Abstract
Heavy metal homeostasis is maintained in plant cells by specialized transporters which compartmentalize or efflux metal ions, maintaining cytosolic concentrations within a narrow range. OsMTP1 is a member of the cation diffusion facilitator (CDF)/metal tolerance protein (MTP) family of metal cation transporters in Oryza sativa, which is closely related to Arabidopsis thaliana MTP1. Functional complementation of the Arabidopsis T-DNA insertion mutant mtp1-1 demonstrates that OsMTP1 transports Zn in planta and localizes at the tonoplast. When heterologously expressed in the yeast mutant zrc1 cot1, OsMTP1 complemented its Zn hypersensitivity and was also localized to the vacuole. OsMTP1 alleviated, to some extent, the Co sensitivity of this mutant, rescued the Fe hypersensitivity of the ccc1 mutant at low Fe concentrations, and restored growth of the Cd-hypersensitive mutant ycf1 at low Cd concentrations. These results suggest that OsMTP1 transports Zn but also Co, Fe, and Cd, possibly with lower affinity. Site-directed mutagenesis studies revealed two substitutions in OsMTP1 that alter the transport function of this protein. OsMTP1 harbouring a substitution of Leu82 to a phenylalanine can still transport low levels of Zn, with an enhanced affinity for Fe and Co, and a gain of function for Mn. A substitution of His90 with an aspartic acid completely abolishes Zn transport but improves Fe transport in OsMTP1. These amino acid residues are important in determining substrate specificity and may be a starting point for refining transporter activity in possible biotechnological applications, such as biofortification and phytoremediation.
Key words: Cation diffusion facilitator, ion selectivity, metal tolerance protein, Oryza sativa, vacuole, zinc transporter.
Introduction
Rice (Oryza sativa) is one of the most widely consumed cereals, being cultivated in ~156 million ha, with ~660 Mt harvested in 2008 (IRRI, 2010). Rice is the main carbohydrate source for more than half the world’s population. It is mainly consumed in its polished form, which is very poor in micronutrients such as iron (Fe) and zinc (Zn). Human Fe nutritional deficiency affects 3 billion people, being the most common mineral nutritional disorder, while Zn deficiency is the second (Welch and Graham, 2004). Therefore, it is important to understand the mechanisms of micronutrient import to the rice grain and develop strategies to increase the levels of these nutrients (Wirth et al., 2009). Although required at relatively low concentrations in plant tissues, heavy metals such as Zn, copper (Cu), and Fe are equally important for the completion of a plant’s life cycle; crop yield is therefore severely impacted by growth on soils with low mineral phytoavailability. Additionally, along with other non-essential heavy metals such as cadmium (Cd) and lead (Pb), these micronutrients are toxic when present in excess, with plants exhibiting symptoms such as chlorosis, necrosis, and growth inhibition (Marschner, 1995). For successful improvement of crops and subsequently human nutrition, it is important to understand the biological processes that govern uptake, homeostasis, and distribution of each ion throughout the plant (Palmgren et al., 2008; Mills et al., 2010, 2012; Mikkelsen et al., 2012).
Zn is important for enzymatic activity and for gene regulation by transcription factors, being described as a cofactor in >300 proteins (Palmgren et al., 2008). Although essential for completing a plant’s life cycle, it is toxic above threshold concentrations and must, therefore, be maintained within a narrow adequate range. This is regulated in plant cells by specialized transporters, controlling the distribution of Zn within the cell. Those at the plasma membrane transport Zn into or out of the cell. Others, positioned at subcellular membranes, compartmentalize Zn when concentrations within the cellular fluid become toxic; this often involves transport to the vacuole.
An array of transporter families exists, allowing plants to cope with varying substrate availabilities, enabling responses to a variety of stress conditions, and meeting specific transport requirements (Hall and Williams, 2003). Cation diffusion facilitators (CDFs) are ubiquitous in all branches of life (Nies and Silver, 1995; Montanini et al., 2007; Gustin et al., 2011). Structural analysis suggests that most family members possess six transmembrane domains (TMDs), with cytoplasmic N- and C-termini (Paulsen and Saier, 1997). Key polar and charged residues conserved within TMDs I, II, V, and VI are likely to be involved in metal transport (Gaither and Eide, 2001; Haney et al., 2005). Some members contain a histidine-rich cytoplasmic loop, thought to be vital for transporter specificity, which might act as a chaperone to determine the identity of metal ions to be transported (Kawachi et al., 2008, 2012; Podar et al., 2012). A signature sequence, proposed by Paulsen and Saier (1997) and modified by Montanini et al. (2007), enables predictions regarding uncharacterized CDF family members. Members of this family cluster phylogenetically according to their main substrate: Zn, Zn and Fe, or manganese (Mn). It has been suggested that metal selectivity of uncharacterized members can be inferred according to their cluster position (Montanini et al., 2007).
In plants, CDFs are known as metal tolerance proteins (MTPs; Montanini et al., 2007). MTP genes have been cloned from a number of plant species and are shown to have a role in heavy metal transport (Blaudez et al., 2003; Kim et al., 2004; Kobae et al., 2004; Delhaize et al., 2007; Peiter et al., 2007; Kawachi et al., 2008; Podar et al., 2012). Twelve MTP genes have been classified in Arabidopsis and 10 in rice (Gustin et al., 2011). The first, identified as ZAT or Zinc Transporter of Arabidopsis (van der Zaal et al., 1999), was renamed AtMTP1. Overexpression in Arabidopsis enhances Zn resistance (van der Zaal et al., 1999) while T-DNA insertion (Kobae et al., 2004) or RNA interference (RNAi)-mediated silencing (Desbrosses-Fonrouge et al., 2005) increases Zn sensitivity. The Zn hyperaccumulator plant Arabidopsis halleri had a pentaplication of the MTP1 gene during the evolutionary process, which is believed to have a role in Zn hypertolerance (Shahzad et al., 2010). AtMTP3 is also thought to be involved in Zn transport (Arrivault et al., 2006), while AtMTP11 transports Mn (Delhaize et al., 2007; Peiter et al., 2007).
The rice orthologue OsMTP1 was recently characterized by Yuan et al. (2012) and Lan et al. (2012). Located on chromosome 5, it is most highly expressed in mature leaves and stem (Yuan et al., 2012). Both overexpression and RNAi-mediated silencing suggest a role for the transporter in Zn, Cd, and nickel (Ni) movement, a hypothesis strengthened by functional complementation of yeast mutants (Yuan et al., 2012). However, there is controversy over OsMTP1 localization, reported at the plasma membrane when expressed in onion epidermal cells (Yuan et al., 2012) or at the vacuole when expressed in Saccharomyces cerevisiae (Lan et al., 2012).
This study aims to clarify the membrane localization of OsMTP1 and investigate the importance of key residues in transport ability and specificity. It is shown here that OsMTP1 is localized to the vacuole when stably expressed in planta. It is further demonstrated that OsMTP1 is a Zn transporter that can also transport cobalt (Co), Fe, and Cd. Moreover, it is shown that single residue substitutions can alter substrate specificity.
Materials and methods
Growth of rice plants (Nipponbare cultivar) for leaf RNA extraction and OsMTP1 amplification
Rice seeds (cultivar Nipponbare) were germinated for 4 d in an incubator at 28 °C, on filter paper soaked with distilled water, and transferred to holders positioned over plastic pots with 5 litres of nutrient solution (16 seedlings per pot). The nutrient solution contained 700 μM K2SO4, 100 μM KCl, 100 μM KH2PO4, 2mM Ca(NO3)2, 500 μM MgSO4, 10 μM H3BO3, 0.5 μM MnSO4, 0.5 μM ZnSO4, 0.2 μM CuSO4, 0.01 μM (NH4)6 Mo7O24, and 100 μM Fe3+-EDTA (as described by Kobayashi et al., 2005). The pH of the nutrient solution was adjusted to 5.4 by addition of 0.5mol l–1 NaOH. Plants were kept at 28±1 °C under a photoperiod of 16h/8h light/dark (150 μmol m–2 s–1) for 10 d. Solutions were replaced every 3–4 d and leaf samples were harvested for RNA extraction (described later).
Growth of Arabidopsis plants
Arabidopsis plants were grown as previously described (Mills et al., 2010; Jaffé et al., 2012) in soil containing equal proportions of vermiculite, Levingtons M2, and John Innes No. 2 compost (Fargro) in 8cm pots; 0.28g l–1 Intercept insecticide (Bayer, Canada) was present and soil was sterilized by autoclaving at 121 °C for 15min at 1 bar pressure. Wild-type (ecotype Wassilewskija), mtp1-1 mutant, and transgenic lines expressing OsMTP1–GFP (green fluorescent protein) were grown in a controlled-environment growth room with a day/night cycle (23 °C 16h light, 120 μmol m–2 s–1; 18 °C 8h dark). The T-DNA knockout mtp1-1 mutant (Kobae et al., 2004) was kindly provided by Professor Masayoshi Maeshima (Nagoya University, Japan).
Metal tolerance assay in Arabidopsis plants
These assays were similar to those previously described (Mills et al., 2008). Arabidopsis thaliana seed from wild-type (ecotype Wassilewskija), mtp1-1 mutant, and transgenic T3 homozygous lines expressing OsMTP1-GFP (lines 1 and 2) were sterilized in 15% (v/v) bleach for 15min and rinsed five times with sterile water. They were then inoculated onto plates containing 0.8% (w/v) agarose (Melford), 1% (w/v) sucrose, and half-strength Murashige and Skoog medium (Murashige and Skoog, 1962) with a range of Zn concentrations (provided as ZnSO4), from 0 to 500 μM. Seeds were stratified at 4 °C for 48h prior to transfer to a controlled-environment cabinet (22 °C, 16h light, 110 μmol m–2 s–1; 18 °C, 8h dark), and plates were incubated vertically. Fresh weight and chlorophyll determinations were performed as described below.
Amplification and cloning of OsMTP1
The cDNA sequence of O. sativa OsMTP1 (accession no. AY266290/LOC_Os05g03780) is found in GenBank™. Total RNA from rice leaves was extracted using the Concert Plant RNA Reagent (Invitrogen) and treated with DNase I (Invitrogen). First-strand cDNA synthesis was performed with oligo(dT) and reverse transcriptase (M-MLV, Invitrogen) using 1 μg of RNA. The OsMTP1 full-length coding sequence was amplified from cDNA with Pfu DNA polymerase (Promega) using forward primer 5′-CACCATGGACAGCCATAACTCAGCA and reverse primer 5′-CTACTCGCGCTCAATCTGAAT. In addition, reverse primer 5′-CTCGCGCTCAATCTGAATG was used to amplify a fragment without the stop codon referred to as non-stop (NS). PCR products were cloned into the entry vector pENTR/D-TOPO (Invitrogen), using Gateway technology. Sequencing confirmed the nucleotide fidelity of OsMTP1 in the entry vector.
Expressing OsMTP1 in the Arabidopsis mtp1-1 mutant
Using Gateway technology, OsMTP1(NS) was transferred from the entry clone to the pMDC83 destination vector (for C-terminal GFP tagging), by LR recombination. Sequencing of expression clones was carried out to confirm correct transfer. Plasmids were transformed into Agrobacterium tumefaciens GV3101 by electroporation. Plants (T-DNA knockout mtp1-1 mutant; Kobae et al., 2004) were transformed using the floral dip method (Clough and Bent, 1998) but including a 3h pre-induction of vir genes by addition of 100 μM acetosyringone to the culture before dipping. Homozygous T3 plants were used for analysis.
Reverse transcription–PCR (RT–PCR) was used to confirm expression of OsMTP1 in Arabidopsis. To do this, RNA was isolated from 11-day-old seedlings grown on 0.5 MS agar plates. cDNA synthesis and RT–PCR were carried out as previously described (Mills et al., 2008) except that the primers used to detect OsMTP1 were forward primer 5′-CACCATGGACAGCCATAACTCAGCA and reverse primer 5′-CTACTCGCGCTCAATCTGAAT.
Fresh weight and chlorophyll measurements
Fresh weight and chlorophyll concentrations were determined for A. thaliana seedlings of wild-type (ecotype Wassilewskija), mtp1-1 mutant, and T3 homozygous lines expressing OsMTP1–GFP (lines 1 and 2), as previously described (Mills et al., 2008). Seedlings were grown (as described above in the metal tolerance assays in Arabidopsis) on six separate plates for a range of Zn concentrations, each plate having four wild-type seedlings, four mtp1-1 mutant seedlings, and four of each transgenic line. Following growth, the seedlings were removed from the plates using forceps and weighed (four seedlings per genotype), and placed in a tube for chlorophyll determination. Chlorophyll was determined after harvesting whole seedlings following extraction in N,N-dimethylformamide (DMF) (Moran and Porath, 1980). The data presented are from a representative experiment (conducted at least twice) and are the means from the six plates ±SE expressed on a per seedling basis.
OsMTP1 constructs for yeast expression, site-directed mutagenesis, and yeast transformation
The entry clones pENTR OsMTP1 full-length and pENTR OsMTP1(NS) (described above) were used to introduce the OsMTP1 cDNAs into the yeast expression vector pAG426-GFP (Alberti et al., 2007) by LR recombination, Gateway technology (Invitrogen). Site-directed mutagenesis using OsMTP1(NS) from the entry vector as the DNA template was performed using the QuikChange® II XL Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer’s instructions. The mutations generated are described in detail in the Results, and the primers used for generating the mutations are presented in Supplementary Table S1 available at JXB online. All mutations were confirmed by DNA sequencing and recombined into pAG426-GFP vector as described above. Constructs were transformed into S. cerevisiae: wild-type BY4741 (MATa, his3-1, leu2-0, met15-0, ura3-0) and zrc1 cot1 double mutant (MATa; his3-1, leu2-0, met15-0, ura3-0, zrc1::natMX cot1::kanMX4) for Zn and Co complementation analyses; wild-type DY150 [MATa ura3-52, leu2-3, 112, trp1-1, his3-11, 15, ade2-1, can1-100(oc)] and ccc1 mutant [MATa ura3-52, leu2-3, 112, trp1-1, his3-11, 15, ade2-1, can1-100(oc) Δccc1::HIS3] for Fe complementation analyses; and wild-type BY4741 and ycf1 mutant (MATa his3-1, leu2-0, lys2-0, ura3-0, YDR135c::kanMX4) for Cd complementation analyses. The Zn-hypersensitive zrc1 cot1 mutant was obtained from Dr U. Kramer (Ruhr University, Bochum), while other mutants were obtained from Euroscarf. Yeast transformation was performed using the LiOAc/PEG method (Gietz et al., 1992). Transformants were selected on SC (synthetic complete) medium without uracil (5g l–1 ammonium sulphate, 1.7g l–1 yeast nitrogen base, 1.92g l–1 yeast synthetic drop-out medium supplement without uracil; Sigma, UK) with 2% glucose (w/v), 2% (w/v) agar (Difco technical) (adjusted to pH 5.3 before addition of agar and prior to autoclaving) (Mills et al., 2010, 2012). Plates were incubated at 30 °C for 3 d.
Metal tolerance assays in yeast
For metal sensitivity tests, yeast cultures were grown overnight at 30 °C in selective liquid medium, SC without uracil (described above). Overnight cultures were centrifuged for 4min at 4000rpm and resuspended in SC galactose liquid medium (pH 5.0) with 2% galactose (w/v) in place of glucose, before further incubation for 4h (30 °C, 200rpm) (Mills et al., 2010, 2012). Yeast cultures were diluted to the same OD600 (~0.4) in SC liquid medium without uracil, 2% (w/v) galactose. Aliquots were inoculated onto SC without uracil, 2% (w/v) agar (Difco technical), 2% (w/v) galactose containing various concentrations of different metals (ZnSO4, CoCl2, FeSO4, CdCl2 and MnCl2, Sigma, UK); adjusted to pH 5.3 before addition of agar and prior to autoclaving. Inoculated plates were incubated at 30 °C for 4–6 d depending on the growth of yeast colonies.
Localization studies in yeast
Subcellular localization of wild-type and mutated proteins in yeast was assessed using in-frame C-terminal fusions with GFP in the pAG426-GFP vector (Alberti et al., 2007). To induce expression of the fusion proteins, yeast cells were pre-grown overnight in SC medium containing glucose to mid-log phase. The cells were switched to SC medium containing galactose and grown for 24h, then were processed for fluorescence microscopy. The GFP fluorescence of yeast was observed using fluorescence microscopy under an Olympus FluoView 1000 confocal laser scanning system. GFP excitation, 488nm; detection, 505–530nm. Images were captured with a high-sensitivity photomultiplier tube detector.
Localization studies in A. thaliana
Seeds of both T3 homozygous lines expressing OsMTP1 with a C-terminal GFP tag (line 1 and 2) and the wild type (ecotype Wassilewskija) were surface sterilized in 15% (v/v) bleach for 15min, rinsed five times with sterile water, and inoculated onto plates containing 0.8% (w/v) agarose (Melford), 1% (w/v) sucrose, and 0.5 MS medium (Murashige and Skoog, 1962) as previously described (Mills et al., 2008). Seed were stratified at 4 °C for 48h prior to transfer to a controlled-environment cabinet (22 °C, 16h light, 110 μmol m–2 s–1; 18 °C, 8h dark) and plates were incubated vertically for 7 d. Whole seedlings were placed on the slide in water and covered with a cover slide for imaging using an Olympus FluoView 1000 confocal laser scanning system. GFP, propidium iodide (PI), and chlorophyll were excited using the 488nm line of an argon ion laser. GFP and PI emission were detected at 505–530nm and 610–650nm, respectively; chlorophyll autofluorescence was detected using an LP580 filter. Cell wall staining was performed with PI (Invitrogen) using a 19.5 μm solution in water. Plants were immersed in the solution for 5min and rinsed three times with sterile water before analysis.
Bioinformatics
OsMTP1 transmembrane domains were predicted using online helix-prediction programmes TMHMM (Krogh et al., 2001) (http://www.cbs.dtu.dk/services/TMHMM/) and Phobius (Kall et al., 2004) (http://phobius.sbc.su.se/). The subsequent topology was created according to their predictions. Sequence alignment and phylogenetic analyses for MTP1 proteins were performed using the MEGA (Molecular Evolutionary Genetics Analysis) 4.1 package (Tamura et al., 2007). Protein multiple alignments were obtained with ClustalW2, and phylogenetic trees were reconstructed with the Neighbor–Joining method and the following parameters: pairwise deletion option, 1000 replicates of bootstrap, and Poisson correction. The consensus tree shows only branches with a bootstrap consensus >50%.
Results
Sequence analysis of OsMTP1
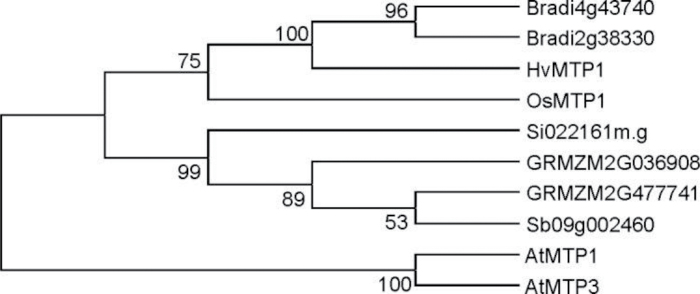
The full-length cDNA fragment of OsMTP1 (accession no. AY266290 /LOC_Os05g03780) was amplified by high-fidelity RT–PCR based on GenBank™ and the Rice Annotation Project information from NCBI (http://www.ncbi.nlm.nih.gov/nuccore). It encodes a protein with 418 amino acids. Percentage identities and similarities between OsMTP1, AtMTP1, and HvMTP1 are shown in Table 1. The highest identity (85%) and similarity (90%) scores at the amino acid level were observed between OsMTP1 and HvMTP1. MTP amino acid sequences were searched for in the genomes of six monocotyledons, Sorghum bicolor, Zea mays, Setaria italica, Hordeum vulgare, O. sativa, and Brachypodium distachyon, and a phylogenetic tree including A. thaliana MTP1 and MTP3 sequences was generated (Fig. 1). In the search, it is clear that O. sativa has only one MTP1 sequence, while Z. mays and B. distachyon have two, indicating that the appearance of new MTP1 sequences occurred after the speciation of each monocot. The multiple sequence alignment of AtMTP1, OsMTP1, and HvMTP1 proteins is shown in Fig. 2; positions with a single, fully conserved residue are highlighted, as are those with conserved similar properties. TMDs predicted by Phobius (http://phobius.sbc.su.se/) are also marked in Fig. 2. The proteins are predicted to possess cytoplasmic N- and C-termini and six TMDs, with a small cytoplasmic loop between TMDs II and III, and a large cytoplasmic, histidine-rich loop between TMDs IV and V. OsMTP1 possesses the loop with the most histidine residues, followed by HvMTP1. The 17 residues of the CDF signature sequence (Montanini et al., 2007) are identical for each sequence, and are highlighted in Fig. 2.
Table 1.
Analyses of protein sequence similarity and identity between AtMTP1, OsMTP1, and HvMTP1. Calculated using the EMBOSS program Matcher. Scores are given as percentage similarity and percentage identity. Accession numbers: AtMTP1, NM130246; OsMTP1, AY266290; HvMTP1, AM286795.
| Similarity | Identity | |
|---|---|---|
| OsMTP1/HvMTP1 | 90 | 85 |
| OsMTP1/AtMTP1 | 77 | 70 |
| HvMTP1/AtMTP1 | 77 | 68 |
Fig. 1.
Phylogenetic tree showing the relationships between MTP1 protein sequences from six monocotyledons. Brachypodium distachyon (Bradi4g43740 and Bradi2g38330), Hordeum vulgare (HvMTP1), Oryza sativa (OsMTP1), Setaria italica (Si022161m.g), Zea mays (GRMZM2G036908 and GRMZM2G477741), and Sorghum bicolor (Sb09g002460), plus Arabidopsis thaliana MTP1 and MTP3 sequences. The tree was generated with MEGA 4.1 software. Bootstrap values from 1000 replicates using the Neighbor–Joining method are indicated at each node when the method agrees with the tree topology.
Fig. 2.
Multiple sequence alignment of OsMTP1, HvMTP1, and AtMTP1 generated by the ClustalW2 program. Black with white letters/asterisk, fully conserved residues between sequences; dark grey with white letters/colon, conservation of residues with strongly similar properties; pale grey with black letters/full stop, conservation of residues with weakly similar properties. Transmembrane domains predicted by Phobius are marked by boxes. The CDF signature sequence is underlined.
OsMTP1 complements the Zn sensitivity of the A. thaliana mtp1-1 mutant
The T-DNA insertion mutant of A. thaliana (Wassilewskija) mtp1-1 is sensitive to elevated Zn, suffering stunted growth and chlorosis compared with the wild type (Fig. 3A). These differences are significant at upward of 100 μM Zn, as indicated by fresh weight and chlorophyll determinations (Fig. 3B). Resistance to Zn is restored when mtp1-1 is transformed with OsMTP1–GFP under the control of the constitutive 35S promoter. Lines 1 and 2 are homozygous T3 lines expressing a C-terminally GFP-tagged version (35S:OsMTP1-GFP:mtp1-1). Similar results were obtained when OsMTP1 without a GFP tag was expressed in the mtp1-1 mutant (results not shown), indicating that GFP tagging does not interfere with the rescue. This also indicates that the localization of OsMTP1 in planta can be investigated using the GFP-tagged construct.
Fig. 3.
Functional complementation of the Arabidopsis knockout mutant mtp1-1 by OsMTP1–GFP. (A) Representative 18-day-old seedlings on low (15 μM) and high (350 μM) Zn-containing media. (B) (i) Bar chart of average fresh weight (mg) per seedling; significance calculated using paired t-test. #significant difference from mtp1-1; *significant difference from the wild type (A. thaliana ecotype Wassilewskija) (P ≤ 0.05); black, wild type; dark grey, mtp1-1; pale grey, 35S:OsMTP1-GFP:mtp1-1 line 1; white, 35S:OsMTP1-GFP:mtp1-1 line 2. (ii) Bar chart of average total chlorophyll content (μg) per seedling; significance calculated using paired t-test. See part (i) for explanation of keys.
OsMTP1 localizes to the vacuolar membrane when stably expressed in A. thaliana
The OsMTP1–GFP-expressing lines that showed rescue of the Arabidopsis mtp1-1 mutant phenotype were used to determine the membrane localization of OsMTP1. Confocal images show that OsMTP1 is localized in the vacuolar membrane of plant cells (Fig. 4, upper). Different cells were analysed, including cells in the root tip region containing small immature vacuoles (Fig. 4, upper A), elongated root cells with a large central vacuole (Fig. 4, upper B–D), and cotyledon cells containing chloroplasts (Fig. 4, upper E). All show the same vacuolar localization pattern.
Fig. 4.
OsMTP1 localizes to the vacuole when expressed in Arabidopsis and yeast. Upper: (A) OsMTP1 vacuolar membrane localization in the A. thaliana mtp1-1 mutant expressing OsMTP1–GFP. The red fluorescence is caused by cell walls stained with propidium iodide (A–D). GFP vacuolar fluorescence (green) in cells of the root tip region, containing small immature vacuoles (A) and in elongated root cells with a large central vacuole (B–D). Scale bar=10 μm. GFP vacuolar fluorescence in cotyledon cells containing chloroplasts (arrow) (E). Scale bar=25 μm. Lower: OsMTP1 localizes to the vacuoles of zrc1 cot1 yeast cells expressing OsMTP1–EGFP, as shown via confocal microscopy. The OsMTP1–EGFP (green) co-localizes with vacuole organelles, which appear as a depression in the DIC image. Scale bar=5 μm. (B) EGFP alone is distributed throughout the cytoplasm. Scale bar=5 μm.
OsMTP1 localizes to the vacuole of S. cerevisiae
In order to study the subcellular localization of OsMTP1, it was cloned into the pAG426-GFP vector, which fuses a GFP tag to the C-terminus of the gene to enable fluorescence localization studies (Alberti et al., 2007). zrc1 cot1 yeast cells expressing the OsMTP1–GFP construct were analysed via confocal microscopy which showed that OsMTP1–GFP co-localizes with the vacuoles, which appear as a depression in the differential interference contrast (DIC) image (Fig. 4, lower). The empty vector gives a GFP signal throughout the cytoplasm (Fig. 4, lower B).
Functional analysis of OsMTP1 in yeast: metal specificity and site-directed mutations
Wild-type and mutant OsMTP1 cDNAs were cloned into the pAG426-GFP vector. OsMTP1 constructs with and without a stop codon were used so that the activity of OsMTP1 with and without a C-terminal GFP tag could be compared when expressed in yeast using the same expression vector. Only the OsMTP1 without a stop codon (indicated by an asterisk in the figures) should produce the OsMTP1–GFP fusion protein. Having such a tag facilitates localization studies and therefore allowed the evaluation of whether particular mutations had an effect on subcellular localization. Although reducing the transport function slightly, GFP tagging did not change the specificity observed in the metal tolerance tests when compared with the non-tagged OsMTP1 protein. These results suggest that OsMTP1 is functionally active with the GFP tag at the C-terminal region. Mutated versions of OsMTP1 with a C-terminal GFP tag also localized to the vacuole, indicating that any differences observed in transport function are not due to altered localization (Supplementary Fig. S1 at JXB online).
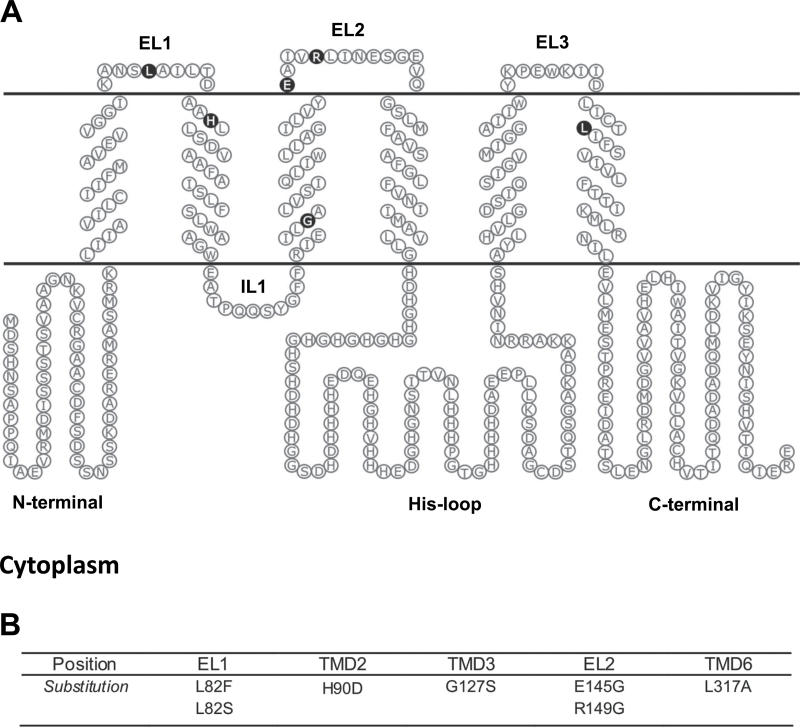
The transmembrane data predicted by Phobius and TMHMM were used to construct the hypothetical membrane topology of OsMTP1 in Fig. 5A. Seven mutations were tested for their influence on the transport function of OsMTP1 (Fig. 5B). The residues were selected based on conservation between CDF family members and also because mutation of some may confer a gain of function (Blaudez et al., 2003; Lin et al., 2009; Hoch et al., 2012). Figure 5A also highlights the substitution sites generated during site-directed mutagenesis, to demonstrate their positions within the protein.
Fig. 5.
(A) Hypothetical membrane topology of OsMTP1 predicted according to the Phobius program. Residues highlighted in black are the sites chosen for OsMTP1 site-directed mutagenesis. EL, extracytosolic loop; IL, intracytosolic loop. (B) Substitutions resulting from site-directed mutations of OsMTP1.
Zn and Co assays in the zrc1 cot1 yeast mutant
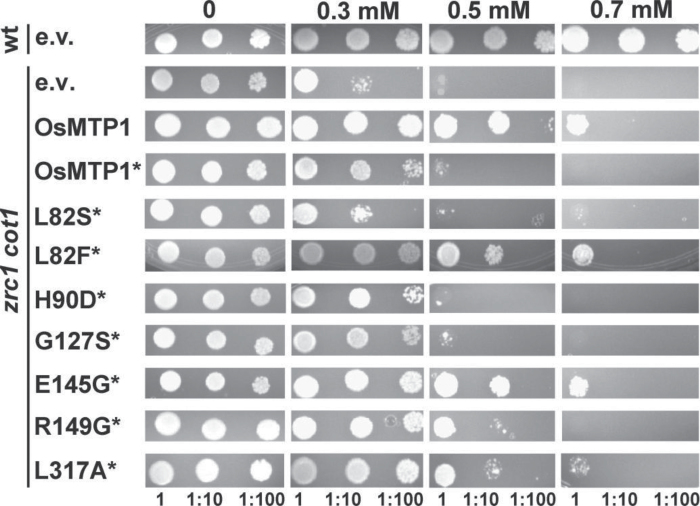
The zrc1 cot1 yeast mutant shows both Zn and Co susceptibility because of lack of the ZRC1 and COT1 genes (Figs 6, 7). Heterologous expression of OsMTP1 fully rescues the mutant phenotype at 10mM Zn (Fig. 6). OsMTP1–GFP also shows rescue of the Zn-sensitive phenotype, with slightly decreased growth at 5mM and 10mM Zn compared with the non-tagged OsMTP1, but was as competent at lower concentrations. Compared with OsMTP1–GFP, all of the mutations reduced the rescue of the Zn-sensitive phenotype to some extent, except R149G. This is obvious in L82S and L82F, but most apparent in H90D, which fails to rescue the Zn sensitivity of zrc1 cot1 on 0.25mM Zn (Fig. 6).
Fig. 6.
Functional analysis of OsMTP1 in the Zn-sensitive Saccharomyces cerevisiae mutant zrc1cot1. BY4741 was transformed with pAG426-GFP empty vector (e.v.); zrc1 cot1 was transformed with pAG426-GFP vector either empty or expressing OsMTP1 with or without stop codon (*). The latter generates an OsMTP1–GFP fusion protein. L82S to L317A refer to site-directed substitutions generated within OsMTP1 without a stop codon (*). Numbers represent serial dilutions of yeast cells in liquid SC galactose without uracil: undiluted (1) OD600=0.4, 1:10, and 1:100, sequentially dropped onto and grown on SC galactose without uracil drop-out medium with different concentrations of ZnSO4. Plates were incubated at 28 °C for 5 d.
In contrast to the results from Yuan et al. (2012), it is shown here that OsMTP1 does have Co transport function. OsMTP1 shows partial rescue of Co sensitivity at all concentrations tested (Fig. 7), but wild-type growth is far from being restored. The GFP tag again impacts the rescuing ability of OsMTP1 (Fig. 7). The rescue of the Co-sensitive phenotype is less obvious than that of Zn. L82S again reduces the rescuing ability of OsMTP1, whereas L82F, E145G, R149G, and L317A improve Co transport relative to the non-mutated OsMTP1–GFP construct.
Fig. 7.
Functional analysis of OsMTP1 in the Co-sensitive Saccharomyces cerevisiae mutant zrc1 cot1. BY4741 was transformed with pAG426-GFP empty vector (e.v.); zrc1 cot1 was transformed with pAG426-GFP vector either empty or expressing OsMTP1 with or without a stop codon (*). L82S to L317A refer to site-directed substitutions generated within OsMTP1 without a stop codon (*). Numbers represent serial dilutions of yeast cells in liquid SC galactose without uracil: undiluted (1) OD600=0.4, 1:10, and 1:100, sequentially dropped onto and grown on SC galactose without uracil drop-out medium with different concentrations of CoCl2. Plates were incubated at 28 °C for 5 d.
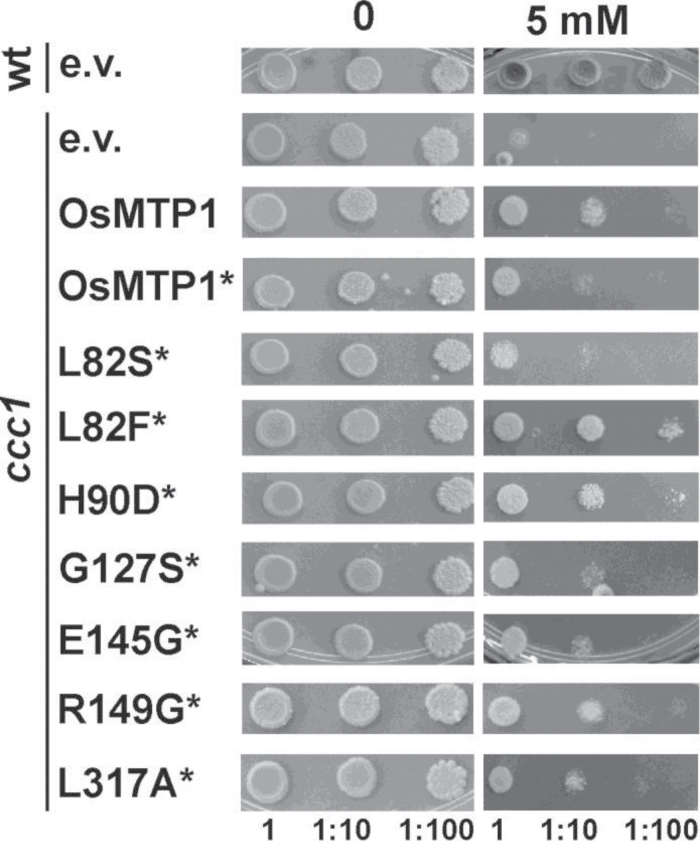
Fe assays in the ccc1 yeast mutant
The Fe transport-deficient ccc1 mutant lacks a vacuolar Fe sequestration function, demonstrating an Fe-sensitive phenotype at high Fe. Growth of ccc1 transformed with an empty pAG426-GFP vector is inhibited on 5mM Fe compared with the wild type (Fig. 8). OsMTP1 shows rescue of ccc1 on 3.5mM (data not shown) and 5mM Fe (Fig. 8), indicating that OsMTP1 can transport iron in yeast. GFP tagging slightly reduces the rescuing ability of OsMTP1 when compared with the non-tagged version of OsMTP1 (Fig. 8). L82F, H90D, and R149G mutations improve Fe transport in ccc1. L82S slightly enhances the sensitive phenotype of ccc1, with poorer growth observed compared with OsMTP1–GFP.
Fig. 8.
Functional complementation of the Fe-sensitive Saccharomyces cerevisiae mutant ccc1. DY150 was transformed with pAG426-GFP empty vector (e.v.); ccc1 was transformed with pAG426-GFP vector either empty or containing OsMTP1 with or without a stop codon (*). L82S to L317A refer to site-directed substitutions generated within OsMTP1 without a stop codon (*). Numbers represent serial dilutions of yeast cells in liquid SC galactose without uracil: undiluted (1) OD600=0.4, 1:10, and 1:100, sequentially dropped onto and grown on SC galactose without uracil drop-out medium with 5mM FeSO4. Plates were incubated at 28 °C for 5 d.
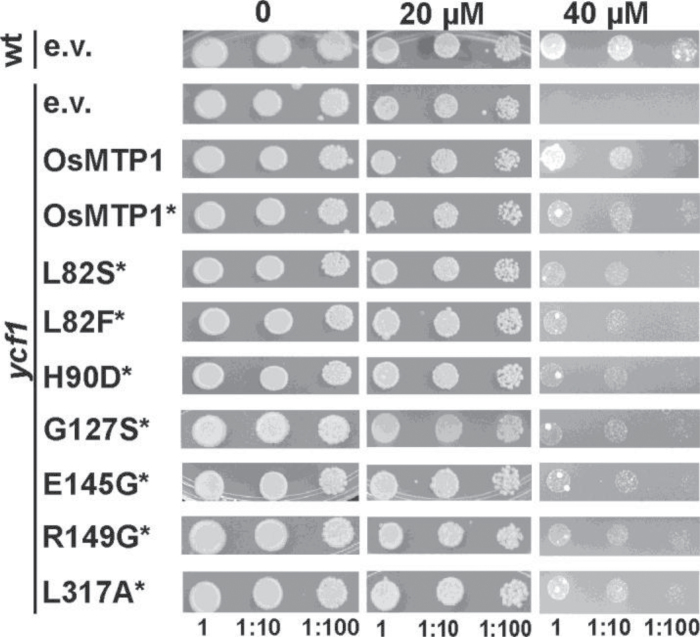
Cd assay in the ycf1 yeast mutant
The Cd-hypersensitive ycf1 mutant transformed with OsMTP1 clearly confers tolerance to low levels of Cd (Fig. 9), indicating that OsMTP1 can transport Cd in yeast. For all metals tested, expressing OsMTP1–GFP consistently reduces transport when compared with the non-tagged construct, and on Cd this reduction is very obvious (Fig. 9). As the mutations were performed in the OsMTP1–GFP construct, their effects on Cd transport are not conclusive, but it seems that none of the mutations have an effect (Fig. 9). To confirm this, L82F and H90D mutations were tested without GFP tagging, but no difference was noticed in ycf1 complementation compared with the non-mutated OsMTP1 (data not shown).
Fig. 9.
Functional complementation of the Cd-sensitive Saccharomyces cerevisiae mutant ycf1. BY4741 was transformed with pAG426-GFP empty vector (e.v.); ycf1 was transformed with pAG426-GFP vector either empty or containing OsMTP1 without a stop codon (*). L82S to L317A refer to site-directed substitutions generated within OsMTP1 without a stop codon (*). Numbers represent serial dilutions of yeast cells in liquid SC galactose without uracil: undiluted (1) OD600=0.4, 1:10, and 1:100, sequentially dropped onto and grown on SC galactose without uracil drop-out medium with different concentrations of CdCl2. Plates were incubated at 28 °C for 5 d.
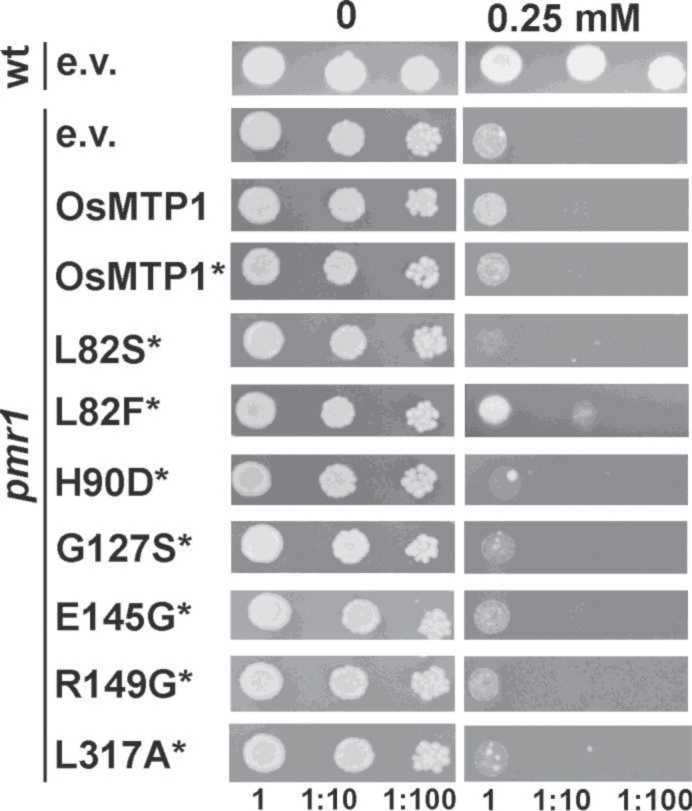
Mn assay in the pmr1 yeast mutant
OsMTP1 does not confer Mn tolerance to the Mn-hypersensitive pmr1 mutant (Fig. 10). Of all mutations tested, it was observed that L82F results in a gain of function for Mn complementation on low Mn concentrations (0.25mM) (Fig. 10).
Fig. 10.
Functional complementation of the Mn-sensitive Saccharomyces cerevisiae mutant pmr1. K601 was transformed with pAG426-GFP empty vector (e.v.); pmr1 was transformed with pAG426-GFP vector either empty or containing OsMTP1 without a stop codon (*). L82S to L317A refer to site-directed substitutions generated within OsMTP1 without a stop codon (*). Numbers represent serial dilutions of yeast cells in liquid SC galactose without uracil: undiluted (1) OD600=0.4, 1:10, and 1:100, sequentially dropped onto and grown on SC galactose without uracil drop-out medium with 0.25mM of MnCl2. Plates were incubated at 28 °C for 5 d.
Summary of the impact of mutations on substrate specificity of OsMTP1
Table 2 summarizes the impact of the mutations on the functional complementation of metal-sensitive yeast mutants by OsMTP1. This includes the positioning of residues within the protein predicted by Phobius and TMHMM, also highlighted in Fig. 2.
Table 2.
Summarizing the effects of site-directed mutations on OsMTP1 substrate selectivity. Amino acid substitutions within the OsMTP1 protein and their position relative to predicted transmembrane domains (TMDs) and loops (EL; extracytosolic loop). Zinc- and cobalt-transporting abilities were assayed by complementation of zrc1cot1 cells on ZnSO4 and CoCl2; iron-transporting ability was assayed by complementation of ccc1 cells on FeSO4; cadmium-transporting ability was assayed by complementation of ycf1 cells on CdCl2; and manganese-transporting ability was assayed by complementation of pmr1 cells on MnCl2.
| Mutation | Location | Complementation of transport defect: | ||||
|---|---|---|---|---|---|---|
| Zn | Co | Fe | Cd | Mn | ||
| L82F | EL1 | xx | ++ | +++ | – | + |
| L82S | EL1 | xxx | x | x | – | – |
| H90D | TMD2 | xxxx | – | ++ | – | – |
| G127S | TMD3 | x | – | – | – | – |
| E145G | TMD3 | x | ++ | – | – | – |
| R149G | TMD3 | – | + | + | – | – |
| L317A | TMD6 | x | + | – | – | – |
x, reduction in growth compared with non-mutated OsMTP1; +, increase in growth; – no difference in growth.
All mutations negatively impact Zn transport ability when compared with the non-mutated OsMTP1–GFP construct, except for R149G; this mutation maintains Zn transport levels and enhances Co and Fe transport, but has no effect on Cd and Mn complementation. L82S shows considerable reduction in Zn, Fe, and Co transport ability. A similar reduction in growth was seen for G127S on Zn-containing media, although no further effect was seen on Co, Fe, Cd, and Mn. All other substitutions have no effect on Cd, but enhance transport of Co or Fe: E145G and L317A both show increased survival on Co. The most notable mutations are L82F and H90D. L82F caused considerable reduction in Zn transport, failing to rescue zrc1cot1 above 1mM Zn (Fig. 6) but resulting in a considerable increase in both Co and Fe transport and a gain of function for Mn. H90D appears to have abolished Zn transport, with certainly no rescue of zrc1cot1 on or above 0.25mM Zn; the substitution shows enhanced survival on Fe.
Discussion
Transport function of OsMTP1
OsMTP1 transports Zn but also Co, Fe, and Cd in yeast, functioning at the vacuole
Expression of OsMTP1 in zrc1 cot1 fully rescues the Zn-sen sitive phenotype, suggesting considerable Zn-transporting ability of OsMTP1. Partial rescue of zrc1 cot1 on Co, ccc1 on Fe, and ycf1 on Cd suggests that while OsMTP1 may primarily transport Zn it can also transport Co, Fe, and Cd in yeast. OsMTP1 was also previously shown to transport Ni (Yuan et al., 2012). The vacuolar localization observed here in yeast is consistent with OsMTP1 functioning at the tonoplast, compartmentalizing primarily Zn, but also Co, Fe, and Cd in the vacuole, and serving as a detoxification system for excess levels of these metals. When removed from the cytosol, these metals are less damaging and the mutant phenotype is partially or fully rescued.
The substrate specificity of OsMTP1 is considerably broader than that of the highly Zn-specific AtMTP1 (Kobae et al., 2004; Desbrosses-Fonrouge et al., 2005) and hybrid poplar PtdMTP1 (Blaudez et al., 2003). CDF family members are classified into three major groups (Zn2+, Fe2+/Zn2+, and Mn2+) based on their substrate specificity (Montanini et al., 2007). OsMTP1 separates phylogenetically with the Zn2+ group, but some members of this group do seem to be capable of transporting other metals. For example, NtMTP1 (Shingu et al., 2005) and HvMTP1 (Podar et al., 2012) have increased Co affinity, while TcMTP1 from the Zn/Cd-hyperaccumulating species Thlaspi caerulescens also transports non-essential Ni and Cd (Kim et al., 2004). Most of the plant MTP1 members characterized to date have not been tested for Fe transport. Here it is shown that OsMTP1 appears to have some Fe transport activity as it is able to reduce the Fe hypersensitivity of the yeast ccc1 mutant. Shingu et al. (2005) suggested that the broad substrate ability of NgMTP1 from hyperaccumulating Nicotiana glauca may be attributed to the number of histidine residues in the loop. OsMTP1 possesses more histidine residues in this loop than AtMTP1, but it remains to be tested whether this contributes to the broader substrate selectivity.
OsMTP1 transports Zn in A. thaliana and is expressed at the tonoplast
The present results suggest that OsMTP1 transports Zn when expressed in Arabidopsis. Consistent with previous findings, it is shown that the Arabidopsis mtp1-1 mutant is sensitive to high Zn concentrations (Kobae et al., 2004; Kawachi et al., 2009). Just as AtMTP1 under the control of the constitutive Cauliflower mosaic virus (CaMV) 35S promoter can be used to rescue this mutant (Kawachi et al., 2009), it is shown that it is also possible with OsMTP1, suggesting conservation of function. OsMTP1–GFP is shown here to localize to the tonoplast in Arabidopsis, and it is proposed that OsMTP1 is functioning in transporting Zn into the vacuole, thereby rescuing the Zn hypersensitivity of the Arabidopsis mtp1-1 mutant. Yuan et al. (2012) ascribed the localization of OsMTP1 to the plasma membrane, but the present results in Arabidopsis and in yeast both support a tonoplast localization, and this is consistent with the vacuolar localization observed for the closely related HvMTP1 from another cereal, barley (Podar et al., 2012).
L82 and H90 play important roles in OsMTP1 substrate specificity
None of the mutations studied here affected targeting, and OsMTP1 localized to the yeast vacuole in all cases. Owing to their reduced or unchanged transporting abilities, G127S and L82S mutants provide little information about selectivity, and loss of function may simply be due to destabilization of the tertiary structure (Lin et al., 2009). L82F, however, maintaining the non-polar environment of leucine with phenylalanine, appears to be a gain-of-function mutant; the protein can still transport low levels of Zn, with an enhanced affinity for Fe and Co and with a gain of function for Mn. L82 falls in the first non-cytoplasmic loop at the beginning of the CDF signature sequence (Montanini et al., 2007). It is equivalent to the L33F mutation that reduces Zn transport but increases Fe and Mn affinity in ScZRC1 (Lin et al., 2009). It may, therefore, be concluded that L82 is an important residue for metal selectivity in plant CDFs as well as in homologous yeast transporters.
Recent work failed to discover a single substitution that entirely shifts the substrate specificity of AtMTP1 from Zn (Kawachi et al., 2012; Podar et al., 2012). It was suggested that CDFs from higher organisms regulate transport specificity more tightly by holding more than one residue responsible for the confinement to specific substrates (Podar et al., 2012). However, it is shown here that the H90D mutation completely abolishes OsMTP1 Zn transport on the lowest concentrations tested (0.25mM) while improving Fe transport. H90, a polar histidine residue, falls within TMD II and the signature CDF sequence of OsMTP1; this residue in AtMTP1 has been proposed to form part of one of the Zn-binding sites (Kawachi et al., 2012). This residue has been mutated in CDFs of other organisms: PtdMTP1 (H89K and H89A; Blaudez et al., 2003); AtMTP1 (H90A; Kawachi et al., 2012), and in the mammalian metal transporters ZnT5 and ZnT8 (H451D and H106D, respectively; Hoch et al., 2012). The mutations abolished Zn transport but the effect on transport of other metals was not investigated in all cases. The H90A mutation in AtMTP1 abolishes Zn transport with no apparent rescue of Co tolerance. In ZnT5 and ZnT8, this mutation allows Cd transport and so it was concluded that this motif was important in discriminating between Zn and Cd, (Hoch et al., 2012). Although this may not be a general feature of CDFs/MTPs, this residue does seem to have a role in selectivity as it enhances Fe transport in OsMTP1.
Three further mutations that cause less extreme substrate changes are E145G, R149G, and L317A. E145G is homologous to E97G, a mutation in ScZRC1 that completely shifts the transported substrate from Zn to Fe and Mn (Lin et al., 2009). In contrast, the corresponding AtMTP1 mutant just extends its transport ability to Co and Mn (Podar et al., 2012) and to Co and Cd when mutated to the non-polar alanine, E145A (Kawachi et al., 2012). In none of the AtMTP1 studies cited above was the effect on Fe hypersensitivity investigated. In contrast to the results with ScZRC1, OsMTP1-E145G continues to transport Zn at a slightly lower level than the non-mutated construct, but with a slight increase in Fe transport and a marked enhancement of Co, demonstrating that this residue can influence OsMTP1 specificity. OsMTP1 containing the R149G mutation does not show a significant difference in Zn tolerance from the wild type; it does, however, enhance Co and Fe tolerance, indicating a distinctive contribution of Arg149 to Zn selectivity over Co and Fe. The homologous mutation in the ScZRC1 protein extends its transport to Fe and Mn (Lin et al., 2009). In AtMTP1, mutation of the homologous residue to cysteine, another non-polar amino acid, conferred tolerance to high levels of Co and Cd (Kawachi et al., 2012).
The leucine-zipper (LZ) motif is conserved in a range of CDF proteins from a number of species. It also appears to be present in OsMTP1, running from L317 to L338 and falling across TMD VI and the C-terminal tail. The OsMTP1 protein containing the L317A mutation continues to transport Zn with slightly lower affinity, while enhancing Co transport. Similarly for AtMTP1, a mutation in L298 (equivalent to L317 in OsMTP1) also has little or no effect in Zn tolerance but conferred Co and Cd gain of function in yeast experiments (Kawachi et al., 2012). This suggests that these leucine residues may have a role in ion selectivity that can be explained by AtMTP1 modelling experiments (Kawachi et al., 2012). The homologous residue in PtdMTP1, L293, falls at the beginning of the LZ motif. Blaudez et al. (2003) serially substituted leucine residues within the motif for alanine, maintaining the non-polar environment but losing the important side chain. These mutations had an increasingly negative effect on the Zn transport ability of PtdMTP1 further along the LZ motif. L293A had little impact, but L314A, the final leucine in the motif, resulted in a severe loss of Zn transport (Blaudez et al., 2003). The last leucine of the LZ motif is highly conserved in the CDF family. In AtMTP1, L319 is essential for protein function, and Kawachi et al. (2012) suggest that it may form a dimerization contact between two protomers, like L205, the corresponding residue of EcYiiP. This study confirms the importance of L317 in the substrate specificity of OsMTP1, and it would be interesting to perform the same mutation on the successive leucines within the sequence, opening up opportunities to explore the importance of the LZ motif.
To conclude, OsMTP1 is a vacuolar transporter that appears to transport Zn, but also Fe, Co, and Cd, perhaps with lower affinity. 35S:OsMTP1:mtp1-1 fully rescues the Zn-sensitive phenotype of the Arabidopsis knockout mutant mtp1-1, and it would be interesting also to test Co- and Fe-transporting abilities in planta. H90 and L82 seem to be important residues for determining the substrate specificity of OsMTP1. Other residues such as E145, R149, and L317 also appear to be related to substrate specificity. These new findings may be useful for defining strategies to generate plants suited for biofortification or phytoremediation applications.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. OsMTP1 site-directed mutations (H90D, L82F, L82S, G127S, E145G, R149, and L317A) with C-terminal GFP tagging were analysed for subcellular localization in zrc1 cot1 yeast cells as shown via confocal microscopy.
Table S1. List of primers used for creating OsMTP1 (NS) site-directed mutations.
Acknowledgements
We gratefully acknowledge Professor Masayoshi Maeshima (Nagoya University, Japan) for mtp1-1 mutant seeds and Professor Ute Kramer (Ruhr University, Bochum) for the zrc1cot1 strain. We thank Mrs Swaha Dhar (University of Southampton, UK) for technical assistance with parts of the project. This research was supported by a CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil), research fellowship to JPF and scholarships to FKR and PKM; by CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brazil), sandwich scholarship to PKM; and partly by funding to LEW from the Gerald Kerkut Trust and European Union Framework Programme 6 as part of the Integrated Project Public Health impact of long-term, low level mixed element exposure in susceptible population strata (PHIME) (contract no FOOD-CT-2006–016253); it reflects only the authors’ views. The Community is not liable for any use that may be made of the information contained therein and had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Glossary
Abbreviations:
- CDFs
cation diffusion facilitators
- DMF
N,N-dimethylformamide
- GFP
green fluorescent protein
- MTPs
metal tolerance proteins
- TMDs
transmembrane domains.
References
- Alberti S, Gitler AD, Lindquist S. 2007. A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast 24, 913–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrivault S, Senger T, Kramer U. 2006. The Arabidopsis metal tolerance protein AtMTP3 maintains metal homeostasis by mediating Zn exclusion from the shoot under Fe deficiency and Zn oversupply. The Plant Journal 46, 861–879. [DOI] [PubMed] [Google Scholar]
- Blaudez D, Kohler A, Martin F, Sanders D, Chalot M. 2003. Poplar metal tolerance protein 1 confers zinc tolerance and is an oligomeric vacuolar zinc transporter with an essential leucine zipper motif. The Plant Cell 15, 2911–2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Delhaize E, Gruber BD, Pittman JK, White RG, Leung H, Miao Y, Jiang L, Ryan PR, Richardson AE. 2007. A role for the AtMTP11 gene of Arabidopsis in manganese transport and tolerance. The Plant Journal 51, 198–210. [DOI] [PubMed] [Google Scholar]
- Desbrosses-Fonrouge AG, Voigt K, Schroder A, Arrivault S, Thomine S, Kramer U. 2005. Arabidopsis thaliana MTP1 is a Zn transporter in the vacuolar membrane which mediates Zn detoxification and drives leaf Zn accumulation. FEBS Letters 579, 4165–4174. [DOI] [PubMed] [Google Scholar]
- Gaither LA, Eide DJ. 2001. Eukaryotic zinc transporters and their regulation. Biometals 14, 251–270. [DOI] [PubMed] [Google Scholar]
- Gietz D, St, Jean A, Woods RA, Schiestl RH. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Research 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin JL, Zanis MJ, Salt DE. 2011. Structure and evolution of the plant cation diffusion facilitator family of ion transporters. BMC Evolutionary Biology 11, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JL, Williams LE. 2003. Transition metal transporters in plants. Journal of Experimental Botany 54, 2601–2613. [DOI] [PubMed] [Google Scholar]
- Haney CJ, Grass G, Franke S, Rensing C. 2005. New developments in the understanding of the cation diffusion facilitator family. Journal of Industrial Microbiology and Biotechnology 32, 215–226. [DOI] [PubMed] [Google Scholar]
- Hoch E, Lin W, Chai J, Hershfinkel M, Fu D, Sekler I. 2012. Histidine pairing at the metal transport site of mammalian ZnT transporters controls Zn2+ over Cd2+ selectivity. Proceedings of the National Academy of Sciences, USA 109, 7202–7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IRRI. 2010. International Rice Research Institute. Accessed 8 December 2012. Available at: http://www.irri.org/
- Jaffé FW, Freschet GE, Valdes BM, Runions J, Terry MJ, Williams LE. 2012. G protein-coupled receptor-type G proteins are required for light-dependent seedling growth and fertility in Arabidopsis. The Plant Cell 24, 3649–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kall L, Krogh A, Sonnhammer EL. 2004. A combined transmembrane topology and signal peptide prediction method. Journal of Molecular Biology 338, 1027–1036. [DOI] [PubMed] [Google Scholar]
- Kawachi M, Kobae Y, Mimura T, Maeshima M. 2008. Deletion of a histidine-rich loop of AtMTP1, a vacuolar Zn(2+)/H(+) antiporter of Arabidopsis thaliana, stimulates the transport activity. Journal of Biological Chemistry 283, 8374–8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawachi M, Kobae Y, Mori H, Tomioka R, Lee Y, Maeshima M. 2009. A mutant strain Arabidopsis thaliana that lacks vacuolar membrane zinc transporter MTP1 revealed the latent tolerance to excessive zinc. Plant and Cell Physiology 50, 1156–1170. [DOI] [PubMed] [Google Scholar]
- Kawachi M, Kobae Y, Kogawa S, Mimura T, Kramer U, Maeshima M. 2012. Amino acid screening based on structural modeling identifies critical residues for function, ion selectivity and structure of Arabidopsis MTP1. FEBS Journal 279, 2339–2356. [DOI] [PubMed] [Google Scholar]
- Kim D, Gustin JL, Lahner B, Persans MW, Baek D, Yun DJ, Salt DE. 2004. The plant CDF family member TgMTP1 from the Ni/Zn hyperaccumulator Thlaspi goesingense acts to enhance efflux of Zn at the plasma membrane when expressed in Saccharomyces cerevisiae. The Plant Journal 39, 237–251. [DOI] [PubMed] [Google Scholar]
- Kobae Y, Uemura T, Sato MH, Ohnishi M, Mimura T, Nakagawa T, Maeshima M. 2004. Zinc transporter of Arabidopsis thaliana AtMTP1 is localized to vacuolar membranes and implicated in zinc homeostasis. Plant and Cell Physiology 45, 1749–1758. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Suzuki M, Inoue H, Itai RN, Takahashi M, Nakanishi H, Mori S, Nishizawa NK. 2005. Expression of iron-acquisition-related genes in iron-deficient rice is coordinately induced by partially conserved iron-deficiency-responsive elements. Journal of Experimental Botany 56, 1305–1316. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of Molecular Biology 305, 567–580. [DOI] [PubMed] [Google Scholar]
- Lan HX, Wang ZF, Wang QH, Wang MM, Bao YM, Huang J, Zhang HS. 2012. Characterization of a vacuolar zinc transporter OZT1 in rice (Oryza sativa L.). Molecular Biology Reports 40, 1201–1210. [DOI] [PubMed] [Google Scholar]
- Lin H, Burton D, Li L, Warner DE, Phillips JD, Ward DM, Kaplan J. 2009. Gain-of-function mutations identify amino acids within transmembrane domains of the yeast vacuolar transporter Zrc1 that determine metal specificity. Biochemical Journal 422, 273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner H. 1995. Mineral nutrition of higher plants. San Diego: Academic Press. [Google Scholar]
- Mikkelsen MD, Pedas P, Schiller M, et al. 2012. Barley HvHMA1 is a heavy metal pump involved in mobilizing organellar Zn and Cu and plays a role in metal loading into grains. PLoS One 7, e49027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills RF, Doherty ML, Lopez-Marques RL, Weimar T, Dupree P, Palmgren MG, Pittman JK, Williams LE. 2008. ECA3, a Golgi-localized P2A-type ATPase, plays a crucial role in manganese nutrition in Arabidopsis. Plant Physiology 146, 116–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills RF, Peaston KA, Runions J, Williams LE. 2012. HvHMA2, a P(1B)-ATPase from barley, is highly conserved among cereals and functions in Zn and Cd transport. PLoS One 7, e42640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills RF, Valdes B, Duke M, Peaston KA, Lahner B, Salt DE, Williams LE. 2010. Functional significance of AtHMA4 C-terminal domain in planta. PLoS One 5, e13388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montanini B, Blaudez D, Jeandroz S, Sanders D, Chalot M. 2007. Phylogenetic and functional analysis of the Cation Diffusion Facilitator (CDF) family: improved signature and prediction of substrate specificity. BMC Genomics 8, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran R, Porath D. 1980. Chlorophyll determination in intact tissues using N,N-dimethylformamide. Plant Physiology 65, 478–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog FA. 1962. A revised medium for a rapid growth and bioassays with tobacco tissues cultures. Plant Physiology 15, 473–479. [Google Scholar]
- Nies DH, Silver S. 1995. Ion efflux systems involved in bacterial metal resistances. Journal of Industrial Microbiology 14, 186–199. [DOI] [PubMed] [Google Scholar]
- Palmgren MG, Clemens S, Williams LE, Kramer U, Borg S, Schjorring JK, Sanders D. 2008. Zinc biofortification of cereals: problems and solutions. Trends in Plant Science 13, 464–473. [DOI] [PubMed] [Google Scholar]
- Paulsen IT, Saier MH., Jr 1997. A novel family of ubiquitous heavy metal ion transport proteins. Journal of Membrane Biology 156, 99–103. [DOI] [PubMed] [Google Scholar]
- Peiter E, Montanini B, Gobert A, Pedas P, Husted S, Maathuis FJ, Blaudez D, Chalot M, Sanders D. 2007. A secretory pathway-localized cation diffusion facilitator confers plant manganese tolerance. Proceedings of the National Academy of Sciences, USA 104, 8532–8537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podar D, Scherer J, Noordally Z, Herzyk P, Nies D, Sanders D. 2012. Metal selectivity determinants in a family of transition metal transporters. Journal of Biological Chemistry 287, 3185–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahzad Z, Gosti F, Frerot H, Lacombe E, Roosens N, Saumitou-Laprade P, Berthomieu P. 2010. The five AhMTP1 zinc transporters undergo different evolutionary fates towards adaptive evolution to zinc tolerance in Arabidopsis halleri. PLoS Genetics 6, e1000911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingu Y, Kudo T, Ohsato S, Kimura M, Ono Y, Yamaguchi I, Hamamoto H. 2005. Characterization of genes encoding metal tolerance proteins isolated from Nicotiana glauca and Nicotiana tabacum. Biochemical and Biophysical Research Communications 331, 675–680. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24, 1596–1599. [DOI] [PubMed] [Google Scholar]
- van der Zaal BJ, Neuteboom LW, Pinas JE, Chardonnens AN, Schat H, Verkleij JA, Hooykaas PJ. 1999. Overexpression of a novel Arabidopsis gene related to putative zinc-transporter genes from animals can lead to enhanced zinc resistance and accumulation. Plant Physiology 119, 1047–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch RM, Graham RD. 2004. Breeding for micronutrients in staple food crops from a human nutrition perspective. Journal of Experimental Botany 55, 353–364. [DOI] [PubMed] [Google Scholar]
- Wirth J, Poletti S, Aeschlimann B, et al. 2009. Rice endosperm iron biofortification by targeted and synergistic action of nicotianamine synthase and ferritin. Plant Biotechnology Journal 7, 631–644. [DOI] [PubMed] [Google Scholar]
- Yuan L, Yang S, Liu B, Zhang M, Wu K. 2012. Molecular characterization of a rice metal tolerance protein, OsMTP1. Plant Cell Reports 31, 67–79. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.