Summary
Protein translocation across the bacterial membrane, mediated by the SecYEG translocon and the SecA ATPase1–4, is enhanced by proton-motive force (PMF)5,6 and membrane-integrated SecDF7–9, which associates with SecYEG. Here, we determined the crystal structure of Thermus thermophilus SecDF at 3.3 Å resolution, which revealed a pseudo-symmetrical, 12-helix transmembrane (TM) domain belonging to the RND superfamily and major periplasmic domains (P1 and P4). Higher resolution analysis of the latter suggested that P1, which proved to bind an unfolded protein, undergoes functionally important conformational changes. In vitro analyses identified an ATP-independent step of protein translocation that requires both SecDF and PMF. Electrophysiological analyses revealed that SecDF conducts protons in a pH- and unfolded protein-dependent fashion, in which conserved Asp and Arg residues at the TM SecD/SecF-interface play essential roles in the movements of protons and preproteins. Therefore, we propose that SecDF functions as a membrane-integrated chaperone, powered by PMF, to achieve ATP-independent protein translocation.
The functional importance of SecDF in protein translocation was previously shown in vivo7,9,10; the SecD/SecF–deficient Escherichia coli mutant secD1 (producing negligible levels of SecD and SecF and exhibiting cold-sensitivity for growth) is severely defective in protein export at any temperature. The role of SecDF has remained unclear, although it was proposed to function in later stages of translocation as well as in membrane protein biogenesis4,11–14. In E. coli, SecD and SecF (ESecD and ESecF, respectively) are encoded by the secD operon (yajC-secD-secF), and form a tight complex8. Each protein contains 6 transmembrane (TM) segments, belonging to the resistance nodulation and cell division (RND) superfamily, and a large periplasmic domain15,16.
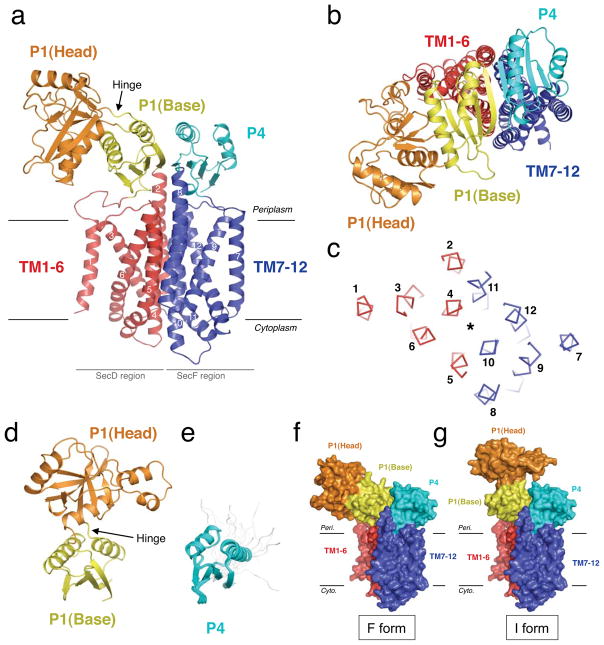
SecDF from Thermus thermophilus HB817 is composed of a single polypeptide chain [TSecDF (MW 80.5 kDa)] containing 12 predicted transmembrane (TM) regions and 6 periplasmic regions (P1–P6), among which P1 (TSecDF31-263) and P4 (TSecDF469-559) are large enough to form distinct domains (Supplementary Fig. 1). We solved the crystal structure of full-length TSecDF at 3.3 Å resolution (Fig. 1a–c,f, Supplementary Fig. 2 and Supplementary Table 1), as well as the crystal structure of P1 (TSecDF36-263) at 2.6 Å resolution and the NMR solution structure of P4 (TSecDF481-557) (Fig. 1d,e and Supplementary Table 2). The TSecDF structure contains twelve TM α-helices, with both termini facing the cytoplasm and P1 and P4 protruding into the periplasm (Fig. 1a,b). The N-terminal (TM1-6) and C-terminal (TM7-12) halves are assembled pseudo-symmetrically, in which TM4 and TM10 form the primary SecD-SecF interface (Fig. 1c). The TM helices are membrane-embedded, except for the ~10 Å extensions of TM2 and TM8 that tether P1 and P4, respectively (Fig. 1a). The tilted TMs 4, 5, 6, 10, 11 and 12 are curved near the cytoplasmic surface.
Figure 1. Structures of T. thermophilus SecDF.
a, b, The crystal structure of full-length SecDF, viewed from the membrane side (a) and the periplasmic side (b). c, TSecDF cross-sectioned at the middle of the TM, viewed from the periplasm. The asterisk indicates the pseudo-symmetrical axis. TMs are numbered. d, Crystal structure of the P1 domain. e, NMR structure of the P4 domain after twenty superimpositions. The disordered regions are shown in gray. f, F form. Crystal structure of full-length SecDF. g, I form. The base subdomain of isolated P1 was docked onto that of the F form, as shown in f.
The NMR structure of P4 revealed a ferredoxin-like fold, consisting of four β-strands flanked by two α-helices (Fig. 1e). P1, protruding by ~40 Å from the membrane, is divided into head (~25 Å periplasmic protrusion) and base subdomains (Fig. 1a,d). The base subdomain of P1 (residues 36-111 and 251-263) is structurally homologous to P4; both form a pseudo-symmetrical 8-stranded anti-parallel β-sheet that covers the TM region (Fig. 1a,b). The crystal structure of isolated P1 (I form) revealed a ~120° rigid-body rotation of its head subdomain toward the periplasmic space, as compared to that observed for P1 in the full-length SecDF (F form) (Supplementary Fig. 3). Thus, SecDF can assume at least two different conformations, F and I, with distinct P1 configurations (Fig. 1f,g). For this conformational change, the two loops connecting the head and the base could act as a hinge (Fig. 1a,d), allowing the head to swing around the base. We built a model of the “full-length I form”, by superimposing the base subdomain of the isolated P1 structure onto that of the full-length SecDF. The in vivo occurrence of the two conformations was supported by disulfide crosslinking experiments, which further suggested that the conformational transition of SecDF is functionally important (Supplementary Fig. 4a–e and Supplementary Discussion).
Despite the vital importance of SecDF in vivo, its specific steps in translocation have not been defined. We examined if SecDF functions in the steps subsequent to the SecA- and ATP-dependent translocation initiation. A proOmpA translocation intermediate was generated, which possessed a 59-residue disulfide-linked loop acting as an obstacle to translocation through the Sec translocon, by subjecting 35S-labeled proOmpA(L59)18 to translocation reactions in the presence of SecA and ATP and with either wild-type inverted membrane vesicles (IMVs) or SecD/SecF-deficient IMVs from the secD1(Cs) mutant (Fig. 2b). ATP was then depleted from the intermediate-bearing IMVs by glucose-hexokinase reaction cycles (Fig. 2a, lanes 1 and 6). The addition of dithiothreitol (DTT) to the wild-type IMVs restored translocation that was PMF-dependent and ATP-independent (compare lanes 2 and 4 for the PMF-dependence). In contrast, the SecD1 IMVs did not support any further translocation of the intermediate, regardless of the presence of a PMF (lane 7). Instead, these IMVs tended to lose the intermediate (lanes 7, 8), probably due to backsliding of the polypeptide19. In the presence of excess ATP, the completion of proOmpA(L59) translocation was not affected by either the secD1 mutation or the lack of PMF (lanes 13, 15, 18 and 20). These results show that SecDF is required for the later, ATP-independent steps of translocation driven by the PMF, while the PMF is thought to accelerate even the initial stage of translocation (Supplementary Discussion). In addition, IMVs bearing inactive SecDF variants (described below) did not support the PMF-dependent completion of proOmpA translocation (Fig. 2c).
Figure 2. SecDF-dependent translocation completion.
a, Identification of a SecDF- and PMF-dependent translocation step. SecDF-deficient (secD1) IMVs were incubated with 35S-labeled proOmpA(L59) to generate translocation intermediates. The protein translocation was continued in the presence or absence of ATP, PMF and DTT. b, Schematic depiction of the translocation intermediate of proOmpA(L59). c, Completion of the proOmpA(L59) translocation using IMVs from the secD1 (Cs) mutant expressing E. coli SecDF derivatives.
To gain structural insights into the PMF-dependence of SecDF function, we compared its structure with that of AcrB20, an RND superfamily proton/multi-drug antiporter16 (Supplementary Fig. 5). AcrB forms a homo-trimer, while SecDF is monomeric. Although the TM segments of SecDF and AcrB share low sequence identity (15%) (Supplementary Fig. 6), the structures of their TM regions are similar, yielding an RMSD of ~2.7 Å for the Cα atoms of the TM helices. By contrast, the structures and functions of the periplasmic regions are quite different between SecDF and AcrB. The TM region of AcrB is thought to participate in proton transport and contains several conserved, charged residues, such as Asp407, Asp408, Lys940 and Arg971, important for the drug export activity21 (Supplementary Fig. 5e). Asp407, Thr978 and Arg971 in AcrB have structural counterparts in TSecDF (Asp340, Thr675 and Arg671, respectively) (Fig. 3a and Supplementary Fig. 5e). The conserved SecDF residues are clustered at the TM SecD-SecF interfaces as well as the periplasmic base region underneath the head (Supplementary Fig. 7). We also note that Asp637 is a highly conserved, membrane-embedded charged residue. Complementation tests indicated that the Asp519Asn mutation in ESecD as well as the Asp213Asn and Arg247Met mutations in ESecF (Fig. 3a) completely abolished the SecDF activity and conferred some dominant-negative phenotypes (Fig. 3b). Thus, these conserved, charged residues are crucial for SecDF function, consistent with the hypothesis that SecDF conducts protons through the conserved TM region.
Figure 3. Functional charged residues and proton conduction of SecDF.
a, Functionally important, conserved residues in the TM regions. The TMs are numbered. b, Complementation of the secD1 (Cs) growth and protein export defects by SecD/F mutants. c, Na+-dependent protein export by VSecDF-1. d, Na+-dependence of export (n=3). e, Single channel currents recorded by patch clamp in membrane patches excised from E. coli spheroplasts containing TSecDF or its mutants, and effects of a pH gradient and casein. f, The channel open probability (n=6 with casein, n=4 without casein).
The halophilic marine bacteria Vibrio use a Na+ gradient, instead of PMF, for some cellular processes22. Vibrio alginolyticus has two sets of secDF genes encoding SecDF-1 and SecDF-2 complexes, respectively. When VSecDF-1 from V. alginolyticus 138-2 was expressed in the E. coli secD1(Cs) mutant, the addition of NaCl, but not KCl, to the medium restored protein export (Fig. 3c lanes 1–3). The VSecDF-1-dependent protein export depended on extracellular Na+ up to 50 mM (Fig. 3d). Such Na+-dependence was not observed with ESecDF. Thus, VSecDF-1 facilitates protein export using a Na+ gradient across the membrane. The Arg240Met and Asp206Asn mutations in VSecF-1, corresponding to ESecF Arg247Met and Asp213Asn, respectively (Fig. 3a), compromised the Na+-dependent protein export activity (Fig. 3c lanes 4–7). These results provide physiological evidence for the cation-coupled protein translocation by SecDF.
To verify that SecDF utilizes the PMF and thus conducts protons, we performed inside-out patch-clamp experiments23, using E. coli giant spheroplasts24 containing TSecDF. Current recordings revealed that TSecDF-containing spheroplasts underwent transient channel openings under both symmetrical and asymmetrical pH conditions, whereas negative-control spheroplasts without TSecDF did not (Supplementary Fig. 8a). The channel activities were markedly enhanced by the imposition of a pH gradient as well as by the addition of a P1-interacting unfolded protein, casein, to the pipette (periplasmic side) solution (Fig. 3e left, Supplementary Fig. 8a–c, Supplementary Discussion). In addition, casein and acidic extracellular conditions increased the probability of opening the SecDF-dependent ion channel (Fig. 3f). Thus, the ion-conduction can be regulated by a proton gradient and by unfolded protein binding to SecDF. The use of a proton-specific fluorescent probe, BCECF, confirmed the SecDF-dependent proton import (Supplementary Fig. 9 and Supplementary Discussion). The TSecDF mutations Asp340Asn and Arg671Met, but not Asp637Asn, abolished the ion channel activity (Fig. 3a,e right), indicating that Asp340 and Arg671 in the TM region are essential for the proton conduction, and might be protonated transiently. Asp637, which is sequestered from the putative main proton pathway in the TM region of both AcrB and SecDF (Supplementary Fig. 5e), might function in subsequent conformational changes required for the enhancement of protein translocation.
In addition to the TM residues, the P1 head domain is a critical element for proton transport. TSecDF(Δ112-248) lacking the head did not show any signals in the patch-clamp assay (Fig. 3e right), and the corresponding ESecDF mutant was defective in protein export (Supplementary Fig. 4f). The conformational flexibility of P1 is also required for proton conduction; the TSecDF double cysteine mutant, Leu106Cys/Leu243Cys (see Supplementary Fig. 4a,b), lacked channel activity in four of the six membrane patches examined. The variable results could be explained by incomplete crosslinking (more than 20%; data not shown). We think it is likely that the putative hinge motion of the P1 domain is coupled with both proton transport and protein export. Taken together, we propose that the proton flow is the driving force for the P1 domain movement and the consequent protein translocation enhancement by SecDF.
We have shown here that SecDF and PMF are required for the post-initiation mode of translocation, which can occur in the absence of ATP and SecA. This function depends on the ability of the periplasmic P1 domain to interact with a substrate and to undergo the structural transition between the I and F forms, which are likely to be in equilibrium. The F state of SecDF may place the tilted P1 head above the translocon pore, enabling it to capture an emerging preprotein (Fig. 4a). The preprotein-bearing F form could then return to the I configuration (Fig. 4b), preventing the backward movement of the substrate and driving the forward movement of the substrate. The release of the bound preprotein from SecDF and the subsequent I to F conversion may be coupled to the proton flow (Fig. 4c). These action cycles will eventually lead to the completion of translocation, in which the substrate is released from the translocon. As the bacterial periplasm lacks ATP, SecDF may utilize the PMF to drive its conformational transition and delivery of substrates. The Asp and Arg residues in the TM region of SecDF could serve as putative proton acceptors in the proton relay pathway (Fig. 4c). The corresponding residues of AcrB (Supplementary Fig. 5e) were proposed to participate in proton conduction, with their side chains assuming different configurations among the asymmetric protomers, which presumably exist in different protonation states20. Likewise, the protonation states of the key charged residues of SecDF could induce the twisting of TM4 and TM10, which would be transmitted to the conserved P1-TM4 linker region (Supplementary Fig. 7) and trigger the conformational transition of the P1 head subdomain. Although direct evidence for the PMF–dependent conformational transition of SecDF will await further structural and functional studies, we have shown that SecDF is a component of the Sec machinery that utilizes the PMF to complete protein translocation after the ATP-dependent SecA function.
Figure 4. A working model of the PMF-driven translocation enhancement by SecDF.

a, F form, capturing state. b, I form, holding state. c, I to F transition and substrate-releasing state. The two essential charged residues of ESecDF are highlighted. SecDF is colored as in Fig. 2. SecYEG, gray; SecA, green; pre-protein, black line; proton movement, white arrow. See the main text discussion for details.
METHODS SUMMARY
The X-ray diffraction analysis of TSecDF was described previously17. The initial phases were determined by the single-wavelength anomalous dispersion method. The initial model was manually built, using the structures of the TM regions of AcrB and the separately determined P1 and P4 domain structures as references (Supplementary Methods). The model was substantially improved by using the zonal scaling25 and methionine-marking26 methods, and was finally refined to Rwork= 29.8% and Rfree = 31.9% at 3.3 Å resolution.
To monitor the later steps of translocation, 35S-labeled proOmpA(L59) and IMVs, prepared as described previously18,27, were incubated to form translocation intermediates, which were treated with proteinase K and then incubated under several conditions. The translocation status of 35S-proOmpA was examined by phosphor imaging after SDS-PAGE.
For the complementation test, E. coli secD1 (Cs) mutant cells carrying plasmids encoding HA-tagged ESecD and ESecF mutants were spotted onto L-agar plates and incubated at 20 °C. The efficiency of protein export in vivo was assessed by the 35S-methionine pulse-labeling procedures.
To monitor the protein transport activity of Vibrio SecDF-1, E. coli secD1 cells expressing VSecD-1 and VSecF-1 were pulse-labeled in the presence or absence of Na+. To detect the single channel activity, electric currents were measured with the inside-out membrane patches excised from TSecDF-containing E. coli giant spheroplasts, using an Axopatch 200B amplifier (Axon CNS, Molecular Devices).
Supplementary Material
Acknowledgments
We thank Y. Akiyama, R. Suno, Y. Morimoto, T. Minamino, K. Namba, K. Inaba, M. Hattori and H. Nishimasu for useful suggestions; T. Sakamoto and A. Kurabayashi for their assistance with sample preparation; R. Yamasaki, M. Sano, K. Mochizuki, K. Yoshikaie, K. Imayoshi and T. Adachi for technical support; M. Homma and S. Kojima for graciously providing the Vibrio genomic DNA; the beamline staff members at BL41XU of SPring-8 (Hyogo, Japan) and at NW12 of KEK PF-AR (Tsukuba, Japan) for technical help during data collection; and M. Ibba for comments on our manuscript. This work was supported by a Grant-in-Aid for Scientific Research (S) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) to O.N., by a CREST grant from JST to K.I., by a BIRD grant from JST to H.M. and R.I., by a grant for the National Project on Protein Structural and Functional Analyses to O.N., by NIH grants to D.G.V., and by grants from MEXT to T.T., H.M., R.I., S.F. and K.I.
Footnotes
Author Contributions
T.T. performed the structural determination and the biochemical experiments with SecDF. H.M. performed the functional analyses of SecDF. Y.E. solved the crystal structure of the SecDF P1 domain and assisted with the functional analysis of SecDF. R.I., S.F., D.G.V. and O.N. assisted with the structural determination. A.D.M. performed patch-clamp and pH fluorescence experiments. T. Tanaka and T.K. solved the structure of the P4 domain by NMR. A.P. and D.G.V. assisted with the crystallization and data collection of SecDF. All authors discussed the results and commented on the manuscript. O.N. and K.I. supervised the work and wrote/edited the manuscript.
Author Information
The coordinates and structure factors have been deposited in the Protein Data Bank, under the accession codes 3AQP for the entire TSecDF protein and 3AQO for the P1 domain. The PDB and BMRB codes for the deposited P4 domain are 2RRN and 11426, respectively. The authors declare no competing financial interests.
References
- 1.van den Berg B, et al. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 2.Zimmer J, Nam Y, Rapoport TA. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature. 2008;455:936–43. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsukazaki T, et al. Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature. 2008;455:988–91. doi: 10.1038/nature07421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.du Plessis DJ, Nouwen N, Driessen AJ. The Sec translocase. Biochim Biophys Acta. 2011;1808:851–65. doi: 10.1016/j.bbamem.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 5.Driessen AJ, Wickner W. Proton transfer is rate-limiting for translocation of precursor proteins by the Escherichia coli translocase. Proc Natl Acad Sci U S A. 1991;88:2471–5. doi: 10.1073/pnas.88.6.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shiozuka K, Tani K, Mizushima S, Tokuda H. The proton motive force lowers the level of ATP required for the in vitro translocation of a secretory protein in Escherichia coli. J Biol Chem. 1990;265:18843–7. [PubMed] [Google Scholar]
- 7.Pogliano JA, Beckwith J. SecD and SecF facilitate protein export in Escherichia coli. EMBO J. 1994;13:554–61. doi: 10.1002/j.1460-2075.1994.tb06293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sagara K, Matsuyama S, Mizushima S. SecF stabilizes SecD and SecY, components of the protein translocation machinery of the Escherichia coli cytoplasmic membrane. J Bacteriol. 1994;176:4111–6. doi: 10.1128/jb.176.13.4111-4116.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hand NJ, Klein R, Laskewitz A, Pohlschroder M. Archaeal and bacterial SecD and SecF homologs exhibit striking structural and functional conservation. J Bacteriol. 2006;188:1251–9. doi: 10.1128/JB.188.4.1251-1259.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nouwen N, Piwowarek M, Berrelkamp G, Driessen AJ. The large first periplasmic loop of SecD and SecF plays an important role in SecDF functioning. J Bacteriol. 2005;187:5857–60. doi: 10.1128/JB.187.16.5857-5860.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuyama S, Fujita Y, Mizushima S. SecD is involved in the release of translocated secretory proteins from the cytoplasmic membrane of Escherichia coli. EMBO J. 1993;12:265–70. doi: 10.1002/j.1460-2075.1993.tb05652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Economou A, Pogliano JA, Beckwith J, Oliver DB, Wickner W. SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell. 1995;83:1171–81. doi: 10.1016/0092-8674(95)90143-4. [DOI] [PubMed] [Google Scholar]
- 13.Arkowitz RA, Wickner W. SecD and SecF are required for the proton electrochemical gradient stimulation of preprotein translocation. EMBO J. 1994;13:954–63. doi: 10.1002/j.1460-2075.1994.tb06340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duong F, Wickner W. The SecDFyajC domain of preprotein translocase controls preprotein movement by regulating SecA membrane cycling. EMBO J. 1997;16:4871–9. doi: 10.1093/emboj/16.16.4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pogliano KJ, Beckwith J. Genetic and molecular characterization of the Escherichia coli secD operon and its products. J Bacteriol. 1994;176:804–14. doi: 10.1128/jb.176.3.804-814.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tseng TT, et al. The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J Mol Microbiol Biotechnol. 1999;1:107–25. [PubMed] [Google Scholar]
- 17.Tsukazaki T, et al. Purification, crystallization and preliminary X-ray diffraction of SecDF, a translocon-associated membrane protein, from Thermus thermophilus. Acta Crystallogr F. 2006;62:376–80. doi: 10.1107/S1744309106007779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uchida K, Mori H, Mizushima S. Stepwise movement of preproteins in the process of translocation across the cytoplasmic membrane of Escherichia coli. J Biol Chem. 1995;270:30862–8. doi: 10.1074/jbc.270.52.30862. [DOI] [PubMed] [Google Scholar]
- 19.Schiebel E, Driessen AJ, Hartl FU, Wickner W. ΔμH+ and ATP function at different steps of the catalytic cycle of preprotein translocase. Cell. 1991;64:927–39. doi: 10.1016/0092-8674(91)90317-r. [DOI] [PubMed] [Google Scholar]
- 20.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443:173–9. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 21.Seeger MA, von Ballmoos C, Verrey F, Pos KM. Crucial role of Asp408 in the proton translocation pathway of multidrug transporter AcrB: evidence from site-directed mutagenesis and carbodiimide labeling. Biochemistry. 2009;48:5801–12. doi: 10.1021/bi900446j. [DOI] [PubMed] [Google Scholar]
- 22.Hase CC, Barquera B. Role of sodium bioenergetics in Vibrio cholerae. Biochim Biophys Acta. 2001;1505:169–78. doi: 10.1016/s0005-2728(00)00286-3. [DOI] [PubMed] [Google Scholar]
- 23.Sasaki M, Takagi M, Okamura Y. A voltage sensor-domain protein is a voltage-gated proton channel. Science. 2006;312:589–92. doi: 10.1126/science.1122352. [DOI] [PubMed] [Google Scholar]
- 24.Hattori M, et al. Mg2+-dependent gating of bacterial MgtE channel underlies Mg2+ homeostasis. EMBO J. 2009;28:3602–3612. doi: 10.1038/emboj.2009.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vassylyev DG, et al. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448:163–8. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- 26.Inaba K, et al. Crystal structure of the DsbB-DsbA complex reveals a mechanism of disulfide bond generation. Cell. 2006;127:789–801. doi: 10.1016/j.cell.2006.10.034. [DOI] [PubMed] [Google Scholar]
- 27.Matsuo E, Mori H, Shimoike T, Ito K. Syd, a SecY-interacting protein, excludes SecA from the SecYE complex with an altered SecY24 subunit. J Biol Chem. 1998;273:18835–40. doi: 10.1074/jbc.273.30.18835. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.