Abstract
Glaucoma is a group of genetically heterogeneous neurodegenerative disorders causing the degeneration of the ganglion neurons of the retina. Increased intraocular pressure (IOP) is a hallmark risk factor promoting the death of ganglion neurons of the retina in glaucoma. Yet, the molecular processes underlying the degeneration of these neurons by increased IOP are not understood. To gain insight into the early molecular events and discover biomarkers induced by IOP, we performed gene and protein expression profiling to compare retinas of eyes with and without high IOP in a rodent model of experimental glaucoma. This pilot study found that the IOP-mediated changes in the transcription levels of a restricted set of genes implicated in peroxisomal and mitochondrial function, modulation of neuron survival and inflammatory processes, were also accompanied by changes in the levels of proteins encoded by the same genes. With the exception of the inflammatory markers, serum amyloid-A1 (SAA1) and serum amyloid-A2 (SAA2), the IOP-induced changes in protein expression were restricted to ganglion neurons of the retina and they were detected also in the vitreous, thus suggesting to an early IOP-mediated loss of ganglion cell integrity. Interestingly, SAA1 and SAA2 were induced in retinal microglia cells, whereas they were reduced in sera of IOP-responsive mice. Hence, this study defines novel IOP-induced molecular processes, biomarkers and sources thereof, and it further validates the extension of the analyses herein reported to other genes modulated by IOP.
Keywords: Neurodegeneration, ganglion neurons, gene expression, retina, glaucoma, pathogenesis, biomarkers, inflammation, stress stimuli, vitreous
Introduction
Glaucoma is the leading cause of irreversible blindness, affecting 66 million people worldwide (1). Glaucoma is a genetically heterogeneous visual and neurodegenerative disorder, which is characterized by progressive optic nerve head cupping and selective loss of retinal ganglion cell bodies and their axons (2, 3). In about 60% of glaucoma patients, glaucoma is triggered by an increase of outflow resistance at the anterior eye that promotes the elevation of intraocular pressure (IOP), which is clinically silent (4–6). Ultimately, this stress insult is a determinant risk factor in inducing the death of ganglion neurons of the retinal tissue lining the posterior eye and ultimately, blindness (2, 3, 7, 8). Hence, the stress insult of IOP sustained by the ganglion cells likely compromises molecular and subcellular processes that are paramount to the function and survival of ganglion neurons (9).
Despite the high prevalence of glaucoma, little is known about the molecular and subcellular mechanisms triggering the death of ganglion cells and the pathogenesis of glaucoma. Likewise, the lack of biomarkers of glaucoma severely hinders the stratification and molecular diagnosis of glaucoma patients before clinical symptoms ensue. Several compounding and confounding factors seem to contribute to the pathogenesis of glaucoma. To this effect, a) to this date very few genes causing glaucoma have been identified (10–16); b) genetic lesions (mutations) isolated in glaucoma patients do not always cosegregate with disease manifestations in family pedigrees (17–20); c) a pharmacological-induced decrease of IOP does not prevent always glaucoma and blindness (21); d) the spatiotemporal segregation between the IOP insult and damage to the ganglion neurons is likely to trigger an array of heterogeneous stress responses by these neurons, as a result of the diversity in genetic background among patients. Collectively, these factors support the idea that phenotypic manifestations linked to glaucoma have variable expressivity and may outlast the stress insult.
Experimental elevation of IOP in animal models (experimental glaucoma), which is known to recapitulate pathological features linked to the development and progression of glaucoma in the human, ultimately causes glaucomatous degeneration of ganglion neurons of the retina (9, 22, 23). In an attempt to understand the molecular processes modulating the expression of glaucoma, gene expression profiling with microarrays were performed in the monkey (24) and various other animal models of experimental glaucoma (4, 25–27). Yet, no consensus exists among such studies as to what genes are modulated by IOP. These apparent disparate outcomes may have arisen from the employment of different experimental parameters that are likely to contribute to confounding outcomes. They include the assessment of genetically distinct forms of glaucoma (e.g. DBA2J vs POAG) (26), the species, age, genetic background, the length and number of experimental procedures to induce IOP, and the timing of the assays to examine the effect of IOP on ganglion neurons of the retina. The objectives of this pilot study are: i) to circumvent many of these limitations by employing a stringent experimental procedure of inducing IOP in an experimental mouse model of glaucoma, ii) to determine differences in gene expression profile between retinas of eyes with and without IOP, iii) to validate some of such changes at protein level with a restricted set of genes and mapping of these changes to ganglion neurons. Collectively, these studies provide proof-of-concept that the approaches employed allow the identification of IOP-induced pathobiological signaling pathways as well as subclinical ocular and systemic biomarkers that may be used also as potential surrogates and stratifiers of disease expression among glaucoma patients.
Materials and Methods
Generation of mice with elevated intraocular pressure
All animal experiments were performed in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research and were approved by the Duke University Animal Care and Use Committee. IOP elevation (experimental glaucoma) was created in 10 week-old inbred C57Bl6/J mice (Jackson Laboratories) by a single and unilateral episcleral vein injection of hypertonic saline in the right eye (22). Non-responders refractory to the single injection treatment were kept through the duration of the study under identical conditions. IOP of mice was measured once a week between 10–12 AM with a handheld TonoLab in mice (28) who were sedated with ketamine/xylazine (ketamine 80mg/kg and xylazine 4mg/kg) as described elsewhere (29). Under this experimental procedure, the elevation of IOP began to rise at week 4 post-injection and remained high until week 7. At the end of week 7, all non- and responder mice were sacrificed between 10–12 AM and ocular tissues and serum were collected for various analyses.
Retinal Ganglion Cell Counts and TUNEL Assays
Whole eyecups were fixed for 4h in 4% paraformaldehyde/phosphate-buffered saline (PBS), pH 7.4. Eyes were transferred to PBS, pH 7.4 and vitreous was removed. Retinas were dissected and immersed in PBS in 24-well tissue culture plates and washed with PBS, pH 7.4. Retinas were permeabilized with 0.3% Triton X-100/PBS, pH 7.4, incubated with Hoechst (1:1000) and washed with PBS, pH 7.4. All retinas were flattened on Superfrost Plus microscope slide (VWR) with the ganglion cell layer upwards, mounted with Prolong Gold Anti-Fade reagent (Invitrogen) and coverslipped (Fisher-Scientific). Pictures of the Hoechst-stained nuclei of ganglion cells layer were acquired with 20×-objective using Nikon C90i upright microscope equipped with epifluorescence. Four image fields of 10,000 μm2 each from each quadrant of the retina were used for analysis of nuclei count. The Metamorph software (Molecular Devices) was used to count the Hoechst-labeled nuclei in each image. TUNEL assays on at least eight sections of the whole eye cup were performed as described elsewhere (30).
Total RNA isolation
After the intraocular pressures became elevated for 3 weeks, the retinas of treated right and untreated left eyes were isolated, singly placed in RNAlater solution (Ambion) and total RNA was purified with RNeasy mini kit (Qiagen). To remove traces of genomic DNA, all sample extracts were subjected to a DNaseI in-column treatment according to the manufacture instructions (Qiagen). RNA purity and concentration was analyzed spectrophometrically (260/280 nm) by electrophoresis on an agarose gel and by microcapillary electrophoresis on an RNA 6000 Nano LabChip kit using an Agilent 2100 bioanalyzer (Agilent Technologies, Palo Alto, CA; IGSP core facility, Duke University).
Microarray analysis
Microarray data were collected at Expression Analysis, Inc. (www.expressionanalysis.com; Durham, NC) using the Affymetrix GeneChip® Mouse 430 2.0 array which contain approximately 39,000 transcripts and variants, including over 34,000 well-characterized mouse genes. The probe sets were selected from sequences derived from GenBank, dbEST, and RefSeq. The sequence clusters were created from the UniGene database (Build 107, June 2002) and then refined by analysis and comparison with the publicly available draft assembly of the mouse genome from the Whitehead Institute Center for Genome Research (MSCG, April 2002).
Target was prepared from mRNA amplified using the NuGEN Ovation® RNA Amplification System V2 following the manufacturer’s specified protocol. In brief, first strand cDNA was prepared from 40 ng of total RNA using a unique first strand DNA/RNA chimeric primer and a reverse transcriptase. The primer has a DNA portion that hybridizes to the 5′ portion of the poly (A) sequence. The resulting cDNA/mRNA hybrid molecule contains a unique RNA sequence at the 5′ end of the cDNA strand. Fragmentation of the mRNA within the cDNA/mRNA complex creates priming sites for DNA polymerase to synthesize a second strand, which includes DNA complementary to the 5′ unique sequence from the first strand chimeric primer. The result is a double stranded cDNA with a unique DNA/RNA heteroduplex at one end. RNase H is used to degrade RNA in the DNA/RNA heteroduplex at the 5′ end of the first cDNA strand. This results in the exposure of a DNA sequence that is available for hybridizing a second DNA/RNA chimeric primer. DNA polymerase then initiates replication at the 3′ end of the primer, displacing the existing strand. The RNA portion at the 5′ end of the newly synthesized strand is again removed by RNase H, exposing part of the unique priming site for initiation of the next round of cDNA synthesis. The process of DNA/RNA primer hybridization, DNA replication, strand displacement and RNA cleavage is repeated, resulting in rapid accumulation of cDNA with sequence complementary to the mRNA. The resultant cDNA product was purified with the Affymetrix cDNA cleanup module (Affymetrix) and 3.75 ug was fragmented and labeled using the NuGEN FL-Ovation™ cDNA Biotin Module V2 following the manufacturer’s specified protocol. The fragmented cDNA was diluted in hybridization buffer (MES, NaCl, EDTA, Tween 20, Herring Sperm DNA, Acetylated BSA) containing biotin-labeled OligoB2 and Eukaryotic Hybridization Controls (Affymetrix). The hybridization cocktail was denatured at 99°C for 5 minutes, incubated at 45°C for 5 minutes and then injected into a GeneChip cartridge. The GeneChip array was incubated at 42°C for at least 16 hours in a rotating oven at 60 rpm. GeneChips were washed with a series of nonstringent (25°C) and stringent (50°C) solutions containing variable amounts of MES, Tween20 and SSPE. The microarrays were then stained with Streptavidin Phycoerythrin and the fluorescent signal was amplified using a biotinylated antibody solution. Fluorescent images were detected in a GeneChip® Scanner 3000 and expression data was extracted using the GeneChip Operating System v 1.1 (Affymetrix). The expression intensities for all genes across the samples were normalized using robust multi-array analysis (RMA) (31), including probe-level quantile normalization and probe summarization, as implemented in the Genespring GX 7.3.1. Differentially expressed genes of retinas from treated and untreated eyes were selected with Volcano (group-wise analysis) with a p-value cutoff of 0.1 and fold change cutoff of 1.5 (Genespring GX 7.3.1) and paired t-test (pair-wise analysis) with a p-value cutoff of 0.05. (Partek 6.3). Hierarchical clustering was performed using Average Linkage with Pearson Correlation as the similarity measure (Genespring GX 7.3.1).
Western blot analyses
Retina and vitreous samples were homogenized in NP-40 buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 1% Nonidet P-40 (NP-40), and Complete protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN, USA). Samples were centrifuged at 10,000g for 15 min and supernatants collected. Protein concentration was measured by Bradford method using BSA as standard. Protein extracts (60 μg unless otherwise noted) were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 10.5–14% Bio-Rad Criterion gradient gel and Western blotting was carried out as described elsewhere (32). Blots were developed with a SuperSignal chemiluminescence substrate (Pierce, Rockford, IL). Primary antibodies used: Mouse anti-Bcl2 (1μg/mL; MBL, Woburn, MA), rabbit anti-catalase (1:8000; Calbiochem), rabbit anti-XIAP (1μg/mL; AnaSpec, San Jose, CA), mouse anti-acetylated tubulin (25 ng/mL; Sigma, St. Louis, MO), rabbit anti-Hsc70 (1:3000; Stressgen, Ann Arbor, MI), rabbit anti-rhodopsin (1:20,000; Affinity Bioreagents, Golden, CO).
Semi-quantitative RT-PCR
Reverse transcription was carried out with 0.5μg of total RNA from each retina sample using SuperScript II Reverse Transcriptase (Invitrogen). Five percent of each resulting cDNA was used for a 50 μl PCR reaction. Differential expression of serum amyloid A1 and A2 (Saa1/Saa2) was validated by semi-quantative RT-PCR as Western blotting failed to detect them. Saa1 and Saa2 gene-specific primers were designed to yield amplicons of 161 bp in length for both SAA1 and SAA2. A common reverse primer (5′-TCAGCCATGGTGTCCTCGTGT) is used for both SAA1 and SAA2 amplification, and specific forward primers were designed to discriminate SAA1 (5′-AAG CTA ACT GGA AAA ACT CA) and SAA2 (5′-AAG CTG GCT GGA AAG ATG GA). The identities of Saa1 and Saa2 amplicons were further confirmed by restriction digestion of these with Hae III, which digests selectively 46 bp from the Saa1 amplicon (Supplemental figure 1). β-actin was used as a loading control.
Immunhistochemistry
Bilateral eyes of three C57Bl6 mice with unilateral pressure elevation of the right eye exactly under the same conditions described previously underwent enucleation. Eyes were then fixed for 4 hours in 4% paraformaldehyde/PBS, pH 7.4 followed by several rinses with PBS, pH 7.4. Tissues were then embedded in OCT compound in cryomolds and stored in −80ºC. Ten μm-thick sections were then cut in a cryostat (Microm HM 5550, Walldorf, Germany), mounted on superfrost slides (Fisher Scientific) and washed in PBS 3 times. Immunochemistry was performed essentially as described elsewhere (29, 33). Briefly, sections were blocked with 5% normal goat serum for 30 minutes at room temperature, incubated with a primary antibody (rabbit anti catalase 1:100, Calbiochem; rabbit anti XIAP, 1:100, AnaSpec, San Jose, CA; mouse anti Bcl2, 1:10, MBL, Woburn, MA; and rat anti SAA, 1:20, R&D Systems, Minneapolis, MN) overnight at 4ºC, rinsed 3× and 5 minutes each in PBS, incubated in secondary antibody for 1 hour at room temperature (goat anti-rabbit Alexa-594, goat anti-mouse Alexa-594, goat anti-rat Alexa-594; Molecular Probes). Then, they were rinsed three times in PBS, stained with Hoechst (1:1000) for 10 minutes followed by three rinses in PBS for 2 minutes each. The laser confocal Nikon C90i microscope with software EZ-C1.3.1 (Nikon, Garden City, NY) was then used to examine and capture immunostained images of retinas of treated and untreated eyes with the same laser power and average number of passes (4).
Hydrogen Peroxide Measurement
Retinas were homogenized in 50 μl de-ionized water per retina on ice with a motorized Kontes Microtube Pellet Pestle Rod. Samples were centrifuged at 10,000g 15 min at 4°C and supernatants collected for hydrogen peroxide measurement. Pellets were dissolved in 50 μl SDS buffer (50 mM Tris-HCl, pH 6.7, 2% SDS, 10% glycerol) and protein contents were measured by BCA method. The hydrogen peroxide levels were assayed using the Amplex Red Hydrogen Peroxide/Peroxidase Assay Kit (Invitrogen) as per manufacturer’s instructions.
Statistical analyses
Whenever applicable, statistical analysis was performed with two-tailed equal or unequal variance t-test.
Results
Generation of mouse models with unilateral increase of intraocular pressure
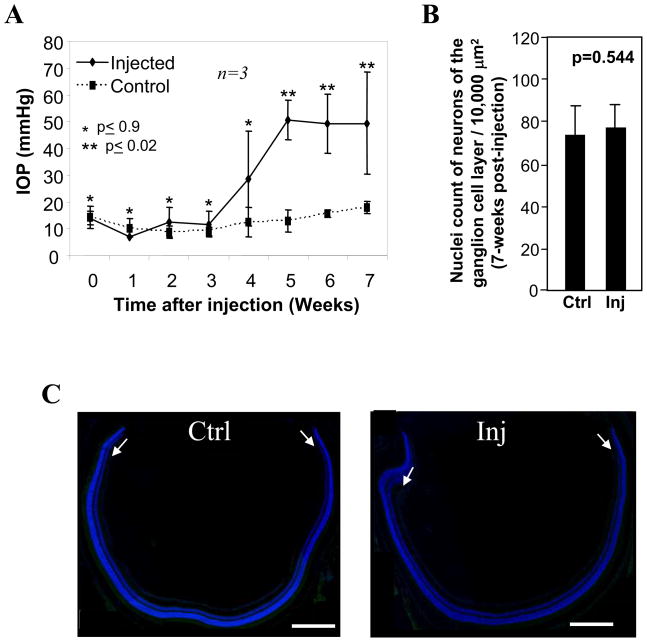
Experimental glaucoma by episcleral vein injection of hypertonic saline leads to the obstruction of the outflow of aqueous humor and elevation of intraocular pressure (IOP) (22, 28). To mitigate the generation of procedural and temporal variations among mice and experimental injection procedures, a batch of age-matched (10 week-old) inbred C57Bl6 mice underwent a single episcleral vein injection of hypertonic saline between 10–12AM. IOPs were subsequently recorded weekly around the same time of the day. Retinas of mice subjected to similar temporal elevation of sustained IOP (3 weeks), in comparison to non-injected retinas, were selected for analyses. As shown in Fig. 1A, the elevation of IOP in injected eyes began to rise at week 4 post-injection and the pressure remained significantly elevated between week 5 and 7. At week 7 (Fig. 1A), mice were sacrificed for isolation of RNA from injected and non-injected retinas and microarray analysis. Retinas of independent set of mice subjected to identical experimental and temporal procedures of induction of IOP were also subjected to further analyses. We were particularly interested in determining changes in gene and protein expression prior to any significant retinal ganglion cell death. Once death has ensued, changes in gene and protein expression related to cell death would skew the data and prevent a clear interpretation of results caused by elevated IOP; instead of comparing similar populations of retinal neurons (the goal of this study), substantially different populations of neurons would be compared. To assess whether our experimental procedure and its timing resulted in cell death, we tallied the number of neurons in the ganglion cell layer and determined if apoptotic nuclei could be observed by the sustained elevation of IOP for 3 weeks. There was not a significant change in the number of neurons of the ganglion cell layer encompassing all four quadrant of the retina (Fig. 1B) and no cell death of neurons of the ganglion cell layer was observed between retinas of eyes with and without elevated IOP (Fig. 1C). Collectively, these procedures help to ensure that IOP-induced changes of gene and protein expression in retinas were derived from a similar number of neurons from which early indicators of changes in biological activity and generation of novel biomarkers by IOP could be discerned and identified.
Figure 1.
Induction of high intraocular pressure (IOP) in a mouse model of experimental glaucoma without neuronal cell loss. A. IOPs begin to increase in injected eyes as compared with the control eyes at week 4 and sustained elevated pressures continue through week 7. Week 0 represents the baseline intraocular pressures just before the hypertonic saline injections were performed. Initially, there were similar pressures in both the injected and control eyes during weeks 0–3. IOPs reached maximal elevation as well as statistical significance at week 5 and they remained elevated throughout the duration of the study (week 7). Data are means of three independent experiments ± s.d. B. No significant neuronal cell loss of the ganglion cell layer of the retina was present between injected eyes at week-7 post-injection (time of sacrifice) and non-injected eyes. Cell loss was measured by comparing Hoechst-positive nuclei of cells of the ganglion cell layer between retinas of non-injected and injected eyes. Data presented as means of sample nuclei counts from all four quadrants of three retinas ± standard deviation. C. TUNEL staining (nucleosomal DNA fragmentation) of retinal sections of eyecups showed no apoptotic cell death in the ganglion cell layer (arrows) or any other nuclear layers of the retina between non-injected (Ctrl; left image) and injected (Inj; right image) eyes. Retinal sections were counterstained with DAPI (blue). Scale bar: 400 μm
Analyses of changes in gene and protein expression profile of retinas induced by IOP
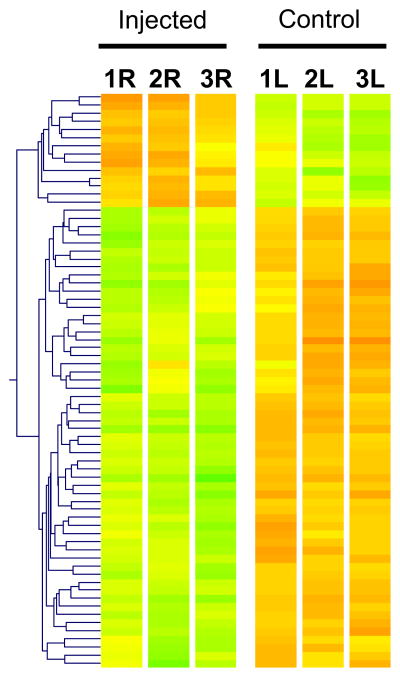
In an effort to define differences in gene expression induced by elevated intraocular pressure, microarray analysis was utilized to compare the transcriptional expression patterns of retinas of three mice subjected to a single episcleral vein injection of hypertonic saline and three weeks of significantly elevated intraocular pressure in their right eyes, and baseline pressure in their left and untreated eyes (Fig. 1A). Pairwise and volcano (group wise) statistical analyses were performed to compare these two groups. We employed a two-tier statistical criteria based on the cut-off of p-values and median ratio of fold-change of gene expression between retinas subjected to non- and elevated IOP found in the pairwise and volcano analyses. This approach identified 10 (fold change ≥2; p≤0.05) and ~50 genes (fold change≥1.5; p≤0.1) up-regulated and ~140 genes down-regulated (fold change≥1.5; p≤0.1) in the volcano analyses, whereas in the pairwise analyses there were 17 and 2 genes down- and up-regulated (fold change≥1.5; p≤0.01), respectively, and 94 and 40 genes down- and up-regulated (fold change≥1.4; p≤0.05), respectively (unpublished data). The overlap of volcano and pairwise analyses represented 71 genes that were modulated by IOP (Fig. 2).
Figure 2.
Overall changes of gene expression by elevated IOP. Hierarchical clustering (heat plot) of 71 retinal genes whose modulation of expression by IOP was shared by volcano and pairwise analyses of untreated (control) left (L) and contralateral treated (injected) right (R) eyes (n=3). Branch lengths of dendrogram on the left depict the relatedness of expression patterns. Genes with higher expression are depicted in red; genes with lower expression are shown in green.
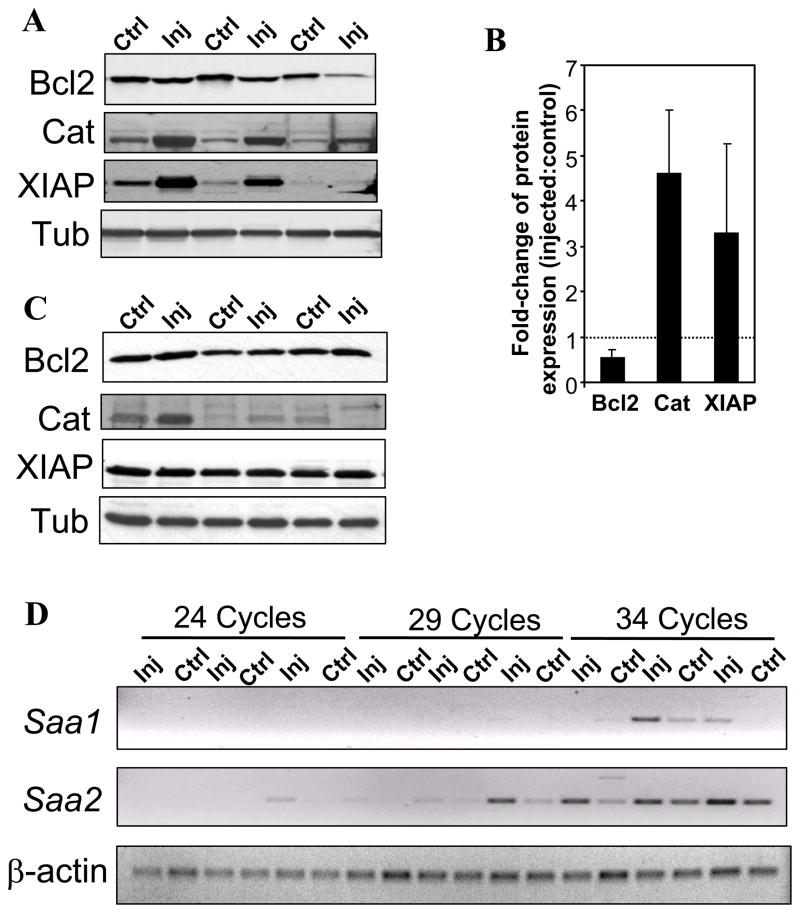
To establish that the outcome of our experimental approach can be employed to identify early indicators of selective IOP-induced changes in protein levels, biological activity, and biomarkers, we focused our pilot analyses on four genes, whose changes in mRNAs or proteins are critical for cell survival or are commonly designated disease biomarkers, but have not been yet reported to be modulated by IOP in the retina (Table 1). These genes were the B-cell leukemia/lymphoma 2 (Bcl2), baculoviral IAP repeat-containing-4 (Birc4/XIAP), catalase (Cat), and serum amyloid A1 (SAA1). Cat, Bcl2, Birc4/XIAP, and SAA1, mediate, respectively, critical peroxisomal, mitochondrial, cell survival, and inflammatory processes. Since changes in the activity of transcriptional expression of the genes identified must lead largely to changes in the levels of proteins encoded by the cognate genes to promote the activation of pathophysiological pathways prior to the onset of clinical glaucoma (ganglion cell death), we probed by immunoblot analysis whether the levels of Bcl2, Birc4/XIAP, Cat, and SAA1, were changed between retinas of eyes with and without elevated IOP. As shown in figures 3A and B, Bcl2 was down-regulated by approximately 50% in injected eyes with IOP, whereas Cat and XIAP were up-regulated by approximately 4.5- and 3-fold, respectively, in the same eyes. The housekeeping protein, tubulin, remained unchanged upon injection, indicating the selective effect of IOP in the modulation of Bcl2, Birc4/XIAP, and Cat of the injected eyes (Fig. 3A, lower panel). In contrast, such changes in Bcl2, Birc4/XIAP and Cat, were not found in the injected eyes of non-responsive (Fig. 3C, lower panel). These observations support that the changes observed were independent of the injection procedure itself and solely dependent on the rise of IOP (Fig. 3C). On the other hand, we were not able to detect changes in the protein levels of SAA1 in the retina by immunoblot analysis. As subsequently described, this observation may reflect changes in protein expression to a very restricted population of neurons or glial cells of the retina. Regardless, we examined whether transcriptional changes occurred between retinas with and without exposure to IOP by semi-quantitative RT-PCR. Serum amyloid A1 and A2 are highly homologous with 90 % nucleotide identity (34, 35). Closer inspection of the microarray employed found that the changes in the SAA1 may reflect changes in SAA1 and SAA2, since probes unique and common to both transcripts are spotted in the array. As shown in Fig. 3D, both SAA1 and SAA2 were induced in retinas of injected eyes, but the induction of latter was more prominent. These results further support that IOP promotes the up-regulation of Saa1 and Saa2 in the retina.
Table 1.
List of selected genes with altered expression between retinas with and without exposure to elevated intraocular pressure
| Gene | Probe set ID | Analysis method | Fold-change | p-value |
|---|---|---|---|---|
| Saa1 | 1419075_s_at | Pairwise | 6.80 | 0.006 |
| Catalase | 1416430_at | Volcano | 1.60 | 0.080 |
| 1416429_a_at | Volcano | 1.54 | 0.087 | |
| XIAP/ | 1450231_a_at | Pairwise | 0.57 | 0.005 |
| 1421394_a_at | Pairwise | 0.55 | 0.018 | |
| 1450231_a_at | Volcano | 0.57 | 0.002 | |
| 1421394_a_at | Volcano | 0.55 | 0.020 | |
| Bcl2 | 1422938_at | Volcano | 0.66 | 0.081 |
| 1427819_at | Volcano | 0.63 | 0.097 |
Figure 3.
Elevated IOP promotes altered expression of proteins in the retina encoded by the genes with IOP-induced change of transcription (as noted in Table 1). A. Western blot analysis of extracts of retinas with (injected; Inj) and without (control, Ctrl) exposure to elevated IOP. When compared to control retinas, Bcl2 levels were decreased in retinas of contralateral injected eyes, whereas the levels of catalase (Cat) and XIAP were increased. Tubulin serves as a loading control. Retinas were exposed to 3-weeks of elevated IOP during the 7-week post-injection treatment as those shown in Fig 1A. B. Fold change of protein expression in retinas of injected eyes compared to control eyes (n=3). Quantitation of protein levels ascertained by Western blot showed an almost 50% reduction in Bcl2 expression, a 4.5-fold increase in catalase expression, and a 3-fold increase in XIAP expression in the eyes with elevated intraocular pressure as compared with the controls. C. Western blot of retinas of injected eyes of non-responders, which were refractory to the single injection treatment (without IOP elevation), and control (non-injected) eyes. The protein expression levels of Bcl2, Cat, and XIAP are comparable in the retinas of both the injected and control eyes. Tubulin served as a loading control. Retinas were collected 7-weeks after the injection treatment. D. Induction of Saa1 and Saa2 gene expression in retinas of eyes with elevated intraocular pressure versus controls using semi-quantitative RT-PCR. Aliquots of RT-CR reactions were taken at 24, 29 and 34 cycles. Saa1 and Saa2 were increased in retinas of eyes exposed to IOP as compared with that of controls with the expression of Saa2 being the most prominent. Retinas were exposed to 3-weeks of elevated IOP during the 7-week post-injection treatment as those shown in Fig 1A. β -actin serves as the loading control.
IOP modulates localized changes in protein expression levels in ganglion neurons and microglial cells of the retina
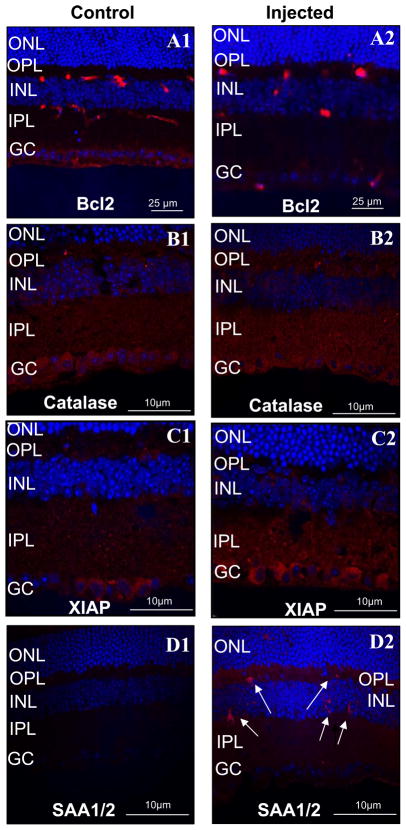
Neurodegeneration of ganglion neurons underlying glaucoma and inflammatory processes are associated to various stages of glaucoma (2, 3, 36, 37). Hence, we examined whether the changes in expression of Cat, Bcl2, Birc4/XIAP, and SAA1, were localized and restricted to ganglion neurons or microglial cells of the retina. As shown in figure 4, changes in expression of Cat, Bcl2 and Birc4/XIAP, were localized by immunocytochemistry to the cell bodies of ganglion neurons or processes thereof (Fig. 4A–C), while changes in SAA1/SAA2 were highly restricted to small cell bodies of microglial cells of the retina (Fig. 4D2, arrows). In particular, decreased levels of Bcl2 were localized to the retinal ganglion cell bodies of injected eyes as compared to the control eyes. In contrast, catalase (Fig. 4B1 and B2) and XIAP (Fig. 4C1 and C2) were prominently increased in the inner plexiform layer and, ganglion cell and inner plexiform layers, of retinas exposed to elevated IOP. SAA1/SAA2 was the only protein without altered protein expression in ganglion neurons, since its expression was induced in either residential or migrating proinflamatory microglial cells of the retina (Fig. 4D1 and D2).
Figure 4.
Immunohistochemistry showing localization of proteins with IOP-induced altered gene expression in retinas of non-injected (A1–D1) and contralateral injected (A2–D2) eyes. Bcl2 expression level in the retina is decreased in the injected eyes (A2), whereas catalase (B2) and XIAP (C2) are increased. These changes in protein expression are localized to the ganglion cells of retinas of injected eyes. In particular, changes in expression of Bcl2, catalase and XIAP, are localized respectively, to the soma, dendritic processes (inner plexiform layer; IPL), and soma and dendritic processes, of ganglion neurons. In contrast, the serum amyloid A1/A2 (SAA1/SAA2) expression was present in scarce microglial cells spread throughout the retina (D2). These SAA1/SAA2-positive cells were not observed in retinas of non-injected eyes (D1). Retinas of injected eyes were exposed to 3-weeks of elevated IOP during the 7-week post-injection treatment as those shown in Fig 1A.
Elevation of intraocular pressure produces biomarkers in non-retinal ocular tissues and extraocularly
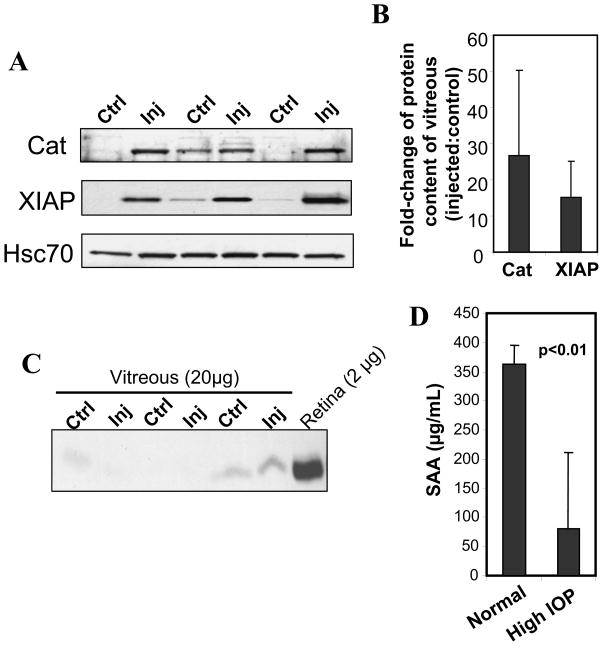
Another goal of this study was to determine whether changes in the protein levels of retinal cells may be detected in extra-retinal tissues and fluids and thus, employed as potential biomarkers of processes linked to glaucoma. To this effect, we examined whether some of the protein changes observed in the retina could be also detected in the vitreous of the eye chamber due to its juxtaposed location to the retina. One of the rationales to this hypothesis was that the vitreous may act as a reservoir of proteins leaked from ganglion neurons due to transitional loss of membrane permeability and leakage of some of its contents as a result of high IOP. We therefore performed Western blots of the vitreous and compared the levels of catalase and XIAP between the injected and control eyes. As shown in Figs. 5A and B, elevated IOP resulted in a 25-fold increase of catalase content in the vitreous compared to that of the non-treated eye, whereas XIAP content increased by 9-fold. There were no significant differences in BCl2 content between the injected and control eyes (data not shown). Interestingly, the increases in the levels of proteins in the vitreous of the injected eyes versus the control eyes seemed greater than those observed in the retina, thus supporting an accumulation of such proteins in the vitreous. This accumulation was not due to contamination of the vitreous with retinal tissue, since reprobing of the vitreous with an antibody against rhodopsin, an extremely abundant protein expressed in the photosensory cells comprising 70% of retinal neurons, did not or just detected traces of this protein (Fig. 5C).
Figure 5.
Detection of altered protein content in the vitreous of eyes with elevated IOP. A. Western blots of catalase (Cat) and XIAP protein levels in the vitreous of injected (Inj) and non-injected (Ctrl) contralateral eyes showed that these are increased only in eyes with elevated IOP. Hsc70 levels (lower panel) are comparable between the two groups of eyes and serves as a loading control. Injected eyes presented 3-weeks of elevated IOP during the 7-week post-injection treatment as those shown in Fig 1A. B. Densitometry analysis of changes the protein levels of catalase and XIAP in the vitreous of injected eye as compared with the control (shown in A). A 25-fold and a 15-fold average increase in the levels of catalase and XIAP were observed in the vitreous of injected eyes as compared with the controls (n=3). C. The vitreous presents no contamination with retina tissue. Western blot analysis of rhodopsin protein levels in the vitreous of injected and control contralateral eyes. The vitreous of injected (Inj) and non-injected (Ctrl) eyes present traces or no detectable rhodopsin, whereas this protein is highly abundant the retina. Note protein content of vitreous extracts loaded is 10-fold higher than the retina. D. Elevated IOP induces a significant decrease of the level of SAA in the serum. Data are presented as mean ± standard deviation (n=6).
SAA1 and SAA2 are hallmark proinflammatory markers generated in response to acute inflammatory stimuli and infection (38–40). In light of the presence of SAA1/SAA2-positive microglial cells in retinas exposed to IOP, we examined whether IOP promotes a change in the circulating levels of SAA1/SAA2 of the serum. Surprisingly, we found that mice with unilateral elevation of IOP presented a substantial decrease of the levels of SAA1/SAA2 in the serum (Figure 5D).
Increased intraocular pressure results in a significant reduction in H2O2 in the retina
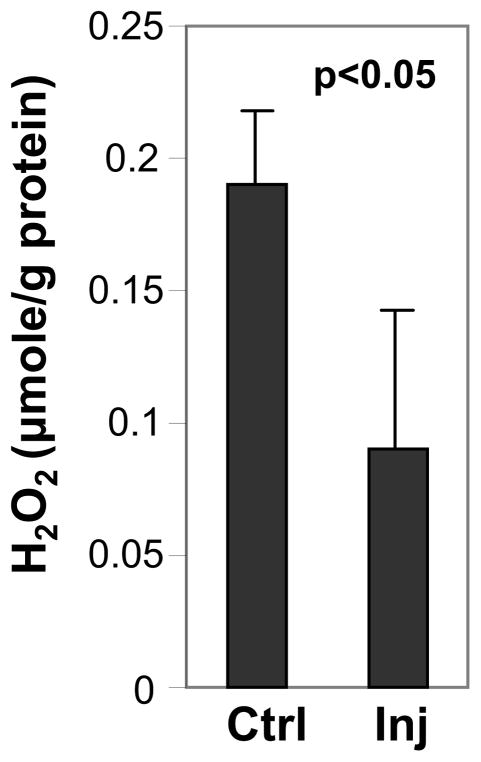
As described previously, we observed an increase of the levels of catalase with increased intraocular pressure. Catalase is critical to the conversion of hydrogen peroxide (H2O2) into H2O and O2 and reactive oxygen species have implicated in processes causing cellular damage in neurodegenerative diseases, such as glaucoma (41, 42). Hence, we examined whether this IOP-induced increase of catalase promotes changes in hydrogen peroxide levels as a result of oxidative stress to the retina. We performed ELISA assays to quantify the levels of hydrogen peroxide in retinas with and without exposure to IOP. We found that increased levels of catalase of retinas exposed to high IOP correlated with significantly lower levels of H2O2. Compared to control eyes, there was a decrease in H2O2 levels by almost 50% (p < 0.05) in eyes under elevated IOP (Fig. 6). These observations support that increased levels of catalase in IOP eyes counteract efficiently any changes in reactive oxygen species potentially produced by IOP.
Figure 6.
The elevation of IOP is associated with a significant decrease in H2O2 in the retina. Elevated IOP induces a significant decrease of the level of H2O2 in the retina of injected eyes (Inj) compared to contralateral non-injected (Ctrl) eyes. Retinas of injected eyes were exposed to 3-weeks of elevated IOP during the 7-week post-injection treatment as those shown in Fig 1A. Data are presented as means ± standard deviation (n=3).
Discussion
The microarray analyses herein reported provide evidence of IOP-induced changes in transcriptional gene expression (Figs. 2 and 3, supplemental Fig. 1) that is reflected and validated by changes in expression of proteins encoded by these genes in the retina (Fig. 3). With the exception of SAA1/SAA2, all changes in protein expression were also localized to ganglion neurons (Fig. 4), thus supporting that IOP has a prominent and selective effect in various facets of ganglion cell function. Furthermore, these protein changes (e.g. catalase) correlated with biological changes in the concentration of their substrates (e.g. H2O2) (Fig. 6). Collectively, these observations validate our experimental approaches to examine (by microarray and related analyses) changes in gene expression modulated by IOP as a tool to identify pathobiological pathways and biomarkers of glaucoma linked to elevated IOP.
Although we limited the scope of this study to the extended analyses of a restricted and novel set of genes selectively modulated by IOP, several important implications and insights into the pathogenesis of glaucoma can be already inferred from this study. First, we observed a positive correlation between a decrease of the transcriptional and protein levels of Bcl2. Bcl2 is a key apoptotic regulator by blocking the pore channel opening of mitochondria and cytochrome c release (43). This biological property confers anti-apoptotic activity to Bcl2. In ganglion neurons, such biological activity of Bcl2 confers neuroprotection to ganglion neurons upon axotomy of their axons (44). Our study supports that IOP down-regulates Bcl2 levels, thus hinting toward the down-modulation of anti-apoptotic activity by the ganglion neurons early on after exposure of these neurons to the IOP stress insult and before cell death ensues. We also found that the transcriptional levels of a member of an inhibitor of apoptosis (IAP) gene family, XIAP (45), to be down-regulated by IOP. In this case however, there was a negative correlation between the transcriptional and protein levels of XIAP upon IOP, supporting the IOP-dependent induction of a post-transcriptional regulatory mechanism in the modulation of XIAP. Similar post-transcriptional regulatory mechanisms have been found in other disorders (46). XIAP is a potent caspase inhibitor (47, 48). The increased levels of XIAP by IOP suggest the stimulation of an intrinsic pathway to prevent the maturation and activity of executioner phase of cell death by apoptotic caspases and that may counteract the effects of decreased protein levels of Bcl2. To this end, ectopic expression of XIAP is known to promote the survival of axons of optic nerve in ganglion neurons to chronic exposure of IOP (49). It is interesting to observe that while changes in Bcl2 expression were confined to the soma of ganglion neurons, changes in XIAP appear limited to dendritic processes of the neurons localized to sublaminar layers of the inner plexiform layer. These observations support a distinct spatial regulation of apoptotic and survival pathways in ganglion neurons. Finally, we observed a strong increase of the levels of catalase with IOP suggesting that IOP promotes the formation of reactive oxygen species (ROS). Instead, we found that the level of the ROS, H2O2, was significantly decreased in retinas exposed to IOP. Although the direct role of oxidative stress in the pathogenesis of the cell death of ganglion neurons upon IOP remains unclear (42, 50–52), the data support that the IOP-induced increase of catalase levels not only counteracts effectively any increase of the steady-state levels of H2O2, but it also promotes a decrease of H2O2 when compared to retinas of normotensive eyes. The impact of low levels of H2O2 on the survival of ganglion neurons exposed to IOP is unclear, but emerging observations suggest that a decline of H2O2 in response to apoptotic stimuli on photoreceptor and ganglion neurons in culture signals cell death (53).
We found the transcriptional levels of SAA1 and SAA2 to be significantly up-regulated in the retina, with SAA2 being most prominently expressed in the retina. Although immunoblots did not detect these proteins in extracts of the retina, immunocytochemistry of retinal sections supports that SAA1/SAA2 expression is confined to a scarce population of small microgial cells typically found throughout the retina. In light of the known robust production and detection in the plasma of SAA1/SAA2 upon acute inflammatory stimuli (38–40), these protein reactants commonly serve as biomarkers of inflammation, but other signaling roles may be associated with the production of SAA1/SAA2, namely in cholesterol metabolism and secretion of pro-inflammatory cytokines (38, 39, 54–56). These events are of significance to glaucoma because inflammatory processes have been linked to glaucoma (57–59). Regardless, it is interesting to note that the induction of SAA2 also occurs in the trabecular meshwork of glaucomatous eyes (60). Altogether, these results suggest that regardless of the function of SAA1/SAA2 in the pathogenesis of glaucoma, these pro-inflammatory reactants are strong candidates for biomarkers of hypertensive glaucoma and stratification of patients with this disorder. To this effect, we extended our analysis of the levels of SAA to the serum of non- and responsive mice to IOP to find surprisingly that these were decreased in mice with unilateral high IOP. It is possible that stimulation of migration of SAA-positive microgial cell to the retina promotes a systemic down-regulation of SAA to prevent the mounting of an immunoresponse toward inflammatory processes focally and restricted to glaucoma. Prior microarray studies in various glaucoma models have demonstrated that neuroinflammatory genes likely play a significant role in the pathophysiology of glaucoma. Microglia, in particular, have been implicated in this process (59) and inflammatory markers such as ceruloplasmin, glial fibrillary acidic protein, vimentin, and chitinase 3-like 1, were found to be up-regulated in several studies (24–27).
To identify additional sources of biomarkers of glaucoma from tissues accessible to potential biopsies in the human, we focused the analyses on the vitreous chamber. The vitreous is found juxtaposed to the retina and it presents low turnover of its contents (61–63); thus, the vitreous may accumulate selective retinal metabolites (64). We postulated the vitreous chamber may act as a reservoir for factors released over time by ganglion cells as a result of transient loss of the membrane permeability or death of these cells. Indeed, when we examined the levels in the vitreous of IOP-modulated proteins previously found in ganglion neurons (e.g. catalase and XIAP), we discovered that these were also significantly increased in the vitreous. The levels of Bcl2 in the vitreous were inconclusive (data not shown) and this outcome may reflect the differential extrusion of factors by ganglion neurons to the vitreous chamber upon stress stimuli. Hence, the data support that the vitreous is an amenable source of biomarkers of glaucoma and the extension of such much needed biopsy procedure to all forms of glaucoma may significantly help to diagnose and stratify the expression of the disease in glaucomatous patients.
Altogether, the outcome of this study and approaches employed provide novel leads to better diagnose and understand the pathogenesis of glaucoma. Further, it validates the extension of similar analyses to the ones herein reported to other changes in gene expression not examined in this study. These changes are likely to identify additional and novel biomarkers and pathogenic processes underlying hypertensive glaucoma and pave the way to comparative studies of the impact of other factors, such as age and various phases of the disease, in the expression of the glaucoma and stratification of its patients.
Supplementary Material
Amplification of Saa1 and Saa2 from mRNAs derived from retinas of high IOP (right) eyes (R) and respective contralateral (left) control eyes (L). A. Alignment of mouse Saa1 and Saa2 sequences. Primers and Hae III site (GGCC, in Saa1) are indicated. The Saa2 reverse primer is also used for Saa1 amplification even though there are two nucleotide mismatches. B. Digestion of Saa1 and Saa2 PCR products with Hae III. The PCR product of Saa1 gives a 46 bp shorter fragment after digestion. The Hae III site is not present in Saa2 sequence. Paired eyes are underlined.
Acknowledgments
We thank Dr. John Morrison for his instruction in the use of the rodent (rat) glaucoma models. This work was supported by NIH K12 grant EY 302-8113 and NIH 2P30-EY005722-21. P.A.F. is the Jules & Doris Stein Research to Prevent Blindness Professor.
References
- 1.Quigley HA. Number of people with glaucoma worldwide. The British journal of ophthalmology. 1996;80:389–393. doi: 10.1136/bjo.80.5.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quigley HA. Neuronal death in glaucoma. Prog Retin Eye Res. 1999;18:39–57. doi: 10.1016/s1350-9462(98)00014-7. [DOI] [PubMed] [Google Scholar]
- 3.Wax MB, Tezel G. Neurobiology of glaucomatous optic neuropathy: diverse cellular events in neurodegeneration and neuroprotection. Mol Neurobiol. 2002;26:45–55. doi: 10.1385/MN:26:1:045. [DOI] [PubMed] [Google Scholar]
- 4.Johnson EC, Jia L, Cepurna WO, Doser TA, Morrison JC. Global changes in optic nerve head gene expression after exposure to elevated intraocular pressure in a rat glaucoma model. Investigative ophthalmology & visual science. 2007;48:3161–3177. doi: 10.1167/iovs.06-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee WR, Grierson I. Relationships between intraocular pressure and the morphology of the outflow apparatus. Transactions of the ophthalmological societies of the United Kingdom. 1974;94:430–449. [PubMed] [Google Scholar]
- 6.Rohen JW, Witmer R. Electron microscopic studies on the trabecular meshwork in glaucoma simplex. Albrecht von Graefes Archiv fur klinische und experimentelle. Ophthalmologie. 1972;183:251–266. doi: 10.1007/BF00496153. [DOI] [PubMed] [Google Scholar]
- 7.Garcia-Valenzuela E, Shareef S, Walsh J, Sharma SC. Programmed cell death of retinal ganglion cells during experimental glaucoma. Exp Eye Res. 1995;61:33–44. doi: 10.1016/s0014-4835(95)80056-5. [DOI] [PubMed] [Google Scholar]
- 8.Quigley HA, Nickells RW, Kerrigan LA, Pease ME, Thibault DJ, Zack DJ. Retinal ganglion cell death in experimental glaucoma and after axotomy occurs by apoptosis. Invest Ophthalmol Vis Sci. 1995;36:774–786. [PubMed] [Google Scholar]
- 9.Quigley HA, Addicks EM. Chronic experimental glaucoma in primates. II. Effect of extended intraocular pressure elevation on optic nerve head and axonal transport. Invest Ophthalmol Vis Sci. 1980;19:137–152. [PubMed] [Google Scholar]
- 10.Monemi S, Spaeth G, DaSilva A, Popinchalk S, Ilitchev E, Liebmann J, Ritch R, Heon E, Crick RP, Child A, Sarfarazi M. Identification of a novel adult-onset primary open-angle glaucoma (POAG) gene on 5q22.1. Human molecular genetics. 2005;14:725–733. doi: 10.1093/hmg/ddi068. [DOI] [PubMed] [Google Scholar]
- 11.Rezaie T, Child A, Hitchings R, Brice G, Miller L, Coca-Prados M, Heon E, Krupin T, Ritch R, Kreutzer D, Crick RP, Sarfarazi M. Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science. 2002;295:1077–1079. doi: 10.1126/science.1066901. [DOI] [PubMed] [Google Scholar]
- 12.Stone EM, Fingert JH, Alward WL, Nguyen TD, Polansky JR, Sunden SL, Nishimura D, Clark AF, Nystuen A, Nichols BE, Mackey DA, Ritch R, Kalenak JW, Craven ER, Sheffield VC. Identification of a gene that causes primary open angle glaucoma. Science. 1997;275:668–670. doi: 10.1126/science.275.5300.668. [DOI] [PubMed] [Google Scholar]
- 13.Wiggs JL. Genetic etiologies of glaucoma. Arch Ophthalmol. 2007;125:30–37. doi: 10.1001/archopht.125.1.30. [DOI] [PubMed] [Google Scholar]
- 14.Bejjani BA, Lewis RA, Tomey KF, Anderson KL, Dueker DK, Jabak M, Astle WF, Otterud B, Leppert M, Lupski JR. Mutations in CYP1B1, the gene for cytochrome P4501B1, are the predominant cause of primary congenital glaucoma in Saudi Arabia. American journal of human genetics. 1998;62:325–333. doi: 10.1086/301725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stoilov I, Akarsu AN, Sarfarazi M. Identification of three different truncating mutations in cytochrome P4501B1 (CYP1B1) as the principal cause of primary congenital glaucoma (Buphthalmos) in families linked to the GLC3A locus on chromosome 2p21. Human molecular genetics. 1997;6:641–647. doi: 10.1093/hmg/6.4.641. [DOI] [PubMed] [Google Scholar]
- 16.Libby RT, Gould DB, Anderson MG, John SW. Complex genetics of glaucoma susceptibility. Annu Rev Genomics Hum Genet. 2005;6:15–44. doi: 10.1146/annurev.genom.6.080604.162209. [DOI] [PubMed] [Google Scholar]
- 17.Petersen MB, Kitsos G, Samples JR, Gaudette ND, Economou-Petersen E, Sykes R, Rust K, Grigoriadou M, Aperis G, Choi D, Psilas K, Craig JE, Kramer PL, Mackey DA, Wirtz MK. A large GLC1C Greek family with a myocilin T377M mutation: inheritance and phenotypic variability. Investigative ophthalmology & visual science. 2006;47:620–625. doi: 10.1167/iovs.05-0631. [DOI] [PubMed] [Google Scholar]
- 18.Raymond V. Molecular genetics of the glaucomas: mapping of the first five “GLC” loci. American journal of human genetics. 1997;60:272–277. [PMC free article] [PubMed] [Google Scholar]
- 19.Sarfarazi M. Recent advances in molecular genetics of glaucomas. Human molecular genetics. 1997;6:1667–1677. doi: 10.1093/hmg/6.10.1667. [DOI] [PubMed] [Google Scholar]
- 20.Hewitt AW, Bennett SL, Dimasi DP, Craig JE, Mackey DA. A myocilin Gln368STOP homozygote does not exhibit a more severe glaucoma phenotype than heterozygous cases. Am J Ophthalmol. 2006;141:402–403. doi: 10.1016/j.ajo.2005.08.073. [DOI] [PubMed] [Google Scholar]
- 21.The Advanced Glaucoma Intervention Study (AGIS): 11. Risk factors for failure of trabeculectomy and argon laser trabeculoplasty. Am J Ophthalmol. 2002;134:481–498. doi: 10.1016/s0002-9394(02)01658-6. [DOI] [PubMed] [Google Scholar]
- 22.Morrison JC, Moore CG, Deppmeier LM, Gold BG, Meshul CK, Johnson EC. A rat model of chronic pressure-induced optic nerve damage. Exp Eye Res. 1997;64:85–96. doi: 10.1006/exer.1996.0184. [DOI] [PubMed] [Google Scholar]
- 23.Morrison JC, Johnson EC, Cepurna W, Jia L. Understanding mechanisms of pressure-induced optic nerve damage. Prog Retin Eye Res. 2005;24:217–240. doi: 10.1016/j.preteyeres.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 24.Miyahara T, Kikuchi T, Akimoto M, Kurokawa T, Shibuki H, Yoshimura N. Gene microarray analysis of experimental glaucomatous retina from cynomologous monkey. Invest Ophthalmol Vis Sci. 2003;44:4347–4356. doi: 10.1167/iovs.02-1032. [DOI] [PubMed] [Google Scholar]
- 25.Ahmed F, Brown KM, Stephan DA, Morrison JC, Johnson EC, Tomarev SI. Microarray analysis of changes in mRNA levels in the rat retina after experimental elevation of intraocular pressure. Invest Ophthalmol Vis Sci. 2004;45:1247–1258. doi: 10.1167/iovs.03-1123. [DOI] [PubMed] [Google Scholar]
- 26.Steele MR, Inman DM, Calkins DJ, Horner PJ, Vetter ML. Microarray analysis of retinal gene expression in the DBA/2J model of glaucoma. Invest Ophthalmol Vis Sci. 2006;47:977–985. doi: 10.1167/iovs.05-0865. [DOI] [PubMed] [Google Scholar]
- 27.Yang Z, Quigley HA, Pease ME, Yang Y, Qian J, Valenta D, Zack DJ. Changes in gene expression in experimental glaucoma and optic nerve transection: the equilibrium between protective and detrimental mechanisms. Invest Ophthalmol Vis Sci. 2007;48:5539–5548. doi: 10.1167/iovs.07-0542. [DOI] [PubMed] [Google Scholar]
- 28.Reitsamer HA, Kiel JW, Harrison JM, Ransom NL, McKinnon SJ. Tonopen measurement of intraocular pressure in mice. Exp Eye Res. 2004;78:799–804. doi: 10.1016/j.exer.2003.11.018. [DOI] [PubMed] [Google Scholar]
- 29.Aslanukov A, Bhowmick R, Guruju M, Oswald J, Raz D, Bush RA, Sieving PA, Lu X, Bock CB, Ferreira PA. RanBP2 modulates Cox11 and hexokinase I activities and haploinsufficiency of RanBP2 causes deficits in glucose metabolism. PLoS genetics. 2006;2:e177. doi: 10.1371/journal.pgen.0020177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho KI, Yi H, Yeh A, Tserentsoodol N, Cuadrado L, Searle K, Hao Y, Ferreira PA. Haploinsufficiency of RanBP2 is neuroprotective against light-elicited and age-dependent degeneration of photoreceptor neurons. Cell death and differentiation. 2009;16:287–297. doi: 10.1038/cdd.2008.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferreira PA. Characterization of RanBP2-associated molecular components in neuroretina. Methods in enzymology. 2000;315:455–468. doi: 10.1016/s0076-6879(00)15861-6. [DOI] [PubMed] [Google Scholar]
- 33.Mavlyutov TA, Cai Y, Ferreira PA. Identification of RanBP2- and kinesin-mediated transport pathways with restricted neuronal and subcellular localization. Traffic (Copenhagen, Denmark) 2002;3:630–640. doi: 10.1034/j.1600-0854.2002.30905.x. [DOI] [PubMed] [Google Scholar]
- 34.Betts JC, Edbrooke MR, Thakker RV, Woo P. The human acute-phase serum amyloid A gene family: structure, evolution and expression in hepatoma cells. Scandinavian journal of immunology. 1991;34:471–482. doi: 10.1111/j.1365-3083.1991.tb01570.x. [DOI] [PubMed] [Google Scholar]
- 35.Sellar GC, Steel DM, Zafiropoulos A, Seery LT, Whitehead AS. Characterization, expression and evolution of mouse beta 2-glycoprotein I (apolipoprotein H) Biochem Biophys Res Commun. 1994;200:1521–1528. doi: 10.1006/bbrc.1994.1623. [DOI] [PubMed] [Google Scholar]
- 36.Tezel G, Wax MB. Glial modulation of retinal ganglion cell death in glaucoma. Journal of glaucoma. 2003;12:63–68. doi: 10.1097/00061198-200302000-00014. [DOI] [PubMed] [Google Scholar]
- 37.Tezel G, Wax MB. The immune system and glaucoma. Curr Opin Ophthalmol. 2004;15:80–84. doi: 10.1097/00055735-200404000-00003. [DOI] [PubMed] [Google Scholar]
- 38.Furlaneto CJ, Campa A. A novel function of serum amyloid A: a potent stimulus for the release of tumor necrosis factor-alpha, interleukin-1beta, and interleukin-8 by human blood neutrophil. Biochem Biophys Res Commun. 2000;268:405–408. doi: 10.1006/bbrc.2000.2143. [DOI] [PubMed] [Google Scholar]
- 39.Yang RZ, Lee MJ, Hu H, Pollin TI, Ryan AS, Nicklas BJ, Snitker S, Horenstein RB, Hull K, Goldberg NH, Goldberg AP, Shuldiner AR, Fried SK, Gong DW. Acute-phase serum amyloid A: an inflammatory adipokine and potential link between obesity and its metabolic complications. PLoS Med. 2006;3:e287. doi: 10.1371/journal.pmed.0030287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malle E, De Beer FC. Human serum amyloid A (SAA) protein: a prominent acute-phase reactant for clinical practice. Eur J Clin Invest. 1996;26:427–435. doi: 10.1046/j.1365-2362.1996.159291.x. [DOI] [PubMed] [Google Scholar]
- 41.Andersen JK. Oxidative stress in neurodegeneration: cause or consequence? Nat Med. 2004;10 (Suppl):S18–25. doi: 10.1038/nrn1434. [DOI] [PubMed] [Google Scholar]
- 42.Izzotti A, Bagnis A, Sacca SC. The role of oxidative stress in glaucoma. Mutat Res. 2006;612:105–114. doi: 10.1016/j.mrrev.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 43.Ferri KF, Kroemer G. Mitochondria--the suicide organelles. Bioessays. 2001;23:111–115. doi: 10.1002/1521-1878(200102)23:2<111::AID-BIES1016>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 44.Bonfanti L, Strettoi E, Chierzi S, Cenni MC, Liu XH, Martinou JC, Maffei L, Rabacchi SA. Protection of retinal ganglion cells from natural and axotomy-induced cell death in neonatal transgenic mice overexpressing bcl-2. J Neurosci. 1996;16:4186–4194. doi: 10.1523/JNEUROSCI.16-13-04186.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deveraux QL, Reed JC. IAP family proteins--suppressors of apoptosis. Genes Dev. 1999;13:239–252. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- 46.Moore DF, Gelderman MP, Ferreira PA, Fuhrmann SR, Yi H, Elkahloun A, Lix LM, Brady RO, Schiffmann R, Goldin E. Genomic abnormalities of the murine model of Fabry disease after disease-related perturbation, a systems biology approach. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8065–8070. doi: 10.1073/pnas.0701991104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eckelman BP, Salvesen GS, Scott FL. Human inhibitor of apoptosis proteins: why XIAP is the black sheep of the family. EMBO Rep. 2006;7:988–994. doi: 10.1038/sj.embor.7400795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salvesen GS, Duckett CS. IAP proteins: blocking the road to death’s door. Nature reviews. 2002;3:401–410. doi: 10.1038/nrm830. [DOI] [PubMed] [Google Scholar]
- 49.McKinnon SJ, Lehman DM, Tahzib NG, Ransom NL, Reitsamer HA, Liston P, LaCasse E, Li Q, Korneluk RG, Hauswirth WW. Baculoviral IAP repeat-containing-4 protects optic nerve axons in a rat glaucoma model. Mol Ther. 2002;5:780–787. doi: 10.1006/mthe.2002.0608. [DOI] [PubMed] [Google Scholar]
- 50.Liu Q, Ju WK, Crowston JG, Xie F, Perry G, Smith MA, Lindsey JD, Weinreb RN. Oxidative stress is an early event in hydrostatic pressure induced retinal ganglion cell damage. Investigative ophthalmology & visual science. 2007;48:4580–4589. doi: 10.1167/iovs.07-0170. [DOI] [PubMed] [Google Scholar]
- 51.Moreno MC, Campanelli J, Sande P, Sanez DA, Keller Sarmiento MI, Rosenstein RE. Retinal oxidative stress induced by high intraocular pressure. Free radical biology & medicine. 2004;37:803–812. doi: 10.1016/j.freeradbiomed.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 52.Mozaffarieh M, Flammer J. Is there more to glaucoma treatment than lowering IOP? Survey of ophthalmology. 2007;52 (Suppl 2):S174–179. doi: 10.1016/j.survophthal.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 53.Mackey AM, Sanvicens N, Groeger G, Doonan F, Wallace D, Cotter TG. Redox survival signalling in retina-derived 661W cells. Cell death and differentiation. 2008;15:1291–1303. doi: 10.1038/cdd.2008.43. [DOI] [PubMed] [Google Scholar]
- 54.Liao F, Lusis AJ, Berliner JA, Fogelman AM, Kindy M, de Beer MC, de Beer FC. Serum amyloid A protein family. Differential induction by oxidized lipids in mouse strains. Arterioscler Thromb. 1994;14:1475–1479. doi: 10.1161/01.atv.14.9.1475. [DOI] [PubMed] [Google Scholar]
- 55.Lindhorst E, Young D, Bagshaw W, Hyland M, Kisilevsky R. Acute inflammation, acute phase serum amyloid A and cholesterol metabolism in the mouse. Biochimica et biophysica acta. 1997;1339:143–154. doi: 10.1016/s0167-4838(96)00227-0. [DOI] [PubMed] [Google Scholar]
- 56.Van Lenten BJ, Hama SY, de Beer FC, Stafforini DM, McIntyre TM, Prescott SM, La Du BN, Fogelman AM, Navab M. Anti-inflammatory HDL becomes pro-inflammatory during the acute phase response. Loss of protective effect of HDL against LDL oxidation in aortic wall cell cocultures. J Clin Invest. 1995;96:2758–2767. doi: 10.1172/JCI118345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sappington RM, Calkins DJ. Contribution of TRPV1 to microglia-derived IL-6 and NFkappaB translocation with elevated hydrostatic pressure. Invest Ophthalmol Vis Sci. 2008;49:3004–3017. doi: 10.1167/iovs.07-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou X, Li F, Kong L, Tomita H, Li C, Cao W. Involvement of inflammation, degradation, and apoptosis in a mouse model of glaucoma. The Journal of biological chemistry. 2005;280:31240–31248. doi: 10.1074/jbc.M502641200. [DOI] [PubMed] [Google Scholar]
- 59.Tezel G, Yang X, Luo C, Peng Y, Sun SL, Sun D. Mechanisms of immune system activation in glaucoma: oxidative stress-stimulated antigen presentation by the retina and optic nerve head glia. Invest Ophthalmol Vis Sci. 2007;48:705–714. doi: 10.1167/iovs.06-0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang WH, McNatt LG, Pang IH, Hellberg PE, Fingert JH, McCartney MD, Clark AF. Increased expression of serum amyloid A in glaucoma and its effect on intraocular pressure. Invest Ophthalmol Vis Sci. 2008;49:1916–1923. doi: 10.1167/iovs.07-1104. [DOI] [PubMed] [Google Scholar]
- 61.Halfter W, Dong S, Dong A, Eller AW, Nischt R. Origin and turnover of ECM proteins from the inner limiting membrane and vitreous body. Eye (London, England) 2008 doi: 10.1038/eye.2008.19. [DOI] [PubMed] [Google Scholar]
- 62.Haddad A, Laicine EM, de Almeida JC, Costa MS. Partial characterization, origin and turnover of glycoproteins of the rabbit vitreous body. Exp Eye Res. 1990;51:139–143. doi: 10.1016/0014-4835(90)90065-3. [DOI] [PubMed] [Google Scholar]
- 63.Laurent UB, Fraser JR. Turnover of hyaluronate in the aqueous humour and vitreous body of the rabbit. Exp Eye Res. 1983;36:493–503. doi: 10.1016/0014-4835(83)90043-x. [DOI] [PubMed] [Google Scholar]
- 64.van Deemter M, Ponsioen TL, Bank RA, Snabel JM, van der Worp RJ, Hooymans JM, Los LI. Pentosidine accumulates in the aging vitreous body: A gender effect. Exp Eye Res. 2009 doi: 10.1016/j.exer.2009.01.004. in press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Amplification of Saa1 and Saa2 from mRNAs derived from retinas of high IOP (right) eyes (R) and respective contralateral (left) control eyes (L). A. Alignment of mouse Saa1 and Saa2 sequences. Primers and Hae III site (GGCC, in Saa1) are indicated. The Saa2 reverse primer is also used for Saa1 amplification even though there are two nucleotide mismatches. B. Digestion of Saa1 and Saa2 PCR products with Hae III. The PCR product of Saa1 gives a 46 bp shorter fragment after digestion. The Hae III site is not present in Saa2 sequence. Paired eyes are underlined.