SUMMARY
Detection of foreign materials is the first step of successful immune responses. Stimulator of interferon genes (STING) was shown to directly bind cyclic diguanylate monophosphate (c-di-GMP), a bacterial second messenger, and to elicit strong interferon responses. Here we elucidate the structural features of the cytosolic c-di-GMP binding domain (CBD) of STING and its complex with c-di-GMP. The CBD exhibits an α + β fold and is a dimer in the crystal and in solution. Surprisingly, one c-di-GMP molecule binds to the central crevice of a STING dimer, using a series of stacking and hydrogen bonding interactions. We show that STING is autoinhibited by an intramolecular interaction between the CBD and the C-terminal tail (CTT) and that c-di-GMP releases STING from this autoinhibition by displacing the CTT. The structures provide a remarkable example of pathogen-host interactions in which a unique microbial molecule directly engages the innate immune system.
INTRODUCTION
Innate immunity is an evolutionarily conserved mechanism that provides the first line of defense against microbial pathogens such as viruses and bacteria. Until fairly recently, innate immunity was considered nonspecific immunity mediated by phagocytosis. The discovery of germline-encoded pattern recognition receptors (PRRs) that recognize conserved pathogen associated molecular patterns (PAMPs) shared by many bacteria, fungi, protozoa, and viruses has greatly advanced the concept of pattern recognition as a prime task of the innate immune system (Medzhitov and Janeway, 2000). After recognition of PAMPs, PRRs trigger signaling pathways that not only alert the immune system to the presence of infection but also help initiate adaptive immune responses through maturation of dendritic cells and antigen presentation (Iwasaki and Medzhitov, 2010; Takaoka and Yanai, 2006).
Several families of PRRs have been identified, most notably transmembrane Toll-like receptors (TLRs) (Akira and Takeda, 2004), cytosolic Nod-like receptors (Inohara et al., 2005), and dsRNA helicases such as RIG-I and MDA5 (Kato et al., 2011). Microbial DNAs represent an important class of PAMPs (Barber, 2011). While unmethylated CpG DNA from bacteria can be detected by TLR9 in the endosome (Hemmi et al., 2000), cytosolic DNA from viral infection is sensed independent of TLR9 (Ishii et al., 2006). Recent studies have revealed the involvement of DAI (Takaoka et al., 2007), AIM2 (Fernandes-Alnemri et al., 2009; Hornung et al., 2009; Roberts et al., 2009), IFI16 (Unterholzner et al., 2010), and p202 (Roberts et al., 2009) as direct cytosolic dsDNA sensors for interferon (IFN) induction and inflammasome formation.
Stimulator of interferon genes (STING) (also known as MITA, MPYS, ERIS, and TMEM173) was first identified as an ER-residing protein relaying signals to IRF activation and IFN transcription from a variety of stimuli, especially cytosolic dsDNAs (Ishikawa and Barber, 2008; Jin et al., 2008; Sun et al., 2009; Zhong et al., 2008). STING activates the IKK-like kinase TBK1, which in turn phosphorylates IRF3, leading to its nuclear translocation and transcription of type I IFNs to exert a potent antiviral state (Ishikawa et al., 2009; Saitoh et al., 2009). STING-deficient cells fail to produce type I IFN in response to transfection with dsDNA or infection with herpes simplex virus 1 (HSV-1) (Ishikawa et al., 2009).
Remarkably, STING was also shown recently to be essential for IFN production in response to another form of nucleic acids known as cyclic dinucleotides such as cyclic diguanylate monophosphate (c-di-GMP) and cyclic diadenylate monophosphate (c-di-AMP) (Sauer et al., 2011). A mutant mouse strain, Goldenticket, that harbors a missense mutation (I199N) in the STING protein, is a nonfunctional allele that fails to produce detectable protein and to activate IFN production in response to c-di-GMP (Sauer et al., 2011). Both c-di-GMP and c-di-AMP are second messengers secreted by bacteria such as the intracellular pathogen Listeria monocytogenes (Sauer et al., 2011). They are known immunostimulatory molecules with potential in novel immunotherapeutics and as vaccination adjuvants (Chen et al., 2010; Karaolis et al., 2007). Using a UV crosslinking assay, STING was identified as the direct binding partner for c-di-GMP, and this interaction was mapped to the cytosolic region and shown to be critical for cellular responses to cyclic dinucleotides (Burdette et al., 2011). In contrast, dsDNA sensing by the STING pathway requires additional components, as STING transfection in HEK293T cells only reconstituted responsiveness to c-di-GMP, but not to dsDNA (Burdette et al., 2011; Sauer et al., 2011).
STING was predicted to contain five transmembrane helices (TM1–TM5) and a sizable cytosolic domain (Figure 1A). The cytosolic domain does not exhibit significant sequence homology to known bacterial c-di-GMP receptors such as those containing the PilZ domain (Tamayo et al., 2007) or bacterial cyclases that generate cyclic dinucleotides (Tamayo et al., 2007), nor to any proteins with known structures. Within the cytosolic domain, a recent study showed that the very C-terminal tail (CTT) (Figure 1A) interacts with and activates TBK1 and IRF3 in vitro (Tanaka and Chen, 2012).
Figure 1. Overall Structure of the STING c-di-GMP Binding Domain.
(A) Domain organization of STING. Five previously suggested transmembrane helices (TMs) are represented with four orange boxes (TM1–4) and a blue box (TM5), followed by the suggested cytosolic domain. The c-di-GMP binding domain (CBD) identified from this study, which contains the previous TM5, is represented by the blue box and the long blue rectangle. The construct used in E. coli expression (139–379) is represented with the dotted box. CTT: C-terminal tail. Starting residue number of each domain is labeled above.
(B) Gel filtration profiles of full-length (F.L., blue) and trypsin-digested (red) cytosolic domain. Elution volumes of protein standards are marked at the top. The black arrow points to high molecular weight species observed only in fulllength cytosolic domain.
(C) Experimental electron density of the α1–α1′ (left) and β4–α3 (right) region contoured at 1.4 σ.
(D) Cartoon representation of one STING protomer. The molecule is in rainbow spectrum with blue at the N terminus and red at the C terminus. N and C termini as well as secondary structure elements are labeled. Disordered regions are drawn as dotted lines.
(E) Topology diagram of the STING CBD. α helices and β strands are represented by cylinders and wide arrows, respectively. Peptide directionality is indicated by black arrows. The molecule is colored in rainbow spectrum as in (D).
Here we report structural characterizations of the STING cytosolic domain. We show that the N-terminal part of the cytosolic domain of STING forms a folded structural entity sufficient for recognition of c-di-GMP, which we here name the c-di-GMP binding domain (CBD) (Figure 1A). The previously identified CTT for TBK1 binding and activation (Tanaka and Chen, 2012) immediately follows this domain. Interestingly, the previously assigned TM5 is not a transmembrane helix but part of the folded, soluble CBD. We show that the CBD of STING is a wing-shaped dimer. Consistent with the lack of significant sequence homology to any proteins with known structures, the CBD exhibits an α + β fold, which to our knowledge was previously unknown. A single c-di-GMP molecule binds at the STING dimerization interface and assumes a dimerically symmetrical conformation. The interaction specifically selects c-di-GMP with high affinity and c-di-AMP with lower affinity and discriminates against other types of nucleotides. We show that STING is autoinhibited via an intramolecular interaction between the CBD and the CTT. The presumed CTT-binding site on CBD must overlap with the c-di-GMP-binding site, leading to the relief of autoinhibition upon c-di-GMP binding and activation of TBK1 and IRF3 for the induction of IFN response. The study provides a remarkable example of pathogen-host interactions in which a unique microbial molecule directly engages the innate immune system at an integrating point in the signaling cascade.
RESULTS
Crystal Structure of the c-di-GMP Binding Domain (CBD)
We set out to elucidate the molecular basis for the functions of the cytosolic region of STING, especially for its ability to recognize c-di-GMP. As a first step, we expressed human STING in E. coli using a construct (139–379) that is equivalent to the mouse STING construct (138–378) previously shown to be sufficient for interaction with c-di-GMP (Burdette et al., 2011). However, the purified recombinant protein eluted in heterogeneous size populations from gel filtration chromatography that are consistent with dimers and higher order oligomers (Figure 1B). Because this heterogeneity may hinder crystallization, we performed limited proteolysis to determine if the protein would become more homogeneous. Indeed, upon digestion by the protease trypsin, the protein eluted from gel filtration chromatography as a single peak that corresponds to dimers of STING (Figure 1B). Since the C-terminal 30 or so residues were predicted to possess mostly random coil structures, we suspected that this region was removed by the proteolysis.
We crystallized the trypsin-digested sample of STING in the P43212 space group. The structure was solved using singlewavelength anomalous diffraction (SAD) of the selenomethionyl protein and was refined at 2.75 Å resolution (Table 1 and Figures 1C and 1D). The ordered region of the structure starts at N154 and ends at K338 (Figures 2 and Figure S1), consistent with our analysis on the region removed during proteolysis. The structure exhibits an α + β fold which contains a central twisted five-stranded β sheet surrounded by three long α helices (α1, α3, and α5) on the convex face and two (α2 and α6) α helices on the concave surface (Figure 1D). A short α4 helix connects α3 and α5. The first helix is very long and kinked at Y167 (α1 and α1′); it overarches over the central β sheet and almost reaches the concave surface. This helix is very hydrophobic and corresponds to the previously predicted TM5 (Figure 1A). The structure has an approximate triangular shape when viewed down the stacks of β strands (Figure 1D). Together with α3 and α5, α1 forms one edge of the triangle while α1′ forms another. The curled β sheet forms the last edge of the triangle. Surprisingly, the overall topology of the subunit (Figure 1E) has not been observed before; DALI structural homology search (Holm and Sander, 1995) only identified low significance matches with Z scores below 4.0 and RMSDs in the range 3–4 Å a and with about half of the Cα positions aligned.
Table 1.
Crystallographic Statistics
| STING | STING/c-di-GMP | |
|---|---|---|
| Constructs | L139–S379 | L139–S379 |
| Structure determination | SAD | MR |
| Data collection | ||
| Beamlines | X25 of NSLS | X29 of NSLS |
| Space group | P43212 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 61.4, 61.4, 118.3 | 60.0, 74.2, 60.0 |
| β (°) | 96.3 | |
| Resolution (Å) | 35.0 – 2.75 | 50.0 – 2.94 |
| Rsym (%) | 4.0 (45.6) | 6.6 (48.5) |
| I/σI | 33.0 (4.3) | 36.2 (4.3) |
| Completeness (%) | 99.9 (100) | 98.9 (100) |
| Redundancy | 8.2 (8.6) | 5.8 (5.9) |
| Refinement | ||
| Resolution (Å) | 35.0 – 2.75 | 38.0 – 2.94 |
| No. reflections | 6,332 | 11,082 |
| Rwork / Rfree (%) | 21.1 / 25.7 | 20.6 / 25.3 |
| No. atoms | ||
| Protein | 1,376 | 2,674 |
| c-di-GMP | 46 | |
| H2O | 21 | |
| Average B (Å2) | ||
| Protein | 68.0 | 92.1 |
| c-di-GMP | 87.8 | |
| H2O | 68.6 | |
| R.m.s deviations Bond lengths (Å) / angles (°) | 0.01 / 1.2 | 0.01 / 1.4 |
| Ramachandran Plot Most favored / allowed (%) | 94.4 / 5.6 | 90.9 / 9.1 |
Values in parenthesis are for highest-resolution shell.
Figure 2. Structural Features of the STING CBD.
Secondary structure elements are labeled on top of the human STING sequence. α helices are shown as green cylinders,β strands as red arrows, and loops as yellow lines. Residues without defined densities are shown as dotted yellow lines. Solvent accessibility is shown underneath each residue by a blue-white gradient. Buried residues are indicated by blue shadings and exposed residues by white shadings. Residues of STING conserved among different species (Figure S1) are highlighted in yellow. Residues at the dimer interface are colored in green, and c-di-GMP contacting residues are in red. Residue T267, involved both in dimerization and c-di-GMP binding, is colored in magenta.
STING Is a Stable Dimer
Although there is only one STING molecule in each crystallographic asymmetric unit of the crystal, the crystallographic two-fold axis generates an intimately associated dimer with the shape of a pair of wings (Figure 3A). Consistently, STING exists in solution as a dimer as well (Figure 1B). The assignment of the dimer structure is conclusive, as the dimerization interface is extraordinarily extensive, burying roughly a total of 1800 Å2 surface area. The dimer interface is mostly formed by the α1 and α3 helices from each protomer (Figure 3B). The two symmetry-related α1helices are nearly orthogonal to each other. While α1 of one protomer interacts with both α1 and α3 of the neighboring protomer, α3 of one protomer only interacts with α1 of the neighboring protomer. The interaction forms an approximate antiparallel four-helix bundle. Peripheral interactions also exist between the short α4 of one protomer and the α5 of the neighboring protomer.
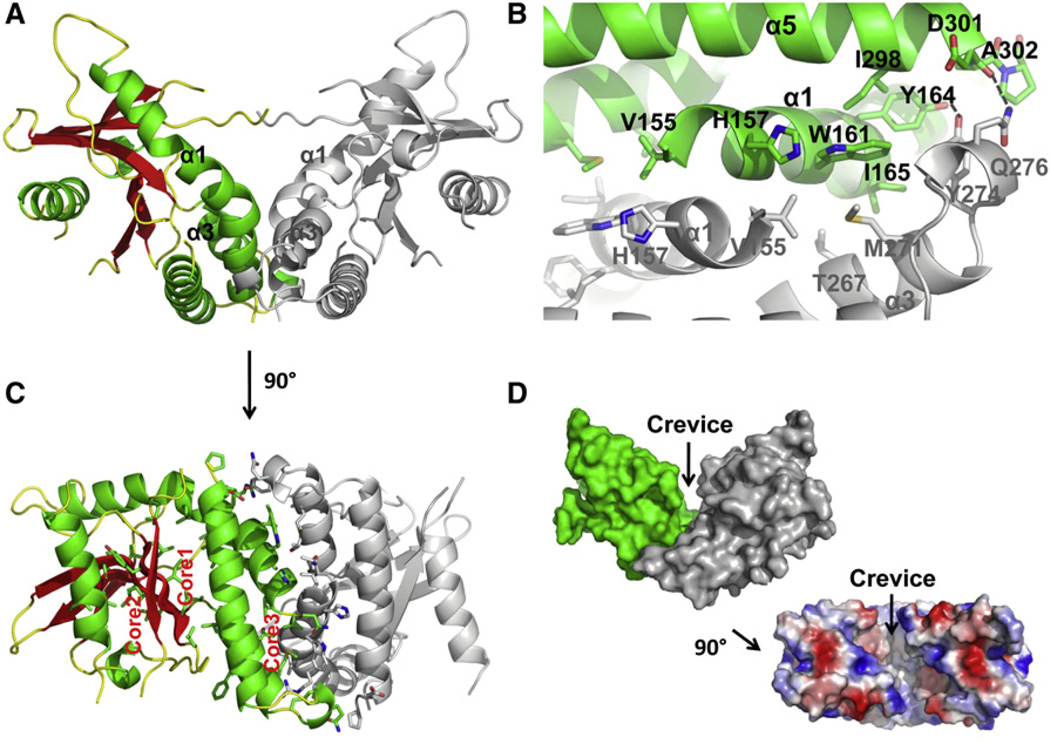
Figure 3. Dimeric Nature of STING CBD.
(A) Cartoon representation of STING dimer. One protomer is colored as in Figure 2 with α helices in green, β strands in red, and loops in yellow. The other protomer is colored in gray. The interacting α1 and α3 are labeled for both protomers.
(B) Detailed interaction at the dimer interface. For clarity, only one of the two symmetrical interfaces is shown. One molecule is colored in green and the other in gray, as in (A). Interfacial residues are shown as sticks. Residues and the helices they belong to are labeled and numbered.
(C) The STING dimer is rotated roughly 90° horizontally from the orientation in (A) and colored in the same way. The three hydrophobic cores are labeled. Residues constituting the hydrophobic cores are shown as sticks. Note core 3 is composed of residues from both protomers.
(D) Surface representation of the STING dimer in a side view, showing the deep crevice across the dimer interface, and the electrostatic surface of the STING dimer shown in a top view.
Residues at the center of the dimerization interface are mostly hydrophobic, including V155, H157, W161, Y164, and I165 from helix α1 and T267, M271, Y274, and Q276 from helix α3 (Figure 3B). Interestingly, these clusters of residues create an additional, third hydrophobic core in the STING dimer (Figure 3C). The STING protomer structure already consists of two somewhat separate hydrophobic cores, one on each face of the central twisted β sheet. The first hydrophobic core is formed mainly from packing of large hydrophobic residues in helices α1, α3, and α5 with those in the convex face of the β sheet. The second hydrophobic core is between large hydrophobic residues in helices α2 and α6 and those in the concave face of the β sheet. The large interface and the hydrophobic nature strongly suggest that the CBD of STING is a stable dimer and that mutations at the interface may expose the hydrophobic surface, leading to aggregation. Consistently, an interfacial double mutant of CBD, W161A/M271A, formed higher order oligomers (Figure S2).
At the more peripheral part of the interface, Y164 from the far end of helix α1 and D301 from helix α5 interact with the tip of the α3– α4 loop (Figure 3B). There are hydrogen bonds between the side chains of Y164 and Y274 and between the main chain of D301 with the side chain of Q276. Strikingly, the surface diagram of the STING dimer has a deep crevice running along the dimerization interface (Figure 3D). The bottom of the crevice is mostly uncharged while the wall of the crevice is decorated with both positive and negative charges (Figure 3D). The prominence of the crevice suggests that it performs certain biological functions.
Crystal Structure of STING in Complex with c-di-GMP
We obtained crystals of STING in complex with c-di-GMP using cocrystallization. The diffraction data were initially processed in space group C2221, and the structure was solved by molecular replacement using the STING alone structure. Difference map calculated without c-di-GMP showed clear density for c-di-GMP (Figure 4A). There is one STING protomer per crystallographic asymmetric unit, but the same STING dimer is generated by a crystallographic two-fold axis. Surprisingly, c-di-GMP sits at a crystallographic two-fold axis, between one STING molecule and its symmetry-related mate (Figure 4B). This is possible because c-di-GMP has an intrinsic dimeric symmetry. In order to build the complete, rather than half of the c-di-GMP molecule in the crystallographic asymmetric unit, we lowered the symmetry to P21 so the asymmetric unit contains one STING dimer and one c-di-GMP molecule (Table 1). The C terminus of STING is more ordered in the complex structure with visible density extending to V343. Binding of c-di-GMP does not induce drastic conformational changes in STING, as both structures superpose well with an RMSD of 0.85 Å. However, if only one protomer of the STING dimer is used for structural superposition, it is clear that the other promoter is slightly plied open, perhaps to accommodate the c-di-GMP molecule precisely (Figure 4C). Supporting the binding ratio of two STING protomers for one c-di-GMP, an isothermal titration calorimetry (ITC) experiment confirmed the stoichiometry of STING to c-di-GMP as 2:1 with a binding dissociation constant of 2.4 ± 0.5 µM (Figure 4D). This binding affinity is consistent with a previous measurement using equilibrium dialysis (Burdette et al., 2011).
Figure 4. Recognition of c-di-GMP by STING.
(A) Difference Fourier (Fo-Fc) map contoured at 2.5 σ at the dimer interface, superimposed with the bound c-di-GMP.
(B) Cartoon representation of STING/c-di-GMP complex. One STING molecule is colored in violet and the other in wheat. The c-di-GMP molecule is shown as a stick model.
(C) Superposition of the STING/c-di-GMP complex with the free STING dimer. The STING protomers in the complex are colored in violet and wheat, while those in the free STING are in green and gray. Red arrow marks the slight outward swing of the STING molecule (wheat) in the complex with c-di-GMP.
(D) Isothermal titration calorimetry measurement for the interaction between STING and c-di-GMP.
(E) Stick representation of c-di-GMP with the intrinsic two-fold symmetry. The orientation is the same as in (C). 2-position nitrogen of the guanine ring, 2′-hydroxyl group of the ribose, and the phosphoryl oxygen atoms are labeled.
(F) Detailed interaction of c-di-GMP with STING. For clarity, only one “half site” is shown. Contacting residues are labeled, and hydrogen bonds are represented by dotted lines.
Mode of Interaction between STING and c-di-GMP
The prominent crevice at the dimerization interface of STING accommodates c-di-GMP. The single c-di-GMP molecule assumes a shape that resembles a curved bow that has been pulled on the string by the arrow (Figure 4E). This curvature fits almost perfectly along the entire 17 Å length of c-di-GMP with the curved crevice floor of the STING dimer that c-di-GMP sits on. Therefore, one side of the c-di-GMP molecule is completely buried in its interaction with STING. The conformation of c-di-GMP bound to STING resembles those in complex with riboswitches (Kulshina et al., 2009; Smith et al., 2009), the PilZ domain (Benach et al., 2007), and the diguanylate cyclase domain (Chan et al., 2004), in which the two guanine bases adopt a cis, nearly parallel orientation. In contrast, in the alternative conformation associated with the EAL domain, the guanine bases are roughly in trans, leading to an extended and rather planar configuration (Barends et al., 2009; Navarro et al., 2009).
The c-di-GMP binding pocket is created by α1, α3, and the loop between β2 and β3 of both protomers of the STING dimer (Figure 4F). The intermolecular interactions are mixed, with a combination of hydrophobic and hydrophilic contacts. The most extensive interaction to c-di-GMP is provided by Y167, the residue in α1 at the kink that marks the boundary with α1′ . The phenol ring of Y167 stacks precisely with the pyrimidine portion of the guanine ring of c-di-GMP in which every atom of the phenol ring makes contacts. The kink is important for making this interaction possible. In addition to Y167, residues S162, Y163, E260, T263, and P264 make multiple van der Waals and hydrogen bonding interactions with c-di-GMP. The carboxylate of E260 points toward N2 of guanine, although this interaction is beyond hydrogen bonding distance. Residues Y167 and E260 appear to properly orient each other through a hydrogen bond between the phenolic hydroxyl of Y167 and the carboxylate of E260. The hydroxyls of S162 of one protomer and T267 of the other protomer are within hydrogen bonding distance to the phosphoryl oxygen, while T263 hydrogen bonds with 2′-hydroxyl of the ribose. Side chains of c-di-GMP contacting residues in STING assume similar orientations in the unbound structure, suggesting that the binding pocket is largely preformed.
ITC measurements revealed that the binding is entirely driven by favorable enthalpy (ΔH = −11.6 ± 0.5 kcal/mol) with unfavorable entropy (−TΔS =3.9 kcal/mol) (Figure 4D). The favorable enthalpy is in agreement with the favorable π–π stacking and hydrogen bonding interactions. Some of the entropy that is lost upon binding may be conformational. The c-di-GMP molecule has several tunable torsion angles and the binding completely restricts this conformation to the bow shape observed in the structure.
Specificity Determinants
The specificity and strength of c-di-GMP recognition by STING are reflected at several structural levels that help to define this cyclic dinucleotide as an optimal ligand in comparison with other nucleotides (Table 2). First of all, STING also senses c-di-AMP, as shown by a recent study that the intracellular bacterial pathogen Listeria monocytogenes stimulates a type I IFN response due to cytosolic detection of bacterially secreted c-di-AMP (Sauer et al., 2011). STING has been shown to interact with c-di-AMP directly and specifically and to reconstitute responsiveness of HEK293T cells to this signaling molecule (Burdette et al., 2011). However, competition experiments suggest that the affinity of STING to c-di-AMP is lower than that to c-di-GMP (Burdette et al., 2011). As a purine, the adenine ring will also be able to stack with Y167. However, in adenine, there is no substitution at the 2-position, losing the interaction with E260 and explaining the apparent lower affinity to STING.
Table 2.
Specificity Determinants in c-di-GMP Recognition
| c-di-GMP | c-di-AMP | c-di-YMP1 | GMP | |
|---|---|---|---|---|
| Stacking with Y167 | Yes | Yes | No | Yes |
| Divalent interaction2 | Yes | Yes | Yes | No |
| H-bond between 2′-OH of ribose and T263 | Yes | Yes | Yes | Yes |
| H-bond between phosphoryl oxygen and S162 and T267 | Yes | Yes | Yes | Yes |
| Interaction between N2 and E260 | Yes | No | No | No |
The interactions seen in the STING/c-di-GMP complex are shown in the left two columns. Potential interactions of STING with other nucleotides, as deduced from the current structures, are shown in the three right columns.
Pyrimidine cyclic dinucleotide.
A single molecule of the nucleotide interacts with a STING dimer.
Second, STING should not sense cyclic pyrimidine dinucleotides. If bound symmetrically at the dimerization interface as c-di-GMP, the single pyrimidine ring in these dinucleotides will not be able to reach Y167 for the favorable stacking interaction, explaining the incapability of the interactions. Third, single GMP also cannot stimulate STING (Burdette et al., 2011). While the “half site” binding may be preserved in the mode of interaction between GMP and STING, the binding energy would have been cut in half without the avidity provided by the dimeric arrangement as in c-di-GMP. Therefore, at least three structural features allow discrimination between c-di-GMP and other nucleotides (Table 2). First, a cognate nucleotide needs to be a cyclic dinucleotide. Second, a cognate nucleotide needs to be a purine nucleotide. Third, guanine has advantage over adenine in its capability to interact with E260 of STING at N2. In addition, it is possible that ribonucleotides are favored over deoxyribonucleotides due to the specific hydrogen bonding interaction at the 2′ -hydroxyl.
Previously Reported STING Mutants Support the c-di-GMP Binding Site
The phenotypes of a number of mutations in mouse STING have been described (Burdette et al., 2011; Sauer et al., 2011). Human and mouse STING are very similar, sharing 80% sequence identity in the CBD (Figure S1). Therefore our structures provide a template for understanding the underlying mechanism in the functional impairment of these mutants. The Goldenticket mouse strain, which fails to produce type I IFNs upon Listeria monocytogenes infection or in response to c-di-GMP, encodes a missense mutation equivalent to I200N in human STING (I199N in mouse STING) (Sauer et al., 2011). Residue I200 is buried in the interior of the STING protomer and I200N would have disrupted the STING structure (Figure 5A and Table 3), explaining the null-phenotype of the mutant.
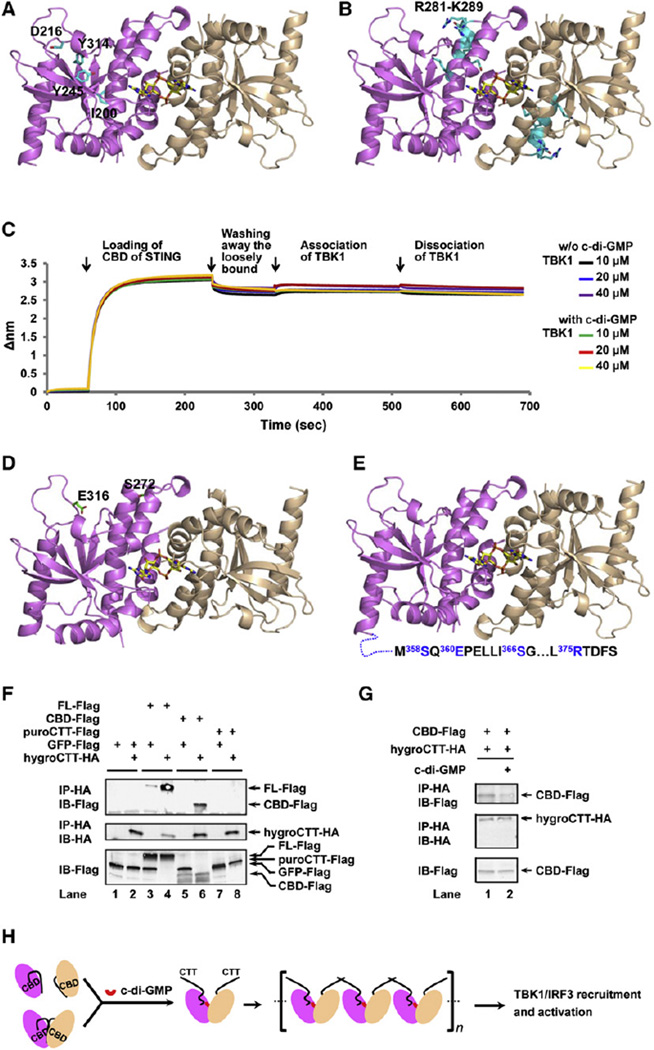
Figure 5. Proposed Mechanism of STING Activation.
(A) Mutants that contain buried residues and fail to interact with c-di-GMP. These buried residues, I200, D216, Y245, and Y314, are shown as sticks on a cartoon representation of the STING/c-di-GMP structure.
(B) A multiply substituted mutant that contains exposed residues and fails to interact with c-di-GMP. These residues,R281–K289,are coloredincyanand shownassticks.
(C) Real time biolayer interferometry measurement of 10, 20, and 40 µM TBK1 to immobilized STING CBD in the absence (black, blue, and purple lines) or presence (green, red, and yellow lines) of c-di-GMP. Biotinylated STING CBD loading and washing as well as TBK1 association and dissociation curves are labeled.
(D) Mutants that bind c-di-GMP, hyperinduce IFN but do not respond to c-di-GMP for further enhancement of IFN production. These residues, E316 and S272, are shown as sticks on the STING/c-di-GMP cartoon representation.
(E) Mutants in the CTT that bind c-di-GMP, induce IFN but do not respond to c-di-GMP for further enhancement of IFN production. These residues, S358/E360/S366 and R375 are highlighted in blue in the appended CTT.
(F) Interaction of CTT with full-length (FL) cytosolic domain of STING and CBD were detected by immunoprecipitation (IP) followed by immunoblotting (IB) in 293 cells transfected with indicated expression constructs.
(G) The interaction between CTT and CBD as detected by coimmunoprecipitation is weakened when cells are stimulated with c-di-GMP.
(H) “Release of autoinhibition” model. In unstimulated cells, STING exists in an autoinhibited status with an intramolecular CBD (violet or wheat ovals)/CTT (black curvy lines) interaction, either as a monomer or dimer. Binding of c-di-GMP at the dimer interface displaces CTT, induces and stabilizes STING dimer, and releases STING from autoinhibition. STING dimers with freed CTT may further oligomerize through CTT. Activated STING then recruits and activates TBK1 and IRF3.
Table 3.
Anticipated Structural Consequences of Previously Described STING Mutations
| Mutant Name | Mutations | Structural Observation | Proposed Mechanism of Deficiency |
|---|---|---|---|
| Mutants that abolish c-di-GMP binding and IFN induction | |||
| goldenticket | I200N | I200 is buried. | Structural perturbation |
| mut8 | D210A, D216A, N218A | D216 is largely buried. | Structural perturbation |
| mut10 | Y240A, S243A, Y245A | Y245 is largely buried. | Structural perturbation |
| mut11 | E260A, Q266A | E260 directly interacts with c-di-GMP and hydrogen bonds with Y167. | Loss of an important interaction; local structural perturbation |
| mut13 | R281A, E282A, D283A, R284A, E286A, Q287A, K289A | These residues pack against α3 at both the dimerization interface and the c-di-GMP binding site. | Local structural perturbation |
| mut22 | Y314A | Y314 is buried. | Structural perturbation |
| Mutants that bind c-di-GMP, hyperinduce IFN, but do not respond to c-di-GMP for further enhanced IFN production | |||
| mut15 | R310A, E316A | The mutational effect most likely comes from the E316A mutation (see next line). | Release of intramolecular interaction-mediated autoinhibition? |
| mut18 | E316A | E316 is exposed and away from the c-di-GMP binding site. | |
| mut19 | E316N | ||
| mut20 | E316Q | ||
| mut34 | S272A | S272 is mostly exposed and away from the c-di-GMP binding site. | |
| Mutants thatbind c-di-GMP, induce IFN, but do not respond to c-di-GMP for further enhanced IFN prduction | |||
| Mut44 | R375A | R375 is in the CTT | Release of intramolecular interaction-mediated autoinhibition plus partial deficiency in TBK1 and IRF3 activation? |
| mut17 | S358A, E360A, S366A | These residues are in the CTT | |
| Mutants that behaves like WT | |||
| Mut24 | E316D | In comparison with E316A, E316N and E316Q, E316D preserves the negative charge. | Preservation of intramolecular interaction-mediated autoinhibition? |
See Burdette et al. (2011). Residue numbers were modified to reflect the equivalent human sequence (residue number in human = residue number in mouse + 1). Deletion mutations were omitted.
Additional mutants have been identified in mouse STING that disrupted c-di-GMP interaction (Burdette et al., 2011). To minimize confusion, we will use the equivalent residues in human STING by adding 1 to the residue numbers in mouse STING. Most of the mutations in this group involve large deletions and nonconservative changes on buried residues in the CBD, including D216, Y245, and Y314 (Figure 5A and Table 3). Like I200N, these changes would be predicted to compromise the folding of the CBD. A multiply substituted mutant on helix α5 (R281A, E282A, D283A, R284A, E286A, Q287A, and K289A) acts as a null likely because these residues pack against the α3 helix for c-di-GMP recognition (Figure 5B) and may cause local structural perturbation in STING. The most informative mutant may be the E260A/Q266A double mutant. Because Q266A is exposed on the surface and does not contact c-di-GMP, the mutational phenotype is likely caused by the E260A mutation. E260 interacts with c-di-GMP (Figure 4F) and may directly compromise c-di-GMP recognition when mutated. In addition, E260 hydrogen bonds with Y167 (Figure 4F), and these residues may be mutually critical in maintaining the productive side chain conformations for c-di-GMP recognition.
The CBD of STING Does Not Interact Measurably with TBK1 in the Presence and Absence of c-di-GMP
Intriguingly, the structures of the CBD domain of STING that we determined do not include the CTT (Figure 1A) that has been shown recently to be necessary and sufficient for TBK1 and IRF3 activation (Tanaka and Chen, 2012). So then how does c-di-GMP binding enhance STING-mediated IFN production? We wondered whether c-di-GMP binding creates a new interact ing surface on the CBD of STING that directly recruits and activates TBK1, in addition to TBK1 and IRF3 activation by CTT. To test this possibility, we measured the direct binding of the CBD of STING with recombinant TBK1 in the presence and absence of c-di-GMP using biolayer interferometry (Figure 5C). The CBD domain of STING was labeled by biotin and bound to streptavidin-coated chips at a 2 µ Mconcentration. After washing away loosely bound materials, biotinylated CBD showed a stable bound level of approximately 3 nm on the chips. Association was initiated by dipping CBD-chips in solutions of recombinant TBK1 at different concentrations and was monitored at real time. Dissociation was initiated by dipping the same chips into buffers and was monitored similarly. Dipping the CBD-bound chips to TBK1 solutions (10–40 µM) did not cause significant change of bound proteins on the chips, either in the presence or absence of c-di-GMP (Figure 5C). These data suggest that the CBD of STING does not contain a TBK1-binding site in its free form or in its c-di-GMP bound form.
STING Is Autoinhibited and c-di-GMP Binding May Unleash Its TBK1-Activating Capability
We wondered if existing mutagenesis data (Burdette et al., 2011) could provide clues in elucidating the potential mechanism of TBK1 activation upon c-di-GMP binding. In this regard, we found that the most informative group of mutants may be those that are still able to interact with c-di-GMP, but do not respond to c-di-GMP for enhanced IFN production. In fact, when overexpressed, these mutants hyperinduce IFN in comparison with the WT STING. Because the CTT is both necessary and sufficient for TBK1 activation and IRF3 phosphorylation, the mutants must have somehow activated the CTT in the absence of c-di-GMP, while the WT STING was not fully activated upon overexpression in the absence of c-di-GMP.
The mutants with the hyperinduction phenotype include R310A/E316A, E316A, E316N, E316Q, and S272A (Burdette et al., 2011) (Table 3). Because the double mutant R310A/ E316A shares the same phenotype as E316A, the key alteration in this mutant should be E316A. When mapped to the CBD structure, residue E316 is exposed on the surface, away from but on the same side of the c-di-GMP binding site (Figure 5D). Similarly, S272 is mapped to an adjacent site to E316 (Figure 5D). The locations of the mutant residues and their phenotypes suggest that in the WT STING, the CTT is autoinhibited through an intramolecular interaction between the CBD and the CTT. This interaction may overlap but is not identical with the c-di-GMP binding site, therefore rendering a capability of c-di-GMP in relieving this autoinhibition. The mutations on E316 or S272 may have compromised this interaction to unleash the CTT for TBK1 and IRF3 interaction and activation, leading to increased IFN production, without interfering with c-di-GMP binding. Interestingly, E316A, E316N, and E316Q all are hyperactive and nonresponsive to c-di-GMP, while E316D behaves like the WT STING (Table 3), suggesting that the negative charge on this residue is important for maintaining the CBD/ CTT interaction.
Further supporting evidence for this hypothesis is that several mutations at the CTT, R375A, and the triple mutant S358A/E360A/S366A (Burdette et al., 2011) (Figure 5E and Table 3) also abolish the responsiveness to c-di-GMP, but induce IFN in levels comparable to WT STING. These mutations may have also compromised the intramolecular interaction that is important for autoinhibition, leading to nonresponsiveness to c-di-GMP; but being at the CTT, they may have also directly affected TBK1 and IRF3 interaction and activation, leading to a comparable level of IFN production with the WT STING that has not been fully activated by c-di-GMP.
To directly test this hypothesis, we cotransfected HA-tagged STING CTT with constructs of FLAG-tagged STING full-length cytoplasmic domain (FL), CBD, and CTT in 293 cells. We then performed anti-HA immunoprecipitation followed by anti-FLAG immunoblotting. While HA-tagged CTT did not pull down control FLAG-tagged GFP, it pulled down both STING FL and CBD (Figure 5F). In addition, the interaction between CTT and CBD was significantly weakened in the presence of c-di-GMP (Figure 5G).
DISCUSSION
Our structural studies of STING uncovered a number of unexpected insights. First, STING is well recognized as an ER-localizing protein with several TM domains (Ishikawa and Barber, 2008; Sun et al., 2009). Our biochemical and structural studies now showed that the last predicted TM helix belongs to the cytosolic region and corresponds to helix α1 and α1′in the compact CBD of STING for c-di-GMP recognition. In fact, this helix provides the most crucial stacking interaction to c-di-GMP as well as participates in STING dimerization. Second, our structure of STING reveals a fold that is distinct from those of known bacterial c-di-GMP receptors. Besides the structural architecture, STING recognizes c-di-GMP uniquely using the dimerization interface and imposes a symmetrical conformation to the bound c-di-GMP. While the current study was under review, Ouyang et al. reported similar crystal structures of STING and its complex with c-di-GMP (Ouyang et al., 2012).
Importantly, our data suggest that STING is autoinhibited due to an intramolecular interaction between the CBD and CTT of STING. Binding of c-di-GMP competes with this interaction and releases the CTT of STING for TBK1 and IRF3 activation and IFN production (Figure 5H). One question that remains to be resolved is whether the autoinhibited form of STING is a monomer or dimer. Although our in vitro data show that STING CBD is a dimer, we do not know if this is because massive overexpression in E. coli during protein production may have already changed the STING conformation to the active one even in the absence of c-di-GMP. The hydrophobic nature of the dimerization interface would suggest that the dimerization is constitutive; however, this interface may have been shielded from solvent by the intramolecular interaction with the CTT. In any case, c-di-GMP binding would release the CTT to generate active STING dimers. Binding of c-di-GMP may well work as a “molecular staple,” locking STING into the active dimer conformation.
Once released, the CTT may mediate further oligomerization (Figure 5H), as the full-length cytosolic domain construct contains higher order oligomers (Figure 1B) and the CTT itself hyperoligomerizes (Tanaka and Chen, 2012). It is also important that STING is a membrane protein and its membrane localization may further facilitate STING oligomerization and activation upon signaling. In this regard, it has been shown that c-di-GMP induced formation of STING homodimers in mouse bone marrow-derived macrophages (Jin et al., 2011) and that forced dimerization of a GyrB gyrase fusion of STING by coumermycin increased luciferase reporter reading (Sun et al., 2009). Formation of large, punctate structures has also been observed for STING (Ishikawa et al., 2009; Saitoh et al., 2009; Zhong et al., 2008).
STING also functions as an adaptor protein in cytosolic dsDNA-induced innate immune response (Ishikawa et al., 2009) in addition to as a direct sensor of bacterial second messengers (Burdette et al., 2011). Several dsDNA sensors upstream of STING such as IFI16 and DDX41 have been identified (Unterholzner et al., 2010; Zhang et al., 2011). However, there seem to be additional intermediaries between these sensors and STING in the pathways. It appears that cyclic dinucleotides and cytosolic dsDNA utilize different structural aspects of STING because a STING mutant inert to c-di-GMP maintains its dsDNA responsiveness (Burdette et al., 2011). Yet, in both cases, they result in STING activation and induction of the same IFN pathway. Is there a general mechanistic basis of STING activation in response to different stimuli? Perhaps regardless of the different triggers, whether it is dsDNA or c-di-GMP, STING activation may result from a release of STING autoinhibition followed by oligomerization and activation of the pathway.
Autoinhibition and ligand-induced release of autoinhibition may also be a general mechanism for self versus non-self discrimination in innate immunity. Activation of RIG-I by dsRNA is an example. In the resting autorepressed state, CARD2 at the N terminus of RIG-I forms extensive interactions with the helical insertion domain (Hel2i), preventing its Polyubiquitination or polyubiquitin binding that is necessary for RIG-I interaction with downstream partners. After activation, dsRNA occupies an overlapping surface on Hel2i and releases CARD2 for free access by ubiquitination machinery (Kowalinski et al., 2011). Given the danger of innate immune activation in the absence of non-self recognition, intramolecular interaction-mediated autoinhibition provides a safety mechanism and a simple molecular switch that can be flipped on specifically by non-self recognition.
EXPERIMENTAL PROCEDURES
Protein Purification
DNA sequence encoding human STING CBD-CTT (139–379) was inserted into pSMT3 vector between BamHI and SalI sites. Protein was expressed in E. coli BL21 DE3 RIPL Codon Plus cell. E. coli cell was induced by0.5mM IPTG when cell density reached 0.5–0.6 and grew at 20°C overnight. Cells were spun down and lysed in lysis buffer (50 mM sodium phosphate [pH 7.4], 300 mM NaCl, 20 mM imidazole, and 5 mM 2-mercaptoethanol). After centrifugation and removal of cell debris, supernatant was incubated with Ni-NTA beads. Ni beads were washed extensively and protein was eluted in lysis buffer with 300 mM imidazole. Eluted protein was incubated with 1/1000 (w/w) Ulp1 protease at 4°C overnight to remove N-terminal SUMO tag. STING protein was further purified by size-exclusion chromatography in running buffer (20 mM Tris HCl [pH 8.0], 150 mM NaCl, and 5 mM DTT). Fraction containing STING protein was pooled, concentrated, and flash frozen for future use. Selenomethionyl protein and mutant proteins were purified in the same way. Samples intended for crystallization were digested with 1/200 (w/w) trypsin at room temperature for 1 hr and purified by a second round of size-exclusion chromatography.
Crystallization, Data Collection, and Structure Determination
Both STING CBD alone and c-di-GMP complex crystals were obtained by hanging drop vapor diffusion methods at 20°C. Selenomethionyl STING CBD at 12.9 mg/ml was mixed with equal volume of reservoir solution of 20%–23% PEG3350, 0.1 M Bis-Tris (pH6.2–6.5), and 0.2 M ammonium sulfate. For complex crystallization, STING CBD was incubated with equimolar of c-di-GMP for 2 hr at 4°C before mixing with equal volume of reservoir solution of 12%–15% PEG8000, 0.08 M sodium cacodylate (pH 6.3–6.5), 0.16 M calcium acetate, and 20% glycerol. Diffraction data was collected at National Synchrotron Light Source (NSLS) beamlines X25 and X29. Data sets were processed using the HKL2000 (Otwinowski and Minor, 1997) and XDS (Kabsch, 2010) softwares. Free STING structure was determined by singlewavelength anomalous dispersion (SAD), while STING/c-di-GMP complex structure was determined by molecular replacement, both using Phenix (Adams et al., 2010). Model building and iterative refinement were achieved using coot (Emsley and Cowtan, 2004), CNS (Brünger et al., 1998), Phenix (Adams et al., 2010), and Refmac (Murshudov et al., 1997). Molecular diagrams were generated using Pymol (Delano, 2002).
Isothermal Titration Calorimetry
Human STING CBD domain and c-di-GMP were dialyzed extensively against running buffer (100 mM NaCl, 10 mM Tris-HCl [pH 8.0]). Protein concentration was measured using absorbance at 280 nm. Prior to titration, both protein and c-di-GMP were centrifuged at 18,000 × g at 25°C for more than 10 min to remove any debris and air bubbles. The calorimetric titrations were carried out at 25°C on a MicroCal ITC200 instrument with 16 successive injections of 2.4 µl (400 µM) c-di-GMP, spaced 180 s apart, into the sample cell containing a solution of 200 µl (40 µM) STING CBD. The data was analyzed using the ORIGIN software. The association constant (Ka), enthalpy change (ΔH), and the stoichiometry (N) were calculated by fitting the thermograms to one set of binding sites. The dissociation constant (Kd), free energy change (ΔG), and the entropy change (ΔS) were calculated using the equations:
Biolayer Interferometry Binding Measurement
Binding between STING CBD and TBK1 in the absence or presence of c-di-GMP was performed using the BLItz system (forteBio Inc.) STING CBD was biotinylated by NHS-PEG4-Biotin following the manufacturer’s instructions (Thermo Scientific). Biotinylated CBD was immobilized onto streptavidin sensors, and the unbound protein was washed off by PBS-based sample dilution buffer (forteBio Inc.). The sensor was then immersed into TBK1 solution of various concentrations for 180 s. Dissociation was carried out by immersion of sensor into sample dilution buffer. For measurements in the presence of c-di-GMP, biotinylated STING CBD was incubated with equimolar of c-di-GMP at 4°C for 2 hr before being immobilized onto streptavidin sensor. Subsequent washing was performed in sample dilution buffer supplemented by 0.5 mM c-di-GMP.
Cell Culture and Transfection
Human embryonic kidney (HEK) 293 cells were cultured in DMEM supplemented with 10% calf serum and antibiotics. Transient transfection was carried out using the calcium phosphate precipitation method. HEK293 cells were plated in 6-well plates and transfected with 1 µg indicated expression constructs.
Lentiviral Vector Mediated Stable Cell Line Construction
Lentivirus transducing vectors (pTY) were constructed to express cDNA driven by Elongation Factor 1a (EF1a) promoter (He and Chang, 2004). The cDNA of interest was fused to the coding sequences for a puromycin-resistant or hygromycin-resistant gene and a picornavirus self-cleaving 2A peptide, which separates the puromycin or hygromycin resistant gene product and the protein of interest.
Stimulation with c-di-GMP
Cells are stimulated with c-di-GMP using the modified reversible digitonin permeabilization method (Girardin et al., 2003). Briefly, cells are incubated with or without 6 µM c-di-GMP in digitonin permeabilization solution (50 mM HEPES [pH7.0], 100 mM KCl, 3 mM MgCl2, 0.1 mM DTT, 85 mM Sucrose, 0.2% BSA, 1 mM ATP, 10 µg/ml Digitonin) for 30 min at 37°C. This solution was then replaced with normal culture media. At 5 hr post stimulation, cells lysates were harvested.
Coimmunoprecipitation
Cells were transfected with indicated expression constructs. Cells were lysed in lysis buffer 36–48 hr after transfection (20 mMTris-Cl [pH 7.4], 100mM NaCl, 0.1% Triton X-100, 25 mM β-glycerophosphate, 1 mM DTT, Roche protease inhibitor cocktail). Cell lysates were incubated with anti-FLAG M2-argarose (Sigma) or HA-affinity gel (Sigma) at 4°C for 2 hr. Beads were then washed in lysis buffer before being analyzed by SDS-PAGE and immunoblotting. For c-di-GMP stimulated samples, 6 µM c-di-GMP was added to lysis buffer and wash buffer.
Supplementary Material
ACKNOWLEDGMENTS
We thank the staff at National Synchrotron Light Source beamlines X25 and X29 for assistance in data collection.
Footnotes
ACCESSION NUMBERS
The coordinates and the structure factors for STING and the STING/c-di-GMP complex have been deposited in the Protein Data Bank with accession codes of 4F9E and 4F9G, respectively.
SUPPLEMENTAL INFORMATION
Supplemental Information includes two figures and can be found with this article online at doi:10.1016/j.molcel.2012.05.029.
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S, Takeda K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- Barber GN. Innate immune DNA sensing pathways: STING, AIMII and the regulation of interferon production and inflammatory responses. Curr. Opin. Immunol. 2011;23:10–20. doi: 10.1016/j.coi.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barends TR, Hartmann E, Griese JJ, Beitlich T, Kirienko NV, Ryjenkov DA, Reinstein J, Shoeman RL, Gomelsky M, Schlichting I. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature. 2009;459:1015–1018. doi: 10.1038/nature07966. [DOI] [PubMed] [Google Scholar]
- Benach J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, Ramos JE, Forouhar F, Neely H, Seetharaman J, Camilli A, Hunt JF. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 2007;26:5153–5166. doi: 10.1038/sj.emboj.7601918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C, Paul R, Samoray D, Amiot NC, Giese B, Jenal U, Schirmer T. Structural basis of activity and allosteric control of diguanylate cyclase. Proc. Natl. Acad. Sci. USA. 2004;101:17084–17089. doi: 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Kuolee R, Yan H. The potential of 3′,5′-cyclic diguanylic acid (c-di-GMP) as an effective vaccine adjuvant. Vaccine. 2010;28:3080–3085. doi: 10.1016/j.vaccine.2010.02.081. [DOI] [PubMed] [Google Scholar]
- Delano WL. The PyMol Molecular Graphics System. CA, USA: DeLano Scientific, San Carlos; 2002. [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardin SE, Boneca IG, Carneiro LA, Antignac A, Jéhanno M, Viala J, Tedin K, Taha MK, Labigne A, Zähringer U, et al. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science. 2003;300:1584–1587. doi: 10.1126/science.1084677. [DOI] [PubMed] [Google Scholar]
- He J, Chang LJ. Functional characterization of hepatoma-specific stem cell antigen-2. Mol. Carcinog. 2004;40:90–103. doi: 10.1002/mc.20019. [DOI] [PubMed] [Google Scholar]
- Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inohara N, Chamaillard M, McDonald C, Nuñez G. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu. Rev. Biochem. 2005;74:355–383. doi: 10.1146/annurev.biochem.74.082803.133347. [DOI] [PubMed] [Google Scholar]
- Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F, Ludwig H, Sutter G, Suzuki K, Hemmi H, et al. A Toll-like receptor-indepen dent antiviral response induced by double-stranded B-form DNA. Nat. Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010;327:291–295. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Waterman PM, Jonscher KR, Short CM, Reisdorph NA, Cambier JC. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol. Cell. Biol. 2008;28:5014–5026. doi: 10.1128/MCB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Hill KK, Filak H, Mogan J, Knowles H, Zhang B, Perraud AL, Cambier JC, Lenz LL. MPYS is required for IFN response factor 3 activation and type I IFN production in the response of cultured phagocytes to bacterial second messengers cyclic-di-AMP and cyclic-di-GMP. J. Immunol. 2011;187:2595–2601. doi: 10.4049/jimmunol.1100088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaolis DK, Means TK, Yang D, Takahashi M, Yoshimura T, Muraille E, Philpott D, Schroeder JT, Hyodo M, Hayakawa Y, et al. Bacterial c-di-GMP is an immunostimulatory molecule. J. Immunol. 2007;178:2171–2181. doi: 10.4049/jimmunol.178.4.2171. [DOI] [PubMed] [Google Scholar]
- Kato H, Takahasi K, Fujita T. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol. Rev. 2011;243:91–98. doi: 10.1111/j.1600-065X.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, Cusack S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- Kulshina N, Baird NJ, Ferré-D’Amaré AR. Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch. Nat. Struct. Mol. Biol. 2009;16:1212–1217. doi: 10.1038/nsmb.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, Janeway C., Jr Innate immunity. N. Engl. J. Med. 2000;343:338–344. doi: 10.1056/NEJM200008033430506. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Navarro MV, De N, Bae N, Wang Q, Sondermann H. Structural analysis of the GGDEF-EAL domain-containing c-di-GMP receptor FimX. Structure. 2009;17:1104–1116. doi: 10.1016/j.str.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Ouyang S, Song X, Wang Y, Ru H, Shaw N, Jiang Y, Niu F, Zhu Y, Qiu W, Parvatiyar K, et al. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity. 2012 doi: 10.1016/j.immuni.2012.03.019. in press. Published online May 10, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc. Natl. Acad. Sci. USA. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer JD, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, Hyodo M, Hayakawa Y, Woodward JJ, Portnoy DA, Vance RE. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect. Immun. 2011;79:688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KD, Lipchock SV, Ames TD, Wang J, Breaker RR, Strobel SA. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat. Struct. Mol. Biol. 2009;16:1218–1223. doi: 10.1038/nsmb.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Li Y, Chen L, Chen H, You F, Zhou X, Zhou Y, Zhai Z, Chen D, Jiang Z. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. USA. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaoka A, Yanai H. Interferon signalling network in innate defence. Cell. Microbiol. 2006;8:907–922. doi: 10.1111/j.1462-5822.2006.00716.x. [DOI] [PubMed] [Google Scholar]
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- Tamayo R, Pratt JT, Camilli A. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu. Rev. Microbiol. 2007;61:131–148. doi: 10.1146/annurev.micro.61.080706.093426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, Shu HB. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.