To develop a culture model for drug development and tissue-engineering human retina, retinal pigment epithelia (RPE) were derived from human embryonic stem cells (hESCs), and their barrier properties were compared with those of a well-regarded model of RPE function, human fetal RPE isolated from 16-week-gestation fetuses (hfRPE). It was found that hESC-derived RPE is highly differentiated but may be less mature than RPE isolated from 16-week fetuses. The study also identified a panel of genes to monitor further maturation of RPE.
Keywords: Retinal pigmented epithelium, Blood-retinal barrier, Claudins, Embryonic stem cells
Abstract
Retinal degenerations are a major cause of impaired vision in the elderly. Degenerations originate in either photoreceptors or the retinal pigment epithelium (RPE). RPE forms the outer blood-retinal barrier and functions intimately with photoreceptors. Animal models and cultures of RPE are commonly used to screen potential pharmaceuticals or explore RPE replacement therapy, but human RPE differs from that of other species. Human RPE forms a barrier using tight junctions composed of a unique set of claudins, proteins that determine the permeability and selectivity of tight junctions. Human adult RPE fails to replicate these properties in vitro. To develop a culture model for drug development and tissue-engineering human retina, RPE were derived from human embryonic stem cells (hESCs). Barrier properties of RPE derived from the H1 and H9 hESC lines were compared with a well-regarded model of RPE function, human fetal RPE isolated from 16-week-gestation fetuses (hfRPE). A serum-free medium (SFM-1) that enhanced the redifferentiation of hfRPE in culture also furthered the maturation of hESC-derived RPE. In SFM-1, the composition, selectivity, and permeability of tight junctions were similar to those of hfRPE. Comparison of the transcriptomes by RNA sequencing and quantitative reverse transcription-polymerase chain reaction revealed a high correlation between the hESCs and hfRPE, but there were notable differences in the expression of adhesion junction and membrane transport genes. These data indicated that hESC-derived RPE is highly differentiated but may be less mature than RPE isolated from 16-week fetuses. The study identified a panel of genes to monitor the maturation of RPE.
Introduction
There is great interest in developing cultures of retinal pigment epithelium (RPE) that can replace RPE damaged by retinal diseases such as age-related macular degeneration—a leading cause of blindness among the elderly. The RPE is unusual, because its apical membrane abuts a tissue layer, the sensory (neural) retina. Both the RPE and sensory retina derive from a diverticulum of neuroepithelium, the optic vesicle. As the two tissues differentiate on opposite sides of the vesicle, the lumen collapses to become a potential space. Consequently, microvilli of the RPE apical membrane interdigitate with the outer segments of photoreceptors [1, 2]. The RPE separates photoreceptors from their blood supply in the choroid. Because the choroidal capillaries are fenestrated, RPE forms the outer blood-retinal barrier to maintain the specialized environment required by photoreceptors [3]. The outer segment of the photoreceptor contains a series of flat disc membranes that resemble a stack of coins. The chromophore, retinal, shuttles back and forth between the RPE and these discs. In the discs, light is transduced into an electrical signal by converting 11-cis retinal to 11-trans retinal. Only the RPE can isomerize retinal back to the cis form. The visual cycle is completed when the retinal is transported back to the photoreceptors [4]. Daily, new discs are added to the base of the outer segments, whereas old discs are shed from the tips and phagocytized by the RPE [5]. These close functional relationships explain why pathology in one tissue often leads to dysfunction or death of the other, as observed in age-related macular degeneration and retinitis pigmentosa [6].
Stem cells are an attractive source of RPE, and based on successes in rodent models of retinal degeneration, phase I clinical trials using stem cell-derived RPE are in progress [7]. In rodents, investigators were encouraged to find that RPE derived from human embryonic stem cells (hESC-RPE) and human induced pluripotent cells (iPS-RPE) formed cobblestone monolayers with melanin granules and tight junctions, expressed RPE signature genes [8, 9], phagocytized rod outer segments, and improved vision [8, 10–13]. A shortcoming of the animal studies was that transplantation was successful only when performed early in the disease and failed to restore vision in late-stage disease. Further, there is no current evidence that transplanted RPE establishes an outer blood-retinal barrier [10]. The barrier function of the tight junctions in hESC- or iPS-RPE has not been thoroughly investigated, because the properties of human RPE junctions have only recently been reported [14–16].
Tight junctions form a partially occluding seal that surrounds each cell of an epithelial monolayer, joining it to its neighbors [3, 17, 18]. The junctions semiselectively retard the diffusion of solutes across the monolayer via the paracellular spaces. Permeability and selectivity are determined by occludin and members of the claudin family [19]. In rodent RPE, claudin-1 is the only detectable claudin, but in chick RPE claudin-20 also plays a major role [18, 20, 21]. In contrast, human RPE expresses predominantly claudin-19, which accounts for the electrophysiological properties of its tight junctions [15, 16]. Further, its absence causes retinal disease [22]. Because the function of tight junctions is coordinated with the plasma membrane pumps and channels that comprise the transcellular contribution to barrier function [3], these differences in composition among the species imply differences in the physiology of the outer blood-retinal barrier.
Just as the choroid and sensory retina differentiate in a gradual, coordinated process, so do the RPE and their tight junctions [1, 2, 23]. During the development of chick RPE, tight junctions develop gradually under a process that is regulated by secretions of the sensory retina [20, 21]. Initially, claudin-5 is the only claudin in evidence. Then, claudin-1 expression rises to become the predominant claudin. In the final stages of development, claudin-20 mRNA comes to be expressed in the highest copy number. Maturation of tight junctions corresponds to the time the choriocapillaris become fully fenestrated, RPE basolateral infoldings are fully elaborated, the intervening Bruch's membrane acquires its five layers of extracellular matrix, and the RPE mechanisms for transcellular transport of glucose are established. In other words, tight junctions mature after other elements of the outer blood-retinal barrier are set in place. The relationship between tight junction and retinochoroidal development has not been studied in human RPE, nor has the expression of claudin-19 or tight junction function been examined in stem cell-derived RPE.
To examine human retinal development in a culture model, it would be useful to have a medium that is compatible with RPE and retinal cells. Such a serum-free medium was devised by Gamm et al. for the culture of RPE and retinal progenitor cells that were isolated from 13-week-gestation (WG) human fetuses (hfRPE) and hESC-derived RPE and retinal progenitors [24, 25]. Recently, we delineated the properties of RPE tight junctions in a well-defined culture model of hfRPE that was derived from 16-WG human fetuses and adapted to the Gamm medium, which we named SFM-1 [16, 26]. We found that SFM-1 promoted the maturation of hfRPE [14–16].
Removing the variable of culture medium allows a direct comparison of hESC-RPE with cultures of hfRPE. An earlier study that compared hESC-RPE with hfRPE used different media to maintain the cells [8]. That study focused on a comparison of hESC-RPE, iPS-RPE, and hfRPE using microarrays to analyze the transcriptome, and left two questions open: (a) these researchers pooled data from hfRPE isolated from 16–21-WG fetuses, which assumes that hfRPE does not mature during fetal life, and (b) they pooled data from HSF1 and H9 hESC lines without examining variability among preparations or how HSF1-RPE and H9-RPE might differ from one another.
To investigate these concerns, we compared the transcriptomes of three independent isolates each from H1-RPE, H9-RPE, and hfRPE cultured from 16-WG human fetuses. Using RNA sequencing, we found that culture medium has a profound effect on gene expression. When maintained in SFM-1, H1-RPE and H9-RPE were quantitatively similar to each other. They were also similar to hfRPE, but significant differences could be found among barrier function-related genes.
Materials and Methods
Cell Culture
For comparison, primary cultures of hfRPE cells were prepared from fetuses of 16 WG, as described previously [15]. Briefly, primary cultures of hfRPE were supplied by the laboratory of Sheldon Miller (National Eye Institute, Bethesda, MD) and subcultured on Transwell or Snapwell culture inserts (1.12 cm2 growth area, 0.4 μm pores; Corning Enterprises, Corning, NY, http://www.corning.com) that were coated with 10 μg of human extracellular matrix (BD Biosciences, Franklin Lakes, NJ, http://www.bdbiosciences.com), according to that laboratory's method [26]. The volumes of medium in the apical chamber and the basolateral chamber were 0.6 and 1.5 ml, respectively for the Transwell, and 0.5 and 2.0 ml for the Snapwell. The cultures were maintained at 37°C in a humidified atmosphere of 95% air/5% CO2. The transepithelial electrical resistance (TER) was monitored using an EVOM2 resistance meter with Endohm electrodes (World Precision Instruments, Sarasota, FL, http://www.wpiinc.com). After 4–6 weeks they became stable, whereupon the cultures were adapted to SFM-1 over the course of 4 weeks [15]. SFM-1 was formulated to culture hfRPE [24] and consisted of 70% Dulbecco's modified Eagle's medium (DMEM) containing 4.5 g/liter d-glucose, 30% F12 nutrient mixture containing l-glutamine, and 1% antibiotic-antimycotic solution, supplemented with 2% B27 (Invitrogen, Carlsbad, CA, http://www.invitrogen.com).
H1-RPE and H9-RPE were derived from the H1 and H9 hESC lines, respectively, by the method of Idelson et al. [27]. Briefly, stem cells were cultured and passaged on Matrigel-coated dishes (BD Biosciences). Embryoid bodies were formed by treating undifferentiated stem cell colonies with 5 mg/ml dispase (StemCell Technologies, Vancouver, BC, Canada, http://www.stemcell.com) and cultured as floating clusters in knockout serum replacement medium (KSR), composed of DMEM/F12 (1:1) medium, 14% knockout serum replacement (Invitrogen), 1% nonessential amino acids (Sigma-Aldrich, St. Louis, MO, http://www.sigmaaldrich.com), 2 mM glutamine (Invitrogen), 50 U/ml penicillin, 50 μg/ml streptomycin (Invitrogen), and 10 mM nicotinamide (Sigma-Aldrich) in six-well ultra-low-attachment cluster plates (Costar, Corning) for 1 week. The embryoid bodies were plated on laminin-coated culture dishes (10 μg/ml) for 6 weeks. During the third and fourth weeks, KSR was supplemented with 140 ng/ml actin A (Peprotech, Rocky Hill, NJ, http://www.peprotech.com). After the sixth week, pigmented epithelial cells were isolated by trypsinization and seeded onto laminin-coated Transwell or Snapwell culture inserts (1.3 × 105 cells per well). Notably, we found that the morphology of the cells was irregular when cultured on the human extracellular matrix-coated filters that promoted the differentiation of hfRPE [26]. On the human matrix, cells formed multilayered clusters or irregularly shaped polygons. Alternatively, differentiated cells were expanded on laminin-coated dishes and purified by weeding unpigmented cells before reseeding on culture inserts. The reseeded cultures were maintained in KSR for 6–8 weeks, when the cultures regained their pigmentation. In some cultures the KSR was then replaced with SFM-1, which had no effect on pigmentation, and the cultures were followed for an additional 4–5 weeks.
Quantitative Reverse Transcription-Polymerase Chain Reaction
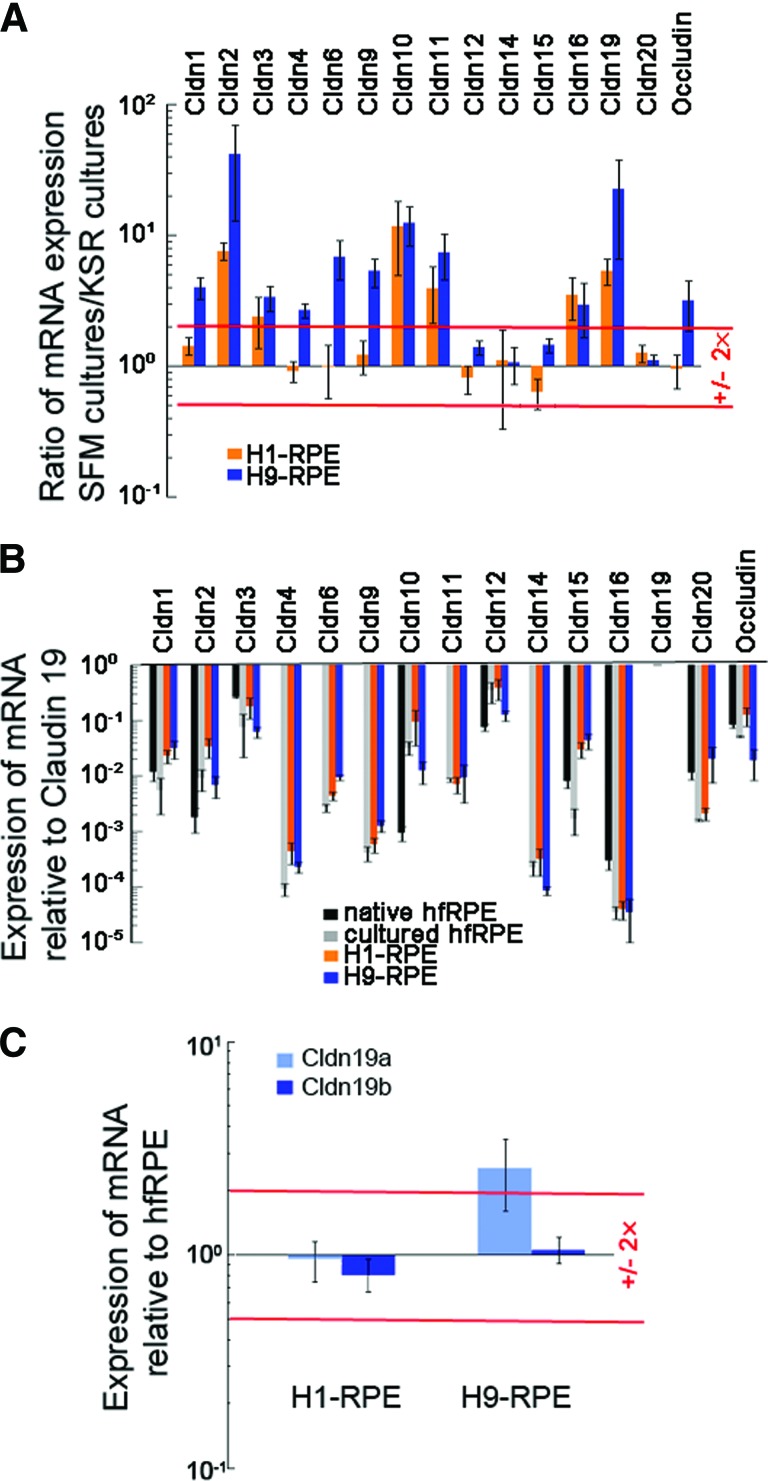
To screen claudin and occludin expression, reverse transcription-polymerase chain reaction was performed as described earlier [14], using 35 cycles of polymerase chain reaction (PCR). Claudins detected by this technique, and other select mRNAs, were further examined by quantitative real-time reverse transcription-polymerase chain reaction (qRT2-PCR) and RNA sequencing as follows. Total RNA was extracted using the RNeasy Mini Kit (Qiagen, Valencia, CA, http://www.qiagen.com). Two micrograms of total RNA was reverse-transcribed to cDNA using the Quantitect Reverse Transcription Kit (Qiagen). The primers used in these experiments are listed in supplemental online Table 1. Experiments were performed in triplicate with a minimum of two biological repeats. Reaction products were analyzed by gel electrophoresis to confirm that a single reaction product of the calculated size was obtained. Relative expression of mRNA was calculated using the 2−ΔΔCT method [28]. Briefly, the data were first normalized to the expression of GAPDH and then to the expression of a reference mRNA. As indicated in Figure 4, the reference mRNA was the same mRNA expressed in hfRPE or in cultures maintained in KSR. Alternatively, the reference mRNA was claudin-19 that was expressed in the same culture. Means and error bars in Figure 4 were calculated from three independent experiments, each performed in triplicate.
Figure 4.
Expression of claudin mRNA increased in SFM-1 to resemble expression in hfRPE. Total RNA was analyzed by quantitative real-time reverse transcription-polymerase chain reaction. A value of 1.0 means that the level of expression equaled that found in the reference sample. (A): Relative to cultures maintained in KSR, expression increased for claudins normally found in hfRPE. In H9-RPE, SFM-1 also increased expression for some claudins absent in native RPE. (B): Expression relative to claudin-19 was similar to that in native and cultured hfRPE. (C): Expression of Cldn-19 isoforms relative to that in hfRPE. Error bars indicate the SE of three independent experiments. Red lines indicate twofold over- or underexpression. Data for hfRPE in (B) were taken from Peng et al. [15]. Abbreviations: Cldn, claudin; hfRPE, human fetal retinal pigment epithelium; KSR, knockout serum replacement medium; RPE, retinal pigment epithelium; SFM, serum-free medium.
RNA Sequencing and Analysis
Total RNA was isolated from RPE that was differentiated and isolated from human embryonic stem cell lines H1 and H9 (three independent isolates from each strain) and compared with RPE that was isolated from three 16-WG human fetuses and expanded in primary culture, as described above. Libraries were prepared according to Illumina's mRNA Sequencing Sample Preparation Guide for mRNA-SeqSample Prep Kit (part no. RS-100-0801) and TruSeq RNA Sample Prep Kit v2 (part no. RS-122-2001) (Illumina Inc., San Diego, CA, http://www.illumina.com). Briefly, poly(A)-containing mRNA molecules were purified from 10 μg of total RNA using poly(T) oligo-attached magnetic beads and then fragmented into small pieces using divalent cations under elevated temperature. The cleaved RNA fragments were copied into first-strand cDNA using reverse transcriptase (SuperScript II; Invitrogen) and random primers followed by second-strand cDNA synthesis using DNA polymerase I and RNaseH. These cDNA fragments then went through an end repair process using a combination of T4 DNA polymerase, Escherichia coli DNA Pol I large fragment (Klenow polymerase), and T4 polynucleotide kinase. The blunt, phosphorylated ends were treated with Klenow fragment (3′ to 5′ exo minus) and dATP to yield a protruding three-A base for ligation of Illumina's adapters, which have a single T base overhang at the 3′ end. These products were then purified and enriched with 15 cycles of PCR to create the final cDNA library. Some library fragments were further purified to remove the adaptor dimer. Insert plus adaptor and PCR primer sequences of 150–350 base pairs were isolated from 2% E-Gel EX Gel (G4020-02; Invitrogen). The purified DNA was captured on an Illumina flow cell for cluster generation. Libraries were sequenced on the Genome Analyzer and HiSeq 2000 following the manufacturer's protocols. Basecalls were performed using Illumina's Consensus Assessment of Sequence and Variation (CASAVA) software. Sequenced reads were imported into the public Galaxy platform (a free online bioinformatics interface available at http://main.g2.bx.psu.edu). The sequences were aligned against the hg19 reference genome using the Tophat for Illumina (version 1.5.0) with default parameters. Transcripts were assembled using Cufflinks (version 0.0.5) with quartile normalization and bias correction. The expression levels for each gene were normalized to reads per kilobase of exon model per million mapped reads to facilitate the comparison of transcripts among samples. Fold change in transcript expression and statistical significance were analyzed using Cuffdiff (version 0.0.5) with false discovery rate of 0.05, minimum alignment count of 100, quartile normalization, and bias correction. Data were deposited in the Gene Expression Omnibus database, accession no. GSE36695. The results for genes of interest were verified by qRT2-PCR.
Immunoblotting and Immunocytochemistry
Cultures were solubilized and prepared for immunoblotting, as described [14]. The level of α-tubulin staining was used as an internal standard to normalize each sample. The following primary antibodies from Invitrogen were used (dilutions in parentheses): rabbit polyclonal anti-claudin-1 (1:100), rabbit polyclonal anti-claudin-2 (1:100), rabbit polyclonal anti-claudin-3 (1:200), mouse monoclonal anti-occludin (1:200), and mouse monoclonal anti-α-tubulin (1:1,500). Mouse anti-E-cadherin (1:5,000) was obtained from BD Biosciences, mouse monoclonal anti-N-cadherin (1:100 dilution) was from Sigma-Aldrich, and rabbit polyclonal anti-claudin-19 (1:200) was a kind gift from Mikio Furuse (Kobe University, Kobe, Japan). The immunoblots were developed using horseradish peroxidase-conjugated secondary antibodies (1:3,000) (Thermo Scientific, Rockford, IL, http://www.thermofisher.com) and SuperSignal West Femto reagent (Thermo Scientific) and were imaged and quantified using a Molecular Imager ChemiDoc XRS System (Bio-Rad, Hercules, CA, http://www.bio-rad.com).
The subcellular distribution of the claudins, ZO-1, and occludin was determined by indirect immunofluorescence, as described [14]. Briefly, cells were fixed in 100% ethanol at 4°C. Besides the antibodies used for immunoblotting, samples were counterlabeled with mouse anti-occludin (1:200) or mouse anti-ZO-1 (1:400) (Invitrogen). The dilutions used for claudin-3 and claudin-19 were reduced to 1:500 and 1:1000, respectively. ML-grade secondary antibodies conjugated with Cy3 or Cy5 dyes (1:200) (Jackson Immunoresearch Laboratories, West Grove, PA, http://www.jacksonimmuno.com). To reveal nuclei and filamentous actin, we used Alexa Fluor 488 phalloidin and 4′,6-diamidino-2-phenylindole. Fluorescence images were acquired with an LSM 410 spinning-disc confocal microscope and processed using AxioVision software (Carl Zeiss, Thornwood, NY, http://www.zeiss.com). Fluorescent channels were captured in gray scale and false-colored using the software, as described in the figure legends.
Estimation of Permeability and Selectivity
Permeability and selectivity were estimated from the TER and transepithelial electrical potential (TEP), as described previously [16]. Briefly, cultures maintained on Snapwell culture inserts were mounted in Ussing chambers (Physiologic Instruments, San Diego, CA, http://www.physiologicinstruments.com) and incubated in a modified Ringer's solution to inhibit membrane transport. Silver chloride electrodes with 3 M KCl agar bridges were used for the current producing and voltage sensing electrodes and were controlled by a model VCC MC6 voltage/current clamp and Acquire & Analyze Revision II software (Physiologic Instruments). The TEP was referenced to the basolateral chamber. For ion selectivity and permeability, dilution and bi-ionic electrical potentials were examined by replacing the solution in the basolateral chamber. Several experiments were performed by replacing the solution in the apical chamber to confirm that the measurements were independent of the direction of the gradient. For dilution potentials, the NaCl in the basolateral chamber was reduced to 75 mM NaCl and the osmolality was balanced with mannitol. For bi-ionic potentials, the NaCl was replaced with 150 mM KCl. The relative ionic permeabilities of the monolayers were calculated using the Goldman-Hodgkin-Katz equation. The individual permeation coefficients for Na+ (PNa), Cl− (PCl), and K+ (PK) were deduced from the method of Kimizuka and Koketsu [29] (see also Yu et al. [30]). Preliminary studies demonstrated that the matrix coating applied to the filters had no effect on permeability or selectivity. Accordingly, bare filters were used for controls in most experiments.
Statistical Analysis
Statistical differences were determined using Student's t test or one-way analysis of variance, except as described above for RNA sequence analysis.
Results
We confirmed that H1-RPE and H9-RPE form confluent, pigmented, low-cuboidal monolayers in KSR, and expressed a number of RPE marker mRNAs including bestrophin, cellular retinaldehyde-binding protein (CRALBP), retinol isomerase (RPE65), pigment epithelium-derived factor (PEDF), microphthalmia-associated transcription factor (MITF), tyrosinase (TYR), and cone-rod homeobox protein (CRX). Adaptation to SFM-1 had minimal effect on pigmentation or the expression of these mRNAs. When expression by hESC-derived RPE was compared with expression by native hfRPE, the levels of expression were similar to those of hfRPE, except for CRX (supplemental online Fig. 1).
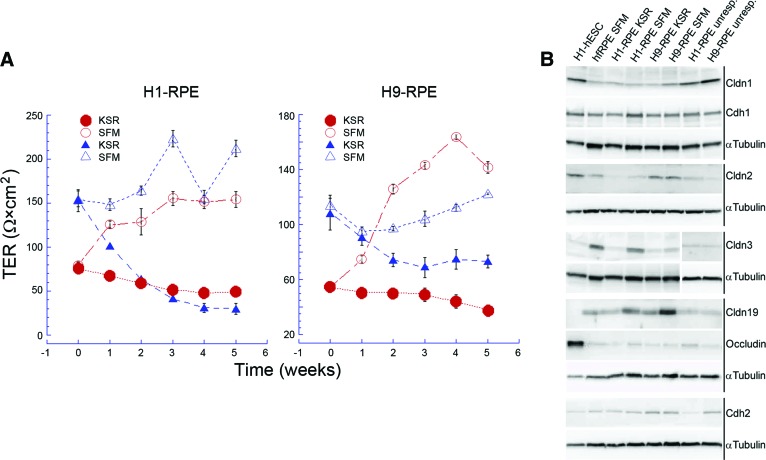
TER was used to assess the integrity of the monolayer and its tight junctions (Fig. 1A). TER varied from preparation to preparation, but SFM-1 narrowed the range of TER values. The cells were differentiated and replated onto filter inserts in KSR. After 6–8 weeks, half of the cultures were transferred to SFM-1 and the control cultures were maintained in KSR. There was no effect on pigmentation or morphology. When the TER was higher at the time of the transfer, 100–150 Ω×cm2, it decreased in control cells but remained high in cultures transferred to SFM-1. When the TER was lower at the time of transfer, ∼60 Ω×cm2, it often increased in the SFM-1 cultures compared with the controls. For reference, the TER of hfRPE in retinal explants of 19–23-WG eyes was reported to be 206 ± 151 Ω×cm2 [31], and that of hfRPE cultured in SFM-1 lay in the range of 250–400 Ω×cm2 [15]. To investigate the effects of SFM-1, we examined the expression and subcellular localization of occludin, ZO-1, and the claudins expressed by native RPE.
Figure 1.
SFM-1 increased and stabilized the TER and increased the expression of claudin-3 and claudin-19. (A): SFM-1 supports a TER that approximates that of hfRPE. At time zero, three cultures were transferred to SFM-1 (open symbols), while three cultures remained in KSR (closed symbols). Error bars indicate the SE and are sometimes smaller than the symbol. (B): Immunoblots demonstrate that steady-state levels of claudin-3 and claudin-19 increased in SFM-1. For comparison, preparations in which TER was unresponsive to SFM-1 are included. Abbreviations: Cdh1, epithelial-cadherin; Cdh2, neural-cadherin; Cldn, claudin; hESC, human embryonic stem cell; hfRPE, human fetal retinal pigment epithelium; KSR, knockout serum replacement medium; RPE, retinal pigment epithelium; SFM, serum-free medium; TER, transepithelial electrical resistance; unresp., unresponsive.
SFM-1 affected the steady-state level of several claudins (Fig. 1B). Of the claudins normally expressed by human RPE, only claudin-1 and claudin-2 were expressed by undifferentiated hESCs along with occludin. Relative to α-tubulin, the expression of both proteins was lower in hfRPE. In H1- and H9-derived RPE, SFM-1 increased steady-state levels of claudin-3 and claudin-19, which previous study demonstrated were the predominant claudins expressed by hfRPE [15]. SFM-1 also increased the steady-state level of claudin-2 in H1-RPE to that of hfRPE, but expression in H9-RPE approximated hfRPE in both media. Some cultures failed to respond to SFM-1 even though the cultures were confluent, pigmented, and displayed a cobblestone morphology. In those cases the TER was low, the steady-state level of claudin-3 and claudin-19 was low, and the steady-state level of claudin-1 was similar to that of hESCs or higher. Those cultures were excluded from further analysis. With regard to claudin and occludin expression, the differences between H1-RPE and H9-RPE were negligible.
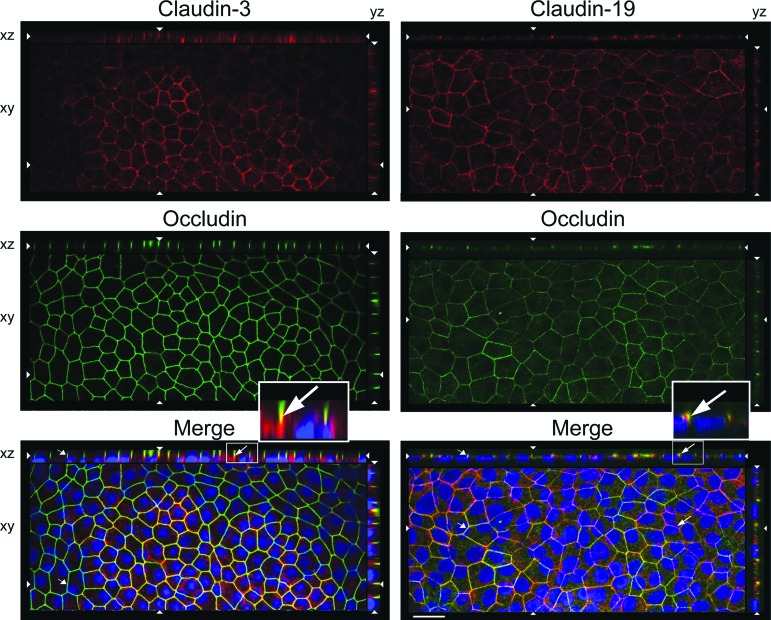
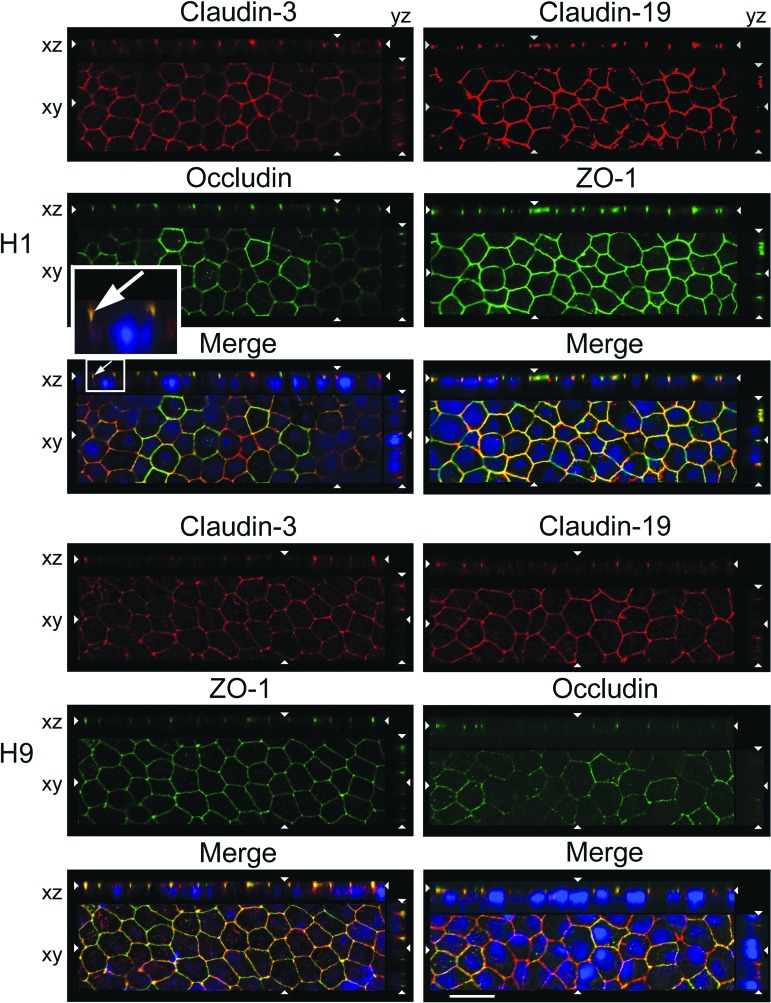
In H1-RPE only, SFM-1 also affected the subcellular localization of claudin-3. Confocal microscopy of KSR cultures reveal that the classic tight junction markers, occludin and ZO-1, were found in the expected location at the apical end of the lateral membranes (Fig. 2). Occludin and claudin-19 colocalized, but whereas occludin was evident in all cells, claudin-19 was undetected in some. Claudin-3 was observed in fewer cells, and there it localized to lateral membranes instead of tight junctions. When the cultures were adapted to SFM-1, both claudins were expressed by all cells and each localized to tight junctions (Fig. 3). The cobblestone morphology of the cultures and polarization of the tight junctions were similar to those in hfRPE [15, 26].
Figure 2.
Claudin-3 was mislocalized in H1-retinal pigment epithelium maintained in knockout serum replacement medium. Confocal images were obtained in the xy, xz, and yz planes. The xy plane includes the tight junctions (revealed by occludin). The apical membrane faces the top (xz) or right (yz). 4′,6-Diamidino-2-phenylindole-labeled nuclei appear blue in the merged image. Long arrows, colocalization (orange) for claudin-19 but not claudin-3; short arrows, absence of claudin; opposing arrowheads, cut-lines for xy, xz, and yz planes. Scale bar = 20 μm.
Figure 3.
Claudin-3 localized to tight junctions in human embryonic stem cell-derived retinal pigment epithelium maintained in serum-free medium 1 (SFM-1). Cultures were adapted to SFM-1 as described in Figure 1A. Images were acquired and labeled as described in the legend to Figure 2. Scale bar = 20 μm.
Claudin-1 and claudin-2 were only detected in subsets of hfRPE [15]. In H1- and H9-RPE, claudin-1 and claudin-2 were found in occasional, distinct colonies within the RPE monolayer. It appeared as though cells that expressed one or the other claudin preferred to self-associate rather than integrate with the rest of the monolayer (supplemental online Fig. 2). Unlike the hfRPE cultures, claudin-1- and claudin-2-positive cells in the hESC cultures had an altered morphology and disorganized actin cytoskeleton. In supplemental online Figure 2, the actin images were overexposed so that melanin granules would become evident. There was no apparent correlation between the level of any claudin's expression and the amount of pigmentation.
The claudins were analyzed by qRT2-PCR and compared with expression in hfRPE in vivo and in culture. SFM-1 increased the expression of many mRNAs >2×, including several that are not normally found in RPE (Fig. 4A). Claudin-19 mRNA increased 5× in H1-RPE and 20× in H9-RPE. To put these increases in perspective, claudin expression in SFM-1 was normalized to claudin-19 and compared with that in hfRPE (Fig. 4B). As in hfRPE, claudin-19 was by far the dominant claudin, followed by claudin-3 and claudin-12 (although the presence of claudin-12 could not be confirmed by immunoblotting even in hfRPE) [15]. Although SFM-1 appeared to have the greatest effect on H9-RPE, the end result was that claudin expression was nearly the same in H1-RPE, H9-RPE, and cultured hfRPE. The exceptions were low-level expression of claudin-4, claudin-6, claudin-9, and claudin-14, which were undetected in hfRPE. Figure 4B also demonstrates that expression in SFM-1 closely resembled expression in freshly isolated hfRPE.
Claudin-19 is the dominant claudin in RPE, because small interfering RNA knockdown of only this claudin is sufficient to eliminate the TER [15]. H1- and H9-RPE differed in their expression of claudin-19 isoforms (Figs. 4C, 5). In cultured hfRPE and H1-RPE, the mRNA for claudin-19b was expressed at 2.5× the level of claudin-19a. For H9-derived RPE, claudin-19a, but not claudin-19b, mRNA was overexpressed relative to hfRPE. Consequently, the ratio of 19b:19a was reduced to 1:1. The difference in these isoforms lies in the short cytoplasmic domain at the C terminus, which interacts with scaffold and regulatory proteins of the tight junction.
Figure 5.
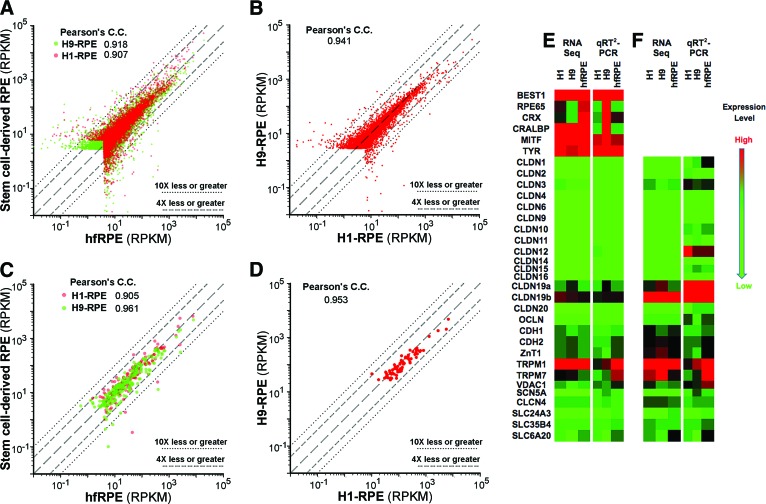
Gene expression levels in human embryonic stem cell-derived RPE correlated with those in hfRPE. RPKM values from RNA sequencing were plotted in two-way comparisons. (A, B): Total transcriptome. (C, D): Signature genes [19]. Long dashes, line of identity; short dashes, 4× over- or underexpression; dots, 10× over- or underexpression. (E, F): Validation of RNA sequencing by qRT2-PCR. The extremes of the color scale were matched to the highest and lowest expressed gene for each gene set. Abbreviations: C.C., correlation coefficient; hfRPE, human fetal retinal pigment epithelium; qRT2-PCR, quantitative real-time reverse transcription-polymerase chain reaction; RPE, retinal pigment epithelium; RPKM, reads per kilobase of exon model per million mapped reads; Seq, sequencing.
The analysis was broadened to include the entire transcriptome by quantitatively sequencing total mRNA (Fig. 5). SFM-1 had a large effect on total gene expression (Table 1; supplemental online Figs. 3–8). The overall effect of SFM-1 was to reduce deviations from the line of identity when hESC-RPE was compared with hfRPE maintained in SFM-1. Aside from increases for claudin-3 and claudin-19 mRNAs, SFM-1 caused few changes in the expression of tight junction-related mRNAs. Comparisons with hfRPE maintained in KSR were not made, because KSR-induced hfRPE to dedifferentiate as noted by a 90% decrease in the TER, distorted morphology, and increased number of apical stress fibers (supplemental online Fig. 9).
Table 1.
Comparison of mRNA expression between hESC-RPE and hfRPE
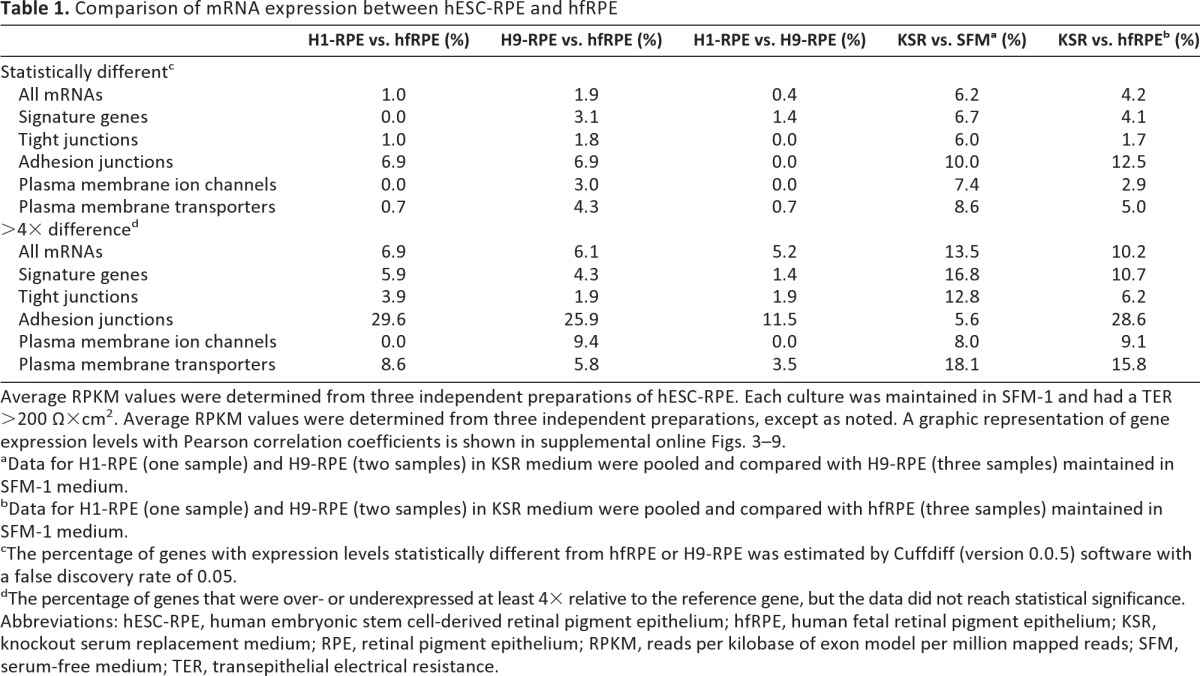
Average RPKM values were determined from three independent preparations of hESC-RPE. Each culture was maintained in SFM-1 and had a TER >200 Ω×cm2. Average RPKM values were determined from three independent preparations, except as noted. A graphic representation of gene expression levels with Pearson correlation coefficients is shown in supplemental online Figs. 3–9.
aData for H1-RPE (one sample) and H9-RPE (two samples) in KSR medium were pooled and compared with H9-RPE (three samples) maintained in SFM-1 medium.
bData for H1-RPE (one sample) and H9-RPE (two samples) in KSR medium were pooled and compared with hfRPE (three samples) maintained in SFM-1 medium.
cThe percentage of genes with expression levels statistically different from hfRPE or H9-RPE was estimated by Cuffdiff (version 0.0.5) software with a false discovery rate of 0.05.
dThe percentage of genes that were over- or underexpressed at least 4× relative to the reference gene, but the data did not reach statistical significance.
Abbreviations: hESC-RPE, human embryonic stem cell-derived retinal pigment epithelium; hfRPE, human fetal retinal pigment epithelium; KSR, knockout serum replacement medium; RPE, retinal pigment epithelium; RPKM, reads per kilobase of exon model per million mapped reads; SFM, serum-free medium; TER, transepithelial electrical resistance.
The remaining analysis focused on comparisons among cultures maintained in SFM-1. The RNA-sequencing data were validated by qRT2-PCR analysis of selected genes related to signature genes or the blood-retinal barrier. To a first approximation, the RNA sequencing data mirrored the qRT2-PCR analysis, although several exceptions could be found (Fig. 5E, 5F).
The transcriptomes of H1- and H9-RPE resembled those of hfRPE, although H1-RPE was nominally a closer match (Table 1; Fig. 5; supplemental online Figs. 3–8). When subsets of genes were examined, H1- and H9-RPE resembled each other more closely than hfRPE, especially for adhesion junctions and plasma membrane transporters. From the categories listed in Table 1, we identified 25 genes that were expressed at significantly different levels from hfRPE, or were over- or underexpressed at least 4× in both H1- and H-9 RPE (supplemental online Table 2).
Unexpectedly, the mRNA for an adherens junction protein, epithelial-cadherin (E-cadherin, Cdh1), was expressed in high amount. In humans, neural-cadherin (N-cadherin, Cdh2) is the predominant cadherin expressed by adult RPE [32]. In other vertebrates, E-cadherin is only found in a subset of RPE, if at all [32, 33]. Immunoblotting confirmed that E-cadherin was present in hfRPE, H1-RPE, and H9-RPE. Like the expression of N-cadherin, the expression of E-cadherin was unaffected by SFM-1 (Fig. 1B).
Because claudins determine the permeability and selectivity of tight junctions, the preceding data suggest that these parameters should be very similar for hESC-RPE and hfRPE. To test this hypothesis, cultures were mounted in an Ussing chamber and bathed with a Ringer's solution that lacked bicarbonate but contained BaCl2, to inhibit bicarbonate transporters and K+ channels, respectively. Under these conditions, transcellular transport was inhibited, as evidenced by the drop in TEP from ∼1 mV to near zero, and the TER approximated the resistance of the tight junctions. Because the junction resistance is lower than the monolayer resistance and dependent on temperature, and because the electrodes are of superior quality, the TER reported in Table 2 is approximately half that measured by Endohm electrodes at ambient temperature. The bare filter was used as a control to assess the effects of liquid junction potentials. The TEP was zero and the electrical resistance of the filter was <10% of the TER for the leakiest monolayer. The bi-ionic and Na+ dilution potentials were used to calculate the permeation coefficients and permeation ratios. The values obtained for the bare filter approximated those predicted from published values [34] and were substantially different from those of the cultures.
Table 2.
The permeability and selectivity of hESC-derived RPE are similar to those of hfRPE in SFM-1
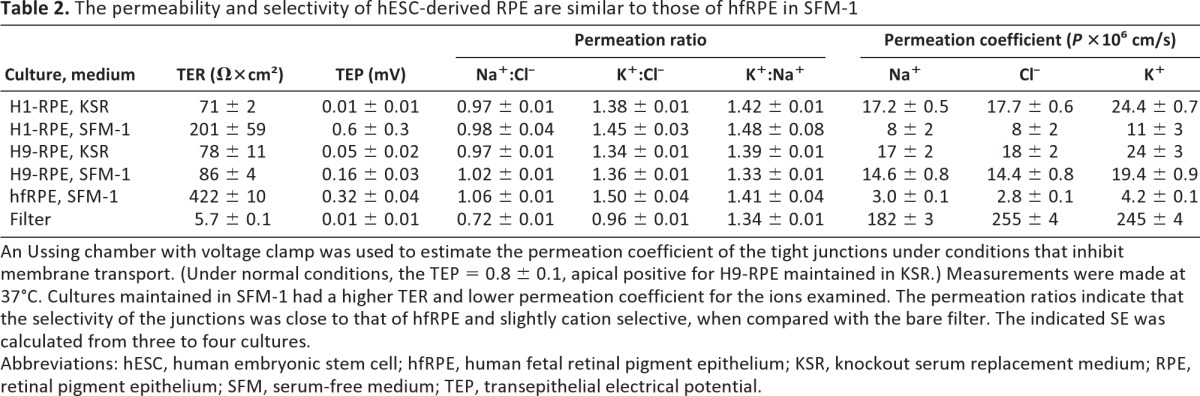
An Ussing chamber with voltage clamp was used to estimate the permeation coefficient of the tight junctions under conditions that inhibit membrane transport. (Under normal conditions, the TEP = 0.8 ± 0.1, apical positive for H9-RPE maintained in KSR.) Measurements were made at 37°C. Cultures maintained in SFM-1 had a higher TER and lower permeation coefficient for the ions examined. The permeation ratios indicate that the selectivity of the junctions was close to that of hfRPE and slightly cation selective, when compared with the bare filter. The indicated SE was calculated from three to four cultures.
Abbreviations: hESC, human embryonic stem cell; hfRPE, human fetal retinal pigment epithelium; KSR, knockout serum replacement medium; RPE, retinal pigment epithelium; SFM, serum-free medium; TEP, transepithelial electrical potential.
SFM-1 increased the TER and decreased ion permeability, as demonstrated by the preparation shown for H1-derived RPE. The data for H9-derived RPE is from a preparation that was minimally responsive to SFM-1 (Table 2). As would be expected from the claudin-expression data, selectivity varied little even though the permeability was inversely proportional to the TER. For cultures that were most responsive to SFM-1, permeability and selectivity of the junctions were nearly the same as observed for hfRPE.
Discussion
A molecular definition of an RPE cell was first proposed by Strunnikova et al. [9], who identified 154 “signature genes” that were either unique to RPE or expressed >10× higher than in other tissues. Liao et al. [8] refined the list by eliminating 67 genes that were also expressed in hESCs. We took another approach that is not restricted to RPE-enriched genes but is suited to addressing the question of whether differentiated RPE can be induced to mature further in culture. The concept is that beyond simple expression, proteins belong to intersecting networks where they must be expressed in balance with other proteins to achieve mature cellular function. The study demonstrated that the expression of many genes and the barrier properties of tight junctions can mature in culture. It also identified a set of 25 candidate genes that will be used to assess future studies on whether interactions with choroid or neural retina might further maturation.
Liao et al. [8] identified a different set of 25 signature genes that were underexpressed by their stem cell-derived RPE relative to hfRPE. SFM-1 increased the expression of eight of these to levels found in hfRPE (supplemental online Table 3). The remaining genes were already expressed at levels found in hfRPE, but note that our cultures were isolated from fetuses that shared the same age and were adapted to SFM-1. In quantitative analyses of the entire transcriptome, SFM-1 narrowed the deviation of hESC-RPE from hfRPE. By contrast, KSR reduced the barrier function of hfRPE to low levels and disrupted morphology. These data suggest the hypothesis that KSR fosters differentiation, SFM-1 furthers maturation, and hfRPE isolated from 16-WG fetuses is the most mature. To examine this hypothesis, consider normal development.
RPE differentiates before other retinal cells and matures in parallel with the differentiation of the choriocapillaris and the neural retina [35, 36]. As detailed in the Introduction, RPE tight junctions of nonprimate vertebrates complete maturation after other elements of the outer-blood retinal barrier are in place. The process is regulated by retinal secretions [20, 21].
Human retina deviates from this pattern [1]. Fenestration of the choriocapillaris (21–22 WG) is completed when photoreceptor inner segments protrude the outer limiting membrane (20–24 WG) [37], which is well before outer segments become evident (24–28 WG). Maturation of RPE tight junctions precedes these milestones. RPE isolated from 13- or 16-WG fetuses exhibit high TER and many other differentiated features [24, 26]. Further, well-developed tight junctions are observed by transmission electron microscopy in 12.5-WG fetuses (Gerald Lutty, Wilmer Eye Institute, personal communication). Tight junction formation corresponds to when the lumen of the choriocapillaris first becomes patent, ∼12–13 WG. In contrast to chick or rodents, the human retina appears to require a tight blood-retinal barrier early in its development.
Given normal human development, it is not surprising that hESC-RPE develop functional tight junctions, and many other RPE-specific properties, in the absence of a choroid or sensory retina. This does not mean that early retinal progenitors cannot play a role in the differentiation of RPE, as the RPE arises in a culture containing many cell types. Isolation of the RPE from that complex milieu leads to dedifferentiation after several cell passages, perhaps because of the loss of epigenetic modifications induced by mixed culture. Regardless, maintenance and maturation of the differentiated state was sensitive to the culture environment. The switch from KSR to SFM-1 led to a higher, more stable TER. Although the effect of on most tight junction mRNAs was minimal, the expression of claudin mRNAs was higher in SFM-1. Claudin expression was nearly the same in H1- and H9-RPE after adaptation to SFM-1 and was similar to that in hfRPE. We found that claudin-19b mRNA was expressed in a greater amount than claudin-19a. Because both isoforms have the same extracellular domains, they would be expected to have the same functional properties, as we observed despite variation in the ratio of these claudins in H1- and H9-RPE. Nonetheless, the different cytoplasmic domains suggest that they might be regulated differently. Despite minimal effects on the expression of scaffold and associated regulatory mRNAs, the functioning of these proteins merits further study.
We did observe a significant difference in adhesion junction-related proteins that we believe should be biologically significant. These are members of signaling complexes that influence a host of cellular properties that include cell morphology, polarity, barrier function, and proliferation [17]. There was a much higher statistical correlation between H1- and H9-RPE than between hESC-RPE and cultured hfRPE. This difference was observed functionally, as hfRPE and hESC-RPE had to be cultured on different substrates. hfRPE was most differentiated when plated on a matrix derived from human placenta [26], which contained mature isoforms of laminin and collagen IV [38–40]. The principal laminin of the placental matrix, laminin 5, was also the most efficacious for culturing several RPE cell lines [41]. In contrast, hESC-RPE failed respond properly to the signals in this mature matrix; it required an embryonic form of laminin, laminin-1, to form uniform monolayers. A related developmental stage-dependent matrix requirement was also observed for chick RPE [20, 42].
Membrane transport mRNAs were the remaining class of barrier-related mRNAs that showed significant differences. Among the signature and blood barrier-related mRNAs, we identified 25 that were expressed in statistically different amounts or more than ±4× relative to hfRPE (supplemental online Table 2). Even though some of these genes are also expressed in other cell types, in RPE they appear to be expressed out of balance with the members of the networks with which they interact. Therefore, this panel would be useful for monitoring environmental factors that might promote the maturation of functional networks within RPE. To explore the effects of environment, coculture experiments are ongoing in several laboratories, including our own.
Conclusion
This culture model provides an advanced platform to study the effects of pharmaceuticals and paracrine signaling from neighboring tissues that could augment or replace transplantation. Retinal progenitor cells mature substantially in the absence of RPE [43, 44], but it is also known that RPE secretes trophic factors that modulate this process (e.g., pigment epithelial-derived factor, brain-derived neurotrophic factor, and vascular endothelial growth factor-A, among others) [45]. Additionally, outer segments fail to form in cultures of retinal progenitors [43, 44]. In preliminary studies, we found that SFM-1 supports the maturation of retinal progenitor cells that were derived from H1 or H9 hESCs (L.J. Rizzolo and S. Peng, unpublished data), which makes coculture experiments a feasible approach to studying how RPE and retinal progenitors foster each other's maturation.
Supplementary Material
Acknowledgments
We thank Drs. Sheldon Miller and Arvydas Maminishkis for helpful suggestions and for providing primary cultures of hfRPE. We also thank Dr. Mikio Furuse for providing antibodies to claudin 19, Lina Li for expert technical assistance, and Peter Zhao for critically reviewing the manuscript. This work was funded by the Research to Prevent Blindness (Yale Department of Ophthalmology), the International Retinal Research Foundation (L.J.R.), Connecticut Stem Cell Research Fund 10SBC02 (L.J.R.), the Leir Foundation (R.A.A.), the Newman's Own Foundation (R.A.A.), the National Natural Science Foundation of China NO: 30772381 (S.P.), and Connecticut Stem Cell Research Fund Core Grant 08SCD-004 (Yale University).
Author Contributions
S.P. and G.G.: conception and design, financial support, collection and assembly of data, data analysis and interpretation; C.Q.: conception and design, data analysis and interpretation, final approval of manuscript; M.Z.: design, collection and assembly of data, data analysis and interpretation; H.A.: collection and assembly of data, data analysis and interpretation; R.A.A.: conception and design, financial support, data analysis and interpretation, final approval of manuscript; L.J.R.: conception and design, financial support, data analysis and interpretation, manuscript writing, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
References
- 1.Barishak YR. Embryology of the Eye and Its Adnexa. Basel, Switzerland: Karger; 2001. 2nd rev. ed. [Google Scholar]
- 2.Stroeva OG, Mitashov VI. Retinal pigment epithelium: Proliferation and differentiation during development and regeneration. Int Rev Cytol. 1983;83:221–293. doi: 10.1016/s0074-7696(08)61689-7. [DOI] [PubMed] [Google Scholar]
- 3.Rizzolo LJ, Peng S, Luo Y, et al. Integration of tight junctions and claudins with the barrier functions of the retinal pigment epithelium. Prog Retin Eye Res. 2011;30:296–323. doi: 10.1016/j.preteyeres.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Kiser PD, Golczak M, Maeda A, et al. Key enzymes of the retinoid (visual) cycle in vertebrate retina. Biochim Biophys Acta. 2012;1821:137–151. doi: 10.1016/j.bbalip.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kevany BM, Palczewski K. Phagocytosis of retinal rod and cone photoreceptors. Physiology. 2010;25:8–15. doi: 10.1152/physiol.00038.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marmor MF, Wolfensberger TJ. The Retinal Pigment Epithelium: Function and Disease. New York, NY: Oxford University Press; 1998. [Google Scholar]
- 7.Schwartz SD, Hubschman JP, Heilwell G, et al. Embryonic stem cell trials for macular degeneration: A preliminary report. Lancet. 2012;379:713–720. doi: 10.1016/S0140-6736(12)60028-2. [DOI] [PubMed] [Google Scholar]
- 8.Liao J-L, Yu J, Huang K, et al. Molecular signature of primary retinal pigment epithelium and stem-cell-derived RPE cells. Hum Mol Genet. 2010;19:4229–4238. doi: 10.1093/hmg/ddq341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strunnikova NV, Maminishkis A, Barb JJ, et al. Transcriptome analysis and molecular signature of human retinal pigment epithelium. Hum Mol Genet. 2010;19:2468–2486. doi: 10.1093/hmg/ddq129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bharti K, Miller SS, Arnheiter H. The new paradigm: Retinal pigment epithelium cells generated from embryonic or induced pluripotent stem cells. Pigment Cell Melanoma Res. 2011;24:21–34. doi: 10.1111/j.1755-148X.2010.00772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vugler A, Carr AJ, Lawrence J, et al. Elucidating the phenomenon of hESC-derived RPE: Anatomy of cell genesis, expansion and retinal transplantation. Exp Neurol. 2008;214:347–361. doi: 10.1016/j.expneurol.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 12.Carr AJ, Vugler A, Lawrence J, et al. Molecular characterization and functional analysis of phagocytosis by human embryonic stem cell-derived RPE cells using a novel human retinal assay. Mol Vis. 2009;15:283–295. [PMC free article] [PubMed] [Google Scholar]
- 13.Carr A-J, Vugler AA, Hikita ST, et al. Protective effects of human iPS-derived retinal pigment epithelium cell transplantation in the retinal dystrophic rat. PLoS One. 2009;4:e8152. doi: 10.1371/journal.pone.0008152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng S, Adelman RA, Rizzolo LJ. Minimal effects of VEGF and anti-VEGF drugs on the permeability or selectivity of RPE tight junctions. Invest Ophthalmol Vis Sci. 2010;51:3216–3225. doi: 10.1167/iovs.09-4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng S, Rao VS, Adelman RA, et al. Claudin-19 and the barrier properties of the human retinal pigment epithelium. Invest Ophthalmol Vis Sci. 2011;52:1392–1403. doi: 10.1167/iovs.10-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng S, Gan G, Rao VS, et al. Effects of proinflammatory cytokines on the claudin-19 rich tight junctions of human retinal pigment epithelium. Invest Ophthalmol Vis Sci. 2012;53:5016–5028. doi: 10.1167/iovs.11-8311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cereijido M, Contreras RG, Shoshani L, et al. Tight junction and polarity interaction in the transporting epithelial phenotype. Biochim Biophys Acta. 2008;1778:770–793. doi: 10.1016/j.bbamem.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Elkouby-Naor L, Ben-Yosef T. Functions of claudin tight junction proteins and their complex interactions in various physiological systems. Int Rev Cell Mol Biol. 2010;279:1–32. doi: 10.1016/S1937-6448(10)79001-8. [DOI] [PubMed] [Google Scholar]
- 19.Van Itallie CM, Anderson JM. Claudins and epithelial paracellular transport. Annu Rev Physiol. 2006;68:403–429. doi: 10.1146/annurev.physiol.68.040104.131404. [DOI] [PubMed] [Google Scholar]
- 20.Rizzolo LJ, Chen X, Weitzman M, et al. Analysis of the RPE transcriptome reveals dynamic changes during the development of the outer blood-retinal barrier. Mol Vis. 2007;13:1259–1273. [PubMed] [Google Scholar]
- 21.Sun R, Peng S, Chen X, et al. Diffusible retinal secretions regulate the expression of tight junctions and other diverse functions of the retinal pigment epithelium. Mol Vis. 2008;14:2237–2262. [PMC free article] [PubMed] [Google Scholar]
- 22.Konrad M, Schaller A, Seelow D, et al. Mutations in the tight-junction gene claudin 19 (CLDN19) are associated with renal magnesium wasting, renal failure, and severe ocular involvement. Am J Hum Genet. 2006;79:949–957. doi: 10.1086/508617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rizzolo LJ. Development and role of tight junctions in the retinal pigment epithelium. Int Rev Cytol. 2007;258:195–234. doi: 10.1016/S0074-7696(07)58004-6. [DOI] [PubMed] [Google Scholar]
- 24.Gamm DM, Melvan JN, Shearer RL, et al. A novel serum-free method for culturing human prenatal retinal pigment epithelial cells. Invest Ophthalmol Vis Sci. 2008;49:788–799. doi: 10.1167/iovs.07-0777. [DOI] [PubMed] [Google Scholar]
- 25.Gamm DM, Wright LS, Capowski EE, et al. Regulation of prenatal human retinal neurosphere growth and cell fate potential by retinal pigment epithelium and Mash1. Stem Cells. 2008;26:3182–3193. doi: 10.1634/stemcells.2008-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maminishkis A, Chen S, Jalickee S, et al. Confluent monolayers of cultured human fetal retinal pigment epithelium exhibit morphology and physiology of native tissue. Invest Ophthalmol Vis Sci. 2006;47:3612–3624. doi: 10.1167/iovs.05-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Idelson M, Alper R, Obolensky A, et al. Directed differentiation of human embryonic stem cells into functional retinal pigment epithelium cells. Cell Stem Cell. 2009;5:396–408. doi: 10.1016/j.stem.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Kimizuka H, Koketsu K. Ion transport through cell membrane. J Theor Biol. 1964;6:290–305. doi: 10.1016/0022-5193(64)90035-9. [DOI] [PubMed] [Google Scholar]
- 30.Yu AS, Cheng MH, Angelow S, et al. Molecular basis for cation selectivity in claudin-2-based paracellular pores: Identification of an electrostatic interaction site. J Gen Physiol. 2009;133:111–127. doi: 10.1085/jgp.200810154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quinn RH, Miller SS. Ion transport mechanisms in native human retinal pigment epithelium. Invest Ophthalmol Vis Sci. 1992;33:3513–3527. [PubMed] [Google Scholar]
- 32.Burke JM. Epithelial phenotype and the RPE: Is the answer blowing in the Wnt? Prog Retin Eye Res. 2008;27:579–595. doi: 10.1016/j.preteyeres.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grunwald GB. Cadherin cell adhesion molecules in retinal development and pathology. Prog Retinal Eye Res. 1996;15:363–392. [Google Scholar]
- 34.Powell DW. Barrier function of epithelia. Am J Physiol. 1981;241:G275–G288. doi: 10.1152/ajpgi.1981.241.4.G275. [DOI] [PubMed] [Google Scholar]
- 35.Braekevelt CR, Hollenberg MJ. Development of the retinal pigment epithelium, choriocapillaris and Bruch's membrane in the albino rat. Exp Eye Res. 1970;9:124–131. doi: 10.1016/s0014-4835(70)80066-5. [DOI] [PubMed] [Google Scholar]
- 36.Greiner JV, Weidman TA. Comparative histogenesis of Bruch's membrane (complexus basalis) Exp Eye Res. 1991;53:47–54. doi: 10.1016/0014-4835(91)90143-3. [DOI] [PubMed] [Google Scholar]
- 37.Lutty GA, Hasegawa T, Baba T, et al. Development of the human choriocapillaris. Eye (Lond) 2010;24:408–415. doi: 10.1038/eye.2009.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gelse K, Poschl E, Aigner T. Collagens: Structure, function, and biosynthesis. Adv Drug Deliv Rev. 2003;55:1531–1546. doi: 10.1016/j.addr.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 39.Nguyen NM, Senior RM. Laminin isoforms and lung development: All isoforms are not equal. Dev Biol. 2006;294:271–279. doi: 10.1016/j.ydbio.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 40.Kim ST, Adair-Kirk TL, Senior RM, et al. Functional consequences of cell type-restricted expression of laminin α5 in mouse placental labyrinth and kidney glomerular capillaries. PLoS One. 2012;7:e41348. doi: 10.1371/journal.pone.0041348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aisenbrey S, Zhang M, Bacher D, et al. Retinal pigment epithelial cells synthesize laminins, including laminin 5, and adhere to them through α3- and α6-containing integrins. Invest Ophthalmol Vis Sci. 2006;47:5537–5544. doi: 10.1167/iovs.05-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rizzolo LJ. Basement membrane stimulates the polarized distribution of integrins but not the Na,K-ATPase in the retinal pigment epithelium. Cell Regul. 1991;2:939–949. doi: 10.1091/mbc.2.11.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phillips MJ, Wallace KA, Dickerson SJ, et al. Blood-derived human iPS cells generate optic vesicle-like structures with the capacity to form retinal laminae and develop synapses. Invest Ophthalmol Vis Sci. 2012;53:2007–2019. doi: 10.1167/iovs.11-9313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eiraku M, Takata N, Ishibashi H, et al. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature. 2011;472:51–56. doi: 10.1038/nature09941. [DOI] [PubMed] [Google Scholar]
- 45.Kolomeyer AM, Sugino IK, Zarbin MA. Characterization of conditioned media collected from aged versus young human eye cups. Invest Ophthalmol Vis Sci. 2011;52:5963–5972. doi: 10.1167/iovs.10-6440. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.