Abstract
Recently, bryophytes, which diverged from the ancestor of seed plants more than 400 million years ago, came into focus in photosynthesis research as they can provide valuable insights into the evolution of photosynthetic complexes during the adaptation to terrestrial life. This study isolated intact photosystem I (PSI) with its associated light-harvesting complex (LHCI) from the moss Physcomitrella patens and characterized its structure, polypeptide composition, and light-harvesting function using electron microscopy, mass spectrometry, biochemical, and physiological methods. It became evident that Physcomitrella possesses a strikingly high number of isoforms for the different PSI core subunits as well as LHCI proteins. It was demonstrated that all these different subunit isoforms are expressed at the protein level and are incorporated into functional PSI–LHCI complexes. Furthermore, in contrast to previous reports, it was demonstrated that Physcomitrella assembles a light-harvesting complex consisting of four light-harvesting proteins forming a higher-plant-like PSI superstructure.
Key words: Photosystem I, photosynthesis, Physcomitrella patens, moss, state transitions, structure
Introduction
Photosystem I (PSI) catalyses the light-driven electron transport from plastocyanin in the thylakoid lumen to NADP+ in the stroma. It is composed of a core complex responsible for charge separation and the first steps of electron transport and a peripheral antenna system, which are involved in light harvesting and transfer of excitation energy to the reaction centre (Jensen et al., 2007; Busch and Hippler, 2011). In plants, PSI is a monomer consisting of at least 16 different proteins to which four peripheral light-harvesting proteins (Lhca1–Lhca4) are attached in a crescent at the PsaF/PsaJ side of the core complex (Amunts and Nelson, 2009).
Throughout the evolution of cyanobacteria towards higher plants, the reaction centre has remained largely conserved with changes only in regard to a few small peripheral subunits. While the subunit PsaX is only present in thermophilic cyanobacteria and PsaM in cyanobacterial and red algal PSI, they are absent in higher plants (Vanselow et al., 2009). In contrast, subunits like PsaG, -H, -N, and -O are acquired in order to accommodate the main structural changes from a trimeric to a monomeric PSI and the acquisition of an external antenna system (Amunts and Nelson, 2009). The extrinsic PSI antenna system is more variable, reflecting the different environmental conditions that photosynthetic organisms thrive in. While the higher plant crystal structure only shows four different light-harvesting proteins associated with PSI (Ben-Shem et al., 2003), two additional Lhca proteins—Lhca5 and Lhca6—have been identified in Arabidopsis thaliana (hereafter Arabidopsis) (Jansson, 1999). However, both are expressed only at a very low level (Klimmek et al., 2006), and solid biochemical data only exist for Lhca5, which has been shown to bind to PSI in substoichiometric amounts in Arabidopsis (Ganeteg et al., 2004). Recently, Lhca5 and Lhca6 have been implicated in NAD(P)H dehydrogenase–PSI supercomplex formation, which is involved in cyclic electron transfer (Peng and Shikanai, 2011).
Research on the structure and function of PSI so far has focused mainly on cyanobacteria, algae (diatoms and green and red algae) and higher plants. Only recently have bryophytes (mosses, liverworts and hornworts), which diverged from the ancestor of seed plants more than 400 million years ago (Falkowski, 2006), come into focus as they represent the transition from aquatic to terrestrial life. Analysing the structure and function of the photosynthetic machinery in a moss such as Physcomitrella patens (hereafter Physcomitrella) can provide valuable insights into the evolution of the system during the adaptation to a terrestrial environment. Together with its evolutionary position, its remarkable resistance against various abiotic stresses such as salt, drought, osmotic stress, and UV radiation have made the moss Physcomitrella an interesting model organism to identify new stress-tolerance genes (Frank et al., 2005; Wolf et al., 2010).
The evolutionary intermediate position of Physcomitrella is clearly reflected within the photosynthetic machinery and was demonstrated recently with the analysis of the non-photochemical quenching mechanism in this moss (Alboresi et al., 2010; Gerotto et al., 2011). Excess absorbed light energy is dissipated thermally, which regulated by an ancient light-harvesting protein (LhcSR) in green algae (Peers et al., 2009). However, during evolution towards higher plants, this protein got lost and was functionally replaced by the PsbS protein (Koziol et al., 2007). As Physcomitrella possesses both proteins (LhcSR and PsbS), it is evident that Physcomitrella represents an intermediate between algae, which employ LhcSR proteins for photoprotection, and higher plants, which rely on the function of PSBS and have lost the LhcSR genes.
Previous analysis of Physcomitrellas photosynthetic machinery focused on the antenna system and their photoprotective mechanisms (Alboresi et al., 2010; Gerotto et al., 2011). Interestingly, a considerably higher number of chlorophyll a/b-binding (Cab) proteins in Physcomitrella compared with Arabidopsis were identified in a bioinformatics study (Alboresi et al., 2008), which confirmed earlier studies by Hofmann et al. (1999). Surprisingly, it was found that an orthologue of Lhca4 was absent in Physcomitrella, which resulted in a PSI–light-harvesting complex I (LHCI) complex with a blue-shifted 77K fluorescence emission spectra due to the lack of the red chlorophyll bound to Lhca4 (Alboresi et al., 2008). Based on results obtained from native gel electrophoresis, it was consequently proposed that Physcomitrella possesses a smaller PSI–LHCI complex compared to Arabidopsis (Alboresi et al., 2008). However, to date, no further evidence for this assumption has been provided. In this report, we have provided a detailed study of the entire PSI–LHCI complex of Physcomitrella including the composition of the core complex. For this, we undertook careful preparation of thylakoids and pigment–protein complexes, which we analysed using mass spectrometry, spectroscopic methods, single-particle electron microscopy imaging, and biochemical assays to provide a comprehensive characterization of the PSI–LHCI structure, composition, and function.
Materials and methods
Plant material and growth conditions
Physcomitrella patens (Hedw.) ecotype Grandsen (Ashton and Cove, 1977) was grown at 22 °C on cellophane discs placed on solid minimal medium (Ashton et al., 1979), supplemented with 0.5g l–1 of ammonium tartrate. Standard irradiance conditions were 50 μmol photons m–2 s–1 white light for 16h and darkness for 8h. Cultures were overlaid with filter foil to shift into state 1 (filter 27, medium red, >600nm, preferentially exciting PSI; Lee Filters, Hampshire, UK) or state 2 (filter 183, moonlight blue, <550nm, preferentially exciting PSII, Lee Filters)
Preparation of thylakoid membranes and isolation of PSI
Protonemata (10–14 d old) was harvested, flash frozen in liquid nitrogen and stored at –80 °C until further use. Crude thylakoids were isolated as described by Haldrup et al. (1999). Protonemal tissue was disrupted in homogenization buffer using a homogenizer (Kinematica Brinkman Polytron PT3000) or a pre-chilled mortar and pestle. Further clean-up of thylakoids followed a modified protocol of Chua and Bennoun (1975). Thylakoids were resuspended in 12ml H3 [1.8M sucrose, 5mM Tricine (pH 7.5), 10mM EDTA], which was overlaid by 12ml H4 [1.3M sucrose, 5mM Tricine (pH 7.5), 10mM EDTA] and 12ml H5 [0.5M sucrose, 5mM Tricine (pH 7.5), 10mM EDTA]. After centrifugation (SW28 rotor, 24000rpm, 1h, 4 °C), the thylakoids accumulated at the H3/H4 interphase. The thylakoids were collected, diluted with 5mM Tricine (pH 7.5) and pelleted (20 000g, 4 °C, 10min). The resulting pellet was resuspended in a small volume of homogenization buffer without ascorbate and BSA but with 20% glycerol (v/v) and stored at –80°C. For analysis of the phosphorylation status of thylakoids, all buffers were supplemented with 10mM NaF and 1mM Na3VO4.
PSI was isolated according to the method of Mullet et al. (1980) with some modifications. The thylakoids were resuspended in solubilization buffer [20mM Tricine (pH 7.5), 10mM EDTA, 0.7% β-d-dodecyl-maltoside] to a final chlorophyll concentration of 0.8 μg μl–1. The samples were incubated on ice for 15min with occasional gentle mixing. After centrifugation (5min, 16 000g, 4 °C), the supernatant was loaded onto a linear sucrose gradient [0.4–0.85M sucrose, 20mM Tricine (pH 7.5), 0.06% β-d-dodecyl-maltoside]. After centrifugation (SW41Ti rotor, 34 000rpm, 16–18h, 4 °C), the PSI was retrieved with a syringe and stored at –80°C. Total chlorophyll was determined in 80% acetone according to the method of Lichtenthaler (1987).
Immunoblot analysis
Criterion XT 12% Bis-Tris gels and MES buffer (Bio-Rad Laboratories, Hercules, CA, USA) were used to separate the proteins and followed by transferring the electrophoresed proteins to nitrocellulose membranes. After immunoblotting, the antibodies were detected using a chemiluminescent detection system (SuperSignal®; Thermo Scientific, Rockford, USA) according to the instructions of the manufacturer. The chemiluminescent signal produced was recorded digitally using a cooled CCD camera with the AC1 AutoChemi System (Ultra-Violet Products, Cambridge, UK) and analysed using LabWorks Analysis software (Ultra-Violet Products). Antibodies against Lhca1–Lhca4 were obtained from Agrisera (Uppsala, Sweden). Anti-phosphothreonine antibodies were obtained from Cell Signaling Technology (MA, USA).
State transition measurements
Photosynthetic state transitions were measured using a DUAL-PAM-100 (Walz; Effeltrich, Germany). After 45min dark adaptation, a saturating flash was applied. Subsequently, samples were illuminated for 20min with the built-in blue light at 44 μE m–2 s–1, followed by a saturating flash to obtain Fm2. The light was switched to built-in far-red light with an intensity of 51 μE m–2 s–1. After 20min of far-red light, a saturating flash was applied to obtain Fm1. State transition was calculated using the equation: (Fm1 – Fm2)/Fm1 (Jensen et al., 2000).
Mass spectrometry
Proteins were separated on a pre-cast NuPAGE® Bis-Tris gel. In-gel-fixed proteins were visualized using colloidal Coomassie blue staining. Protein bands were excised from the gel. Cysteines were reduced and S-alkylated prior to in-gel trypsin digestion, peptide extraction, and analysis by liquid chromatography-tandem mass spectrometry (LC-MS/MS) (Jensen et al., 1998). Mass spectra were acquired in positive-ion mode applying an automatic data-dependent switch between one Orbitrap survey MS scan in the mass range of 300–1650 m/z followed by fragmentation and detection of the seven most intense ions observed from the MS scan. The target value in the Orbitrap for the MS scan was 1 000 000 ions at a resolution of 50 000 at m/z 400, with a higher-energy collisional dissociation-normalized collision energy of 40. The ion selection threshold was set to 15 000 counts. The Orbitrap was coupled to a ThermoFisher/Proxeon EasyLC system. Solvent A was 0.1% formic acid in water and solvent B was 0.1% formic acid in acetonitrile. Peptides were trapped on a homemade 1cm C18, 150 μm internal-diameter column (ReproSil Pur C18 AQ 3 μm sorbent) and separated on a homemade 15cm, 75 μm internal-diameter C18 analytical column (particle size 3 μm). The run time was 113min per sample. Raw files were processed and searched using Proteome Discoverer version 1.3.0.339 (Thermo Fisher Scientific, Bremen, Germany). Tandem mass spectra were converted to peak lists and searched against a Physcomitrella protein sequence database (https://www.cosmoss.org/physcome_project/wiki/Genome_Annotation/V1.6). Database searches were performed with the following parameters: precursor mass tolerance at 10 ppm and MS/MS mass tolerance at 0.1Da. Variable modifications included methionine oxidation and deamidation of asparagine and glutamine. Carbamidomethylation on cysteine was set as a fixed modification.
Analysis of Physcomitrella PSI complex by single-particle electron microscopy
Samples from the sucrose gradient were dialysed (12–14kDa molecular weigh cut-off) against buffer [5mM Tricine (pH 7.8), 5mM MgCl2, 0.03% β-d-dodecyl-maltoside) overnight to reduce the sucrose content and thus improve the contrast in the electron microscope. Samples were then stained with 2% uranyl acetate on glow-discharged carbon-coated copper grids. Electron microscopy was performed on a Philips CM120 electron microscope equipped with a LaB6 filament operating at 120kV. Images (2048×2048 pixels) were recorded with a Gatan 4000 SP 4 K slow-scan CCD camera at 130 000× magnification at a pixel size (after binning the images) of 2.3 Å at the specimen level with GRACE software for semi-automated specimen selection and data acquisition (Oostergetel et al., 1998). Single-particle analysis was performed using GRIP software including multireference and non-reference alignments, multivariate statistical analysis, and classification, as described by Boekema et al. (1999).
NADP+ photoreduction
The NADP+ photoreduction activities of thylakoids from Arabidopsis and Physcomitrella were determined as described by Jensen et al. (2011).
The total P700 content was determined as described by Markwell et al. (1980) from the ferricyanide-oxidized minus ascorbate-reduced difference spectrum using an extinction coefficient of 64 000 M–1 cm–1 at 700nm (Hiyama and Ke, 1972; Sonoike and Katoh, 1988). The thylakoids were solubilized in 0.2% Triton X-100, and the measurements were repeated four to five times on several independent thylakoid preparations.
Results
Photosynthetic genes in Physcomitrella are highly redundant
To elucidate the number of genes encoding PSI subunits in the genome of Physcomitrella, the entire set of Arabidopsis PSI genes was taken from Jensen et al. (2007) and used as reference for a BLAST search on the most current moss genomic database (version 1.6; see Methods). To avoid ambiguities during the BLAST search, the putative chloroplast signal sequence was determined using ChloroP (Emanuelsson et al., 1999) and the signal sequence was removed from the Arabidopsis reference sequences before the search was performed. To our surprise, Physcomitrella contained a large number of isoforms for the different PSI subunits (Table 1). For the chloroplast-encoded subunits PsaA, -B, -C, -I, and –J, only one copy was found in the chloroplast genome; however, the nuclear-encoded subunits showed a large number of isoforms ranging from two copies of PsaG, -H, and -O up to four different gene entries for PsaD and PsaF. Additionally, we assessed the number of available moss expressed sequence tags (ESTs) moss genomic database (version 1.6) for each of the nuclear-encoded gene copies, assuming that the number of available ESTs indicates a relative expression level of the respective genes. In addition to the total number of ESTs, we also assessed their number in the different tissue types: protonema, gametophore, and sporophyte (Table 1). To obtain a consistent nomenclature, we ranked the isoforms according to their number of ESTs and numbered them starting with the isoform with the highest number of ESTs.
Table 1.
Identification of PSI–LHCI subunits in Physcomitrella. The protein names are indicated, together with where the proteins are encoded [chloroplast (C) or nucleus (N)], the gene copy number in Arabidopsis, the hits found by BLAST analysis in moss, a proposed gene designation in moss, and the total number of ESTs found for each gene with the numbers found for the different tissue types indicated in parentheses (protonema+gametophore+sporophyte). Proteins that were identified (+) or not (–) by MS are indicated. NA, not applicable.
| Protein | Encoded in: | No. copies in Arabidopsis | BLAST hits in Physcomitrella | Proposed designation | No. ESTs | Identified with MS/MS |
|---|---|---|---|---|---|---|
| Core proteins | ||||||
| PsaA | C | 1 | PhpapaCp039 | PpPsaA | NA | + |
| PsaB | C | 1 | PhpapaCp040 | PpPsaB | NA | + |
| PsaC | C | 1 | PhpapaCp076 | PpPsaC | NA | + |
| PsaD | N | 2 | Pp1s4_321V6.1 | PpPsaD-1 | 190 (177+4+9) | + |
| Pp1s77_69V6.1 | PpPsaD-2 | 102 (88+7+7) | + | |||
| Pp1s1_788V6.1 | PpPsaD-3 | 90 (80+6+4) | +a | |||
| Pp1s107_188V6.1 | PpPsaD-4 | 12 (8+4+0) | – | |||
| PsaE | N | 2 | Pp1s334_17V6.1 | PpPsaE-1 | 60 (60+0+0) | + |
| Pp1s319_36V6.1 | PpPsaE-2 | 53 (51+0+2) | + | |||
| Pp1s101_2V6.1 | PpPsaE-3 | 37 (33+2+2) | + | |||
| PsaF | N | 1 | Pp1s19_276V6.1 | PpPsaF-1 | 223 (203+11+9) | – |
| Pp1s345_25V6.1 | PpPsaF-2 | 150 (139+3+8) | + | |||
| Pp1s121_54V6.1 | PpPsaF-3 | 108 (70+36+2) | + | |||
| Pp1s80_23V6.1 | PpPsaF-4 | 37 (33+3+1) | – | |||
| PsaG | N | 1 | Pp1s83_246V6.1 | PpPsaG-1 | 183 (138+18+27) | + |
| Pp1s78_205V6.1 | PpPsaG-2 | 18 (18+0+0) | + | |||
| PsaH | N | 2 | Pp1s89_62V6.1 | PpPsaH-1 | 42 (26+2+14) | + |
| Pp1s206_11V6.1 | PpPsaH-2 | 20 (14+2+4) | + | |||
| PsaI | C | 1 | PhpapaCp029 | PpPsaI | NA | – |
| PsaJ | C | 1 | PhpapaCp017 | PpPsaJ | NA | – |
| PsaK | N | 1 | Pp1s74_135V6.1 | PpPsaK-1 | 72 (66+2+4) | – |
| Pp1s5_384V6.1 | PpPsaK-2 | 27 (19+4+4) | – | |||
| Pp1s5_396V6.1 | PpPsaK-3 | 0 | – | |||
| PsaL | N | 1 | Pp1s54_165V6.1 | PpPsaL-1 | 201 (153+22+26) | + |
| Pp1s15_408V6.1 | PpPsaL-2 | 75 (69+1+5) | – | |||
| Pp1s41_267V6.1 | PpPsaL-3 | 71 (66+4+1) | – | |||
| PsaM | C | 0 | PhpapaCp051 | PpPsaM | NA | – |
| PsaN | – | 1 | – | – | – | – |
| PsaO | N | 1 | Pp1s248_60V6.1 | PpPsaO-1 | 111 (100+3+8) | – |
| Pp1s248_61V6.1 | PpPsaO-2 | 49 (48+1+0) | + | |||
| Light-harvesting proteins* | ||||||
| Lhca1 | Pp1s158_109V6.1 | Lhca1.1 | + | |||
| Pp1s161_32V6.1 | Lhca1.2 | + | ||||
| Pp1s247_7V6.1 | Lhca1.3 | + | ||||
| Lhca2 | Pp1s32_1V6.1 | Lhca2.1 | – | |||
| Pp1s330_37V6.1 | Lhca2.2 | + | ||||
| Pp1s241_66V6.1 | Lhca2.3 | + | ||||
| Pp1s651_2V6.1 | Lhca2.4 | + | ||||
| Lhca3 | Pp1s214_86V6.1 | Lhca3.1 | – | |||
| Pp1s429_33V6.2 | Lhca3.2 | + | ||||
| Pp1s197_123V6.1 | Lhca3.3 | + | ||||
| Pp1s214_87V6.1 | ||||||
| Pp1s214_87V6.2 | Lhca3.4 | – | ||||
| Lhca5 | Pp1s284_6V6.1 | Lhca5 | + | |||
a Unique peptide that was only identified using version 1.2 of the moss genome database.
* annotation according to Alboresi et al., 2008.
EST support could be found for almost all nuclear-encoded genes with the exception of PpPsaK-3 (Pp1s5_396V6.1). However, it became clear that the apparent expression levels differed for the different isoforms of a protein. While, for example, PpPsaD-4 (Pp1s107_188V6.1) had only 12 ESTs, the PpPsaD-1 (Pp1s4_321V6.1) isoform was heavily supported by 190 ESTs, indicating that this might be the major PsaD isoform. For the other PSI subunits, it was also evident that at least one isoform appeared to be the dominating form with at least a two-fold higher number of ESTs compared with the other isoforms. With respect to the different tissue types, most of the EST support stemmed from protonemal tissue. However, the distribution of abundance within the gametophores and sporophyte followed the number of ESTs in the protonema. One notable exception was PpPsaF-3 (Pp1s121_54V6.1), which was only the third most abundant isoform in the protonemal tissue but was the dominating isoform in the gametophore (Table 1). However, this result has to be taken with caution as the sample size of the EST collection was too small for robust quantitative conclusions.
It was of note that the chloroplast genome of Physcomitrella also encoded the cyanobacterial PsaM subunit, which is not present in vascular plants (Table 1). In contrast, no PsaN homologue could be identified in the Physcomitrella genome.
Additional thylakoid clean-up is required for efficient biochemical assays
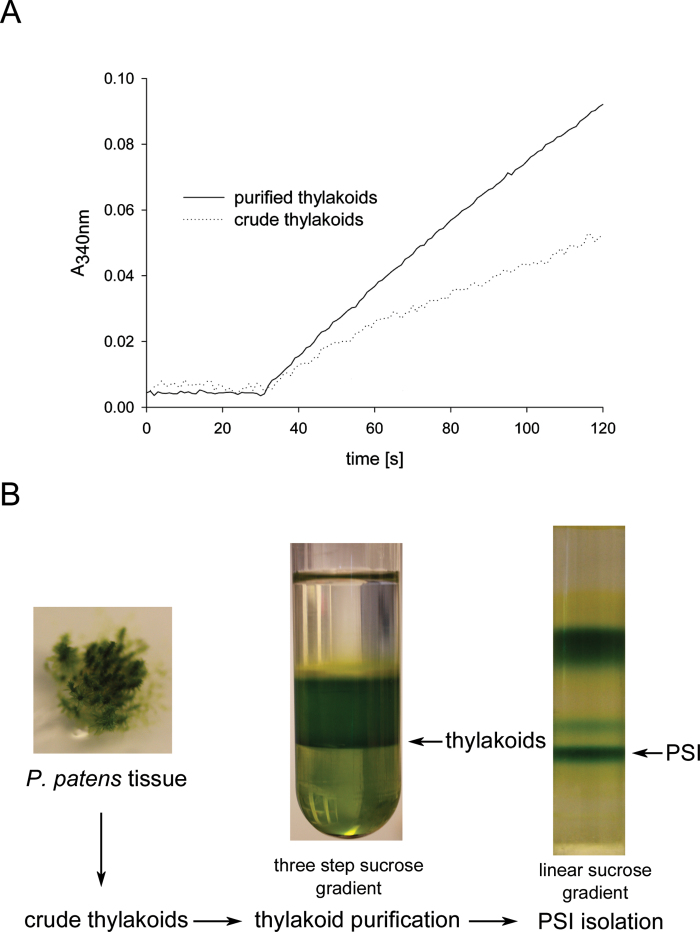
Standard plant protocols for thylakoid isolation (Haldrup et al., 1999) used on Physcomitrella gave a weak and noisy signal when functional measurements such as NADP+ photoreduction (Fig. 1A, dotted trace) were performed. Hence, an additional clean-up step was tested to obtain more homogenous thylakoids for proper biochemical assays. We subjected isolated crude thylakoids to an additional sucrose density centrifugation on a discontinuous sucrose gradient according to the method of Chua and Bennoun (1975). After centrifugation, the thylakoids were collected at the 1.8/1.3M interphase (Fig. 1B), while insoluble components such as starch formed a pellet. Additionally, a white layer could be observed at the 1.3/0.5M interphase, which probably represents additional membranes such as the plasmalemma, endoplasmic reticulum and/or Golgi. We compared the performance of crude thylakoid extract and purified thylakoids in a NADP+ photoreduction assay using equivalent amounts of chlorophyll (Fig. 1A). It was clear that additional thylakoid purification enhanced the quality of the signal significantly (Fig. 1A, solid trace). Hence, we used sucrose density gradient-purified thylakoids for all subsequent experiments.
Fig. 1.
Isolation of thylakoids from Physcomitrella. (A) Absorption trace (at 340nm) of in vitro NADP+ photoreduction assay using crude thylakoids (dotted trace) or purified thylakoids (solid trace) of Physcomitrella. (B) Physcomitrella tissue was disrupted and a crude thylakoid extract prepared, which was subjected to a further clean-up step via a discontinuous sucrose gradient (1.8, 1.3, and 0.5M sucrose layered on top of each other). After centrifugation, the thylakoids were collected at the 1.8/1.3M sucrose interphase. The thylakoids were used for biochemical assays or were solubilized using β-d-dodecyl-maltoside and subjected to a centrifugation on a linear sucrose gradient to obtain intact the PSI–LHCI complex.
Different PSI subunits can be detected by immunoblotting
An extensive collection of plant PSI-subunit specific antibodies has been established (Jensen et al., 2007). In order to elucidate whether these antibodies can be used to study Physcomitrella PSI–LHCI complexes, Physcomitrella thylakoids and isolated PSI were separated by SDS-PAGE, transferred to a nitrocellulose membrane and probed with the different antibodies (Fig. 2A). Isolated thylakoids of Arabidopsis served as a reference for the individual antibodies. It was evident that for subunits PsaA, -C, -D, -F, -H, -K, and -L, bands that corresponded to Arabidopsis could be detected both in thylakoids and in PSI complexes of Physcomitrella. In contrast, antibodies directed towards the PsaE, -G, -N, and -O subunits gave no detectable signal in either Physcomitrella thylakoids or the PSI samples.
Fig. 2.
Immunoblot analysis of PSI subunits in Physcomitrella. Physcomitrella thylakoids and isolated PSI were separated by SDS-PAGE, blotted, and analysed with the indicated antibodies against different PSI core subunits (A) or light-harvesting proteins (B). Arabidopsis thylakoids served as a reference. Samples were loaded according to equal amounts of chlorophyll (5 μg of chlorophyll for thylakoids and 3 μg for isolated PSI).
Interestingly, for PsaD, two bands possibly representing different isoforms could be detected on the immunoblot (Fig. 2A). This assumption was supported by alignment of the four isoforms of PpPsaD (Supplementary Fig. S1 at JXB online), which showed that the isoforms PpPsaD-1 and PpPsaD-2 both had an additional insert of 7 aa (DQKPDAK), making the mature protein approximately 1kDa larger than the other two isoforms.
Another interesting observation was the band pattern obtained for PpPsaF. While for isolated Arabidopsis thylakoids only one band was visible, which corresponded to the predicted size of 17.5kDa (Jensen et al., 2007), the Physcomitrella thylakoids and isolated PSI showed two bands (Fig. 2A). The higher band migrated at around 20kDa and the lower band was slightly higher than the corresponding Arabidopsis band. When aligning the PsaF sequences of Arabidopsis and Physcomitrella, a 7 aa C-terminal extension (NITVSPR) within the Physcomitrella sequence was apparent (Supplementary Fig. S2 at JXB online). This extension added approximately 0.8kDa to the protein size, which could explain the slightly lower electrophoretic mobility of Physcomitrella PsaF. Interestingly, this extension can also be found in cyanobacteria (Synechocystis sp. PCC 6803), red algae (Galdieria sulphuraria) and green algae (Chlamydomonas reinhardtii) (Supplementary Fig. S2). However, the nature of the second band with higher molecular weight remains unclear.
To study the composition of the PSI-associated light-harvesting complexes, commercially available antibodies against Lhca1–4 were likewise used to probe protein blots of thylakoids and PSI isolated from Physcomitrella (Fig. 2B). Again, Arabidopsis thylakoid proteins served as a reference. Antibodies against Lhca1 and -3 could detect proteins at the size that correspond to the respective sizes of Arabidopsis Lhca1 and -3 (Fig. 2B), indicating that the antibodies recognized PpLhca1 and -3. In contrast, the antibody against Lhca2 produced two major bands with the Physcomitrella thylakoids, with the lower one running slightly higher than Lhca2 of Arabidopsis. Interestingly, the high-molecular-weight band for Lhca2 in Physcomitrella was not present in isolated PSI, indicating that it represented non-specific binding of the antibody to proteins present in Physcomitrella thylakoids. Again, the Lhca2 band in Physcomitrella PSI indicated a slightly larger protein than that in Arabidopsis, which is in agreement with the predicted sizes of the mature Lhca2 (Arabidopsis ~23.3kDa, Physcomitrella ~24.4kDa). Surprisingly, Lhca4 gave a signal in the isolated thylakoids and PSI–LHCI of Physcomitrella despite the fact that Physcomitrella does not encode an Lhca4-like subunit (Alboresi et al., 2008). Hence, the obtained signal most likely stemmed from non-specific binding of the antibody to other Lhc proteins.
Clearly, immunoblot-based analysis of isolated PSI–LHCI from Physcomitrella was not able to resolve the existence of all the different isoforms of PSI subunits encoded by Physcomitrella. Therefore, the isolated Physcomitrella PSI–LHCI complex was characterized further by proteome analysis by MS to resolve its subunit composition in more detail.
Different isoforms are clearly identified at the protein level
For MS analyses, isolated Physcomitrella PSI samples were digested with trypsin and separated on an LC system before MS/MS analysis was carried out on an LTQ Orbitrap system. With this approach, we aimed to identify unambiguously the different isoforms for the respective PSI subunits at the protein level. The obtained results clearly showed that the diversity of isoforms for the different PSI subunits encoded by Physcomitrella (Table 1) was also reflected at the protein level (Table 1 and Supplementary Table S1 at JXB online).
Using MS analysis, we were able to identify the following PSI core subunits: PsaA, -B, -C, -D, -E, -F, -G, -H, -K, -L, and -O (Table 1 and Supplementary Table S1). Only the small subunits PsaI, PsaJ, and PsaM could not be identified unambiguously with unique peptides. This might be due to the fact that these small proteins do not always yield tryptic peptides suitable for MS analysis due to size or charge issues. Alternatively, the peptide sequences that were detected could not be assigned unambiguously because they were not unique.
With respect to the different isoforms of the subunits encoded by Physcomitrella (Table 1), we could uniquely identify at least one isoform for each of the PSI core subunits with the exception of PsaK (Table 1 and Supplementary Table S1), while for PsaE (three isoforms), -G (two isoforms), and -H (two isoforms) we could identify all possible isoforms. For PsaD (four isoforms) and PsaO (two isoforms), one isoform remained unidentified, and for PsaF (four isoforms) and PsaL (three isoforms), two isoforms remained unidentified. It is of note that, for PpPsaD-3, a unique peptide could only be identified using version 1.2 of the moss database. Although we could identify three distinct peptides uniquely identifying PsaK (Supplementary Table S1), we were not able to match them to any of the three different isoforms of PsaK, as the peptides were derived from a highly conserved region of PsaK (Supplementary Fig. S3 at JXB online and Supplementary Table S1). One peptide uniquely identified PsaK-2, but manual inspection of the fragmented peptide could not rule out other sequence combinations.
With regard to the members of the light-harvesting complex of PSI, we followed the annotation of Alboresi et al. (2008). We were able to identify Lhca1, -2, -3, and -5 and most of their different isoforms (Table 1 and Supplementary Table S1). For Lhca1 (three isoforms) and Lhca2 (four isoforms), all isoforms except Lhca2.1 were detected by unique peptides. For Lhca3, two out of four isoforms could be detected, and Lhca5 was present only in one isoform, which also could be identified.
Chl/P700 and in vitro NADP+ photoreduction
After characterizing the PSI–LHCI complex at the protein level, we aimed to study the function of Physcomitrella PSI further using spectroscopic as well as biochemical methods.
The number of chlorophylls associated with both photosystems but normalized per P700 reaction centre was assessed in thylakoids using chemical oxidation. For Physcomitrella thylakoids, we measured 693±66 (n=5) chlorophylls per P700 reaction centre (Table 2), which was only marginally more than the 666±24 chlorophylls per P700 reaction centre that were reported previously for Arabidopsis wild-type thylakoids (Jensen et al., 2002).
Table 2.
In vitro NADP+ photoreduction with different electron donor systems. Values are means ±standard deviation.
| Arabidopsis | Physcomitrella | |
|---|---|---|
| Chlorophyll/P700 | 666±24a | 693±66 (n=5) |
| NADP+ photoreduction (μmol NADPH s–1 per P700) | ||
| 2 μM plastocyanin | 24.35±2.65 (n=3) | 11.92±0.85 (n=3) |
| 2 μM cytc6 | 1.61±0 (n=3) | 0.09±0.12 (n=3) |
| 4 μM cytc6 | 3.04±0.31 (n=3) | 2.05±0.32 (n=3) |
a Value taken from Jensen et al. (2002).
An in vitro NADP+ photoreduction assay was used to measure and assess the activity of Physcomitrella PSI. With isolated thylakoids of wild-type Arabidopsis, we obtained an activity of 24.35±2.65 μmol s–1 per P700 reaction centre of NADPH (n=3; Table 2), which was in good agreement with previously published activities (Jensen et al., 2002). Using the same set-up, we also measured an activity of 11.92±0.85 μmol s–1 per P700 reaction centre of NADPH (n=3; Table 2) with isolated Physcomitrella thylakoids.
Although in some algae cytochrome can be used as alternative electron donor to PSI when copper becomes a limiting factor (Wood, 1978), higher plants are dependent on the presence of plastocyanin (Weigel et al., 2003). To test the ability of Physcomitrella PSI to use cytochrome c6 (cytc6) as an alternative electron donor, the 2 μM plastocyanin was substituted with 2 μM cyanobacterial cytc6 in the NADP+ photoreduction assay (Table 2). For comparison, we also performed the assay with Arabidopsis thylakoids and, as expected, an almost complete loss of activity was recorded. While for Arabidopsis thylakoids an activity of 1.61±0 μmol s–1 per P700 reaction centre of NADPH (n=3) was recorded, for Physcomitrella thylakoids no activity could be seen (0.09±0.12 μmol s–1 per P700 reaction centre of NADPH; n=3) (Table 2). Also, a doubling of the cytc6 concentration did not dramatically increase the activity (Table 2). This clearly showed that neither Arabidopsis nor Physcomitrella PSI is able to utilize cytc6 as an alternative electron acceptor.
State 1–state 2 transitions
We also tested the ability of Physcomitrella to induce state transition, the reversible phosphorylation and movement of LHCII to PSI upon overexcitation of PSII (Haldrup et al., 2001). Protonemal tissue was cultivated under blue light (state 2 light) or far-red light (state 1 light) for 1h. Subsequently, thylakoids were isolated from both states and the phosphorylation pattern of the proteins was analysed by immunoblotting using antibodies against phosphothreonine (Fig. 3). Three major bands at ~40, ~30, and ~25kDa were visible whose phosphorylation increased under blue light compared with samples derived from far-red light conditions, which was in line with results obtained from thylakoid samples from Arabidopsis grown under comparable conditions (Fig. 3). Interestingly, the band at 25kDa, which represents LHCII proteins (Rintamaki et al., 2000), was not as pronounced in the Physcomitrella state 2 conditions as in the Arabidopsis state 2, where it represented the strongest band (Fig. 3). This suggested a lower degree of LHCII phosphorylation in Physcomitrella compared with Arabidopsis. However, these results have to be judged with caution, as the signal intensity is very dependent on the brand of antibody and plant material used (Turkina et al., 2006). The extent of state 1–state 2 transition was additionally assessed by measuring chlorophyll a fluorescence on protonema tissue grown on agar plates and compared with intact Arabidopsis plants (Supplementary Fig. S4 at JXB online). The samples were dark adapted for 45min and the maximum fluorescence yield was determined. After 20min of blue light illumination, which favours PSII, the maximum fluorescence in state 2 (Fm2) was determined. Subsequently, the light was changed to far-red light, which favours PSI and induces state 1. Again, after 20min, the maximum fluorescence (Fm1) was determined. The ability to perform state transitions can be expressed as a ratio of Fm1 and Fm2 [(Fm1 – Fm2)/Fm2], where a smaller ratio indicates a lower state transition (Jensen et al., 2000). For Physcomitrella, a ratio of 0.127±0.004 (n=4) was obtained, while for Arabidopsis a ratio of 0.167±0.006 (n=3) was measured. These values were significantly different (Student’s t-test, P <0.001). Hence, Physcomitrella indeed performs state transition; however, it has a slightly smaller capability to do so, in agreement with the lower ability to phosphorylate LHCII (Fig. 3). Additionally, standard photosynthetic parameters were determined (Supplementary Table S2 at JXB online).
Fig. 3.
Analysis of phosphorylation pattern during state transitions. Thylakoids from Physcomitrella and Arabidopsis under state 1 (S1) and state 2 (S2) conditions were isolated, separated by SDS-PAGE, blotted, and analysed with anti-phosphothreonine antibodies to visualize changes in phosphorylation patterns. The equivalent of 12 and 3 μg of chlorophyll from thylakoids was loaded per lane for Physcomitrella and Arabidopsis, respectively.
Topology of the PSI–LHCI complex
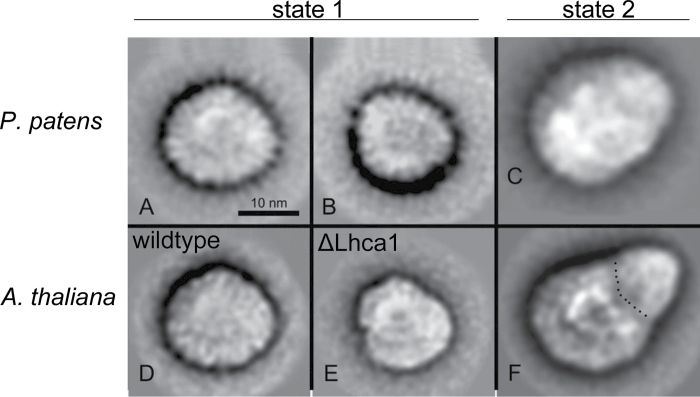
Based on the higher mobility of isolated the PSI–LHCI complex on native gels, it was suggested by earlier studies that the PSI–LHCI complex of Physcomitrella is smaller than that of Arabidopsis and Chlamydomonas (Alboresi et al., 2008). This might reflect the absence of the Lhca4 in Physcomitrella, which in turn leads to a smaller LHCI complex. However, to date, no further evidence has been presented on this matter. Therefore, purified and homogeneous preparations of PSI–LHCI complexes of Physcomitrella were analysed using transmission electron microscopy and single-particle analysis. Classification of single-particle projection maps resulted in two maps of different size, indicating that the PSI–LHCI complex is present in two forms differing in size (Fig. 4A, B), with larger complex as the dominant form with a ratio of ~3:1. This larger projection map (Fig. 4A) stemmed from 921 particles, and although these particles lacked a distinct outline and features making them hard to align, they resembled the structure of PSI–LHCI complexes isolated from wild-type Arabidopsis (Fig. 4D) (Wientjes et al., 2009). The smaller projection (Fig. 4B) represented a sum of only 395 top projections and hence the image resolution was low. However, it is important to note that these smaller projections were found repeatedly in different independent biological and technical replicates.
Fig. 4.
Transmission electron microscopy and single-particle analysis of the isolated PSI–LHCI complex of Physcomitrella. Projection maps of PSI from Physcomitrella in state 1 summed from 921 particles (A) and from 395 particles (B), and in state 2 summed from 613 (C), in comparison with Arabidopsis PSI maps from state 1 (D, E) and from state 2 (F). The dotted line indicates the border between the core part and the extra antenna moiety attached under state 2. Parts (D) and (E) have been reproduced from Wientjes et al. (2009); part (F) was reproduced from Galka et al. (2012).
When analysing PSI–LHCI particles isolated from state 2, an additional density in the PSI–LHCI complex became evident, and a sum of 613 projections is shown in Fig. 4C. This projection looked very similar to PSI–LHCI–LHCII complexes isolated from higher plants under state 2 (Fig. 4F) (Kouril et al., 2005; Wientjes et al., 2009). This indicated that, under state 2 conditions, additional LHCII proteins attach to PSI in Physcomitrella, supporting the results obtained with chlorophyll a fluorescence measurements.
Discussion
Subunit composition of the moss PSI–LHCI complex
Intact PSI–LHCI complexes were isolated from Physcomitrella and its subunit composition was studied in detail. It was evident that the evolutionary intermediate position of Physcomitrellas is also reflected in the subunit composition of the PSI core. While still retaining the cyanobacterial PsaM subunits, it also contains the plant-specific PsaG, -H, and -O subunits. However, a PsaN homologue could not be identified (Table 1). In addition, the protein sequence of PsaF is an example that Physcomitrella still retains a more ancient version of a protein compared with higher plants. Physcomitrella PsaF still contains a 7 aa C-terminal extension, which can also be found in cyanobacteria and in red and green algae but not in higher plants (Supplementary Fig. S2). This extension has been proposed to be involved in the interaction with phycobilisomes (Vanselow et al., 2009); however, due to the fact that it is still present in green algae and moss, which do not possess phycobilisomes, the extension might serve a different purpose.
For all of the nuclear-encoded PSI core subunits, as well as for the light-harvesting proteins, Physcomitrella encodes numerous isoforms (Table 1) (Alboresi et al., 2008). Interestingly, this diversity could also be detected at the protein level within native PSI–LHCI complexes (Table 1 and Supplementary Table S1). This striking wealth of isoforms raises the question of their biological significance.
Molecular heterogeneity of PSI and the presence of two isoforms for PsaD, -E, and -H is well known in higher plants such as Nicotiana and Arabidopsis (Obokata et al., 1993; Jensen et al., 2007). In Nicotiana sylvestris, which encodes two PsaD isoforms, a differential expression pattern during leaf development has been shown. While the PsaD-1 level is highest in developing leaves, the PsaD-2 protein level peaks in mature leaves (Yamamoto et al., 1993). In Arabidopsis, PsaD-1 has been shown to be the dominating form; however, it can be functionally replaced by overexpressing the almost identical PsaD-2 isoform (Ihnatowicz et al., 2004). To date, no distinct function has been attributed to either of the PsaD isoforms. It has been speculated that duplicated genes have been retained to allow the same subunits to respond to multiple signal transduction pathways (Ihnatowicz et al., 2004).
Physcomitrella underwent whole-genome duplication approximately 45 million years ago, leading to a wealth of multiple genes (Rensing et al., 2007). One effect of gene duplication is that it alleviates the pressure to maintain a single important gene, introducing the potential for development of new genes. It is tempting to speculate that Physcomitrella retained the high number of isoforms and specialized them to fine-tune the structure and function of PSI during different developmental stages or stress conditions. Expression data from GENEVESTIGATOR (http://www.genevestigator.com) indicated an isoform-specific gene regulation during development (Supplementary Fig. S5 at JXB online). For example, the PsaD and PsaL isoforms show differential expression during the different development stages. Also, for the isoforms of PsaF, we could see differences in expression pattern based on EST data. PpPsaF-3 is the dominating form in the gametophore while being only the third most abundant form in the protonemal stage (Table 1). However, it remains to be shown experimentally that the different isoforms have a distinct functional role.
Structure of the PSI–LHCI complex
Based on native gel electrophoresis, it has been proposed previously that the PSI–LHCI complex of Physcomitrella is smaller than its plant counterpart (Alboresi et al., 2008). Our data obtained by electron micrscopy imaging analysis of isolated PSI particles did not support these findings. Interestingly, we found two populations of PSI complexes. The larger particles (Fig. 4A), which were present in the majority, represented a PSI with four Lhca proteins at the PsaF/PsaJ side of the PSI core, as seen in higher plants (Fig. 4D) (Boekema et al., 2001; Ben-Shem et al., 2003). This raises the question of which Lhca protein is replacing Lhca4 in the complex, as Physcomitrella does not have an Lhca4 homologue (Alboresi et al., 2008).
It has been shown in Arabidopsis that deletion of an Lhca protein leads to a hole in the PSI–LHCI complex, as each individual Lhca protein has a fixed position (Ballottari et al., 2007; Wientjes et al., 2009). However, for Lhca4, a notable exception was found. In ΔLhca4 plants (Fig. 4E), a small fraction of complete complex containing four Lhca proteins could be isolated. Further analysis suggested that Lhca5 occupies the Lhca4 position, thereby forming a complete complex (Wientjes et al., 2009). One could envision a similar scenario in Physcomitrella. However, semi-quantitative analysis of our proteomics data revealed that the Lhca5 protein level was much lower than that of Lhca1, -2, and -3, which are present in a nearly 1:1:1 stoichiometry (Supplementary Fig. S6 at JXB online). This was further supported by a preliminary analysis of transcript level derived from RNAseq data of Physcomitrella (Supplementary Fig. S7 at JXB online). Also here Lhca5 showed a much lower expression compared with the other Lhca proteins. Hence, one of the Lhca1–Lhca3 isoforms has to occupy the Lhca4 position in order to form the half-moon-shaped LHCI complex. However, which of the Lhca proteins undertakes this remains to be elucidated in a future study.
It is of note that we were able to find two different sizes of PSI particles in Physcomitrella (Fig. 4A, B). Given the fact that we found this size distribution in numerous independent PSI preparations and that smaller PSI particles have been reported previously (Alboresi et al., 2008), it is unlikely that these different sizes are mere preparation artefacts. It is possible that the two forms represent a dynamic equilibrium that responds to different light intensities or qualities. The analysis is further complicated by the fact that PSI does not possess distinct outlines, which makes it technically challenging to obtain high-quality images. In contrast, for PSII–LHCII particles, which have a much more distinct outline, only a few images are necessary to obtain good-quality pictures (Supplementary Fig. S8 at JXB online).
Function of the PSI–LHCI complex
As Physcomitrella also encodes cytc6, which could potentially be an alternative electron donor to PSI, we tested this possibility in an in vitro NADP+ photoreduction assay. However, under the assay conditions used, it was clear that Physcomitrella was not able to accept electrons from cytc6 (Table 2), showing a similar behaviour to that of Arabidopsis. It should be noted that the cytc6 used efficiently donates electrons to cyanobacterial PSI (data not shown). It has been proposed previously that, while cytc6 can serve as an electron donor in green algae such as Chlamydomonas, it has acquired novel functions not related to electron transport to PSI throughout the evolution of higher plants (Weigel et al., 2003). Following this idea, we analysed the phylogeny of different cytc6 sequences including the one from Physcomitrella (Supplementary Fig. S9 at JXB online). Clearly, the Physcomitrella cytc6 sequence grouped with the cytc6 of higher plants, indicating that already in moss cytc6 has acquired a novel function.
With respect to state transition, we showed that Physcomitrella is able to perform state transitions. However, based on the state transition coefficient we obtained [(Fm1 – Fm2)/Fm2] (Jensen et al., 2000), it appeared that the movement of the PSII antenna towards PSI happens to a smaller extent than in Arabidopsis under the same conditions. Recently, Lhcb9, a light-harvesting protein that associates with PSII, was characterized in Physcomitrella (Alboresi et al., 2011). This Lhcb9 protein, unlike all other known LHCII protein, harbours a red-shifted chlorophyll leading to changes in absorption properties of PSII. Hence, the excitation pressure on PSII in Physcomitrella might be different from that in Arabidopsis under the state 2 light-inducing conditions we applied in our experiments.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Sequence comparison of PsaD isoforms of Physcomitrella.
Supplementary Fig. S2. Alignment of full-length PsaF proteins from different species.
Supplementary Fig. S3. Alignment of the three different isoforms of PsaK of Phscomitrella.
Supplementary Fig. S4. Traces from the state transition experiment.
Supplementary Fig. S5. Expression data from GENEVESTIGATOR for selected isoforms.
Supplementary Fig. S6. Label-free quantification of PSI-LHCI proteins in Physcomitrella.
Supplementary Fig. S7. Expression profile of light-harvesting proteins in Physcomitrella.
Supplementary Fig. S8. Transmission electron microscopy and single-particle analysis of isolated PSII–LHCII complexes of Physcomitrella.
Supplementary Fig. S9. Phylogenetic tree based on the protein sequence of cytochrome c6 sequences.
Supplementary Table S1. Identified proteins and corresponding peptides.
Supplementary Table S2. Photosynthetic parameters derived from chlorophyll a fluorescence measurements.
Acknowledgements
The authors gratefully acknowledge financial support from the Villum Foundation to the research centre ‘Pro-Active Plants’, from the ‘Center of Synthetic Biology’ funded by the UNIK research initiative of the Danish Ministry of Science (09-064047), and from the ‘Nutriefficient’ project (10-093498) funded by the Danish Strategic Research Council. A.B. is grateful for support from the Deutsche Forschungsgemeinschaft (BU2563/1-1).
Glossary
Abbreviations:
- cytc6
Cytochrome c6
- EST
expressed sequence tag
- LC
liquid chromatography
- LHC
light-harvesting complex
- MS
mass spectrometry
- PS
photosystem.
References
- Alboresi A, Caffarri S, Nogue F, Bassi R, Morosinotto T. 2008. In silico and biochemical analysis of Physcomitrella patens photosynthetic antenna: identification of subunits which evolved upon land adaptation. PLoS One 3, e2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alboresi A, Gerotto C, Cazzaniga S, Bassi R, Morosinotto T. 2011. A red-shifted antenna protein associated with photosystem II in Physcomitrella patens . Journal of Biological Chemistry 286, 28978–28987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alboresi A, Gerotto C, Giacometti GM, Bassi R, Morosinotto T. 2010. Physcomitrella patens mutants affected on heat dissipation clarify the evolution of photoprotection mechanisms upon land colonization. Proceedings of the National Academy of Sciences U S A 107, 11128–11133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amunts A, Nelson N. 2009. Plant photosystem I design in the light of evolution. Structure 17, 637–650. [DOI] [PubMed] [Google Scholar]
- Ashton NW, Cove DJ. 1977. Isolation and preliminary characterization of auxotrophic and analog resistant mutants of moss, Physcomitrella patens . Molecular & General Genetics 154, 87–95. [Google Scholar]
- Ashton NW, Grimsley NH, Cove DJ. 1979. Analysis of gametophytic development in the moss, Physcomitrella patens, using auxin and cytokinin resistant mutants. Planta 144, 427–435. [DOI] [PubMed] [Google Scholar]
- Ballottari M, Dall’Osto L, Morosinotto T, Bassi R. 2007. Contrasting behavior of higher plant photosystem I and II antenna systems during acclimation. Journal of Biological Chemistry 282, 8947–8958. [DOI] [PubMed] [Google Scholar]
- Ben-Shem A, Frolow F, Nelson N. 2003. Crystal structure of plant photosystem I. Nature 426, 630–635. [DOI] [PubMed] [Google Scholar]
- Boekema EJ, Jensen PE, Schlodder E, van Breemen JF, van Roon H, Scheller HV, Dekker JP. 2001. Green plant photosystem I binds light-harvesting complex I on one side of the complex. Biochemistry 40, 1029–1036. [DOI] [PubMed] [Google Scholar]
- Boekema EJ, Van Roon H, Van Breemen JF, Dekker JP. 1999. Supramolecular organization of photosystem II and its light-harvesting antenna in partially solubilized photosystem II membranes. European Journal of Biochemistry 266, 444–452. [DOI] [PubMed] [Google Scholar]
- Busch A, Hippler M. 2011. The structure and function of eukaryotic photosystem I. Biochimica et Biophysica Acta 1807, 864–877. [DOI] [PubMed] [Google Scholar]
- Chua NH, Bennoun P. 1975. Thylakoid membrane polypeptides of Chlamydomonas reinhardtii: wild-type and mutant strains deficient in photosystem II reaction center. Proceedings of the National Academy of Sciences U S A 72, 2175–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G. 1999. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Science 8, 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkowski PG. 2006. Evolution. Tracing oxygen’s imprint on earth’s metabolic evolution. Science 311, 1724–1725. [DOI] [PubMed] [Google Scholar]
- Frank W, Ratnadewi D, Reski R. 2005. Physcomitrella patens is highly tolerant against drought, salt and osmotic stress. Planta 220, 384–394. [DOI] [PubMed] [Google Scholar]
- Galka P, Santabarbara S, Khuong TT, Degand H, Morsomme P, Jennings RC, Boekema EJ, Caffarri S. 2012. Functional analyses of the plant photosystem I–light-harvesting complex II supercomplex reveal that light-harvesting complex II loosely bound to photosystem II is a very efficient antenna for photosystem I in state II. Plant Cell 24, 2963–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganeteg U, Klimmek F, Jansson S. 2004. Lhca5—an LHC-type protein associated with photosystem I. Plant Molecular Biology 54, 641–651. [DOI] [PubMed] [Google Scholar]
- Gerotto C, Alboresi A, Giacometti GM, Bassi R, Morosinotto T. 2011. Role of PSBS and LHCSR in Physcomitrella patens acclimation to high light and low temperature. Plant, Cell & Environment 34, 922–932. [DOI] [PubMed] [Google Scholar]
- Haldrup A, Jensen PE, Lunde C, Scheller HV. 2001. Balance of power: a view of the mechanism of photosynthetic state transitions. Trends in Plant Science 6, 301–305. [DOI] [PubMed] [Google Scholar]
- Haldrup A, Naver H, Scheller HV. 1999. The interaction between plastocyanin and photosystem I is inefficient in transgenic Arabidopsis plants lacking the PSI-N subunit of photosystem I. The Plant Journal 17, 689–698. [DOI] [PubMed] [Google Scholar]
- Hiyama T, Ke B. 1972. Difference spectra and extinction coefficients of P700. Biochimica et Biophysica Acta 267, 160–171. [DOI] [PubMed] [Google Scholar]
- Hofmann AH, Codon AC, Ivascu C, Russo VE, Knight C, Cove D, Schaefer DG, Chakhparonian M, Zryd JP. 1999. A specific member of the Cab multigene family can be efficiently targeted and disrupted in the moss Physcomitrella patens . Molecular and General Genetics 261, 92–99. [DOI] [PubMed] [Google Scholar]
- Ihnatowicz A, Pesaresi P, Varotto C, Richly E, Schneider A, Jahns P, Salamini F, Leister D. 2004. Mutants for photosystem I subunit D of Arabidopsis thaliana: effects on photosynthesis, photosystem I stability and expression of nuclear genes for chloroplast functions. The Plant Journal 37, 839–852. [DOI] [PubMed] [Google Scholar]
- Jansson S. 1999. A guide to the Lhc genes and their relatives in Arabidopsis . Trends in Plant Science 4, 236–240. [DOI] [PubMed] [Google Scholar]
- Jensen K, Jensen PE, Moller BL. 2011. Light-driven cytochrome p450 hydroxylations. ACS Chemical Biology 6, 533–539. [DOI] [PubMed] [Google Scholar]
- Jensen ON, Larsen MR, Roepstorff P. 1998. Mass spectrometric identification and microcharacterization of proteins from electrophoretic gels: strategies and applications. Proteins: Structure, Function, and Bioinformatics 33 (Suppl. 2 ), 74–89. [DOI] [PubMed] [Google Scholar]
- Jensen PE, Bassi R, Boekema EJ, Dekker JP, Jansson S, Leister D, Robinson C, Scheller HV. 2007. Structure, function and regulation of plant photosystem I. Biochimica et Biophysica Acta 1767, 335–352. [DOI] [PubMed] [Google Scholar]
- Jensen PE, Gilpin M, Knoetzel J, Scheller HV. 2000. The PSI-K subunit of photosystem I is involved in the interaction between light-harvesting complex I and the photosystem I reaction center core. Journal of Biological Chemistry 275, 24701–24708. [DOI] [PubMed] [Google Scholar]
- Jensen PE, Rosgaard L, Knoetzel J, Scheller HV. 2002. Photosystem I activity is increased in the absence of the PSI-G subunit. Journal of Biological Chemistry 277, 2798–2803. [DOI] [PubMed] [Google Scholar]
- Klimmek F, Sjodin A, Noutsos C, Leister D, Jansson S. 2006. Abundantly and rarely expressed Lhc protein genes exhibit distinct regulation patterns in plants. Plant Physiology 140, 793–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouril R, Zygadlo A, Arteni AA, de Wit CD, Dekker JP, Jensen PE, Scheller HV, Boekema EJ. 2005. Structural characterization of a complex of photosystem I and light-harvesting complex II of Arabidopsis thaliana . Biochemistry 44, 10935–10940. [DOI] [PubMed] [Google Scholar]
- Koziol AG, Borza T, Ishida K, Keeling P, Lee RW, Durnford DG. 2007. Tracing the evolution of the light-harvesting antennae in chlorophyll a/b-containing organisms. Plant Physiology 143, 1802–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK. 1987. Chlorophylls and carotenoids—pigments of photosynthetic biomembranes. Methods in Enzymology 148, 350–382. [Google Scholar]
- Markwell JP, Thornber JP, Skrdla MP. 1980. Effect of detergents on the reliability of a chemical assay for P-700. Biochimica et Biophysica Acta 591, 391–399. [DOI] [PubMed] [Google Scholar]
- Mullet JE, Burke JJ, Arntzen CJ. 1980. Chlorophyll proteins of photosystem I. Plant Physiology 65, 814–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obokata J, Mikami K, Hayashida N, Nakamura M, Sugiura M. 1993. Molecular heterogeneity of photosystem I. psaD, psaE, psaF, psaH, and psaL are all present in isoforms in Nicotiana spp. Plant Physiology 102, 1259–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostergetel GT, Keegstra W, Brisson A. 1998. Automation of specimen selection and data acquisition for protein electron crystallography. Ultramicroscopy 74, 47–59. [Google Scholar]
- Peers G, Truong TB, Ostendorf E, Busch A, Elrad D, Grossman AR, Hippler M, Niyogi KK. 2009. An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature 462, 518–521. [DOI] [PubMed] [Google Scholar]
- Peng L, Shikanai T. 2011. Supercomplex formation with photosystem I is required for the stabilization of the chloroplast NADH dehydrogenase-like complex in Arabidopsis . Plant Physiology 155, 1629–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing S, Ick J, Fawcett J, Lang D, Zimmer A, Van de Peer Y, Reski R. 2007. An ancient genome duplication contributed to the abundance of metabolic genes in the moss Physcomitrella patens . BMC Evolutionary Biology 7, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rintamaki E, Martinsuo P, Pursiheimo S, Aro EM. 2000. Cooperative regulation of light-harvesting complex II phosphorylation via the plastoquinol and ferredoxin-thioredoxin system in chloroplasts. Proceedings of the National Academy of Sciences U S A 97, 11644–11649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoike K, Katoh S. 1988. Effects of sodium dodecyl sulfate and methyl viologen on the differential extinction coefficient of P-700—a band shift of chlorophyll a associated with oxidation of P-700. Biochimica et Biophysica Acta (BBA)—Bioenergetics 935, 61–71. [Google Scholar]
- Turkina MV, Kargul J, Blanco-Rivero A, Villarejo A, Barber J, Vener AV. 2006. Environmentally modulated phosphoproteome of photosynthetic membranes in the green alga Chlamydomonas reinhardtii . Molecular & Cellular Proteomics 5, 1412–1425. [DOI] [PubMed] [Google Scholar]
- Vanselow C, Weber AP, Krause K, Fromme P. 2009. Genetic analysis of the Photosystem I subunits from the red alga, Galdieria sulphuraria . Biochimica et Biophysica Acta 1787, 46–59. [DOI] [PubMed] [Google Scholar]
- Weigel M, Varotto C, Pesaresi P, Finazzi G, Rappaport F, Salamini F, Leister D. 2003. Plastocyanin is indispensable for photosynthetic electron flow in Arabidopsis thaliana . Journal of Biological Chemistry 278, 31286–31289. [DOI] [PubMed] [Google Scholar]
- Wientjes E, Oostergetel GT, Jansson S, Boekema EJ, Croce R. 2009. The role of Lhca complexes in the supramolecular organization of higher plant photosystem I. Journal of Biological Chemistry 284, 7803–7810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf L, Rizzini L, Stracke R, Ulm R, Rensing SA. 2010. The molecular and physiological responses of Physcomitrella patens to ultraviolet-B radiation. Plant Physiology 153, 1123–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood PM. 1978. Interchangeable copper and iron proteins in algal photosynthesis. Studies on plastocyanin and cytochrome c-552 in Chlamydomonas . European Journal of Biochemistry 87, 9–19. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Tsuji H, Obokata J. 1993. Structure and expression of a nuclear gene for the Psi-D subunit of Photosystem-I in Nicotiana sylvestris . Plant Molecular Biology 22, 985–994. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.