fig. 1A to C; see also Fig. S1). Consistent with these results, the highly CUG-mistranslating cells were more adherent than wild-type cells to all tested abiotic and biotic substrates (Fig. 1A and C). While the greatest difference in adherence was found for polystyrene (more than a 4-fold increase), the increase of CUG mistranslation also enhanced C. albicans adherence to biotic substrates: 1.5-fold to fibronectin, 2.6-fold to vitronectin, and 2.25-fold to gelatin (Fig. 1A). Collectively, our results clearly indicate that enhancing the leucine incorporation at CUG sites results in increased adherence to a variety of substrates. The increased adherence of the highly CUG-mistranslating strain could also be due to the altered surface expression or function of adhesin proteins. To test whether the level of CUG mistranslation influenced surface expression of adhesins, we analyzed Als1 and Als3, which mediate adherence to a variety of host substrates (20). The level of surface expression of Als1 and Als3 proteins was quantified by flow cytometry using a polyclonal antibody that recognizes both Als proteins (21). No difference in Als protein expression was found between the wild-type and highly CUG-mistranslating strains (see Fig. S2), suggesting that the increased adherence of the CUG-mistranslating strain is likely due to altered function of adhesins rather than increased surface expression of these adhesins.
FIG 1 .
CUG mistranslation increases adherence and cell surface hydrophobicity. (A) Percentage of yeast cells of the indicated strains with at least one bead attached. (B) Representative images of the wild-type strain and the highly CUG-mistranslating strain of C. albicans, demonstrating that the highly CUG-mistranslating strain (blue) binds to more polystyrene beads (which fluoresce red). (C) Mean fluorescence intensity (MFI) of the indicated strains. (D) Cell surface hydrophobicity was assessed by partitioning into n-hexadecane. Results in panels A and C represent the means ± standard deviations of five experiments, each analyzing 50,000 cells. *, P < 0.05 compared to the wild-type strain. Abbreviations: PS, polystyrene; FN, fibronectin; VN, vitronectin; GEL, gelatin. See also Fig. S1 and S2 in the supplemental material.
Cell surface hydrophobicity is known to be important for the adherence of C. albicans (22). Because leucine is hydrophobic and serine is polar, it is likely that the increased leucine content in the cell surface proteins of the highly CUG-mistranslating strain enhances the hydrophobicity of this strain. Cell surface hydrophobicity was quantified using a classical approach based upon microbial adherence to hydrocarbons (23). Cells of the highly CUG-mistranslating strain were significantly more hydrophobic than those of the wild-type strain (Fig. 1D). These results suggest that serine/leucine exchange in CUG translation contributes significantly to the promotion of cell surface hydrophobicity.
The CUG-encoded residues of Als proteins are predicted to be surface exposed.
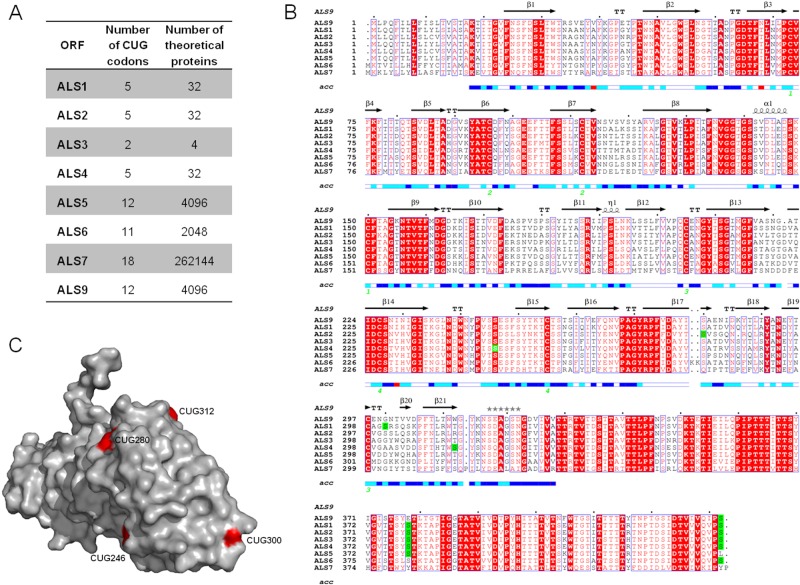
If serine/leucine switching in CUG decoding alters C. albicans adherence, where are CUG codons located in cell wall-coding genes? To gain insight into this topic, we analyzed the ALS gene family, which is composed of 8 genes. Als proteins are composed of four domains: a C-terminal domain, containing a site for covalent attachment of the glycosylphosphatidylinositol (GPI) anchor and rich in serine/threonine amino acid sequences; tandem repeats of highly conserved 108-bp units; an amyloid-forming region; and an N-terminal domain which contains the substrate-binding region (24). The coding sequences of ALS genes contain multiple CUG codons, and their mistranslation increases the number of proteins synthesized from each mRNA transcript by a factor of 2n (Fig. 2A), depending on the arbitrary serine/leucine incorporation at CUG positions. Based on the number of CUG codons and their localization, ALS genes can be divided into two groups: ALS1 to ALS4 and ALS5 to ALS9 (Fig. 2A and B). Members of the ALS1 to -4 group contain 2 to 5 CUG codons, located mainly at the 5′ end of the mRNA sequence that codes for the N-terminal region (Fig. 2B). Members of the ALS5 to -9 group possess 5 to 18 CUG codons, which are predominantly located in the 3′ end of the mRNA, corresponding to the C terminus of the protein. Since the crystal structure of the Als9 N-terminal domain was recently solved and the residue identity is 41 to 84% among Als sequences (25), we did a multiple protein sequence alignment in order to predict the location of CUG-encoded residues in this domain. The solvent accessibility of each residue was scored (Fig. 2B). The three-dimensional structure of the Als9 N-terminal domain (Protein Data Bank [PDB] code 2Y7N) showed that serine/leucine residues encoded by CUG codons are totally or partially exposed to the solvent (Fig. 2C), where both residues can be accommodated with no protein folding disruption. However, the aliphatic side chain of leucine lacks the chemical reactivity of the serine hydroxyl group, which can act as a hydrogen donor. Hence, the variable incorporation of leucine or serine in the N-terminal domain should create functional and structural diversity at the Alsp molecular surface, which is exposed in the cell wall. The Als9 crystal structure does not include residues 379 and 433 (encoded by CUG codons); however, the multiple sequence alignment demonstrated that these residues are mostly encoded by CUG codons among members of the ALS gene family, suggesting that they have an important role in Alsp function (Fig. 2B).
FIG 2 .
Number of CUG codons and their localization in the 5′ domain (N-terminal) coding sequence of the ALS gene family. (A) Coding sequences of ALS1, ALS2, ALS3, ALS4, ALS5, ALS6, ALS7, and ALS9 were retrieved from the Candida Genome Database. The number of CUG codons (n) present in each ORF and the number of theoretical proteins (2n) resulting from its translation, depending on probabilistic serine-versus-leucine incorporation during CUG translation. (B) Multiple sequence alignment of N-terminal domains of Als proteins in which the CUG-encoded serines are highlighted in green. Strictly conserved residues among Als proteins are indicated in red letters against a white background, while conserved residues are represented by white letters boxed in red. Secondary elements from the C. albicans ALS9 crystal sequence structure (PDB code 2Y7N) are displayed above the alignment. The solvent accessibility for each residue is indicated by a bar at the bottom of the sequence (blue, accessible; cyan, intermediate; white, buried; red, no correspondence between ALS9 crystal structure and protein sequence). (C) Distribution of CUG positions (red residue; ALS9 protein numbering) on the molecular surface of the ALS N-terminal domain (expecting residues 379 and 433).
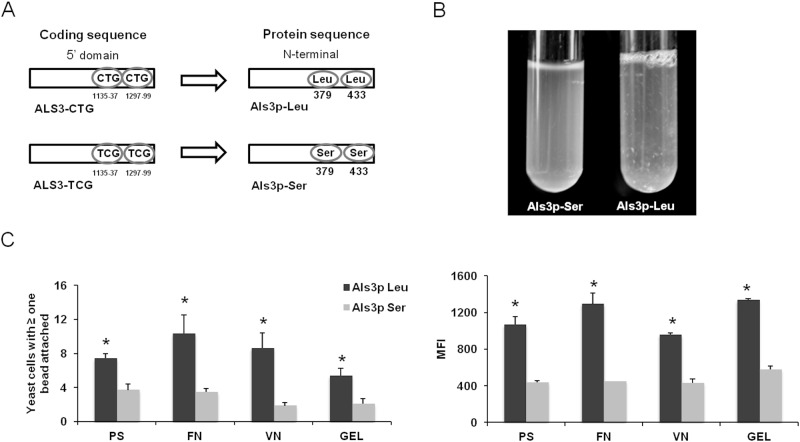
S. cerevisiae expressing Als3p-Leu has increased adherence.
We used a heterologous expression approach to investigate the functional effects of incorporating serine versus leucine at residues 379 and 433 in Als3p, a key C. albicans adhesin. Two different isoforms of C. albicans ALS3 were expressed in the normally nonadherent S. cerevisiae. One was the wild-type allele, which, when expressed in S. cerevisiae, resulted in the production of Als3p containing leucine at residues 379 and 433 (Als3p-Leu). In the other allele, the 2 CUG codons were mutated to UCG, so that the resulting protein contained serine at these positions (Als3p-Ser) (Fig. 3A). When grown in liquid medium, S. cerevisiae expressing Als3p-Leu showed greater flocculation than did the strain expressing Als3p-Ser (Fig. 3B), indicating that the variable translation of the CUG codon significantly influences cell-cell adhesion. Substitution of leucine for serine in Als3p also altered its capacity to bind to different substrates. S. cerevisiae cells expressing Als3p-Leu had significantly greater adherence to polystyrene, fibronectin, vitronectin, and gelatin than did cells expressing Als3p-Ser, as determined both by the number of cells with beads attached and by the mean fluorescence intensity (Fig. 3C). Again, these phenotypes were not due to differences in the amount of Als3p expressed on the fungal cell surface (see Fig. S3 in the supplemental material). Overall, our findings that the Als3p-Ser and Als3p-Leu isoforms exhibit markedly different adherence properties indicate that CUG mistranslation has significant functional consequences.
FIG 3 .
C. albicans Als3p-Leu and Als3p-Ser isoforms confer distinct adhesion profiles when expressed in the normally nonadherent S. cerevisiae. (A) Location of the two Leu-CUG codons in ALS3 that were mutated to Ser-UCG codons. (B) Images of broth cultures of S. cerevisiae expressing Als3p-Ser (left) or Als3p-Leu (right). S. cerevisiae expressing Als3p-Ser did not flocculate, whereas the strain expressing Als3p-Leu exhibited strong flocculation. (C) Adherence of S. cerevisiae expressing Als3p-Leu or Als3p-Ser to either bare polystyrene beads or beads coated with the indicated substrates. Adherence was measured as the percentage of cells with at least one bead attached (left panel) or as the mean fluorescence intensity (right panel). Results in panel C represent the means of five flow cytometry experiments, each analyzing 50,000 cells *, P < 0.05 compared to Als3p-Leu. Abbreviations: PS, polystyrene; FN, fibronectin; VN, vitronectin; GEL, gelatin. See also Fig. S3 in the supplemental material.
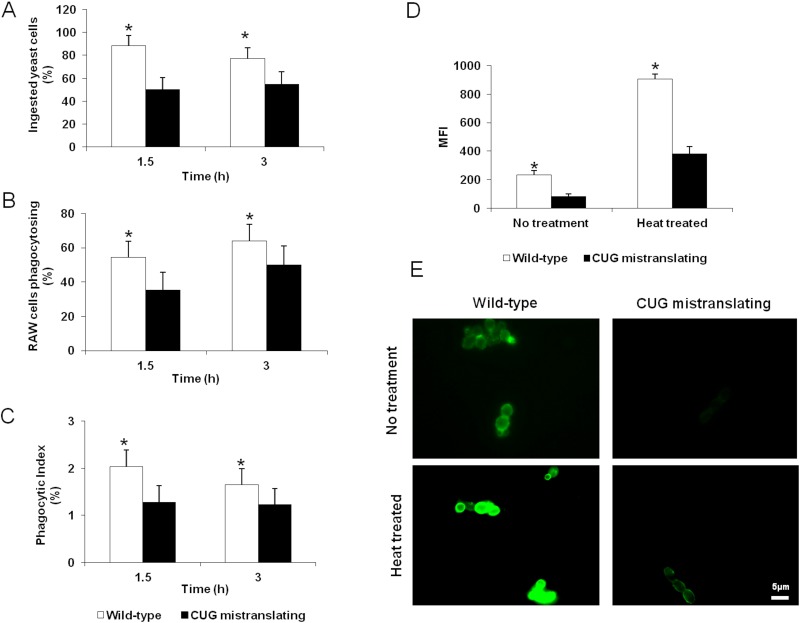
CUG mistranslation reduces susceptibility to macrophage phagocytosis.
Next, we investigated the influence of CUG mistranslation on the phagocytosis of C. albicans by macrophages. RAW 264.7 macrophages were challenged with the wild-type and highly CUG-mistranslating C. albicans strains (pUA12 and pUA15, respectively). The CUG-mistranslating cells were more resistant than the wild-type strain to macrophage phagocytosis, as indicated by a reduced number of phagocytosed yeasts and fewer phagocytosed yeasts per macrophage (Fig. 4A to C). This resistance to phagocytosis was most evident after 1.5 h of incubation, and it decreased by 3 h. The reduced phagocytosis of the highly CUG-mistranslating cells could be due to the reduced surface expression of cell wall components that are recognized by macrophage pattern recognition receptors. One of these receptors is dectin-1, which recognizes β-glucans exposed on the fungal cell surface and triggers the phagocytosis of C. albicans (26–29). We hypothesized that the reduced phagocytosis of the highly CUG-mistranslating strain was due to reduced cell surface exposure of β-glucans. To test this hypothesis, we assessed the level of surface-exposed β-glucan on wild-type C. albicans and the highly CUG-mistranslating strain with an anti-β-1,3-glucan monoclonal antibody using flow cytometry (30). We found that the highly CUG-mistranslating strain had 2- to 3-fold less exposure of β-glucans on its surface than did wild-type cells (Fig. 4D and E). This reduced exposure was seen in viable cells as well as heat-killed cells. Collectively, these data suggest that increased CUG mistranslation results in decreased β-glucan exposure on the fungal surface, which in turn reduces susceptibility to macrophage phagocytosis.
FIG 4 .
CUG mistranslation reduces C. albicans phagocytosis by macrophages and reduces the surface exposure of β-glucans. (A) Percentages of wild-type and CUG-mistranslating cells ingested by a murine macrophage cell line after 1.5 h and 3 h of incubation. (B) Percentages of macrophages phagocytosing one or more yeast cells of the indicated strains after 1.5 h and 3 h of incubation. (C) Phagocytic index of macrophages (calculated as the number of ingested yeasts divided by the number of phagocytosing macrophages) incubated with wild-type and CUG-mistranslating strains. (D) Surface β-glucan exposure in the indicated strains quantified by flow cytometry using an anti-β-1,3 glucan antibody. (E) Highly mistranslating strain display reduced β-glucan exposure comparably to the wild-type strain even after heat treatment. Images of fluorescence microscopy of C. albicans cells incubated with an anti-β-1,3 glucan antibody. Results are means ± standard deviations of three experiments, each performed in triplicate. *, P < 0.05 compared to the wild-type strain.
DISCUSSION
C. albicans has evolved multiple mechanisms to alter its surface characteristics. These mechanisms include white-opaque switching and yeast-to-hypha transition (31). Our data demonstrate that CUG mistranslation through the variable incorporation of polar serine or hydrophobic leucine residues is another mechanism for expanding the structural and functional diversity of cell surface proteins (Fig. 5). Genes encoding these proteins are particularly enriched in CUG codons (58% of cell wall proteome), suggesting that the protein diversity generated by CUG mistranslation may play an important role in the interactions of C. albicans with host cells (17). We found that increasing the rate of CUG mistranslation enhanced the adherence of C. albicans to polystyrene and host constituents. This increase in adherence is mediated by at least two mechanisms. First, CUG mistranslation enhances cell surface hydrophobicity, which is known to play a role in C. albicans adherence to both plastic and host cells (32). Second, CUG mistranslation increases the substrate avidity of adhesins such as the Als proteins. Indeed, expression of Als3p-Leu in S. cerevisiae conferred significantly greater adherence to some substrates than did the expression of Als3p-Ser.
FIG 5 .
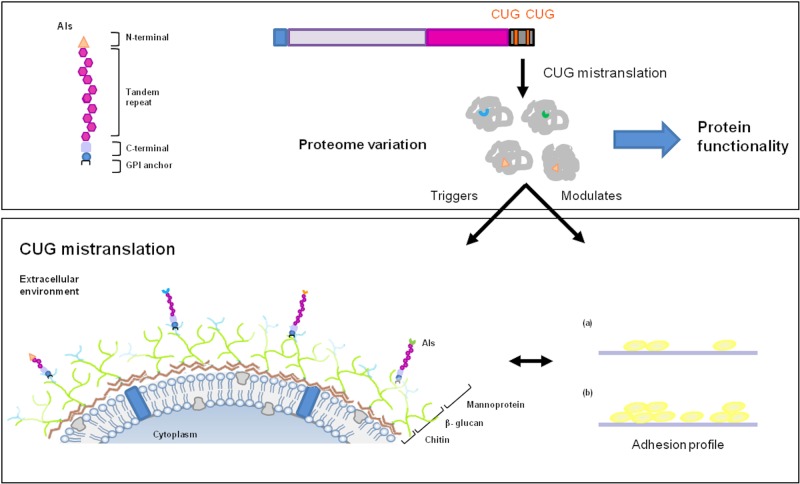
Cell surface variability in C. albicans originated by CUG mistranslation. Since more than 50% of the cell wall ORFeome possesses at least 1 CUG codon, its mistranslation, either as serine or as leucine, creates a natural proteome expansion. Protein isoforms, resulting from a single ORF decoding, constitute a basis for cell surface variability in C. albicans and modulation of its functionality/activity. For example, the ALS3 gene, containing 2 CUG codons in its coding sequence, can originate 4 different protein isoforms (Als-Leu versus Als3-Ser), whose expression confers differential fungal adherence profiles (a and b).
The highly conserved location of CUG codons in the regions of ALS1 to ALS4 that encode the substrate-binding domain suggests that CUG mistranslation likely influences the adhesive function of other Als proteins besides Als3p. Furthermore, C. albicans possesses additional adhesins such as Eap1 that are not members of the Als protein family (33). The EAP1 gene also contains 2 CUG codons. Thus, CUG mistranslation might enhance the structural diversity and modulate the adherence function of other proteins, as well. This functional variability may broaden the range of host substrates to which C. albicans can bind by changing the ligand interaction region. Recently, it has been reported that Als proteins can coalesce into amyloid-like aggregates that result in the formation of adhesive nanodomains and increased cell-cell adherence (24, 34, 35). The CUG codons in ALS1 to ALS4 are located near the regions of these genes that are predicted to encode amyloid-forming domains. Therefore, it is also possible that CUG mistranslation might modulate the amyloid profile of Als proteins.
Even though the highly CUG-mistranslating strain of C. albicans had increased adherence to host constituents, we found that it was phagocytosed poorly by macrophages. Macrophages possess multiple receptors that recognize various components of the C. albicans cell wall (36). CR3 and dectin-1 both recognize β-glucans exposed on the surface of C. albicans, and both of these receptors mediate the phagocytosis of yeast-phase organisms (26, 29, 36). We found that the highly CUG-mistranslating strain of C. albicans had lower surface exposure of β-glucans than did the wild-type strain. This reduced β-glucan exposure may contribute to the reduced macrophage phagocytosis of the highly CUG-mistranslating strain. Although Ser and Asp are the residues for O-glycosylation and 59% of the open reading frames (ORFs) associated with cell wall organization and biogenesis are CUG-containing ORFs (17), N-linked and O-linked mannan appear not to be affected by CUG mistranslation. In contrast, the mannan outer layer is effectively masking β-glucans in the CUG-mistranslating strain comparably to the wild type. A defect in N- and O-glycosylation would imply an increase in C. albicans uptake by macrophages (37, 38). The lower phagocytosis rate observed in the CUG-mistranslating strain may also be related to a depletion of phosphomannan cell wall, as happens in the pmr1 mutant (38). Interestingly, the PMR1 gene has a single CUG codon encoding a strictly conserved serine (17). The impact on Pmr1p functionality of the replacement of a serine by a leucine resulting from CUG decoding is unknown; however, if this serine-to-leucine switching implies a reduction in Pmr1p activity, a defect in phosphomannan cell wall may occur. It was notable that the highly CUG-mistranslating strain had the same susceptibility to the β-1,3-glucan synthase inhibitor caspofungin as did the wild-type strain. This result suggests that the total amounts of β-1,3-glucan in the cell walls of the two strains might be similar (39).
It was previously demonstrated that the level of leucine misincorporation at CUG codons is variable and depends upon external conditions, namely, temperature and pH (15). In addition, a disparity in activity and stability between two protein isoforms of the C. albicans seryl-tRNA synthetase (SerRS) was found. When the only CUG codon in SerRS is translated as leucine, the activity of this enzyme is increased, thereby raising the percentage of tRNACAG that is serylated (17). Besides SerRS and through structural studies, authors suggest that other proteins associated with the pathogenesis process may display functional changes resulting from serine/leucine incorporation at CUG sites (17).
Although they are relatively uncommon, similar mistranslation events are known to occur in other biological systems and enhance resistance to stress. For example, when mammalian cells are subjected to oxidative stress, the population of tRNAs misacylated with methionine increases remarkably (40). The presence of methionine in some proteins protects them from damage caused by reactive oxygen species. Similarly, the mistranslation of the UGA stop codon as selenocysteine protects neurons from oxidative stress (41) and is required for the normal function of macrophages (42). However, an important difference between mammalian and C. albicans cells is that approximately 20 selenoproteins have been described in mammalian cells (43), whereas 3,998 C. albicans genes contain at least one CUG codon.
Organisms able to generate statistical proteins are thought to be in the origin of life (44). Their improved adaptability is granted by the formation of protein variants, resulting from the same coding sequence, which possess enhanced activity but reduced specificity (45). Selective growth advantages conferred by ambiguous codes, similar to the natural CUG mistranslation system but artificially originated, have been demonstrated in S. cerevisiae (46), Escherichia coli (45), and Acinetobacter baylyi (47).
C. albicans has lived as a commensal with mammalian hosts for millions of years. Prerequisites for a successful commensal organism include the capacity to adhere to host constituents and to avoid being cleared by the host immune response. Thus, it is likely that CUG mistranslation may constitute an advantage by conferring increased capability of cells to adhere to a broad range of host substrates while minimizing interactions with immune effector cells. CUG mistranslation has the capacity not only to provide greater diversity in the cell surface proteins of C. albicans but also to expand the repertoire of metabolic responses of this organism (Fig. 5). Thus, CUG mistranslation likely expands the host microniches in which C. albicans can flourish.
MATERIALS AND METHODS
Strains, growth conditions, and cell culture conditions.
The two C. albicans strains, wild type (pUA12) and highly CUG mistranslating (pUA15) (18), were grown in minimal medium lacking uridine (MM-uri) (0.67% [wt/vol] yeast nitrogen base without amino acids, 2% [wt/vol] glucose, and 100 µg/ml of the required amino acids). For hydrophobicity and adherence assays, yeast cells were grown overnight at 30°C in a shaking incubator. For use in the phagocytosis experiments, the yeast cells were grown on MM-uri agar at 30°C for 48 h.
Escherichia coli strain DH5α was used as a host to carry out ALS3 CUG codon mutagenesis (described below), and it was grown in LB with 100 µg ml−1 ampicillin.
S. cerevisiae strain S150-2B (leu2 his3 trp1 ura3) was used for heterologous expression of C. albicans ALS3-Leu and ALS3-Ser genes (20). The Als3p-expressing clones were grown overnight at 30°C in minimal medium supplemented with appropriate amino acids.
The RAW 264.7 murine macrophage cell line was obtained from Sigma-Aldrich (catalog no. ECACC 91062702) and maintained in RPMI 1640 medium (Sigma), containing 10% heat-inactivated fetal bovine serum and 2 mM l-glutamine. All mammalian cells were grown in 5% CO2 at 37°C.
Flow cytometric adhesion assay.
The adherence of C. albicans and S. cerevisiae to polystyrene and host substrates was measured by flow cytometry (19). Carboxylated highly fluorescent polystyrene beads (1 µm; F-8823; Invitrogen) were covalently linked with fibronectin (0.5 mg ml−1), vitronectin (10 µg ml−1), or gelatin (5 mg ml−1) (Sigma-Aldrich) by the carbodiimide method, according to the manufacturer’s instructions. C. albicans yeast-form cells from an overnight culture were harvested, washed with ice-cold phosphate-buffered saline (PBS), and resuspended in PBS at 2 × 106 cells ml−1. To acid-washed glass tubes was added 200 µl of the yeast suspension and an equal volume of PBS containing 2 × 107 beads ml−1. The mixture was equilibrated at room temperature for 2 min, vortexed vigorously for 30 s, and then incubated on a rotary shaker (150 rpm) for 30 min at room temperature. The mixture was then vortexed briefly and then analyzed with a FACSCalibur (BD Biosciences, Sydney, Australia) flow cytometer. Cell-associated fluorescence was read in the FL3 (LP, 670 nm) fluorescence channel, and 50,000 events were analyzed per condition. Controls containing yeast cells alone and beads alone were also analyzed. The adherence results were expressed as the percentage of yeast cells with at least one bead attached, the mean fluorescence intensity (MFI). Each experiment was performed in triplicate at least five times.
Microscopic visualization of yeast-bead attachment.
For visualization of yeast cell-bead attachment, an aliquot of the yeast cell suspension prepared for the flow cytometric adhesion assay was stained with calcofluor white (0.05% [vol/vol]) (Sigma-Aldrich) and mixed with fluorescent beads. Images were taken with a Zeiss Axioplan microscope coupled with an AxioVision image acquisition system (Zeiss).
Cell surface hydrophobicity determination.
The cell surface hydrophobicity of the two strains of C. albicans was analyzed as described previously (23). Yeast cells were harvested at stationary growth phase and washed twice with PBS, pH 7.0. Then, yeasts were resuspended to an optical density at 600 nm (OD600) of 0.4 to 0.5 in PBS (A0). Next, 3 ml of the yeast suspension was added to acid-washed spectrophotometer glass vials and overlaid with 0.4 ml of n-hexadecane. After vigorous vortexing, the phases were allowed to separate for 10 min at 30°C and the OD600 of the aqueous phase was measured (A1). The percentage of hydrophobicity was calculated as follows: hydrophobicity (%) = [1 − (A1/A0)] × 100. All assays were performed in triplicate.
Sequence analysis.
The amino acid sequences of the N termini of C. albicans Als proteins were retrieved from the Candida Genome Database (http://www.candidagenome.org/). The position of CUG-encoded residues in the corresponding nucleotide coding sequence (bp 1 to 1299) was determined using CodonPlot (48). Multiple sequence alignments of the N-terminal domains of Als1 to Als9 were carried out using the ClustalW algorithm (49) and displayed with ESPript (similarity score matrix, BLOSUM62) (50).
ALS3 CUG site-directed mutagenesis.
The plasmid pALS3 (20) was used as the template to mutate the two CUG codons located in the 5′ domain of the coding sequence of ALS3, namely, CUGs encoding amino acids 379 and 433. CUG codons were mutated to Ser-UCG codons sequentially, using the indicated primers (ALS3_Ser379_For, 5′ GTTGGTGTGACTACTTCCTACTCGACCAAAACTGCACC 3′; ALS3_Ser379_Rev, 5′ GGTGCAGTTTTGGTCGAGTAGGAAGTAGTCACACCAAC 3′; ALS3_Ser433_For, 5′ CTGTCATTGTACAAGTTCCATCGCCAAACCCAACTGTTAC 3′; ALS3_Ser433_Rev, 5′ GTAACAGTTGGGTTTGGCGATGGAACTTGTACAATGACAG 3′) and the QuikChange site-directed mutagenesis kit (Stratagene). Plasmids pALS3 and pALS3-UCG379/433 were transformed into S. cerevisiae strain S150-2B by the lithium acetate method.
Macrophage phagocytosis assay.
The phagocytosis of the two strains of C. albicans, wild type and CUG mistranslating, by the RAW 264.7 macrophage cell line was quantified as described previously (51). Briefly, yeast cells suspended in PBS were labeled with Oregon Green 488 (1 µM; Invitrogen) in the dark for 1 h at 30°C. Afterward, the yeast cells were washed twice with PBS-100 mM glycine and resuspended in PBS to a final yeast cell concentration of 5 × 107 cells ml−1. RAW 264.7 macrophages were added to coverslips in a 6-well-tissue culture plate (1 × 106 cells well−1) and incubated with 100 µl of yeast cell suspension, for 1.5 h and 3 h. The cells were then rinsed with ice-cold PBS and fixed with 4% paraformaldehyde in the dark for 45 min at 4°C. The nonphagocytosed yeasts were stained with calcofluor white M2R (2.5 µM; Sigma-Aldrich) in the dark for 5 min at 4°C and then rinsed to remove the unincorporated dye. The cells were dehydrated by immersion in graded ethanol solutions (70%, 90%, and 100%; 20 s each), cleared with xylene for 20 s, and mounted on glass slides using DPX mounting medium (Fluka BioChemika, Germany). The numbers of phagocytosed yeast cells (green fluorescent), noningested yeast cells (blue fluorescent), and macrophages were quantified by phase-contrast and fluorescence microscopy. Digital images were taken and analyzed using ImageJ software. At least three clones of each yeast strain were analyzed. For each clone, four slides were prepared and at least two experiments were carried out in consecutive days. The data were expressed as the percentage of yeast cells phagocytosed, as the percentage of RAW 264.7 cells phagocytosing at least one yeast, and as a phagocytosis index (the number of yeast phagocytosed by each macrophage).
Detection of β-glucan surface expression.
The amount of surface-exposed β-glucan on the two strains of C. albicans, wild type and CUG mistranslating, was determined (30). Briefly, viable and heat-killed (90°C for 20 min) yeast cells were incubated with an anti-β-1,3-glucan mouse monoclonal antibody (Biosupplies) for 15 min at 4°C. After being rinsed, the cells were incubated with a fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG secondary antibody (Sigma) for 15 min at 4°C. Yeasts were then fixed with 0.4% paraformaldehyde, and 20,000 events were analyzed in the FL1 (BP, 530/30 nm) fluorescence channel using a FACSCalibur flow cytometer. Each strain was tested in triplicate. The experiment was repeated two times. Quantitative flow cytometry assays were confirmed by epifluorescence microscopy using a Zeiss Axioplan microscope coupled with an AxioVision image acquisition system (Zeiss).
SUPPLEMENTAL MATERIAL
Comparative histogram of the adherence profiles of wild-type C. albicans cells (dark line) and cells of the highly CUG-mistranslating strain (gray line). The intensity of fluorescence (FL3-H) is plotted against the number of cells (counts), and the results are from the analysis of 50,000 cells. The arrow indicates cells with no beads attached, and numbered peaks indicate the fluorescence of cells with 1, 2, or 3 beads attached. Download
CUG mistranslation does not influence the amount of surface-exposed Als1p and Als3p. Flow cytometric quantification of Als protein expression in wild-type (dark line) and highly CUG-mistranslating (gray line) cells, using an anti-Als3 polyclonal antibody that recognizes both Als3 and Als1. Download
Flow cytometric measurement of Als3p surface expression on S. cerevisiae expressing Als3p-Leu or Als3p-Ser. Download
ACKNOWLEDGMENTS
This study was supported by project POCI/SAU-IMI/61598/2004 financed by the Fundação para Ciência e Tecnologia (FCT). I.M. is supported by FCT Ciência 2008 and the European Social Fund. A.S.-D. is supported by an FCT Ph.D. grant (SFRH/BD/44896/2008). S.G.F. was supported in part by grants R01AI054928 and R01DE017088 from the National Institutes of Health, United States.
Footnotes
Citation Miranda I, Silva-Dias A, Rocha R, Teixeira-Santos R, Coelho C, Gonçalves T, Santos MAS, Pina-Vaz C, Solis NV, Filler SG, Rodrigues AG. 2013. Candida albicans CUG mistranslation is a mechanism to create cell surface variation. mBio 4(4):e00285-13. doi:10.1128/mBio.00285-13.
REFERENCES
- 1. Stringer JR, Keely SP. 2001. Genetics of surface antigen expression in Pneumocystis carinii. Infect. Immun. 69:627–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Deitsch KW. 2005. Malaria virulence genes controlling expression through chromatin modification. Cell 121:1–2 [DOI] [PubMed] [Google Scholar]
- 3. Rando OJ, Verstrepen KJ. 2007. Timescales of genetic and epigenetic inheritance. Cell 128:655–668 [DOI] [PubMed] [Google Scholar]
- 4. Verstrepen KJ, Reynolds TB, Fink GR. 2004. Origins of variation in the fungal cell surface. Nat. Rev. Microbiol. 2:533–540 [DOI] [PubMed] [Google Scholar]
- 5. Verstrepen KJ, Jansen A, Lewitter F, Fink GR. 2005. Intragenic tandem repeats generate functional variability. Nat. Genet. 37:986–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Halme A, Bumgarner S, Styles C, Fink GR. 2004. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell 116:405–415 [DOI] [PubMed] [Google Scholar]
- 7. Krinos CM, Coyne MJ, Weinacht KG, Tzianabos AO, Kasper DL, Comstock LE. 2001. Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature 414:555–558 [DOI] [PubMed] [Google Scholar]
- 8. De Las Peñas A, Pan SJ, Castaño I, Alder J, Cregg R, Cormack BP. 2003. Virulence-related surface glycoproteins in the yeast pathogen Candida glabrata are encoded in subtelomeric clusters and subject to RAP1- and SIR-dependent transcriptional silencing. Genes Dev. 17:2245–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boothroyd CE, Dreesen O, Leonova T, Ly KI, Figueiredo LM, Cross GA, Papavasiliou FN. 2009. A yeast-endonuclease-generated DNA break induces antigenic switching in Trypanosoma brucei. Nature 459:278–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sundstrom P. 2002. Adhesion in Candida spp. Cell. Microbiol. 4:461–469 [DOI] [PubMed] [Google Scholar]
- 11. Nobile CJ, Andes DR, Nett JE, Smith FJ, Yue F, Phan QT, Edwards JE, Filler SG, Mitchell AP. 2006. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2:e63. 10.1371/journal.ppat.0020063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Phan QT, Myers CL, Fu Y, Sheppard DC, Yeaman MR, Welch WH, Ibrahim AS, Edwards JE, Jr, Filler SG. 2007. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol. 5:e64. 10.1371/journal.pbio.0050064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Almeida RS, Brunke S, Albrecht A, Thewes S, Laue M, Edwards JE, Filler SG, Hube B. 2008. The hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin. PLoS Pathog. 4:e1000217. 10.1371/journal.ppat.1000217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Santos MA, Gomes AC, Santos MC, Carreto LC, Moura GR. 2011. The genetic code of the fungal CTG clade. C. R. Biol. 334:607–611 [DOI] [PubMed] [Google Scholar]
- 15. Gomes AC, Miranda I, Silva RM, Moura GR, Thomas B, Akoulitchev A, Santos MA. 2007. A genetic code alteration generates a proteome of high diversity in the human pathogen Candida albicans. Genome Biol. 8:R206. 10.1186/gb-2007-8-10-r206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Santos MA, Keith G, Tuite MF. 1993. Non-standard translational events in Candida albicans mediated by an unusual seryl-tRNA with a 5′-CAG-3 (leucine) anticodon. EMBO J. 12:607–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rocha R, Pereira PJ, Santos MA, Macedo-Ribeiro S. 2011. Unveiling the structural basis for translational ambiguity tolerance in a human fungal pathogen. Proc. Natl. Acad. Sci. U. S. A. 108:14091–14096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Miranda I, Rocha R, Santos MC, Mateus DD, Moura GR, Carreto L, Santos MA. 2007. A genetic code alteration is a phenotype diversity generator in the human pathogen Candida albicans. PLoS One 2:e996. 10.1371/journal.pone.0000996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Silva-Dias A, Miranda IM, Rocha R, Monteiro-Soares M, Salvador A, Rodrigues AG, Pina-Vaz C. 2012. A novel flow cytometric protocol for assessment of yeast cell adhesion. Cytometry A 81:265–270 [DOI] [PubMed] [Google Scholar]
- 20. Sheppard DC, Yeaman MR, Welch WH, Phan QT, Fu Y, Ibrahim AS, Filler SG, Zhang M, Waring AJ, Edwards JE., Jr. 2004. Functional and structural diversity in the Als protein family of Candida albicans. J. Biol. Chem. 279:30480–30489 [DOI] [PubMed] [Google Scholar]
- 21. Fu Y, Ibrahim AS, Sheppard DC, Chen YC, French SW, Cutler JE, Filler SG, Edwards JE., Jr. 2002. Candida albicans. Als1p: an adhesin that is a downstream effector of the EFG1 filamentation pathway. Mol. Microbiol. 44:61–72 [DOI] [PubMed] [Google Scholar]
- 22. Rauceo JM, Gaur NK, Lee KG, Edwards JE, Klotz SA, Lipke PN. 2004. Global cell surface conformational shift mediated by a Candida albicans adhesin. Infect. Immun. 72:4948–4955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rosenberg M, Gutnick D, Rosenberg E. 1980. Adherence of bacteria to hydrocarbons: a simple method for measuring cell-surface hydrophobicity. FEMS Microbiol. Lett. 9:29–33 [Google Scholar]
- 24. Ramsook CB, Tan C, Garcia MC, Fung R, Soybelman G, Henry R, Litewka A, O’Meally S, Otoo HN, Khalaf RA, Dranginis AM, Gaur NK, Klotz SA, Rauceo JM, Jue CK, Lipke PN. 2010. Yeast cell adhesion molecules have functional amyloid-forming sequences. Eukaryot. Cell 9:393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Salgado PS, Yan R, Taylor JD, Burchell L, Jones R, Hoyer LL, Matthews SJ, Simpson PJ, Cota E. 2011. Structural basis for the broad specificity to host-cell ligands by the pathogenic fungus Candida albicans. Proc. Natl. Acad. Sci. U. S. A. 108:15775–15779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gow NA, Netea MG, Munro CA, Ferwerda G, Bates S, Mora-Montes HM, Walker L, Jansen T, Jacobs L, Tsoni V, Brown GD, Odds FC, Van der Meer JW, Brown AJ, Kullberg BJ. 2007. Immune recognition of Candida albicans beta-glucan by dectin-1. J. Infect. Dis. 196:1565–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rubin-Bejerano I, Abeijon C, Magnelli P, Grisafi P, Fink GR. 2007. Phagocytosis by human neutrophils is stimulated by a unique fungal cell wall component. Cell Host Microbe 2:55–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Galán-Díez M, Arana DM, Serrano-Gómez D, Kremer L, Casasnovas JM, Ortega M, Cuesta-Domínguez A, Corbí AL, Pla J, Fernández-Ruiz E. 2010. Candida albicans beta-glucan exposure is controlled by the fungal CEK1-mediated mitogen-activated protein kinase pathway that modulates immune responses triggered through dectin-1. Infect. Immun. 78:1426–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gantner BN, Simmons RM, Underhill DM. 2005. Dectin-1 mediates macrophage recognition of Candida albicans yeast but not filaments. EMBO J. 24:1277–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Martínez-Esparza M, Sarazin A, Poulain D, Jouault T. 2009. A method for examining glycans surface expression of yeasts by flow cytometry. Methods Mol. Biol. 470:85–94 [DOI] [PubMed] [Google Scholar]
- 31. Berman J, Sudbery PE. 2002. Candida albicans: a molecular revolution built on lessons from budding yeast. Nat. Rev. Genet. 3:918–930 [DOI] [PubMed] [Google Scholar]
- 32. Glee PM, Cutler JE, Benson EE, Bargatze RF, Hazen KC. 2001. Inhibition of hydrophobic protein-mediated Candida albicans attachment to endothelial cells during physiologic shear flow. Infect. Immun. 69:2815–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li F, Palecek SP. 2008. Distinct domains of the Candida albicans adhesin Eap1p mediate cell-cell and cell-substrate interactions. Microbiology 154:1193–1203 [DOI] [PubMed] [Google Scholar]
- 34. Otoo HN, Lee KG, Qiu W, Lipke PN. 2008. Candida albicans Als adhesins have conserved amyloid-forming sequences. Eukaryot. Cell 7:776–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Alsteens D, Garcia MC, Lipke PN, Dufrêne YF. 2010. Force-induced formation and propagation of adhesion nanodomains in living fungal cells. Proc. Natl. Acad. Sci. U. S. A. 107:20744–20749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Netea MG, Brown GD, Kullberg BJ, Gow NAR. 2008. An integrated model of the recognition of Candida albicans by the innate immune system. Nat. Rev. Microbiol. 6:67–78 [DOI] [PubMed] [Google Scholar]
- 37. Lewis LE, Bain JM, Lowes C, Gillespie C, Rudkin FM, Gow NA, Erwig LP. 2012. Stage specific assessment of Candida albicans phagocytosis by macrophages identifies cell wall composition and morphogenesis as key determinants. PLoS Pathog. 8:e1002578. 10.1371/journal.ppat.1002578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McKenzie CG, Koser U, Lewis LE, Bain JM, Mora-Montes HM, Barker RN, Gow NA, Erwig LP. 2010. Contribution of Candida albicans cell wall components to recognition by and escape from murine macrophages. Infect. Immun. 78:1650–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wheeler RT, Kombe D, Agarwala SD, Fink GR. 2008. Dynamic, morphotype-specific Candida albicans beta-glucan exposure during infection and drug treatment. PLoS Pathog. 4:e1000227. 10.1371/journal.ppat.1000227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Netzer N, Goodenbour JM, David A, Dittmar KA, Jones RB, Schneider JR, Boone D, Eves EM, Rosner MR, Gibbs JS, Embry A, Dolan B, Das S, Hickman HD, Berglund P, Bennink JR, Yewdell JW, Pan T. 2009. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature 462:522–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Morley SJ, Willett M. 2009. Kinky binding and SECsy insertions. Mol. Cell 35:396–398 [DOI] [PubMed] [Google Scholar]
- 42. Carlson BA, Yoo MH, Sano Y, Sengupta A, Kim JY, Irons R, Gladyshev VN, Hatfield DL, Park JM. 2009. Selenoproteins regulate macrophage invasiveness and extracellular matrix-related gene expression. BMC Immunol. 10:57. 10.1186/1471-2172-10-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ambrogelly A, Palioura S, Söll D. 2007. Natural expansion of the genetic code. Nat. Chem. Biol. 3:29–35 [DOI] [PubMed] [Google Scholar]
- 44. Ribas de Pouplana L, Schimmel P. 2000. A view into the origin of life: aminoacyl-tRNA synthetases. Cell. Mol. Life Sci. 57:865–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pezo V, Metzgar D, Hendrickson TL, Waas WF, Hazebrouck S, Döring V, Marlière P, Schimmel P, De Crécy-Lagard V. 2004. Artificially ambiguous genetic code confers growth yield advantage. Proc. Natl. Acad. Sci. U. S. A. 101:8593–8597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Santos MA, Cheesman C, Costa V, Moradas-Ferreira P, Tuite MF. 1999. Selective advantages created by codon ambiguity allowed for the evolution of an alternative genetic code in Candida spp. Mol. Microbiol. 31:937–947 [DOI] [PubMed] [Google Scholar]
- 47. Bacher JM, Waas WF, Metzgar D, de Crécy-Lagard V, Schimmel P. 2007. Genetic code ambiguity confers a selective advantage on Acinetobacter baylyi. J. Bacteriol. 189:6494–6496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stothard P. 2000. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 28:1102-1104 [DOI] [PubMed] [Google Scholar]
- 49. Thompson JD, Higgins DG, Gibson TJ. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gouet P, Robert X, Courcelle E. 2003. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31:3320–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fernández-Arenas E, Cabezón V, Bermejo C, Arroyo J, Nombela C, Diez-Orejas R, Gil C. 2007. Integrated proteomics and genomics strategies bring new insight into Candida albicans response upon macrophage interaction. Mol. Cell. Proteomics 6:460–478 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparative histogram of the adherence profiles of wild-type C. albicans cells (dark line) and cells of the highly CUG-mistranslating strain (gray line). The intensity of fluorescence (FL3-H) is plotted against the number of cells (counts), and the results are from the analysis of 50,000 cells. The arrow indicates cells with no beads attached, and numbered peaks indicate the fluorescence of cells with 1, 2, or 3 beads attached. Download
CUG mistranslation does not influence the amount of surface-exposed Als1p and Als3p. Flow cytometric quantification of Als protein expression in wild-type (dark line) and highly CUG-mistranslating (gray line) cells, using an anti-Als3 polyclonal antibody that recognizes both Als3 and Als1. Download
Flow cytometric measurement of Als3p surface expression on S. cerevisiae expressing Als3p-Leu or Als3p-Ser. Download