Abstract
Flowering time in the model plant Arabidopsis thaliana is regulated by both external environmental signals and internal developmental pathways. Natural variation at the FLOWERING H (FLH) locus has previously been described, with alleles present in the Cape Verde Islands accession causing early flowering, particularly after vernalization. The mechanism of FLH-induced early flowering is not understood. Here, the integration of FLH activity into the known flowering time pathways is described using molecular and genetic approaches. The identification of molecular markers that co-segregated with the FLH locus allowed the generation of multiple combinations of FLH alleles with mutations in flowering time genes in different flowering pathways. Combining an early flowering FLH allele with mutations in vernalization pathway genes that regulate FLC expression revealed that FLH appears to act in parallel to FLC. Surprisingly, the early flowering allele of FLH requires the floral integrator FD, but not FT, to accelerate flowering. This suggests a model in which some alleles of FLH are able to affect the FD-dependent activity of the floral activator complex.
Key words: FLH, flowering time, quantitative trait loci, vernalization.
Introduction
The transition from a vegetative to reproductive phase of development in flowering plants is tightly regulated by a complex network of control mechanisms that sense environmental signals (Andres and Coupland, 2012). In Arabidopsis thaliana, four major pathways containing hundreds of genes have been identified – the autonomous, photoperiod, gibberellic acid, and vernalization pathways. A smaller number of genes function as floral integrators and respond to these multiple pathways to regulate the transition to flowering.
The difference between late and early flowering varieties of Arabidopsis is partly due to natural allelic variation in two genes with winter annual plants having active alleles of FLOWERING LOCUS C (FLC) and FRIGIDA (FRI) (Clarke and Dean, 1994; Koornneef et al., 1994; Lee et al., 1994a; Lee and Amasino, 1995; Gazzani et al., 2003; Shindo et al., 2006). FLC is a MADS-box transcription factor which delays flowering by repressing the expression of the floral integrators SUPRESSOR OF OVER-EXPRESSION OF CO (SOC1) (Lee et al., 2000; Onouchi et al., 2000), the RAF kinase inhibitor-like/phosphatidylethanolamine binding family encoding gene FLOWERING LOCUS T (FT) and the bZIP transcription factor FD (Kardailsky et al., 1999; Kobayashi et al., 1999; Searle et al., 2006). FLC expression is determined by the RNA Polymerase associated complex (Paf1C) and the coil–coil protein FRI via interactions with SUPPRESSOR OF FRIGIDA4 (SUF4), FRIGIDA-LIKE1 (FRL1) and FRIGIDA ESSENTIAL1 (FES1) (Michaels et al., 2004; Schmitz et al., 2005; Kim and Michaels, 2006; Kim et al., 2006).
The core polycomb repressive complex 2 (PRC2) is associated with FLC chromatin prior to, during and after a cold exposure (De Lucia et al., 2008). Vernalization accelerates flowering through stable repression of FLC through the increased transcription of the antisense FLC transcript COOLAIR (Swiezewski et al., 2009) and the non-coding COLDAIR sense transcript from a cryptic promoter with the first intron of FLC, with COLDAIR thought to recruit the PHD proteins VNR5, VIN3, and VEL1 to form a complex with PRC2 (to produce the PHD–PRC2 complex) at the FLC locus (Sung and Amasino, 2004; Sung et al., 2006b; Greb et al., 2007; De Lucia et al., 2008; Heo and Sung, 2011). These proteins induce the trimethylation of lysine 27 of histone 3 (H3K27me3) that maintain FLC in a repressed state upon the return to warm conditions (Bastow et al., 2004). Once the vernalized state is established, it is subsequently epigenetically maintained by the activity of VERNALIZATION 1 (VRN1) (Levy et al., 2002) and LIKE-HETEROCHROMATIN PROTEIN 1 (LHP1) (Mylne et al., 2006; Sung et al., 2006a).
Although FLC is the primary regulator of flowering in response to vernalization in winter-annual varieties of Arabidopsis, analysis of flc null mutants has demonstrated that an FLC-independent pathway also exists (Michaels and Amasino, 2001; Sung and Amasino, 2004; Alexandre and Hennig, 2008). Two MADS-box genes promote flowering in response to vernalization independently of FLC; AGAMOUS-LIKE 19 (AGL19) (Schonrock et al., 2006) and AGAMOUS-LIKE 24 (AGL24) (Michaels et al., 2003a). Similarly to FLC, AGL19 is maintained in a transcriptionally repressed state by polycomb proteins (Schonrock et al., 2006) and this repression is alleviated by vernalization via a mechanism requiring VIN3, but which is independent of VRN2 (Schonrock et al., 2006). Once activated, AGL19 expression induces flowering by upregulating LFY and APETALA11, but not SOC1 (Schonrock et al., 2006). In contrast, AGL24 has a complex interaction with SOC1, as both genes are able to positively regulate the expression of the other, and overexpression of one without the other has a minimal effect (Michaels et al., 2003a). Like AGL19, AGL24 is thought to promote flowering by upregulating LFY (Yu et al., 2002).
When released from FLC repression, the FT protein is transported from the leaves to the shoot apical meristem where it interacts with the bZIP transcription factor FD (An et al., 2004; Abe et al., 2005; Wigge et al., 2005; Corbesier et al., 2007; Ikeda et al., 2007). In rice, this interaction is mediated by the 14-3-3 GF14c protein (Taoka et al., 2011), which forms a hexameric florigen activation complex (FAC), composed of two molecules each of Hd3a (the rice FT orthologue), FD, and GF14c (Taoka et al., 2011). In this model, FD anchors the FAC to regulatory regions of FAC target genes through the bZip DNA binding domain of FD. Consistent with this model, similar pairwise interactions have been described in tomato (Pnueli et al., 2001). In Arabidopsis, FD binds the promoters of several SQUAMOSA PROMOTER BINDING PROTEIN LIKE (SPL) genes, including SPL3, SPL4, SPL5, and SOC1 (Teper-Bamnolker and Samach, 2005; Jung et al., 2012). SOC1 also binds the promoters of SPL3, SPL4, and SPL5 (Jung et al., 2012), and SPL3, SPL4, and SPL5 can directly activate LFY expression (Yamaguchi et al., 2009; Jung et al., 2012). FD also, probably indirectly, activates AP1 expression (Abe et al., 2005; Wigge et al., 2005; Benlloch et al., 2011). The TERMINAL FLOWER 1 (TFL) protein, an FT paralogue with opposite function, normally represses flowering perhaps by competing with FT for binding to FD or the FAC (Hanzawa et al., 2005; Ahn et al., 2006).
Although several studies have investigated natural variation in vernalization response in Arabidopsis, several of these studies have revealed variation at FRI, FLC, or loci interacting with these loci accounted for much of the observed variation (Shindo et al., 2006; Strange et al., 2011; Coustham et al., 2012; Sanchez-Bermejo et al., 2012). While some additional loci have been identified (Sanchez-Bermejo et al., 2012), little is known about the identity or function of other loci or how these loci function in the context of existing vernalization pathways. One such locus FLOWERING H (FLH) was identified in recombinant inbred lines (RILs) derived from crosses of the Landsberg erecta (Ler) accession with the Cape Verdi Islands (CVI) accession (Alonso-Blanco et al., 1998). Plants carrying CVI alleles of FLH flower earlier than Ler, particularly after vernalization, but responded similarly to photoperiod (Alonso-Blanco et al., 1998).
This study further characterizes the FLH flowering time locus and shows that FLH is likely to represent a novel flowering time gene. Furthermore, the ability of CVI alleles of FLH to confer early flowering after vernalization operates in parallel to the PHD-PRC2 mediated repression of FLC. This study also demonstrates that the earliness conferred by CVI alleles of FLH is dependent on the presence of FD, but does not require FT, suggesting that FLH may act as a modifier of the FD-specific activity of the FAC.
Materials and methods
Plant growth conditions, vernalization treatment, and flowering time analysis
Seeds of Landsberg erecta (Ler, NW20), ft-1 and fd-1 (Koornneef et al., 1991), ap1-1 (Mandel et al., 1992), and ap1-1/cal1-1 (Ditta et al., 2004), all in the Ler background, were obtained from the Arabidopsis Biological Resource Collection (ABRC, Columbus, Ohio, USA). Seeds of near-isogenic line 1 (NIL1) containing the CVI allele at FLH (Alonso-Blanco et al., 1998) were provided by C. Alonso-Blanco. Seeds of flc-5, vrn5-1, vin3-7 (Greb et al., 2007), and vrn1-2 (Levy et al., 2002) in the Ler background were provided by C. Dean. Seeds of soc1-2 in Ler (Melzer et al., 2008) were provided by G. Coupland.
Seeds were sown on moist soil (Debco seed raising mix/vermiculite, 4:1) before being vernalized in the dark for 3 weeks in the dark at 4 °C and then transferred to a controlled-environment growth room at 22 °C under an 8/16 light/dark regime (short day condition, SD) with cool white fluorescent light (Sylvania Luxline Plus F36W/840). Control, non-vernalized plants were stratified at in the dark at 4 °C for 2 days prior to transfer to SD or long day (LD) conditions. For plants grown in LD, the photoperiod was 16/8 light/dark under the same illumination conditions. Plant were grown for 21 days (SD) or 7 days (LD) before being transferred to individual wells of a 48-well tray. Flowering time was measured by counting the total number of rosette leaves and cauline leaves present on the main stem.
Genetic and physical mapping of FLH
The FLH gene was initially mapped by genotyping F2 plants from a cross of Ler and FLH-CVI (NIL1). Genomic DNA was extracted from 648 plants according to the method described by Klimyuk et al. (1993), and plants with recombination events between the markers CER455033 (www.arabidopsis.org; last accessed 23 April 2013) and a CAPS marker derived from the AFLP marker SM78–320 marker (Peters et al., 2001) were selected. The FLH genotype of recombinant plants was determined by analysing the flowering time of 24–48 plants in the F3 generation. Subsequent fine mapping was performed using a selection of new markers (Supplementary Table S1, available at JXB online) on high-quality genomic DNA extracted from recombinants according to the method by Dellaporta et al. (1983).
Mutant alleles were detected as previously described for flc-5, vrn5-1 (Greb et al., 2007), and vrn1-1 (Levy et al., 2002) or using the markers described in Supplementary Table S2.
Reverse-transcription PCR analysis
For semi-quantitative reverse-transcription (RT)-PCR, whole seedlings were harvested at 10 days post germination or vernalization every 10 days for 40 days after being germinated on MS plates without sucrose. Total RNA was extracted from pooled seedlings at each stage with and without vernalization using the RNeasy Plant Kit (Qiagen) as per the manufacturer’s instructions. The optional on-column DNAse step was also included. The yield and RNA purity was determined spectrophotometrically (NanoDrop ND-1000) and visualized by gel electrophoresis. cDNA was synthesized using Superscript III (Invitrogen) as per the manufacturer’s instructions. Total RNA (500ng) was used per cDNA synthesis reaction in an Eppendorf Mastercycler. cDNA concentration across samples was normalized using ACTIN7 as an internal control and visualized on a 1% (w/v) agarose gel using ethidium bromide staining. The primer sequences are described in Supplementary Table S3.
For quantitative RT (qRT) PCR, total RNA was extracted from material enriched in shoot apices (Lopez-Juez et al., 2008) collected from pools of 24 plants each of FLH-CVI and Ler grown for at 15–20 short days post germination/vernalization (i.e. at the same developmental stage). Three biological replicates of each genotype and treatment were used. RNA was purified using the Qiagen RNeasy Kit as per the manufacturer’s instructions. Total RNA (2 µg) was DNase treated with the Promega RQ1 DNase Kit as per the manufacturer’s instructions to remove any genomic DNA contamination. Quantitative real-time PCR was performed using the Bio-RAD iCycler and the iScript One Step RT-PCR Kit with SYBR Green as per the manufacturer’s instructions. PCR conditions consisted of a reverse-transcription step at 50 °C for 10 minutes, a reverse transcription inactivation step at 95 °C for 5 minutes, and 40 cycles of 95 °C for 10 seconds followed by 64.5 °C for 30 seconds. Results were visualized using the BioRad iQ5 Optical System Software. Expression of FD and FT was normalized relative to the expression of UBC_21 previously validated as a reference gene for qRT-PCR (Czechowski et al., 2005). Details of qRT-PCR primers are described in Supplementary Table S4. Transcript abundance was calculated using the Pfaffl model for relative quantification with efficiency correction (Pfaffl, 2001), and statistical analysis was performed using a Student’s t-test.
Results
CVI alleles of FLH accelerate flowering compared to Ler alleles
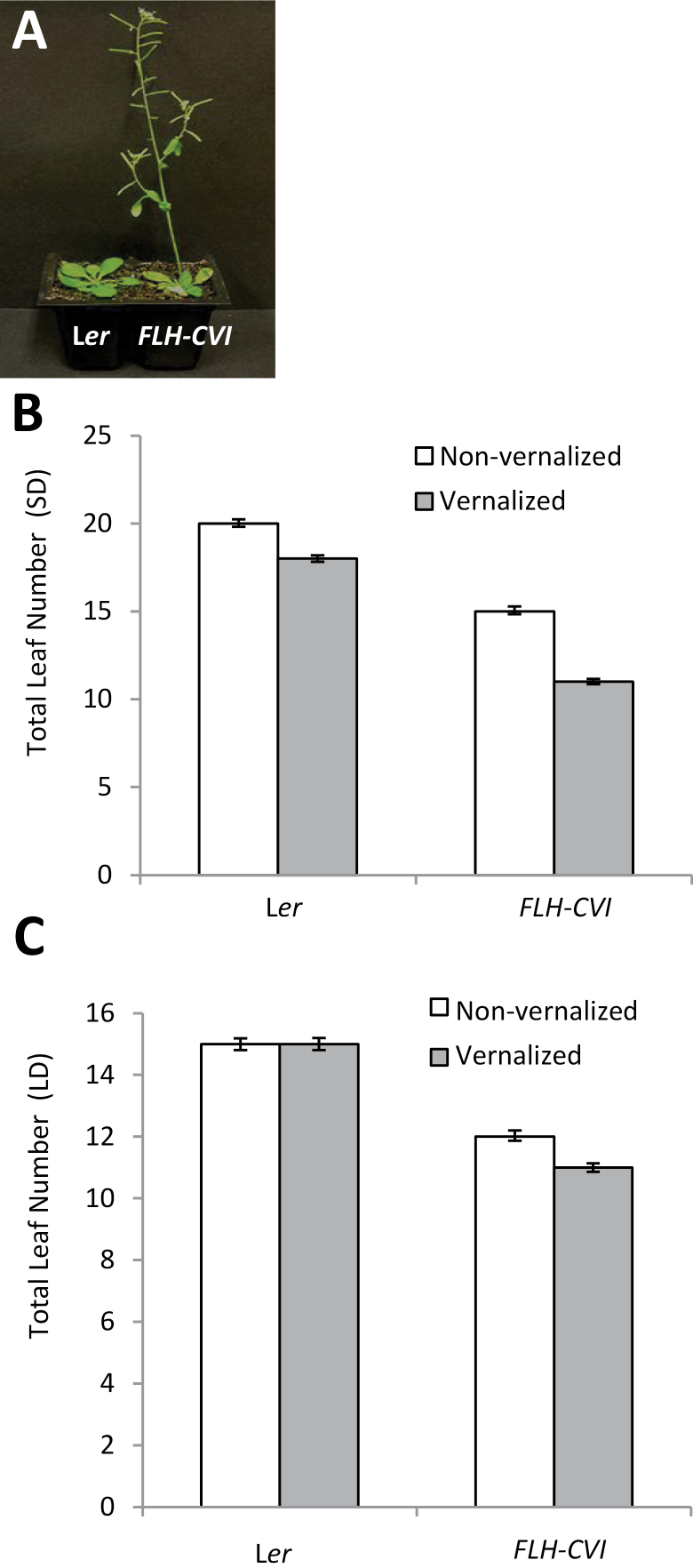
The presence of the alleles at FLH originating from the CVI accession in an otherwise Ler near-isogenic line reduced flowering time when grown in a SD photoperiod, particularly in response to vernalization (Fig. 1A; Alonso-Blanco et al., 1998). FLH-CVI plants appeared otherwise normal and did not exhibit any additional other phenotypes associated with early flowering, and produced normal flowers and cauline leaves (Fig. 1A). In the absence of a vernalization treatment, FLH-CVI plants flowered with approximately five fewer leaves than Ler in SD conditions (Fig. 1B). CVI alleles of FLH also conferred slightly earlier flowering in LD conditions, with FLH-CVI plants flowering with approximately three fewer leaves in the absence of vernalization and with four fewer leaves than Ler after a vernalization treatment (Fig. 1C).
Fig. 1.
CVI alleles of FLH accelerate flowering compared to Ler alleles. (A) Comparison of Ler and FLH-CVI (NIL1) grown for 30 short days after 3 weeks of vernalization. (B, C) Flowering time of Ler and FLH-CVI in short days (B) and long days (C) as measured by total leaf number in response to 3 weeks of vernalization at 4 °C (n = 48). Bars are standard errors.
In order to further analyse the function of FLH in response to vernalization, this study measured the flowering time of FLH-CVI and Ler after exposure to different durations of vernalization as measured by total leaf number after growth in SD conditions (Fig. 2). A vernalization response was detected in both FLH-CVI and Ler after 3-week vernalization, with vernalized FLH-CVI plants flowering significantly earlier than Ler. Exposure to a longer vernalization treatment of 6 weeks did not lead to significantly earlier flowering, suggesting that saturation of the vernalization response occurs at or before 3 weeks in plants with a CVI allele of FLH.
Fig. 2.
Dosage sensitivity of FLH alleles to increasing durations of vernalization. Flowering time of Ler and FLH-CVI as measured by total leaf number in response to various durations of vernalization (n = 24). Bars are standard errors.
CVI of FLH alleles differentially affects gene expression
In order to gain an understanding of where FLH may fit into the current model of floral induction in Arabidopsis, semi-quantitative RT-PCR was performed on genes located throughout the vernalization-induced flowering pathway (FLC, AGL24, and AGL19), floral integrator genes (FT, FD, SOC1, and LFY), and the meristem identify gene AP1, all of which are known to be regulated either directly or indirectly by vernalization. The transcript levels of these genes were examined in Ler and FLH-CVI plants every 10 days for 40 days in both control (non-vernalized) plants and in plants exposed to 3-week vernalization (Fig. 3). Transcript levels in vernalized and non-vernalized samples were then compared to identify those transcripts with altered abundance in FLH-CVI relative to Ler. After applying these criteria, only AP1 appeared to show any significant change in abundance in FLH-CVI compared to Ler, relative to its levels when non-vernalized at the same stages of development. In the FLH-CVI early-flowering plant line, AP1 transcript abundance was increased, particularly at 40 days of growth. This increase in AP1 expression was not accompanied with an increase in SOC1, LFY, FT, or FD expression, implying that CVI alleles of FLH may be capable of upregulating AP1 independently of these genes. The semi-quantitative RT-PCR data also implied that FLH does not regulate any of the other vernalization-induced flowering time genes investigated, including the major repressor FLC.
Fig. 3.
Semi-quantitative RT-PCR analysis of flowering time genes in the presence of CVI alleles of FLH, showing transcript levels of flowering time genes in seedlings of Ler relative to FLH-CVI at 10-day increments with and without vernalization. Days is days of growth in SD conditions post stratification or post vernalization.
FLH is a novel flowering time locus
To further investigate the mechanism by which CVI alleles of FLH confer early flowering, combinations of CVI alleles of FLH with mutants in other flowering time genes were generated. In order to accurately determine which progeny of various crosses contained the CVI alleles of FLH, this study sought to identify a molecular marker closely linked to FLH. This approach also allowed a more precise determination of the chromosomal location of the FLH locus, as the first step towards cloning the FLH quantitative trait locus.
The FLH locus was originally identified by Alonso-Blanco et al. (1998) and further characterized in a FLH-CVI near-isogenic line derived from introgression of CVI alleles into Ler. The FLH-CVI line contains an approximately 10 centimorgan region of CVI-derived DNA at the bottom of the long arm of chromosome 5 in a Ler background (Fig. 4A; Alonso-Blanco et al., 1998). The Ler-CVI polymorphic marker G2368 was previously used as a marker for FLH in the FLH NIL1. G2368 maps to 109 cM on the Ler-CVI genetic map (Alonso-Blanco et al., 1998) and is located at approximately 25.8Mb on chromosome 5 of the Columbia accession reference genome, close to the MAF2–MAF5 genes (Ratcliffe et al., 2003). NIL1 has previously been shown to contain Ler alleles at EG7F2 (24.6Mb) and CVI alleles at markers T6B16 (24.8Mb) and MQB2 (25.2Mb) (Swarup et al., 1999) thus delimiting containing FLH to a region of approximately 2.4Mb between 24.6Mb and the end of chromosome 5 at 27.0Mb.
Fig. 4.
Identification of markers that co-segregate with FLH. (A) Location of CVI-derived DNA on chromosome 5 in the FLH-CVI NIL is indicated by the filled area. (B) Segregation of flowering time in the F2 progeny of a cross between Ler and the FLH-CVI NIL (n = 48); filled triangles indicate the mean flowering time of each of the parents and horizontal lines indicate the range of flowering times (n ≥ 35). (C) Mapping of FLH; filled triangles indicate the positions of markers used to map FLH. Recombinants were identified in 648 F2 progeny of a cross between Ler and the FLH-CVI NIL.
To determine the degree of linkage between G2368 and FLH, NIL1 was first crossed to Ler and the F2 progeny were analysed for flowering time, which confirmed that early flowering conferred by the CVI alleles of FLH was dominant (Fig. 4B). Plants carrying recombination events between markers CER455033 and PLOP3 were identified, and the F3 progeny were tested for segregation of late flowering, indicating that the F2 parent was homozygous for the Ler allele of FLH. F2 plants with recombinants events between CER455033 and SM78-320 were then further analysed using additional polymorphic markers (Fig. 4C). In a population of 648 F2 plants, 11 recombinants between G2368 and FLH could be detected. Furthermore, FLH co-segregated with the marker 2662, and thus the region containing FLH was delimited by markers K1F13D and 2672. Therefore, marker 2662 was used for all further genotyping of FLH alleles. Interestingly, a reduction in recombination was detected distal to FLH (i.e. towards the telomere), suggesting that recombination is suppressed in this region in the Ler × NIL1 cross.
This rough mapping of FLH also suggests that FLH is likely to represent a new flowering time locus, as only a single gene with a flowering time effect, DDM1, is localized to this region (Jeddeloh et al., 1999). However, it seems unlikely that FLH is an allele of DDM1, as mutations in DDM1, which encodes a SWI-SNF2-like protein, produces plants with exhibit unstable pleiotropic phenotypes that become progressively more severe with each generation (Kakutani et al., 1996) – phenotypes which are not shared with plants carrying CVI alleles of FLH.
CVI alleles of FLH act independently of VIN3, VNR5, and FLC to accelerate flowering
In order to identify the role and interaction that CVI alleles of FLH have with known vernalization genes within the vernalization flowering time pathway, the FLH-CVI NIL1 was crossed with various mutants that regulate FLC expression. In order to avoid any effects on flowering time due to mixing of genetic backgrounds, this study only used mutations that were in the Ler genetic background, which contains a weakly active allele of FLC (Koornneef et al., 1994; Lee et al., 1994b; Gazzani et al., 2003; Michaels et al., 2003b). F2 progeny were genotyped at FLH using the 2662 marker, and F3 plants doubly homozygous for CVI alleles at FLH and the mutation of interest were subjected to flowering time analysis.
Mutations in VIN3 lead to late flowering, particularly after vernalization, as upregulation of VIN3 during the cold is required for the initial repression of FLC (Sung and Amasino, 2004; Greb et al., 2007). vin3 mutant plants were slightly late flowering in the absence of a vernalization treatment and were completely unresponsive to a vernalization response and subsequent growth in SD conditions (Fig. 5A and B). When CVI alleles of FLH were combined with a vin3 mutation, the resulting FLH-CVI vin3 plants flowered earlier than the vin3 mutant but later than FLH-CVI plants and did not respond to vernalization, suggesting that the earliness conferred by CVI alleles of FLH does not require VIN3 activity. Combinations of CVI alleles of FLH with mutations in VRN5, which forms a dimer with VIN3, behaved similarly. FLH-CVI vrn5 plants flowered earlier than vrn5 mutants, but not as early as FLH-CVI plants, and FLH-CVI vrn5 plants retained a weak response to vernalization (Fig. 5A and B). FLH-CVI vrn1 plants flowered almost as early as FLH-CVI plants, but did not exhibit a vernalization response (Fig. 5A and B).
Fig. 5.
Genetic analysis of FLH-CVI interaction with vernalization pathway genes. (A) Vernalized wildtype Ler, flc-5, vrn5-1, vin3-7, and vrn1-2 mutants and FLH-CVI combinations grown in for 21 short days after 3-week vernalization. (B) Comparison of flowering time between vernalized treatments of wild-type Ler, FLH-CVI, various known vernalization mutants flc-5, vin5–1, vin3-7, and vrn1-2, and FLH-CVI in the mutant backgrounds grown in short days (n ≥ 48). (C) Comparison of flowering time between vernalized treatments of wild-type Columbia and Ler, FLH-CVI, and FLH-CVI introgressed into Col (FLH-CVI in Col) grown in short days (n ≥ 48). Bars are standard errors.
To determine if there is an interaction between CVI alleles of FLH and active alleles of FLC, the CVI allele of FLH was introgressed into Columbia and selected plants with CVI alleles at FLH using the molecular marker 2662. CVI alleles of FLH were able to confer earlier flowering, particularly after vernalization in the Columbia background; however, the flowering time was more similar to Columbia (Fig. 5C). This suggests that CVI alleles of FLH confer earlier flowering than Col alleles of FLH and that CVI alleles of FLH are not able to completely overcome the lateness caused by active alleles of FLC.
CVI alleles of FLH require FD but not FT to accelerate flowering
To establish how CVI alleles of FLH may be effecting the transcript levels of AP1 through the activitivy of floral integrators, CVI alleles of FLH were introduced by crossing into floral integrator mutants in a Ler background. Combinations of CVI alleles of FLH with ap1 and ap1/cal mutations produced plants with a flowering time that was intermediate between the two parents, suggesting FLH is able to promote flowering partially independently of AP1 and CAL (Fig. 6B and C). When CVI alleles of FLH were combined with mutations in soc1, the resulting FLH-CVI soc1 plants were slightly earlier flowering than soc1 plants (Fig. 6B), suggesting that the earlinesss conferred by CVI allels of FLH is partially dependent on, or acts additively with, SOC1.
Fig. 6.
Genetic analysis of the interaction of FLH-CVI with floral integrator genes. (A) Vernalized wildtype Ler, FLH-CVI, fd-1, ft-1, and FLH-CVI combinations grown in short days for 30 days after 3-week vernalization. (B) Comparison of flowering time between vernalized treatments of wild-type Ler, FLH-CVI, and various mutants and FLH-CVI combinations grown in SD conditions (n ≥ 48). (C) Comparison of flowering time between vernalized treatments of wild-type Ler, FLH-CVI, and various mutants and FLH-CVI combinations grown in LD conditions (n ≥ 48). Bars are standard errors.
The most striking results were observed when CVI alleles of FLH were combined with mutations in FD and FT. FLH-CVI ft plants flowered with an average of 13 fewer leaves compared to the ft single mutant control in both vernalized and non-vernalized treatments (Fig. 6), which was similar to the number of leaves in wildtype Ler plants. CVI alleles of FLH were thus able to completely overcome the loss of FT and to restore flowering time comparable to that of wild type Ler. However, FLH-CVI ft plants did not flower as early as FLH-CVI plants, suggesting that the ability of FLH to accelerate flowering is partially dependent on FT.
In contrast, CVI alleles of FLH were unable to overcome the late flowering due to the loss of FD, in both non-vernalized and vernalized treatments. FLH-CVI fd plants exhibiting a flowering time similar to that of the fd single mutant in both treatments, suggesting that fd mutations are completely epistatic to CVI alleles of FLH (Fig. 6A and B). This suggests that the ability of CVI alleles of FLH to accelerate flowering is completely dependent on FD activity.
To determine if CVI alleles of FLH can accelerate flowering when the photoperiod pathway is active, the flowering time measurements were repeated using plants grown in LD conditions. CVI alleles of FLH behaved similarly in combination with most mutations in long days as they did in short days. Mutations in ap1 and fd prevented the vernalization response of CVI alleles of FLH (Fig. 6C), and mutation of soc1 reduced the ability of CVI alleles of FLH to confer earliness.
FD expression is increased by CVI alleles of FLH in the absence of vernalization
As the results of the genetic analysis suggested that FLH requires FD to confer early flowering, this study examined the possibility that CVI alleles of FLH may lead to the increased expression of FD in the shoot meristem. Therefore, material enriched in shoot apices were collected from Ler and FLH-CVI plants grown in SD conditions and FD expression was examined by qRT-PCR (Fig. 7A). Consistent with previous reports, the levels of FD expression was higher in plants that had been vernalized (Searle and Coupland, 2004). FD mRNA levels were slightly elevated (by approximately 2-fold) in FLH-CVI apices compared to Ler plants in non-vernalized control plants, but FD expression was not different between vernalized Ler and FLH-CVI plants. This suggests that CVI alleles of FLH can increase FD expression under some conditions. Consistent with the genetic analysis, no substantial differences in FT expression between FLH-CVI and Ler plants were detected (Fig. 7B).
Fig. 7.
FT and FD expression in FLH-CVI. Expression of FD (A) and FT (B) in shoot apical meristem-enriched tissues, as measured by quantitative RT-PCR relative to the internal control UBC. Data are means and standard errors of three biological replicates. Different letters indicate significant differences in relative expression levels (P ≤ 0.05).
Discussion
Natural variation in vernalization responses in Arabidopsis has been investigated using a variety of approaches, and several QTLs have been identified (Lempe et al., 2005; Sanchez-Bermejo et al., 2012). In some cases, genes responsible for these QTLs have also been identified (Shindo et al., 2006). This study characterized the activity of the FLOWERING H locus and demonstrated that alleles from the Cape Verde Islands accession accelerate flowering in an FD-dependent manner.
FLH was identified in the Ler/CVI recombinant inbred line population, as a locus which conferred early flowering, particularly in response to vernalization, with CVI alleles at FLH conferring early flowering compared to Ler alleles (Alonso-Blanco et al., 1998). This suggests that the CVI allele of FLH may be a gain-of-function polymorphism, or that the Ler allele of FLH may represent a loss-of-function allele. The direct comparison of CVI and Columbia alleles of FLH (Fig. 5) revealed that Columbia appeared to have a similar allele of FLH as Ler, as plants with CVI alleles of FLH were earlier flowering than those with Col alleles. Flowering time analysis of RILs derived from crosses of Ler to Col did not identify a flowering time QTL at the FLH locus, suggesting that Ler and Col have similar FLH alleles (Jansen et al., 1995). The present analysis of CVI alleles of FLH introgressed into Columbia is also consistent with a previous QTL analysis performed on a Col × CVI RIL population when a weak flowering time QTL was detected close to FLH in the Col × CVI RIL, with Columbia alleles conferring later flowering than CVI alleles (Simon et al., 2008).
This situation is similar to that observed in an analysis of natural variation of CRYPTOCHROME 2 (CRY2). The allele of CRY2 present in CVI was identified as the underlying cause of the EARLY DAYLENGTH INSENSITIVE (EDI) QTL in the Ler/CVI RIL population (Alonso-Blanco et al., 1998; El-Din El-Assal et al., 2001). CRY2-CVI confers dominant, day-length-insensitive early flowering due to the increased accumulation of CRY2-CVI (El-Din El-Assal et al., 2001). Although the CVI allele of CRY2 appears to be unique, and other functional variants in CRY2 have not been described, this example highlights the use of natural variation to uncover novel alleles that shed light on gene function. Similarly, identification of natural variation in an advanced multiparent population uncovered a previously unknown function in the control of short architecture for the AGAMOUS-LIKE6 gene (Huang et al., 2012).
As several genes are located close to the previously reported location of FLH, it was important to be able to exclude these genes as FLH. The fine mapping of FLH confirms that FLH is not a novel allele of MAF2, MAF3, MAF4, or MAF5, which lie close to FLH. MAF2 is involved in vernalization response (Ratcliffe et al., 2003) and natural variation in MAF2 and MAF3 has been described (Rosloski et al., 2010). One FLH candidate with a role in flowering time that could not be eliminated by fine mapping has been described, the SWI/SNF2-like protein DECREASED DNA METHYLATION (DDM1) (Jeddeloh et al., 1999). The role of DDM1 in maintenance of ‘global’ chromatin methylation is also not consistent with the subtle effect of FLH on flowering time. ddm1 mutants exhibit pleiotropic defects and are unstable (Jeddeloh et al., 1999), phenotypes that are not observed in multiple generations of crosses and propagation of the FLH-CVI NIL. Furthermore, preliminary sequence analysis does not support the hypothesis that FLH is DDM1 (A. Dinsdale, unpublished).
CVI alleles of FLH did not affect the expression of any of the tested genes differently compared to Ler alleles of FLH, except AP1 and FD. This suggests that FLH may be acting very late in a floral promoting pathway to activate transcription or is acting post-transcriptionally. CVI alleles of FLH can still accelerate flowering in the absence of AP1, particularly in short days, suggesting at least a partial independence from AP1. The observation that CVI alleles of FLH do not affect the expression of FT, and only weakly increase FD expression in the absence of vernalization, suggests that the CVI allele of FLH may act as a weak transcriptional activator of FD expression.
One flowering time gene with a similar phenotype to FLH-CVI is TERMINAL FLOWER1 (TFL1) (Shannon and Meeks-Wagner, 1991; Alvarez et al., 1992). The FLH-CVI phenotype is similar to that conferred by the overexpression of a TFL1-VP16 fusion protein, in which the TFL repressor is converted to a strong transcriptional activator. The early flowering of 35S::TFL-VP16 is strongly dependent on FD (Hanano and Goto, 2011), similar to that observed with CVI alleles of FLH. Furthermore, the early flowering tfl mutant phenotype is suppressed by a fd mutation, similarly to the phenotype observed in the FLH-CVI fd plants. The dominance and activity of CVI alleles of FLH has some similarity to that of FWA, but dominant late flowering alleles of FWA are due to the misexpression of FWA (Soppe et al., 2000), which interacts with FT, and FWA may therefore be competing for the FT-binding site on the 14-3-3 molecule in the FAC (Ikeda et al., 2007).
Taken together, this suggests several models for FLH activity. CVI alleles of FLH may increase the expression or widen the expression domain of FD, resulting in earlier flowering. However, the increase in FD expression in FLH-CVI plants is subtle and only occurs prior to vernalization. In wild-type plants, FD expression dramatically increases by approximately 10-fold in the shoot apex upon the transition to flowering (Wigge et al., 2005), so it seems unlikely that the small increase in FD expression observed in FLH-CVI plants could entirely account for the observed earliness. Alternatively, CVI alleles of FLH may result in the misexpression, either by expression in a wider range of cells or by increased levels of expression, of a protein that can interact and cooperate directly with FD, perhaps as a transcriptional co-activator as part of the FAC. Alternatively, the protein encoded by CVI alleles of FLH may itself interact with the FAC, with FLH proteins encoded by the Ler and Columbia alleles of FLH may be unable to bind to the FAC or may bind at reduced affinity compared to the FLH protein encoded by CVI alleles. The ability of CVI alleles of FLH to rescue the late flowering of ft mutants also supports the notion that FLH may be able to substitute for FT as part of the FAC. However, none of the predicted genes in the FLH region of the genome encodes an FT- or TFL-like protein suggesting that if FLH does interact with the FAC, it may represent a novel interaction.
Supplementary material
Supplementary data are available at JXB online.
Supplementary Table S1. Mapping primers
Supplementary Table S2. Genotyping primers
Supplementary Table S3. Semi-quantitative RT-PCR primers
Supplementary Table S4. Quantitative RT-PCR primers
Acknowledgements
This work was supported by the Australian Research Council (grant DP0449651 to ARG) and a La Trobe University Postgraduate Research Scholarship to NS. We thank Carlos Alonso-Blanco, Caroline Dean, George Coupland, and the ABRC for providing seed stocks.
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. 2005. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056. [DOI] [PubMed] [Google Scholar]
- Ahn JH, Miller D, Winter VJ, Banfield MJ, Lee JH, Yoo SY, Henz SR, Brady RL, Weigel D. 2006. A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. The EMBO Journal 25, 605–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandre CM, Hennig L. 2008. FLC or not FLC: the other side of vernalization. Journal of Experimental Botany 59, 1127–1135. [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, El-Assal SE, Coupland G, Koornneef M. 1998. Analysis of natural allelic variation at flowering time loci in the Landsberg erecta and Cape Verde Islands ecotypes of Arabidopsis thaliana. Genetics 149, 749–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez J, Guli CL, Yu XH, Smyth DR. 1992. terminal flower: a gene affecting inflorescence development in Arabidopsis thaliana. The Plant Journal 2, 103–116. [Google Scholar]
- An H, Roussot C, Suarez-Lopez P, et al. 2004. CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131, 3615–3626. [DOI] [PubMed] [Google Scholar]
- Andres F, Coupland G. 2012. The genetic basis of flowering responses to seasonal cues. Nature Reviews Genetics 13, 627–639. [DOI] [PubMed] [Google Scholar]
- Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C. 2004. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427, 164–167. [DOI] [PubMed] [Google Scholar]
- Benlloch R, Kim MC, Sayou C, Thevenon E, Parcy F, Nilsson O. 2011. Integrating long-day flowering signals: a LEAFY binding site is essential for proper photoperiodic activation of APETALA1. The Plant Journal 67, 1094–1102. [DOI] [PubMed] [Google Scholar]
- Clarke JH, Dean C. 1994. Mapping FRI, a locus controlling flowering time and vernalization response in Arabidopsis thaliana. Molecular and General Genetics 242, 81–89. [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, et al. 2007. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033. [DOI] [PubMed] [Google Scholar]
- Coustham V, Li P, Strange A, Lister C, Song J, Dean C. 2012. Quantitative modulation of polycomb silencing underlies natural variation in vernalization. Science 337, 584–587. [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W-R. 2005. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology 139, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lucia F, Crevillen P, Jones AM, Greb T, Dean C. 2008. A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proceedings of the National Academy of Sciences, USA 105, 16831–16836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta S, Wood J, Hicks J. 1983. A plant DNA minipreparation: version II. Plant Molecular Biology Reporter 1, 19–21. [Google Scholar]
- Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF. 2004. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Current Biology 14, 1935–1940. [DOI] [PubMed] [Google Scholar]
- El-Din El-Assal S, Alonso-Blanco C, Peeters AJ, Raz V, Koornneef M. 2001. A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nature Genetics 29, 435–440. [DOI] [PubMed] [Google Scholar]
- Gazzani S, Gendall AR, Lister C, Dean C. 2003. Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiology 132, 1107–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greb T, Mylne JS, Crevillen P, Geraldo N, An H, Gendall AR, Dean C. 2007. The PHD finger protein VRN5 functions in the epigenetic silencing of Arabidopsis FLC. Current Biology 17, 73–78. [DOI] [PubMed] [Google Scholar]
- Hanano S, Goto K. 2011. Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. The Plant Cell 23, 3172–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzawa Y, Money T, Bradley D. 2005. A single amino acid converts a repressor to an activator of flowering. Proceedings of the National Academy of Sciences, USA 102, 7748–7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JB, Sung S. 2011. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331, 76–79. [DOI] [PubMed] [Google Scholar]
- Huang X, Effgen S, Meyer RC, Theres K, Koornneef M. 2012. Epistatic natural allelic variation reveals a function of AGAMOUS-LIKE6 in axillary bud formation in Arabidopsis. The Plant Cell 24, 2364–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y, Kobayashi Y, Yamaguchi A, Abe M, Araki T. 2007. Molecular basis of late-flowering phenotype caused by dominant epi-alleles of the FWA locus in Arabidopsis. Plant and Cell Physiology 48, 205–220. [DOI] [PubMed] [Google Scholar]
- Jansen RC, Van, Ooijen JW, Stam P, Lister C, Dean C. 1995. Genotype-by-environment interaction in genetic mapping of multiple quantitative trait loci. Theoretical and Applied Genetics 91, 33–37. [DOI] [PubMed] [Google Scholar]
- Jeddeloh JA, Stokes TL, Richards EJ. 1999. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nature Genetics 22, 94–97. [DOI] [PubMed] [Google Scholar]
- Jung JH, Ju Y, Seo PJ, Lee JH, Park CM. 2012. The SOC1-SPL module integrates photoperiod and gibberellic acid signals to control flowering time in Arabidopsis. The Plant Journal 69, 577–588. [DOI] [PubMed] [Google Scholar]
- Kakutani T, Jeddeloh JA, Flowers SK, Munakata K, Richards EJ. 1996. Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proceedings of the National Academy of Sciences, USA 93, 12406–12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. 1999. Activation tagging of the floral inducer FT. Science 286, 1962–1965. [DOI] [PubMed] [Google Scholar]
- Kim S, Choi K, Park C, Hwang HJ, Lee I. 2006. SUPPRESSOR OF FRIGIDA4, encoding a C2H2-Type zinc finger protein, represses flowering by transcriptional activation of Arabidopsis FLOWERING LOCUS C. The Plant Cell 18, 2985–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Michaels SD. 2006. SUPPRESSOR OF FRI 4 encodes a nuclear-localized protein that is required for delayed flowering in winter-annual Arabidopsis. Development 133, 4699–4707. [DOI] [PubMed] [Google Scholar]
- Klimyuk VI, Carroll BJ, Thomas CM, Jones JD. 1993. Alkali treatment for rapid preparation of plant material for reliable PCR analysis. The Plant Journal 3, 493–494. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. 1999. A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Blankestijn-de Vries H, Hanhart C, Soppe W, Peeters T. 1994. The phenotype of some late-flowering mutants is enhanced by a locus on chromosome 5 that is not effective in the Landsberg erecta wild-type. The Plant Journal 6, 911–919. [Google Scholar]
- Koornneef M, Hanhart CJ, van der Veen JH. 1991. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Molecular and General Genetics 229, 57–66. [DOI] [PubMed] [Google Scholar]
- Lee H, Suh SS, Park E, Cho E, Ahn JH, Kim SG, Lee JS, Kwon YM, Lee I. 2000. The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes and Development 14, 2366–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Amasino RM. 1995. Effect of vernalization, photoperiod, and light quality on the flowering phenotype of Arabidopsis plants containing the FRIGIDA gene. Plant Physiology 108, 157–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Aukerman MJ, Gore SL, Lohman KN, Michaels SD, Weaver LM, John MC, Feldmann KA, Amasino RM. 1994a. Isolation of LUMINIDEPENDENS: a gene involved in the control of flowering time in Arabidopsis. The Plant Cell 6, 75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Michaels SD, Masshardt AS, Amasino RM. 1994b. The late-flowering phenotype of FRIGIDA and mutations in LUMINIDEPENDENS is suppressed in the Landsberg erecta strain of Arabidopsis. The Plant Journal 6, 903–909. [Google Scholar]
- Lempe J, Balasubramanian S, Sureshkumar S, Singh A, Schmid M, Weigel D. 2005. Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genetics 1, 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy YY, Mesnage S, Mylne JS, Gendall AR, Dean C. 2002. Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science 297, 243–246. [DOI] [PubMed] [Google Scholar]
- Lopez-Juez E, Dillon E, Magyar Z, Khan S, Hazeldine S, de Jager SM, Murray JA, Beemster GT, Bogre L, Shanahan H. 2008. Distinct light-initiated gene expression and cell cycle programs in the shoot apex and cotyledons of Arabidopsis. The Plant Cell 20, 947–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF. 1992. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360, 273–277. [DOI] [PubMed] [Google Scholar]
- Melzer S, Lens F, Gennen J, Vanneste S, Rohde A, Beeckman T. 2008. Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nature Genetics 40, 1489–1492. [DOI] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. 2001. Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. The Plant Cell 13, 935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Bezerra IC, Amasino RM. 2004. FRIGIDA-related genes are required for the winter-annual habit in Arabidopsis. Proceedings of the National Academy of Sciences, USA 101, 3281–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Ditta G, Gustafson-Brown C, Pelaz S, Yanofsky M, Amasino RM. 2003a. AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. The Plant Journal 33, 867–874. [DOI] [PubMed] [Google Scholar]
- Michaels SD, He YH, Scortecci KC, Amasino RM. 2003b. Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proceedings of the National Academy of Sciences, USA 100, 10102–10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylne JS, Barrett L, Tessadori F, Mesnage S, Johnson L, Bernatavichute YV, Jacobsen SE, Fransz P, Dean C. 2006. LHP1, the Arabidopsis homologue of HETEROCHROMATIN PROTEIN1, is required for epigenetic silencing of FLC. Proceedings of the National Academy of Sciences, USA 103, 5012–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onouchi H, Igeno MI, Perilleux C, Graves K, Coupland G. 2000. Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. The Plant Cell 12, 885–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JL, Constandt H, Neyt P, Cnops G, Zethof J, Zabeau M, Gerats T. 2001. A physical amplified fragment-length polymorphism map of Arabidopsis. Plant Physiology 127, 1579–1589. [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnueli L, Gutfinger T, Hareven D, Ben-Naim O, Ron N, Adir N, Lifschitz E. 2001. Tomato SP-interacting proteins define a conserved signaling system that regulates shoot architecture and flowering. The Plant Cell 13, 2687–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe OJ, Kumimoto RW, Wong BJ, Riechmann JL. 2003. Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. The Plant Cell 15, 1159–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosloski SM, Jali SS, Balasubramanian S, Weigel D, Grbic V. 2010. Natural diversity in flowering responses of Arabidopsis thaliana caused by variation in a tandem gene array. Genetics 186, 263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Bermejo E, Mendez-Vigo B, Pico FX, Martinez-Zapater JM, Alonso-Blanco C. 2012. Novel natural alleles at FLC and LVR loci account for enhanced vernalization responses in Arabidopsis thaliana. Plant, Cell and Environment 35, 1672–1684. [DOI] [PubMed] [Google Scholar]
- Schmitz RJ, Hong L, Michaels S, Amasino RM. 2005. FRIGIDA-ESSENTIAL 1 interacts genetically with FRIGIDA and FRIGIDA-LIKE 1 to promote the winter-annual habit of Arabidopsis thaliana. Development 132, 5471–5478. [DOI] [PubMed] [Google Scholar]
- Schonrock N, Bouveret R, Leroy O, Borghi L, Kohler C, Gruissem W, Hennig L. 2006. Polycomb-group proteins repress the floral activator AGL19 in the FLC-independent vernalization pathway. Genes and Development 20, 1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle I, Coupland G. 2004. Induction of flowering by seasonal changes in photoperiod. The EMBO Journal 23, 1217–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle I, He Y, Turck F, Vincent C, Fornara F, Krober S, Amasino RA, Coupland G. 2006. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes and Development 20, 898–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon S, Meeks-Wagner DR. 1991. A mutation in the Arabidopsis TFL1 gene affects inflorescence meristem development. The Plant Cell 3, 877–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo C, Lister C, Crevillen P, Nordborg M, Dean C. 2006. Variation in the epigenetic silencing of FLC contributes to natural variation in Arabidopsis vernalization response. Genes and Development 20, 3079–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M, Loudet O, Durand S, Berard A, Brunel D, Sennesal FX, Durand-Tardif M, Pelletier G, Camilleri C. 2008. Quantitative trait loci mapping in five new large recombinant inbred line populations of Arabidopsis thaliana genotyped with consensus single-nucleotide polymorphism markers. Genetics 178, 2253–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppe WJJ, Jacobsen SE, Alonso-Blanco C, Jackson JP, Kakutani T, Koornneef M, Peeters AJM. 2000. The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Molecular Cell 6, 791–802. [DOI] [PubMed] [Google Scholar]
- Strange A, Li P, Lister C, Anderson J, Warthmann N, Shindo C, Irwin J, Nordborg M, Dean C. 2011. Major-effect alleles at relatively few loci underlie distinct vernalization and flowering variation in Arabidopsis accessions. PLoS ONE 6, e19949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S, Amasino RM. 2004. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427, 159–164. [DOI] [PubMed] [Google Scholar]
- Sung S, He Y, Eshoo TW, Tamada Y, Johnson L, Nakahigashi K, Goto K, Jacobsen SE, Amasino RM. 2006a. Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires LIKE HETEROCHROMATIN PROTEIN 1. Nature Genetics 38, 706–710. [DOI] [PubMed] [Google Scholar]
- Sung S, Schmitz RJ, Amasino RM. 2006b. A PHD finger protein involved in both the vernalization and photoperiod pathways in Arabidopsis. Genes and Development 20, 3244–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup K, Alonso-Blanco C, Lynn JR, Michaels SD, Amasino RM, Koornneef M, Millar AJ. 1999. Natural allelic variation identifies new genes in the Arabidopsis circadian system. The Plant Journal 20, 67–77. [DOI] [PubMed] [Google Scholar]
- Swiezewski S, Liu F, Magusin A, Dean C. 2009. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462, 799–802. [DOI] [PubMed] [Google Scholar]
- Taoka K, Ohki I, Tsuji H, et al. 2011. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476, 332–335. [DOI] [PubMed] [Google Scholar]
- Teper-Bamnolker P, Samach A. 2005. The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. The Plant Cell 17, 2661–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. 2005. Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309, 1056–1059. [DOI] [PubMed] [Google Scholar]
- Yamaguchi A, Wu MF, Yang L, Wu G, Poethig RS, Wagner D. 2009. The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Developmental Cell 17, 268–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Xu Y, Tan EL, Kumar PP. 2002. AGAMOUS-LIKE 24, a dosage-dependent mediator of the flowering signals. Proceedings of the National Academy of Sciences, USA 99, 16336–16341. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.