Abstract
Rice seed storage proteins glutelin and α-globulin are synthesized in the endoplasmic reticulum (ER) and deposited in protein storage vacuoles (PSVs). Sar1, a small GTPase, acts as a molecular switch to regulate the assembly of coat protein complex II, which exports secretory protein from the ER to the Golgi apparatus. To reveal the route by which glutelin and α-globulin exit the ER, four putative Sar1 genes (OsSar1a/b/c/d) were cloned from rice, and transgenic rice were generated with Sar1 overexpressed or suppressed by RNA interference (RNAi) specifically in the endosperm under the control of the rice glutelin promoter. Overexpression or suppression of any OsSar1 did not alter the phenotype. However, simultaneous knockdown of OsSar1a/b/c resulted in floury and shrunken seeds, with an increased level of glutelin precursor and decreased level of the mature α- and β-subunit. OsSar1abc RNAi endosperm generated numerous, spherical, novel protein bodies with highly electron-dense matrixes containing both glutelin and α-globulin. Notably, the novel protein bodies were surrounded by ribosomes, showing that they were derived from the ER. Some of the ER-derived dense protein bodies were attached to a blebbing structure containing prolamin. These results indicated that OsSar1a/b/c play a crucial role in storage proteins exiting from the ER, with functional redundancy in rice endosperm, and glutelin and α-globulin transported together from the ER to the Golgi apparatus by a pathway mediated by coat protein complex II.
Key words: Endoplasmic reticulum, OsSar1, protein body, protein intracellular transport, protein storage vacuole, rice storage proteins.
Introduction
Rice seed storage proteins are mainly composed of three groups with distinct solubility profiles: alcohol-soluble prolamin, acid- and alkaline-soluble glutelin, and saline-soluble α-globulin. The prolamins are synthesized in the endoplasmic reticulum (ER) and retained in the ER lumen, forming a spherical protein body PB-I (Bechtel and Juliano, 1980; Tanaka et al., 1980; Yamagata and Tanaka, 1986). Glutelins are initially synthesized on the ER as 57kDa precursors and are transported to the protein storage vacuole (PSV; also called PB-II) through or bypassing the Golgi apparatus. These precursors are then cleaved to 37–39kDa acidic and 19–20kDa basic subunits and stored in the PSV (Yamagata et al., 1982; Krishnan et al., 1986; Yamagata and Tanaka, 1986). The α-globulin is deposited together with glutelin in the PSV.
Many studies of glutelin intracellular transportation have focused on factors related to protein folding, movement from the Golgi apparatus to the PSV, and processing in the PSV. Protein disulphide isomerase (PDI) plays an essential role in proglutelin folding and the segregation of proglutelin and prolamin polypeptides within the ER lumen. In mutants with loss of function of PDI1-1 (esp2), glutelin heterotypically interacts with prolamin polypeptides within the ER lumen, which leads to the formation of new PBs containing both glutelin and prolamin in the ER (Takemoto et al., 2002; Satoh-Cruz et al., 2010). OsRab5a, a small GTPase, is implicated in the intracellular transport of proglutelin from the Golgi to the PSV, and helps maintain the general structure of the endomembrane system in developing rice seeds. In OsRab5a mutants (gpa1 and glup4), some of the glutelin and α-globulin is not delivered to the PSV but is secreted extracellularly to form unique paramural bodies (Wang et al., 2010; Fukuda et al., 2011). The vacuolar processing enzyme (VPE) is essential for proper PSV structure and compartmentalization of rice storage proteins in the PSV. In mutants of VPE (W379 and glup3), glutelin is deposited in the PSV as a precursor and the PSV lacks the typical crystalline lattice structure (Wang et al., 2009; Kumamaru et al., 2010). In addition, independent mutants such as glup1, 2, 5, 6, and 7 have been reported, which suggests that many genes participate in rice storage protein targeting, transport, and deposition (Ueda et al., 2010), although the genes have not been identified. However, little is known about how rice storage proteins are exported from the ER.
Golgi-dependent and -independent pathways are involved in storage protein transport from the ER to the PSV. The direct ER–PSV pathway involving precursor-accumulating (PAC) vesicles has been reported in developing pumpkin seeds (Hara-Nishimura et al., 1998) and in rice (Takahashi et al., 2005). However, most storage proteins travel through the Golgi apparatus to the PSV via dense vesicles (DVs) (Jolliffe et al., 2005; Vitale and Hinz, 2005). In rice, glutelin and α-globulin have been detected in Golgi-associated DVs in developing endosperm cells, which suggests that the Golgi apparatus plays an important role in rice storage protein transport to the PSV (Krishnan et al., 1986, 1992; Fukuda et al., 2011; Washida et al., 2012).
Protein is exported from the ER to the Golgi apparatus via the coat protein complex II (COPII) (Barlowe, 2002). The COPII consists of five proteins: secretion-associated, Ras-related protein 1 (Sar1), and Sec23, Sec24, Sec13, and Sec31. Sar1 is a small GTPase, and Sec23/24 and Sec13/31 are two structural heterodimers that form the inner and outer COPII coat (Barlowe et al., 1994). Sar1 plays a crucial role in initiating the COPII assembly on the ER. Sar1-GTP activated by Sec12 binds to the ER membrane and recruits Sec23/24 and then Sec13/31 heterodimers to form the COPII (Gurkan et al., 2006). Sar1 also participates in cargo selection, membrane curvature, and COPII vesicle fission, and controls membrane constriction to regulate ER export in mammalian cells (Giraudo and Maccioni, 2003; Lee et al., 2005; Sato and Nakano, 2005, 2007; Long et al., 2010). Sar1 is highly conserved in eukaryotes, and its homologues have been identified in Arabidopsis (d’Enfert et al., 1992; Bar-Peled and Raikhel, 1997; Vernoud et al., 2003), Brassica campestris (Kim et al., 1997), and tobacco (Takeuchi et al., 1998). The Sar1 dominant-negative mutant in the GTP-restricted form inhibits protein transport from the ER to the Golgi apparatus, which results in destroyed Golgi integrity in tobacco and Arabidopsis cells (Takeuchi et al., 2000; Phillipson et al., 2001; daSilva et al., 2004; Yang et al., 2005; Hanton et al., 2008; Osterrieder et al., 2009). Arabidopsis contains several Sar1 isoforms. Despite highly identical amino acid sequences, AtSARA1a and AtSARA1b show different localization and different levels of inhibition of soluble α-amylase secretion, which suggests functional heterogeneity among Sar1 isoforms in plants (Hanton et al., 2008). Functional studies of the Sec24 isoforms in Arabidopsis also suggested diverse functions of plant COPII components (Faso et al., 2009; Nakano et al., 2009; Conger et al., 2011). However, the components of the COPII vesicle have not been reported in rice.
In this study, the function of Sar1 in rice was identified by stable endosperm-specific knockdown transformation. Deficiency of OsSar1 in rice endosperm blocked the exit of glutelin and α-globulin from the ER, forming novel electron-dense PBs. The data indicated that the rice storage proteins glutelin and α-globulin were exported from the ER to the Golgi apparatus by a COPII-mediated pathway. The identification and characterization of OsSar1 can help in understanding the detailed mechanism of intracellular transportation of seed storage proteins.
Materials and methods
Isolation of OsSar1a/b/c/d and their promoters
Total RNA from rice was isolated by the TRIpure reagent method (Bioteke), and first-strand cDNA was generated as a template for amplification of OsSar1a/b/c/d coding sequences. Promoters of each OsSar1 were amplified by PCR with rice genomic DNA used as a template. All primer sequences are listed in Supplementary Table S1 available at JXB online. The PCR products were cloned into a pMD18-T vector (Takara), then inserts were sequenced.
GFP and mCherry fusion constructs for transient expression in rice protoplasts
Green fluorescent protein (GFP) or mCherry was fused to the C-terminus of Sar1a/b/c/d (Sar1–GFP or Sar1–mCherry). The ER marker (mCherry–HDEL) was created by combining the signal peptide of AtWAK2 at the N-terminus of mCherry and the ER retention signal His–Asp–Glu–Leu at its C-terminus. The Golgi marker (GmMan1–mCherry) was created by combining the cytoplasmic tail and transmembrane domain (first 49 amino acids) of GmMan1 at the N-terminus of mCherry. The chimeric genes were subcloned into pBI221 under the control of the Cauliflower mosaic virus (CaMV) 35S promoter to obtain transient expression vectors, which were co-transformed into rice protoplasts as described (Chen et al., 2006). Transformed cells were examined under a confocal microscope (Leica TCS SP5) and digital images were recorded. The images were presented as stacks of neighbouring sections according to the cell diameter. The three-dimensional reconstruction functions were employed to compute projections of serial confocal sections.
The soluble and membrane fractions of transformed protoplasts were prepared as described (Hanton et al., 2008).
Construction of the binary vectors and rice transformation
To construct the OsSar1a/b/c/dpro-GUS vector, promoters were introduced upstream of the gusA gene, which encodes β-glucuronidase (GUS) in the binary vector of pCAMBIA1300. To construct the gene overexpression vector, the OsSar1a/b/c/d fragments were inserted in the binary vector pGPTV-GluC-GUS-35S-HPT (Qu et al., 2008) containing the endosperm-specific GluC promoter by replacing the GUS gene. To construct the RNA interference (RNAi) vector, the specific sequence was amplified and ligated to the GluA2-pTCK303 vector, with the endosperm-specific GluA2 promoter replacing the ubiquitin promoter (Wang et al., 2004). All primer sequences used above are listed in Supplementary Table S1 at JXB online.
The binary vectors were introduced into rice (Oryza sativa cv. Kitaake) by Agrobacterium tumefaciens-mediated transformation as described (Qu et al., 2005). Successful transformants were verified by PCR and grown in a greenhouse as described (Qu et al., 2008).
Analysis of GUS expression
Histochemical analysis of GUS expression was conducted as described (Jefferson et al., 1987). Different tissues of OsSar1a/b/c/dpro-GUS transgenic lines were vacuum-infiltrated for 15min in GUS staining buffer and then incubated at 37 °C for 12h. The organs were destained in 70% ethanol until chlorophyll was removed.
Real-time quantitative PCR
Total RNA extraction and first-strand cDNA synthesis was as described above. Real-time quantitative PCR (qRT-PCR) involved the LightCycler480 real-time PCR system (Roche). The rice Actin-1 gene was used as the endogenous control, and all experiments involved at least three biological replicates. All primer sequences used for qRT-PCR are given in Supplementary Table S1 at JXB online.
Protein extraction, SDS–PAGE, and western blot analysis
Total protein was extracted from seeds with use of 0.125M TRIS-HCl, pH 6.8, 4% SDS, 4M urea, and 2% β-mercaptoethanol. Proteins were resolved and separated by 13.6% SDS–PAGE, then transferred onto a polyvinylidene difluoride (PVDF) membrane, which was blocked with non-fat milk and incubated with antibodies, then alkaline phosphatase-conjugated secondary antibodies for immunodetection.
Transmission electron microscopy and immunogold localization
Transverse sections of developing seeds were fixed in PIPES buffer (pH 7.2) as described (Kumamaru et al., 2010). Samples were embedded in Spurr’s low-viscosity resin and sectioned into semi-thin and ultra-thin sections. Semi-thin sections were prepared for immunofluorescence analysis. Ultra-thin sections were stained with uranyl acetate and lead citrate solution, then observed by transmission electron microscopy (TEM; JEM-1230, Hitachi).
For immunogold localization, ultra-thin sections were blocked with 1% bovine serum albumin (BSA) in TRIS-buffered saline (TBS) buffer, then incubated with diluted antibodies. With non-specifically bound antibodies rinsed off, sections were incubated with gold-conjugated secondary antibodies (15nm or 5nm gold particles) and stained with uranyl acetate, then observed by TEM.
Immunofluorescence microscopy
Semi-thin sections were treated with blocking buffer containing 0.9% BSA in phosphate-buffered saline (PBS), then incubated with diluted antibody in blocking buffer. After non-specifically bound antibodies were washed off, sections were incubated with secondary antibody with fluorescein isothiocyanate (FITC) or rhodamine red (Invitrogen), then examined by confocal microscopy (Zeiss 510).
Antibodies
For OsSar1 antibody production, OsSar1c cDNA was cloned into the pGEX-2T vector, and the glutathione S-transferase (GST)-tagged OsSar1 fusion protein was purified. Polyclonal antibody against OsSar1 was then raised in mice and diluted 1:1000. Antibody against glutelin basic subunits was raised in mice and diluted 1:5000. Antibodies against α-globulin, 14kDa prolamin, and OsBip1 were raised in rabbits and diluted 1:1000. Antibodies against α-tubulin (Sigma) and OsCBL (Beijing protein innovation, China) were diluted 1:1500.
Accession numbers
OsSar1a (Os01g23620), OsSar1b (Os12g37360), OsSar1c (Os01g15010), OsSar1d (Os06g12090), AtSar1a (At1g09180), AtSar1b (At1g56330), NtSar1a (BAA13463), NtSar1b (AAF17254), BcSar1a (AAC49716), BcSar1b (AAC49717), LeSar1 (AAA34168), ScSar1 (CAA35978), CeSar1 (CCD73074), MmSar1a (AAH05549), MmSar1b (AAH82550), HsSar1a (AAH03658), and HsSar1b (AAH02847).
Results
Isolation of OsSar1 genes from rice
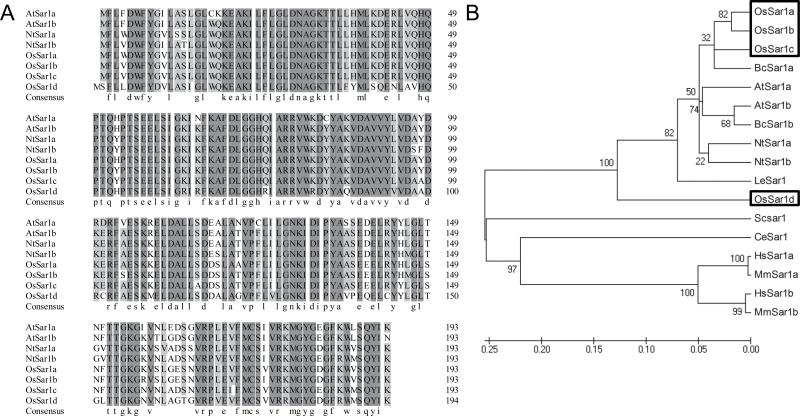
A survey of the rice (O. sativa) genome revealed four Sar1 homologues, named OsSar1a, OsSar1b, OsSar1c, and OsSar1d. The first three homologues encode a polypeptide with 193 amino acids and the latter encodes a polypeptide with 194 amino acids. The amino acid sequences of OsSar1a, b, and c are nearly 90% identical to that of AtSar1 and NtSar1. The amino acid sequence of OsSar1a shares 96, 93, and 78% identity with those of OsSar1b/c/d, respectively (Fig. 1A). A Neighbor–Joining phylogenetic tree was constructed for the 17 Sar1 proteins from plants, animals, and yeast (Fig. 1B). Although Sar1 is highly conserved in eukaryotes, 17 proteins could be classified into two clades, with all of the plant Sar1 proteins in the same clade. OsSar1a, b, and c belong to a group with AtSar1a and AtSar1b, while OsSar1d does not. The phylogenetic analysis suggests that Sar1 proteins are evolutionarily conserved in plants, and OsSar1a, b, and c may function similarly to AtSar1.
Fig. 1.
Multiple sequence alignment of Sar1. (A) Amino acid sequence alignment of OsSar1a/b/c/d by the DNAMAN program. (B) Phylogenetic tree of Sar1. Species designations: animals, Ce, Caenorhabditis elegans; Mm, Mus musculus; Hs, Homo sapiens; yeast, Sc, Saccharomyces cerevisiae; plants, At, Arabidopsis thaliana; Bc, Brassica campestris; Le, Lycopersicon esculentum (Solanum lycopersicum); Nt, Nicotiana tabacum; Os, Oryza sativa. The phylogenetic tree was constructed by use of the MEGA program (Tamura et al., 2011) and used full-length protein sequences by the Neighbor–Joining method with the following parameters: bootstrap (1000 replicates; random seed), pair-wise deletion, and Poisson correction.
OsSar1 isoforms were distributed on the ER and ER export sites (ERES) in rice protoplasts
To investigate the subcellular localization of OsSar1, GFP was fused to the C-terminus of the four Sar1 isoforms and the constructs were transiently expressed in rice protoplasts. Co-expression of each Sar1 isoform with the ER marker mCherry–HDEL showed that the green fluorescence of OsSar1–GFP was partly merged with the red fluorescence of mCherry–HDEL, which indicated that the OsSar1 proteins distributed on the ER. In addition to the reticular structures of OsSar1 proteins, multiple punctate structures were observed (Supplementary Fig. S1A at JXB online).
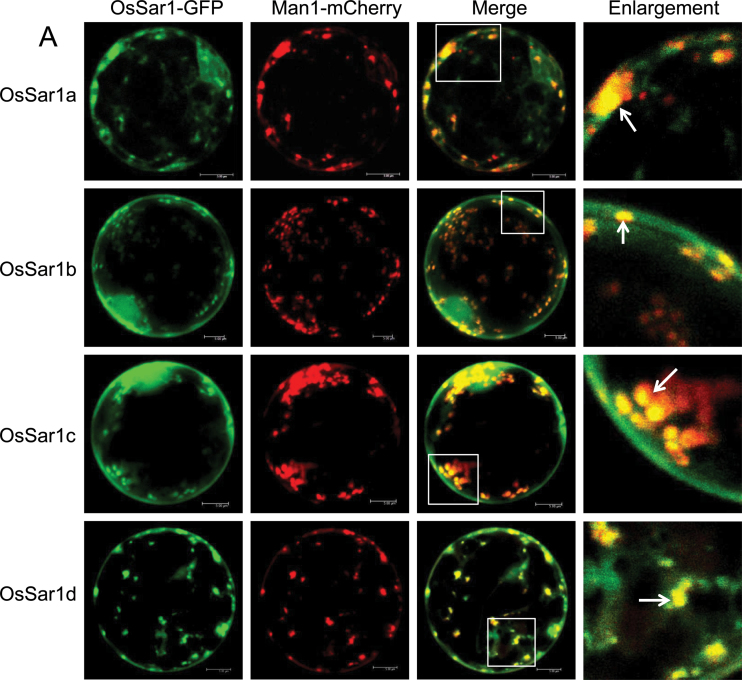
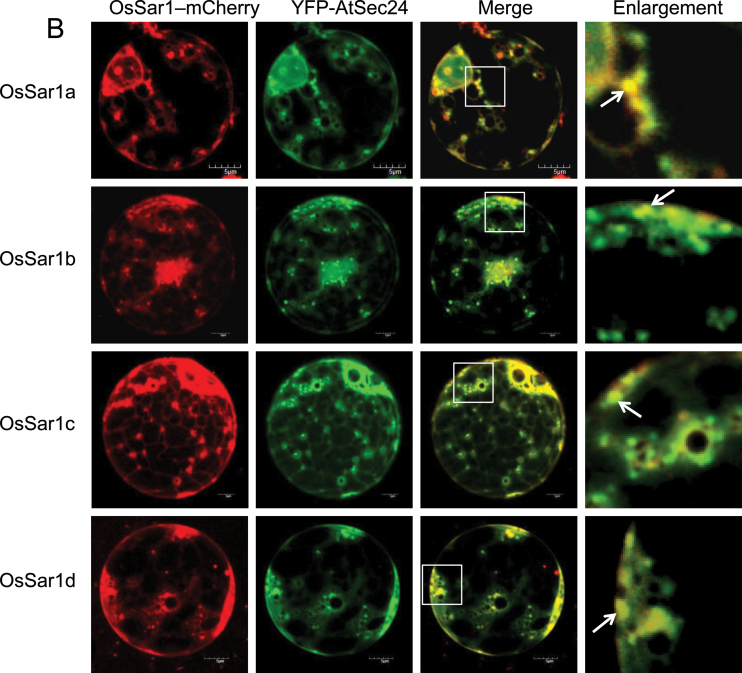
To determine whether the OsSar1 punctate structures were similar to those of NtSar1 and AtSar1 (daSilva et al., 2004; Hanton et al., 2008), each OsSar1–GFP was co-expressed with the Golgi marker Man1–mCherry. In cells transformed with both OsSar1–GFP and Man1–mCherry, most of the punctate structures of OsSar1–GFP co-localized with the Golgi marker (Fig. 2A). To examine whether the OsSar1 was distributed at ERES, OsSar1 was fused with mCherry (OsSar1–mCherry) and co-expressed with yellow fluorescent protein (YFP)–AtSec24, which labels ERES (Hanton et al., 2009), in rice protoplast cells. The punctate structures labelled with OsSar1–mCherry coincided with YFP–AtSec24 fluorescence (Fig. 2B), and the distribution pattern of the four Sar1 proteins was similar. These data indicated that the OsSar1 isoforms were distributed at ERES, at the ER, and in the cytosol.
Fig. 2.
Subcellular localization of Sar1 in rice protoplasts. Rice protoplasts were co-transformed by OsSar1a/b/c/d–GFP with GmMan1–mCherry (A) or OsSar1a/b/c/d–mCherry with YFP–AtSec24 (B). Confocal images of GFP/YFP signal (green), mCherry signal (red), and merged signal. The white arrow indicates the merged punctate structures. Bars=5 μm.
In Arabidopsis, AtSARA1b is more membrane associated than is AtSARA1a (Hanton et al., 2008). To determine the intracellular partitioning of OsSar1, soluble and membrane proteins were extracted from OsSar1–GFP-expressing protoplasts for western blot analysis with antibody against GFP. Each Sar1 isoform was detected in soluble and membrane fractions, but the expression of all isoforms was higher in the soluble than in the membrane fractions (Supplementary Fig. S1B at JXB online). This subcellular distribution of OsSar1 isoforms was similar to that of AtSar1a but differed from that of AtSar1b (Hanton et al., 2008).
OsSar1a, b, and c are expressed universally in rice
qRT-PCR of each OsSar1 revealed that OsSar1a, b, and c were expressed in all examined tissues; however, the expression of OsSar1d was barely detectable (Supplementary Fig. S2 at JXB online). OsSar1a was highly expressed in vegetative tissues such as seedlings, leaf sheaths, and leaves. Expression was lower for OsSar1b than for OsSar1a in most tissues, but higher in leaves and leaf sheaths than in other tissues. OsSar1c expression was abundant in leaves, leaf sheaths, developing embryo, and endosperm.
The expression pattern of OsSar1 homologues was also verified by a stable transgenic strategy using GUS as a reporter gene driven by the native promoter of each OsSar1. GUS signals were detected in callus, young leaves, rootlet, sheath, mature leaves, stems, nodes, glume, stamen, pistil, and seeds of constructs containing the promoter for OsSar1a/b/c (Supplementary Fig. S3 at JXB online). OsSar1d GUS signal was not detected in any tissues because of its low expression. These results were consistent with the qRT-PCR results and indicate that OsSar1a, b, and c are ubiquitously expressed in plants, with OsSar1a mainly expressed in vegetative tissues and OsSar1c abundant in seeds.
No phenotype alteration with endosperm-specific overexpression or knockdown of a single OsSar1
To reveal the function of each OsSar1 in ER export of rice storage proteins in vivo, transgenic rice were generated with OsSar1 specifically overexpressed or suppressed in endosperm under the control of glutelin GluC and GluA-2 promoters, respectively. Successful transformants were screened by RT-PCR. At least five independent transgenic lines were obtained for each construct. The transcription of each OsSar1 was stronger in OsSar1a/b/c/d-overexpressing transformants than in the wild type (Supplementary Fig. S4 at JXB online). The phenotypes of OsSar1-overexpressing seeds were normal. The protein profiles of OsSar1a/b/c/d-overexpressing transformants did not differ from that of the wild type, especially the 57kDa glutelin precursor (Supplementary Fig. S5A). This implied that wild-type seeds contained enough OsSar1 for COPII assembly to transport the storage proteins.
An endosperm-specific RNAi knockdown vector was constructed using the 3′-untranslated regions (UTRs) of OsSar1a, b, or c because of the high similarity of OsSar1 coding sequences. The expression of OsSar1a, b, or c was significantly knocked down in seeds of RNAi transformants (Supplementary Fig. S6 at JXB online). The phenotype of OsSar1a, b, or c RNAi transformant seeds was identical to that of the wild type, with no difference between the wild type and transformants in seed storage protein profiles (Suppelementay Fig. S5B). Therefore, deficiency of any OsSar1 isoform did not affect storage protein transport, and functional redundancy may exist among OsSar1a/b/c in rice endosperm. Because the transcription of OsSar1d was low and GUS signal was not detectable in seeds, OsSar1d RNAi knockdown was not performed.
OsSar1abc RNAi produces abnormal seeds and accumulates proglutelin
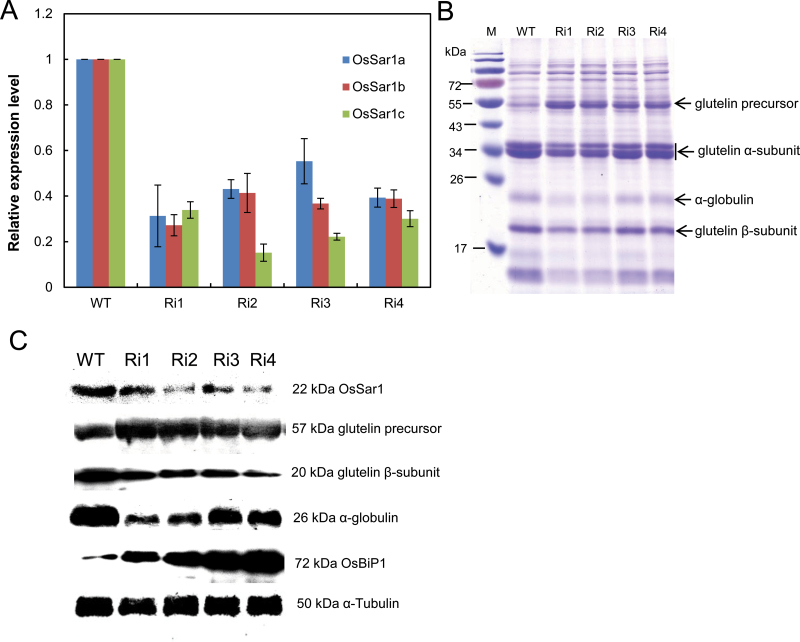
To elucidate the function of OsSar1 in endosperm, transgenic rice were produced with OsSar1a, b, and c knocked down simultaneously by RNAi in endosperm. An OsSar1abc RNAi vector was constructed by using a conserved part of the OsSar1c coding sequence fragment (427bp) under the control of the endosperm-specific promoter GluA-2. Eighteen independent transformants were produced and the expression of OsSar1a, b, and c in the endosperm was measured by qRT-PCR. The mRNA levels of OsSar1a, b, and c were lower in RNAi transgenic lines than in the wild type (Fig. 3A). The western blot analysis revealed that the protein level of OsSar1 was greatly decreased in transgenic endosperm (Fig. 3C).
Fig. 3.
Expression of OsSar1 in OsSar1abc RNAi seed. (A) Quantitative RT-PCR (qRT-PCR) analysis of mRNA levels of OsSar1a, b, and c at 15 DAF in seeds of transformants. (B) SDS–PAGE analysis of seed storage protein in OsSar1a/b/c RNAi transformant seeds. (C) Western blot analysis of protein levels of OsSar1, glutelin precursor and basic subunit, α-globulin, and OsBiP1 in OsSar1abc RNAi transformant seeds. The α-tubulin was used as a loading control. (This figure is available in colour at JXB online.)
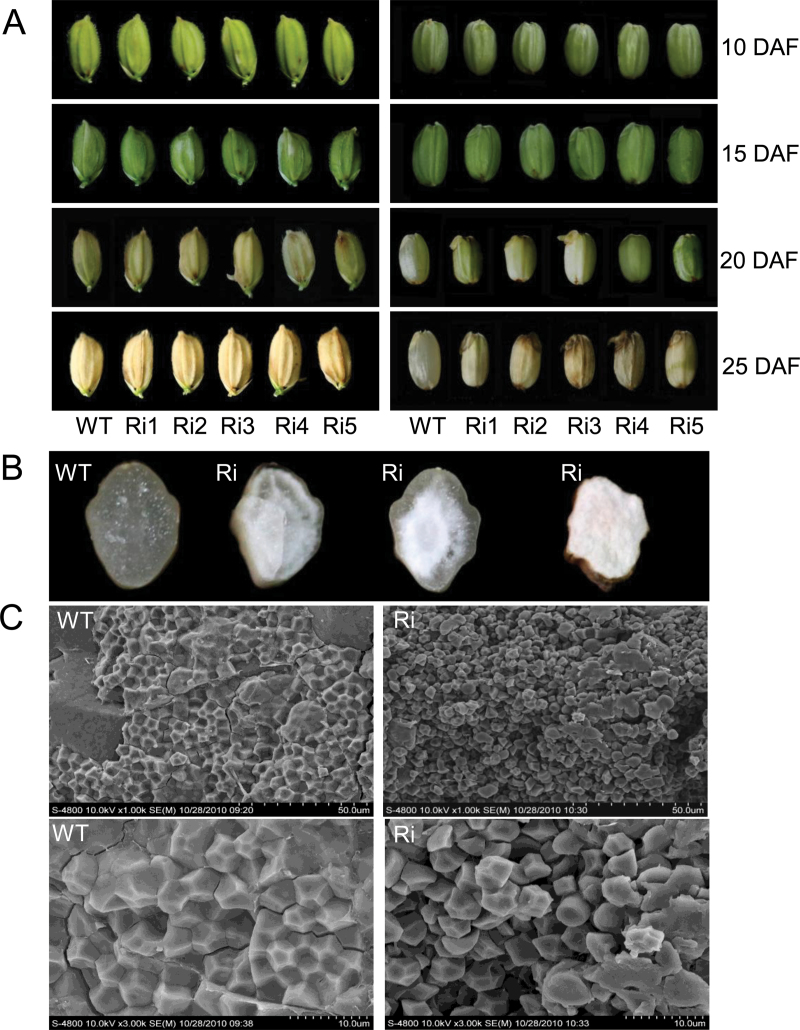
The growth and development of OsSar1abc RNAi lines were almost identical to those of the wild type, because OsSar1a, b, and c were simultaneously knocked down in seeds but not other tissues. However, most mature seeds of the OsSar1abc RNAi lines were opaque, with a floury and shrunken appearance, and failed to germinate. Furthermore, some seeds were even pre-harvest sprouting (Fig. 4A, B). Because the features of seeds were significantly altered in the OsSar1abc RNAi transformants, starch granules were observed in the seeds of transformants by scanning electron microscopy (SEM). In the wild type, the endosperm appeared to be tightly packed with irregularly polyhedral starch granules (Fig. 4C). In contrast, endosperm in the OsSar1abc RNAi transformants appeared to be a loosely packed and fragile structure with spherical starch granules. These morphological changes in the starch granules may be one of the reasons for the floury and shrunken features of the OsSar1abc RNAi transformants.
Fig. 4.
Phenotype analysis of OsSar1abc RNAi transgenic seeds. (A) Morphology of OsSar1abc RNAi seeds at 10, 15, 20, and 25 days after flowering (DAF). The right panel shows the seeds on the left without glumes. (B) Transverse sections of grains. (C) Scanning electron microscopy of endosperm. WT, wild type, Ri, OsSar1abc RNAi lines. (This figure is available in colour at JXB online.)
SDS–PAGE and western blot analyses of developing seeds at 15 days after flowering (DAF) were used to examine whether suppression of OsSar1a/b/c affected accumulation of proteins. The level of the 57kDa glutelin precursor was greatly increased in OsSar1abc RNAi transgenic plants, while the levels of the acidic and basic subunits were greatly reduced (Fig. 3B). In addition, the levels of 26kDa α-globulin and prolamin were reduced. Western blot analysis revealed significant accumulation of the 57kDa glutelin precursor, with the level of the basic subunit lower than that of the wild type. Furthermore, accumulation of the 26kDa α-globulin was suppressed in transformants. However, the level of OsBiP was markedly increased in OsSar1abc RNAi seeds (Fig. 3C). These results implied that the intracellular transport of storage proteins was inhibited and the 57kDa glutelin precursor was blocked in the ER in OsSar1abc RNAi seeds, which enhanced the level of the glutelin precursor in the ER and decreased that of mature glutelin acidic and basic subunits.
Novel ER-derived electron-dense protein bodies formed in OsSar1abc RNAi endosperm
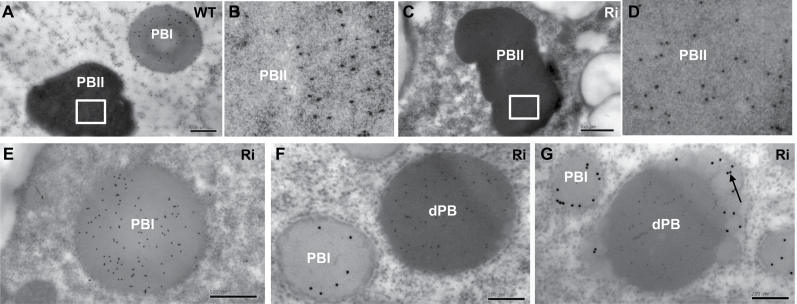
Because the profile of seed storage protein was greatly altered in OsSar1abc RNAi transformants, the intracellular structures of developing endosperm cells in transformants was observed by immunofluorescence microscopy and TEM. Developing rice endosperm has two types of PBs: spherical, electron-lucent PB-I containing prolamin; and irregularly shaped, electron-dense PB-II containing glutelin and α-globulin. Therefore, PB-I was detected by anti-prolamin antibody and PB-II by anti-glutelin antibody. Prolamin and glutelin in the wild type were packaged into separate PBs, with prolamin in a red, spherical, smaller, dot-like PB-I (Supplementary Fig. S7B at JXB online) and glutelin in a green, irregularly shaped, larger PB-II (Supplementary Fig. S7A). In OsSar1abc RNAi endosperm, PB-I was identical to that in the wild type; however, in addition to the normal PB-II, many novel, spherical, smaller glutelin PBs were observed, which were separated with PB-I (Supplementary Fig. S7H).
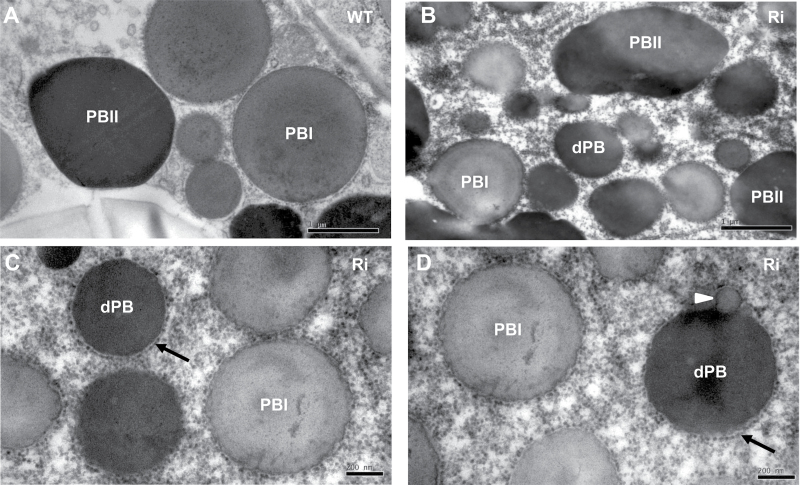
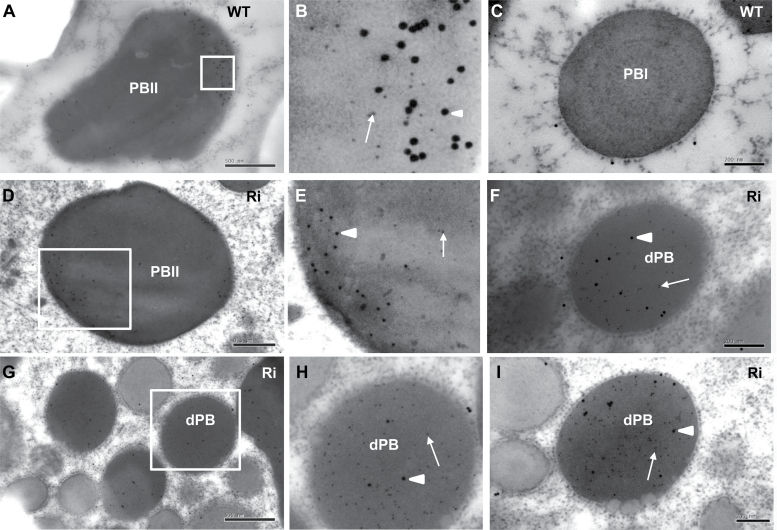
TEM of the endosperm of 15 DAF wild-type seeds revealed that the ER-derived prolamin-containing PB-I was spherical, electron-lucent, and surrounded by rough ER membrane with ribosomes, whereas the glutelin- and α-globulin-containing PB-II was irregularly shaped with high electron density (Fig. 5A). In endosperm cells of OsSar1abc RNAi transformants, in addition to the normal PB-I and PB-II (Fig. 5B), many smaller and spherical novel structures with high electron density and surrounded by ribosomes were observed (Fig. 5C, D). The attachment of the ribosomes on the surface of the novel structures indicated that they were derived from the ER, so these novel structures were designated as ER-derived electron-dense PBs (dPBs). Furthermore, electron-lucent blebbing structures were attached to some of the dPBs (Fig. 5D).
Fig. 5.
Transmission electron microsopy (TEM) of protein bodies in developing endosperm. (A) Wild type; (B–D) OsSar1abc RNAi transformants. The black arrow indicates the ER-derived dense protein body (dPB) surrounded by rough ER membrane with ribosomes. The white arrowhead indicates some dPBs with blebbing structures. Scale bars=1 μm in (A, B) and 200nm in (C, D).
Export of proglutelin and α-globulin from the ER was affected in OsSar1abc RNAi endosperm cells
To determine whether the formation of dPBs was due to the accumulation of proglutelin in the ER, the intracellular localization of glutelin was examined. In wild-type endosperm cells, glutelin was detected in PB-II (Fig. 6A). In OsSar1abc RNAi endosperm cells, glutelin was detected in normal PB-II (Fig. 6B) and in dPBs (Fig. 6C), but not in the electron-lucent blebbing structures (Fig. 6D). These results confirmed that when the expression of OsSar1 was suppressed, a large part of glutelin precursors were not able to transport to the PB-II but were blocked in the ER and formed dPBs, which suggested that the glutelin transport from the ER to PSV depended on COPII.
Fig. 6.
Immunocytochemistry of glutelin in protein bodies. (A) Glutelin in PB-II in the wild type. (B, C) Glutelin localized on the normal PB-II and dPB in OsSar1abc RNAi lines. The black arrow indicates the dPB surrounded by rough ER membrane with ribosomes. (D) The white arrowhead indicates that glutelin did not accumulate in the blebbing structures of dPBs. Gold particles (10nm) indicate the reaction of glutelin antibody. Scale bars=0.5 μm in (A) and 200nm in (B–D).
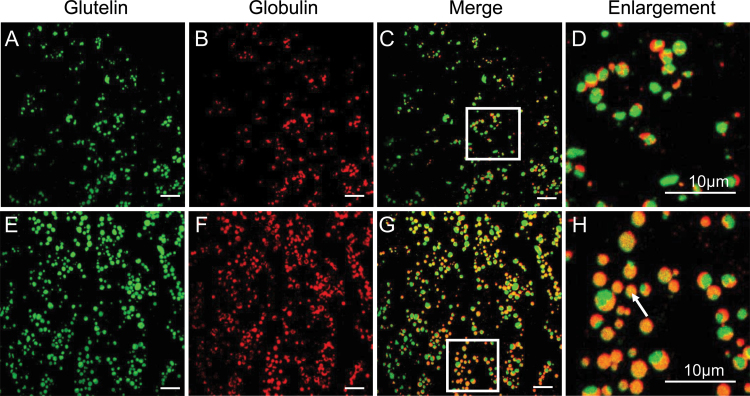
Rice 26kDa α-globulin is also synthesized in the ER and deposited in PB-II. To determine whether the export of α-globulin from the ER was affected in OsSar1abc RNAi endosperm cells, the subcellular co-localization of α-globulin and glutelin was examined by immunofluorescence and immunoelectron microscopy. In the wild type, α-globulin and glutelin were packaged together in irregularly shaped, larger PB-II, with the red fluorescence of α-globulin mainly distributed in the peripheral, amorphous matrix area, and the green fluorescence of glutelin located in the bulky crystalloid regions. The red and green fluorescence were almost not overlapping in the wild type (Fig. 7A–D). However, in OsSar1abc RNAi endosperm cells, in addition to distribution in the normal PB-II, the red fluorescence of α-globulin and the green fluorescence of glutelin were uniformly distributed in dPBs, with the expression of both proteins merged completely. The appearance of yellow fluorescence in dPBs implied no spatial partitioning of α-globulin and glutelin within the dPBs (Fig. 7E–H).
Fig. 7.
Immunofluorescence analysis of glutelin and α-globulin in rice endosperm. Secondary antibodies labelled with FITC (green, A and E) and rhodamine (red, B and F) were used to visualize glutelin and α-globulin, respectively, in wild-type (A–D) and OsSar1abc RNAi endosperm (E–H). (C and G) Merged images. (D and H) Enlarged images of areas inside the white boxes in (C, G). A white arrow indicates α-globulin and glutelin merged in dPBs. Bars=10 μm.
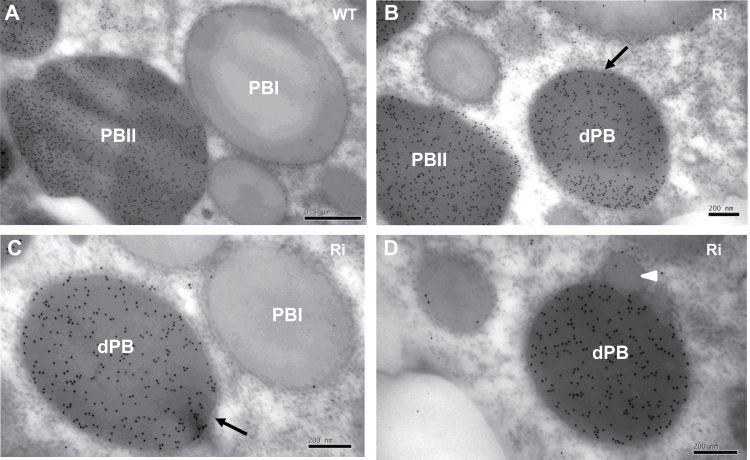
Immunocytochemistry revealed that in the wild type, α-globulin accumulated mainly in the peripheral regions of PB-II, whereas glutelin was mainly distributed in the inner region (Fig. 8A, B). The α-globulin and glutelin were not detected in the PB-I of the wild type (Fig. 8C). Their distribution in normal PB-II was similar in OsSar1abc RNAi and wild-type cells (Fig. 8D, E). However, α-globulin was not distributed in the peripheral regions of dPBs but rather randomly in the inner areas with glutelin (Fig. 8F–H). The α-globulin and glutelin were not detected in the blebbing structures, with low electron density of dPBs (Fig. 8I). Therefore, a large proportion of proglutelin and α-globulin was not transported to PSVs but accumulated in the ER and was sequestered in the smaller, spherical dPBs.
Fig. 8.
Immunocytochemistry of glutelin and α-globulin in protein bodies. (A–C) Wild type; (D–I) OsSar1abc RNAi transformants. (B, E, H) Enlarged images of areas inside the boxes in (A, D, G), respectively. Glutelin and α-globulin antibodies are labelled with 5nm (white arrow) and 15nm (white arrowhead) immunogold particles, respectively. Scale bars=500nm in (A, D, G) and 200nm in (C, F, I).
To make sure that the abnormal accumulation of glutelin and α-globulin in ER is due to the specificity of COPII assembly malfunction, and not a result of the secondary effect from ER stress response, OsCBL, a homologue of AtCBL6 whose transport to the vacuole was COPII independent (Batistic et al., 2010; Bottanelli et al., 2011), was selected as a negative control. The results of western blot analysis showed that the expression levels of OsCBL in OsSar1abc RNAi seeds did not differ from that of the wild type (Supplementary Fig. S8 at JXB online), indicating that the abnormal accumulation of glutelin and α-globulin in the ER was caused by COPII deficiency.
Blebbing structures of dPBs were PB-I derivatives
OsSar1abc RNAi endosperm cells showed no distribution of α-globulin and glutelin in electron-lucent blebbing structures of dPBs. Therefore, if prolamin was distributed in the blebbing structures, it may have caused the low electron density. To support this assumption, the co-localization of prolamin and glutelin was examined, and glutelin was found to be distributed in irregularly shaped PB-II of the wild type and normal PBs of OsSar1abc RNAi, with high electron density (Fig. 9A–D), whereas prolamin was detected only in the spherical PB-I with low electron density (Fig. 9A, E). Most dPBs showed no prolamin, and only glutelin was detected in the spherical dPBs (Fig. 9F). For the blebbing dPBs, glutelin accumulated in the inner areas of dPBs, with high electron density, and some prolamin was in the blebbing structures, with low electron density (Fig. 9G). Therefore, the blebbing structure was an ER-derived PB-I derivative.
Fig. 9.
Immunocytochemistry of glutelin and prolamin in protein bodies. (A, B) Wild type; (C–G) OsSar1abc RNAi transformants. (B, D) Enlarged images of areas inside the boxes in (A, C). Glutelin and prolamin antibodies were labelled with 5nm and 15nm immunogold particles, respectively. The black arrow indicates prolamin accumulated in the blebbing structures of dPBs. Scale bars=500nm in (A, C, E) and 200nm in (F, G).
Discussion
Rice glutelin is synthesized in the ER and then transported to PSVs through or bypassing the Golgi apparatus for final processing to mature acid and basic subunits. Research into glutelin intracellular transportation has focused on factors such as chaperones that affect protein folding, factors affecting glutelin transport from the Golgi to PSVs, and glutelin processing in PSVs (Takemoto et al., 2002; Wang et al., 2009, 2010; Satoh-Cruz et al., 2010; Kumamaru et al., 2010; Fukuda et al., 2011). Little is known about the factors regulating glutelin export from the ER to the Golgi apparatus.
Subcellular distribution of OsSar1 and functional redundancy in rice endosperm
We focused on the function of Sar1 GTPases in storage protein transport in rice because Sar1 plays an essential role in the formation of COPII vesicles in ER to Golgi traffic in yeast, mammalian, and plant cells (Takeuchi et al., 2000; Hanton et al., 2006; Marti et al., 2010). Different subcellular localization of Sar1 isoforms in plant cells has been reported in tobacco and Arabidopsis, which suggests that Sar1 proteins might be functionally distinct in plant cells. In tobacco leaf epidermal cells, NtSar1 is distributed predominantly in the cytosol but is visibly recruited to ERES on co-expression of membrane cargo proteins destined for the Golgi apparatus (daSilva et al., 2004). In Arabidopsis, AtSARA1a and AtSARA1b are distributed at multiple punctate structures and localize at ERES, independently of co-expression of Golgi-destined membrane cargo (Hanton et al., 2008). In this study, we describe four Sar1 isoforms with high amino acid sequence identity in rice. Transient expression of all OsSar1 isoforms revealed a punctate pattern in rice protoplast cells. On co-expression of OsSar1–GFP with the cis-Golgi marker GmMan1–mCherry, the punctate structures labelled with OsSar1 were associated with the Golgi area, while co-expression of OsSar1–mCherry with YFP–AtSec24 confirmed that OsSar1 localized at ERES (Fig. 2). The localization of OsSar1 at ERES is similar to that of AtSARA1, because punctate structures were also observed in the absence of Golgi cargo. Furthermore, the two AtSar1 isoforms show different subcellular distribution and activity despite near-identical protein sequences. However, the subcellular distribution of the four Sar1 isoforms in rice was consistent with that of AtSARA1a but differed from that of AtSARA1b, since they mainly existed in the soluble fraction (Supplementary Fig. S1 at JXB online). This may be because the amino acid residue at the C-terminus of all OsSar1 isoforms is lysine (K), which is conserved in AtSARA1a (Hanton et al., 2008). Although the C-termini of Sar1 may contribute to the membrane association of the protein, the exchange of GDP for GTP on Sar1 triggers the exposure of its N-terminal amphipathic α-helix, which constitutes a membrane anchor to the ER, and then initiates the COPII coat assembly (Bielli et al., 2005; Lee and Miller, 2007). Therefore, GFP/mCherry should be fused to the C-termini of Sar1 for subcellular distribution similar to AtSARA1 (Hanton et al., 2008).
The similar subcellular localization and distribution of the four OsSar1 isoforms raised the question of tissue specificity and redundancy for OsSar1 isoforms in rice. qRT-PCR and stable transgenic analysis with the GUS reporter gene revealed OsSar1a, b, and c expressed universally in rice without tissue specificity (Supplementary Figs S2, S3 at JXB online). To avoid a detrimental effect on growth and development, transgenic plants were produced by endosperm-specific overexpression or suppression of the OsSar1 genes. Endosperm-specific suppression of any OsSar1 meant that rice endosperm could not accumulate proglutelin in the ER, but suppression of the three OsSar1 genes simultaneously caused abnormal seeds and proglutelin accumulation. These results suggest functional redundancy of OsSar1a, b, and c in rice endosperm, which is different from AtSec24, another COPII component. AtSec24 isoforms have incompletely overlapping functions, because a missense point mutation of AtSec24A induced the formation of clusters of the ER and Golgi apparatus, and total loss of AtSec24A function was lethal; the phenotypes could not be complemented by AtSec24B and AtSec24C (Faso et al., 2009; Nakano et al., 2009; Conger et al., 2011). The difference in functional redundancy of OsSar1 and AtSec24 may be related to their different functions. Sar1 acts as a molecular switch to control the assembly of COPII (Memon, 2004), whereas Sec24 selects and captures cargo via specific binding sites of ER export motifs (Sato and Nakano, 2007). The different AtSec24 isoforms may interact with specific cargo that participates in different pathways in cells. The present data suggest that the OsSar1 isoforms are functionally redundant in rice endosperm, but they might have different roles in other tissues.
Co-suppression of OsSar1abc blocked the storage proteins in the ER, forming novel ER-derived electron-dense PBs
Rice glutelins are synthesized on the ER as 57kDa precursors, and finally transported to the PSV, where they are processed into mature acidic and basic subunits. If any step in glutelin transport and processing is defective, the glutelin will remain as 57kDa precursors (Takemoto et al., 2002; Wang et al., 2009, 2010; Fukuda et al., 2011). The accumulation of most mature glutelin, α-globulin, and some prolamin was severely inhibited in OsSar1abc RNAi transformants. Much glutelin was deposited as precursors, and post-translational processing into mature subunits was suppressed (Fig. 3B, C). The intracellular structures of endosperm cells were significantly disrupted by the formation of spherical electron-dense PBs, many of which were surrounded by ribosomes that were never observed in the wild type (Fig. 5). Similar ER-derived PBs were observed in transgenic soybean seeds in which the expression of α and α’ subunits of β-conglycinin was co-suppressed (Kinney et al., 2001). Of note, the dPBs were smaller than the PB-II, and some dPBs were connected to electron-lucent blebbing structures (Figs 5, 6, 8, 9). The highly electron-dense matrix of dPBs contained glutelin and α-globulin, and the low electron-dense blebbing structures contained prolamin (Figs 6, 8, 9). Therefore, dPBs were derived from the ER, and dPBs with blebbing structures may be formed by the adjacent area of the PB ER and cisternal ER, with the blebbing structures as PB-I derivatives.
The dPBs formed in the OsSar1abc RNAi lines contained glutelin and α-globulin but not prolamin. The dPBs differ from those in the rice PDI1-knockout mutant esp2 and BiP-overexpressing lines, which contain both glutelin and prolamin (Takemoto et al., 2002; Yasuda et al., 2009). In esp2, because of loss function of PDI1-1, the glutelin precursor could not fold correctly but, instead aggregated with prolamin via interachain disulphide bonds (Takemoto et al., 2002; Satoh-Cruz et al., 2010). In BiP-overexpressing lines, the high concentrations of BiP might perturb entry into the ER and the folding and sorting of secreted proteins (Yasuda et al., 2009). In OsSar1abc RNAi lines, proglutelin could fold correctly and separate with prolamin but could not be transported from the ER perhaps because of COPII deficiency. ER-derived PBs (MAG bodies) were also found in Arabidopsis mag2 and mag4 mutants, with reduced efficiency of protein transport from the ER to the Golgi complex (Li et al., 2006; Takahashi et al., 2010). However, the distribution of storage proteins in dPBs differed from that in MAG bodies. Within the MAG bodies, two major storage proteins were separately localized with 2S albumin in the core and 12S globulin in the matrix region, whereas in dPBs, α-globulin and glutelin were distributed uniformly without spatial partitioning (Figs 7, 8).
The glutelins and α-globulins are transported to PSVs by a Golgi-dependent pathway in rice
Two pathways were reported for storage protein transport from the ER to PSVs, the Golgi-dependent pathway in which the storage proteins travel through the Golgi apparatus into the PSV via DVs, and the Golgi-independent machinery with the storage proteins aggregated within the ER and transported directly to PSVs by PAC. PAC has been reported in pumpkin (Hara-Nishimura et al., 1998), but the report in rice remains unique (Takahashi et al., 2005) since these vesicles have not been verified in other ultrastructural studies of rice PB biogenesis (Fukuda et al., 2011). Moreover the detection of glutelin and α-globulin in DVs suggested that the Golgi apparatus may mediate the rice storage protein transport (Krishnan et al., 1986, 1992; Fukuda et al., 2011; Washida et al., 2012). Using endosperm-specific RNAi technology, it was confirmed that rice storage protein transport from the ER to PSV depended on COPII. The deficiency of OsSar1 may perturb the formation of COPII, which results in blocking glutelin precursors in the ER and in the formation of dPBs in the ER lumen (Figs 6, 8, 9). Some of the dPBs were connected to but not surrounded by prolamin-containing electron-lucent blebbing structures (Fig. 9), which differed from PAC-like structures reported in rice (Takahashi et al., 2005). It is not surprising that some glutelins and α-globulin were exported from the ER and transported to the PSV in OsSar1abc RNAi lines because RNAi is a knockdown but not knockout technology. The results, together with glutelin and α-globulin being detected in Golgi-associated DVs in developing endosperm cells, indicated that the rice storage protein transport to the PSV is a Golgi-dependent pathway. Further investigation on how the rice storage proteins selectively packaged into the vesicles will be required. The ER export signal and vacuolar sorting determinants in rice glutelin and α-globulin are being identified.
Rice α-globulin RNAs, like those of prolamin, are targeted to the PB ER, whereas glutelin RNAs are enriched on the adjacent cisternal ER (Choi et al., 2000; Crofts et al., 2004, 2005; Washida et al., 2012). However, α-globulin is deposited together with glutelin in the PSV, whereas prolamin is deposited in the ER-derived PB-I. The α-globulin and glutelin are mainly sequestered in the matrix and the crystalloid regions of PSVs, respectively (Kumamaru et al., 2010). Previous studies suggested that α-globulin might be exported directly from the PB ER to the Golgi apparatus because α-globulin was detected only on the PB ER and not on the cisternal ER (Washida et al., 2012). In the OsSar1abc RNAi transformants, α-globulin could not exit from the ER but was distributed uniformly with glutelin in the smaller, spherical ER-derived dPBs without spatial partitioning (Fig. 8). The newly synthesized α-globulin might move from the PB ER lumen to export sites on the cisternal ER and then be transported to PSVs together with glutelin via COPII. How α-globulin moves from the PB ER lumen to export sites on the cisternal ER remains to be investigated.
Deficiency of OsSar1 results in ER stress response
Secretory proteins must move from the ER to reach a final destination for their proper function. When the exit of proteins is disturbed, the proteins will accumulate in the ER, where the high concentrations will cause ER stress. Severe ER stress induces cell death in plant cells (Iwata and Koizumi, 2005). BiP is involved in a signalling pathway as a sensor of the ER stress response and an indicator of ER stress. Overexpression of BiP in transgenic rice seed resulted in an opaque phenotype and produced ER-derived PB-like compartments filled with both prolamin and glutelin (Yasuda et al., 2009; Wakasa et al., 2011). The increased expression of BiP and the formation of dPBs in the OsSar1abc RNAi lines (Figs 3, 5) suggested that the cell might sense a severe stressful condition in the ER because the secretory proteins were blocked in the ER. OsSar1abc RNAi showed reduced levels of prolamin compared with the wild type, similar to that in the proglutelin-accumulating mutants esp2, glup4, glup5, and glup6 (Ueda et al., 2010), which might be a general phenomenon of ER stress response. Deficiency of OsSar1 in rice endosperm induced physiological damage leading to an opaque phenotype, even pre-harvest sprouting. OsSar1abc RNAi lines showed inhibited sorting of glutelin and α-globulin from the ER to PB-II and inhibited normal transport of other secretory proteins. Accumulation of secretory proteins in the ER resulted in ER stress, thus leading to changes to an opaque seed phenotype, with floury and shrunken features (Fig. 4). The seed morphological features of OsSar1abc RNAi lines was similar to that of ER stress lines caused by BiP overexpression (Yasuda et al., 2009), whereas that of the dPBs differed from that of PB-like structures in BiP-overexpressing transformants. The appearance of pre-harvest sprouting suggested that the transport of some proteins involved in seed germination and dormancy was defective in OsSar1abc RNAi seeds, but the identity of the specific proteins is not clear.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Distribution of OsSar1 in rice protoplasts.
Figure S2. The qRT-PCR analysis of relative mRNA expression of OsSar1a, b, c, and d in rice tissue.
Figure S3. Expression pattern of OsSar1 in rice.
Figure S4. RT-PCR analysis of mRNA expression of OsSar1 in wild-type (WT) and overexpressng transgenic seeds.
Figure S5. SDS–PAGE of total seed protein level in OsSar1a overexpression lines (A) and OsSar1c RNAi lines (B).
Figure S6. RT-PCR analysis of mRNA expression of OsSar1a (A), b (B), and c (C) in seeds of relative RNAi transgenic lines.
Figure S7. Immunofluorescence analysis of glutelin and prolamin in rice endosperm.
Figure S8. Western blot analysis of protein levels of OsCBL in OsSar1abc RNAi transformant seeds.
Table S1. Primer sequences used in this study.
Acknowledgements
We thank Dr Kang Chong (Institute of Botany, the Chinese Academy of Sciences, China) for providing the pTCK303 vector. We also thank Dr Liwen Jiang (Centre for Cell and Developmental Biology, the Chinese University of Hong Kong, China) for the YFP–Sec24 construct. This work was supported by the Natural Science Foundation of China (nos 30971562 and 31171368), the National Program of Transgenic Variety Development of China (2013ZX08001–006), and the National High Technology Research and Development Program (863) (2011AA100604).
References
- Barlowe C. 2002. COPII-dependent transport from the endoplasmic reticulum. Current Opinion in Cell Biology 14, 417–422. [DOI] [PubMed] [Google Scholar]
- Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M, Schekman R. 1994. COPII: a membrane coat formed by sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell 77, 895–907. [DOI] [PubMed] [Google Scholar]
- Bar-Peled M, Raikhel NV. 1997. Characterization of AtSec12 and AtSar1: proteins likely involved in endoplasmic reticulum and Golgi transport. Plant Physiology 114, 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistic O, Waadt R, Steinhorst L, Held K, Kudla J. 2010. CBL-mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. The Plant Journal 61, 211–222. [DOI] [PubMed] [Google Scholar]
- Bechtel DB, Juliano BO. 1980. Formation of protein bodies in the starchy endosperm of rice (Oryza sativa L.): a re-investigation. Annals of Botany 45, 503–509. [Google Scholar]
- Bielli A, Haney CJ, Gabreski G, Watkins SC, Bannykh SI, Aridor M. 2005. Regulation of Sar1 NH2 terminus by GTP binding and hydrolysis promotes membrane deformation to control COPII vesicle fission. Journal of Cell Biology 171, 919–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottanelli F, Foresti O, Hanton S, Denecke J. 2011. Vacuolar transport in tobacco leaf epidermis cells involves a single route for soluble cargo and multiple routes for membrane cargo. The Plant Cell 23, 3007–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Tao L, Zeng L, Vega-Sanchez ME, Umemura K, Wang GL. 2006. A highly efficient transient protoplast system for analyzing defence gene expression and protein–protein interactions in rice. Molecular Plant Pathology 7, 417–427. [DOI] [PubMed] [Google Scholar]
- Choi SB, Wang C, Muench DG, Ozawa K, Franceschi VR, Wu Y, Okita T. 2000. Messenger RNA targeting of rice seed storage proteins to specific ER subdomains. Nature 407, 765–767. [DOI] [PubMed] [Google Scholar]
- Conger R, Chen Y, Fornaciari S, Faso C, Held MA, Renna L, Brandizzi F. 2011. Evidence for the involvement of the Arabidopsis SEC24A in male transmission. Journal of Experimental Botany 62, 4927–4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crofts AJ, Washida H, Okita TW, Ogawa M, Kumamaru T, Satoh H. 2004. Targeting of proteins to endoplasmic reticulum-derived compartments in plants. The importance of RNA localization. Plant Physiology 136, 3414–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crofts AJ, Washida H, Okita TW, Satoh M, Ogawa M, Kumamaru T, Satoh H. 2005. The role of mRNA and protein sorting in seed storage protein synthesis, transport and deposition. Biochemistry and Cell Biology 83, 728–737. [DOI] [PubMed] [Google Scholar]
- daSilva LL, Snapp EL, Denecke J, Lippincott-Schwartz J, Hawes C, Brandizzi F. 2004. Endoplasmic reticulum export sites and Golgi bodies behave as single mobile secretory units in plant cells. The Plant Cell 16, 1753–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Enfert C, Gensse M, Gaillardin C. 1992. Fission yeast and a plant have functional homologues of the Sar1 and Sec12 proteins involved in ER to Golgi traffic in budding yeast. EMBO Journal 11, 4205–4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faso C, Chen YN, Tamura K, et al. 2009. A missense mutation in the Arabidopsis COPII coat protein Sec24A induces the formation of clusters of the endoplasmic reticulum and Golgi apparatus. The Plant Cell 21, 3655–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M, Satoh-Cruz M, Wen L, et al. 2011. The small GTPase Rab5a is essential for intracellular transport of proglutelin from the Golgi apparatus to the protein storage vacuole and endosomal membrane organization in developing rice endosperm. Plant Physiology 157, 632–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudo CG, Maccioni HJ. 2003. Endoplasmic reticulum export of glycosyltransferases depends on interaction of a cytoplasmic dibasic motif with Sar1. Molecular Biology of the Cell 14, 3753–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurkan C, Stagg SM, Lapointe P, Balch WE. 2006. The COPII cage: unifying principles of vesicle coat assembly. Nature Reviews Molecular Cell Biology 7, 727–738. [DOI] [PubMed] [Google Scholar]
- Hanton SL, Chatre L, Matheson LA, Rossi M, Held MA, Brandizzi F. 2008. Plant Sar1 isoforms with near-identical protein sequences exhibit different localisations and effects on secretion. Plant Molecular Biology 67, 283–294. [DOI] [PubMed] [Google Scholar]
- Hanton SL, Matheson LA, Brandizzi F. 2006. Seeking a way out: export of proteins from the plant endoplasmic reticulum. Trends in Plant Science 11, 335–343. [DOI] [PubMed] [Google Scholar]
- Hanton SL, Matheson LA, Chatre L, Brandizzi F. 2009. Dynamic organization of COPII coat proteins at endoplasmic reticulum export sites in plant cells. The Plant Journal 57, 963–974. [DOI] [PubMed] [Google Scholar]
- Hara-Nishimura I, Shimada T, Hatano K, Takeuchi Y, Nishimura M. 1998. Transport of storage proteins to protein storage vacuoles is mediated by large precursor-accumulating vesicles. The Plant Cell 10, 825–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y, Koizumi N. 2005. An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proceedings of the National Academy of Sciences, USA 102, 5280–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: betaglucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO Journal 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolliffe NA, Craddock CP, Frigerio L. 2005. Pathways for protein transport to seed storage vacuoles. Biochemical Society Transactions 33, 1016–1018. [DOI] [PubMed] [Google Scholar]
- Kim WY, Cheong NE, Je DY, Kim MG, Lim CO, Bahk JD, Cho MJ, Lee SY. 1997. The presence of a Sar1 gene family in Brassica campestris that suppresses a yeast vesicular transport mutation Sec12-1. Plant Molecular Biology 33, 1025–1035. [DOI] [PubMed] [Google Scholar]
- Kinney AJ, Jung R, Herman EM. 2001. Cosuppression of the α-subunits of β-conglycinin in transgenic soybean seeds induces the formation of endoplasmic reticulum-derived protein bodies. The Plant Cell 13, 1165–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan HB, Franceschi VR, Okita TW. 1986. Immunochemical studies on the role of the Golgi complex in protein-body formation in rice seeds. Planta 169, 471–480. [DOI] [PubMed] [Google Scholar]
- Krishnan HB, White JA, Pueppke SG. 1992. Characterization and location of rice (Oryza sativa L.) seed globulins. Plant Science 81, 1–11. [Google Scholar]
- Kumamaru T, Uemura Y, Inoue Y, Takemoto Y, Siddiqui SU, Ogawa M, Hara-Nishimura I, Satoh H. 2010. Vacuolar processing enzyme plays an essential role in the crystalline structure of glutelin in rice seed. Plant and Cell Physiology 51, 38–46. [DOI] [PubMed] [Google Scholar]
- Lee MC, Miller EA. 2007. Molecular mechanisms of COPII vesicle formation. Seminars in Cell and Developmental Biology 18, 424–434. [DOI] [PubMed] [Google Scholar]
- Lee MC, Orci L, Hamamoto S, Futai E, Ravazzola M, Schekman R. 2005. Sar1p N-terminal helix initiates membrane curvature and completes the fission of a COPII vesicle. Cell 122, 605–617. [DOI] [PubMed] [Google Scholar]
- Li L, Shimada T, Takahashi H, Ueda H, Fukao Y, Kondo M, Nishimura M, Hara-Nishimura I. 2006. MAIGO2 is involved in exit of seed storage proteins from the endoplasmic reticulum in Arabidopsis thaliana . The Plant Cell 18, 3535–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long KR, Yamamoto Y, Baker AL, Watkins SC, Coyne CB, Conway JF, Aridor M. 2010. Sar1 assembly regulates membrane constriction and ER export. Journal of Cell Biology 190, 115–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti L, Fornaciari S, Renna L, Stefano G, Brandizzi F. 2010. COPII-mediated traffic in plants. Trends in Plant Science 15, 522–528. [DOI] [PubMed] [Google Scholar]
- Memon AR. 2004. The role of ADP-ribosylation factor and SAR1 in vesicular trafficking in plants. Biochimica et Biophysica Acta 1664, 9–30. [DOI] [PubMed] [Google Scholar]
- Nakano RT, Matsushima R, Ueda H, Tamura K, Shimada T, Li L, Hayashi Y, Kondo M, Nishimura M, Hara-Nishimura I. 2009. GNOM-LIKE1/ERMO1 and SEC24a/ERMO2 are required for maintenance of endoplasmic reticulum morphology in Arabidopsis thaliana . The Plant Cell 21, 3672–3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterrieder A, Hummel E, Carvalho CM, Hawes C. 2009. Golgi membrane dynamics after induction of a dominant-negative mutant Sar1 GTPase in tobacco. Journal of Experimental Botany 61, 405–422. [DOI] [PubMed] [Google Scholar]
- Phillipson BA, Pimpl P, daSilva LL, Crofts AJ, Taylor JP, Movafeghi A, Robinson DG, Denecke J. 2001. Secretory bulk flow of soluble proteins is efficient and COPII dependent. The Plant Cell 13, 2005–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu LQ, Xing YP, Liu WX, Xu XP, Song YR. 2008. Expression pattern and activity of six glutelin gene promoters in transgenic rice. Journal of Experimental Botany 59, 2417–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu LQ, Yoshihara T, Ooyama A, Goto Y, Takaiwa F. 2005. Iron accumulation does not parallel the high expression level of ferritin in transgenic rice seeds. Planta 222, 225–233. [DOI] [PubMed] [Google Scholar]
- Sato K, Nakano A. 2005. Dissection of COPII subunit–cargo assembly and disassembly kinetics during Sar1p-GTP hydrolysis. Nature Structural and Molecular Biology 12, 167–174. [DOI] [PubMed] [Google Scholar]
- Sato K, Nakano A. 2007. Mechanisms of COPII vesicle formation and protein sorting. FEBS Letters 581, 2076–2082. [DOI] [PubMed] [Google Scholar]
- Satoh-Cruz M, Crofts AJ, Takemoto-Kuno Y, Sugino A, Washida H, Crofts N, Okita TW, Ogawa M, Satoh H, Kumamaru T. 2010. Protein disulfide isomerase like 1-1 participates in the maturation of proglutelin within the endoplasmic reticulum in rice endosperm. Plant and Cell Physiology 51, 1581–1593. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Saito Y, Kitagawa T, Morita S, Masumura T, Tanaka K. 2005. A novel vesicle derived directly from endoplasmic reticulum is involved in the transport of vacuolar storage proteins in rice endosperm. Plant and Cell Physiology 46, 245–249. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Tamura K, Takagi J, Koumoto Y, Hara-Nishimura I, Shimada T. 2010. MAG4/Atp115 is a Golgi-localized tethering factor that mediates efficient anterograde transport in Arabidopsis . Plant and Cell Physiology 51, 1777–1787. [DOI] [PubMed] [Google Scholar]
- Takemoto Y, Coughlan SJ, Okita TW, Satoh H, Ogawa M, Kumamaru T. 2002. The rice mutant esp2 greatly accumulates the glutelin precursor and deletes the protein disulfide isomerase. Plant Physiology 128, 1212–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi M, Tada M, Saito C, Yashiroda H, Nakano A. 1998. Isolation of a tobacco cDNA encoding Sar1 GTPase and analysis of its dominant mutations in vesicular traffic using a yeast complementation system. Plant and Cell Physiology 39, 590–599. [DOI] [PubMed] [Google Scholar]
- Takeuchi M, Ueda T, Sato K, Abe H, Nagata T, Nakano A. 2000. A dominant negative mutant of Sar1 GTPase inhibits protein transport from the endoplasmic reticulum to the Golgi apparatus in tobacco and Arabidopsis cultured cells. The Plant Journal 23, 517–525. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony method. Molecular Biology and Evolution 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Sugimoto T, Ogawa M, Kasai Z. 1980. Isolation and characterization of 2-types of protein bodies in the rice endosperm. Agricultural Biology and Chemistry 44, 1633–1639. [Google Scholar]
- Ueda Y, Satoh-Cruz M, Matsusaka H, Takemoto-Kuno Y, Fukuda M, Okita TW, Ogawa M, Satoh H, Kumamaru T. 2010. Gene–gene interactions between mutants that accumulate abnormally high amounts of proglutelin in rice seed. Breeding Science 60, 568–574. [Google Scholar]
- Vernoud V, Horton AC, Yang Z, Nielsen E. 2003. Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiology 131, 1191–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale A, Hinz G. 2005. Sorting of proteins to storage vacuoles: how many mechanisms? Trends in Plant Science 10, 316–323. [DOI] [PubMed] [Google Scholar]
- Wakasa Y, Yasuda H, Oono Y, Kawakatsu T, Hirose S, Takahashi H, Hayashi S, Yang L, Takaiwa F. 2011. Expression of ER quality control-related genes in response to changes in BiP1 levels in developing rice endosperm. The Plant Journal 65, 675–689. [DOI] [PubMed] [Google Scholar]
- Wang Z, Chen C, Xu Y, Jiang R, Han Y, Xu Z, Chong K. 2004. A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L). Plant Molecular Biology Reporter 22, 409–417. [Google Scholar]
- Wang Y, Ren Y, Liu X, et al. 2010. OsRab5a regulates endomembrane organization and storage protein trafficking in rice endosperm cells. The Plant Journal 64, 812–824. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhu S, Liu S, et al. 2009. The vacuolar processing enzyme OsVPE1 is required for efficient glutelin processing in rice. The Plant Journal 58, 606–617. [DOI] [PubMed] [Google Scholar]
- Washida H, Sugino A, Doroshenk KA, Satoh-Cruz M, Nagamine A, Katsube-Tanaka T, Ogawa M, Kumamaru T, Satoh H, Okita TW. 2012. RNA targeting to a specific ER sub-domain is required for efficient transport and packaging of α-globulins to the protein storage vacuole in developing rice endosperm. The Plant Journal 70, 471–479. [DOI] [PubMed] [Google Scholar]
- Yamagata H, Sugimoto T, Tanaka K, Kasai Z. 1982. Biosynthesis of storage proteins in developing rice seeds. Plant Physiology 70, 1094–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata H, Tanaka K. 1986. The site of synthesis and accumulation of storage proteins. Plant and Cell Physiology 27, 135–145. [Google Scholar]
- Yang YD, Elamawi R, Bubeck J, Pepperkok R, Ritzenthaler C, Robinson DG. 2005. Dynamics of COPII vesicles and the Golgi apparatus in cultured Nicotiana tabacum BY-2 cells provides evidence for transient association of Golgi stacks with endoplasmic reticulum exit sites. The Plant Cell 17, 1513–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda H, Hirose S, Kawakatsu T, Wakasa Y, Takaiwa F. 2009. Overexpression of BiP has inhibitory effects on the accumulation of seed storage proteins in endosperm cells of rice. Plant and Cell Physiology 50, 1532–1543. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.