Abstract
Pheophorbide a oxygenase (PaO) is a key enzyme in chlorophyll catabolism that is known to suppress cell death in maize and Arabidopsis. The catalytic activity of PaO in chlorophyll degradation has been clearly demonstrated, but the function of PaO in the regulation of cell death and plant–microbe interactions is largely unknown. In this study, we characterized a PaO homologue in wheat of the lethal leaf-spot 1 gene, TaLls1, that was induced in leaves infected by Puccinia striiformis f.sp. tritici (Pst) and wounding treatment. The TaLls1 protein contains a conserved Rieske [2Fe-2S] motif and a mononuclear iron-binding site typical of PaOs. Silencing of TaLls1 by virus-induced gene silencing in wheat led to leaf cell death without pathogen attacks, possibly due to the accumulation of pheophorbide a (upstream substrate of PaO), indicating a suppressor role of TaLls1, while overexpression of TaLls1 also triggered cell death in both tobacco and wheat leaves, probably owing to the accumulation of the red chlorophyll catabolite (downstream product of PaO). Further deletion mutant analysis showed that the conserved Rieske domain, but not the iron-binding site, was essential for cell death induction. These results thus suggest a threshold for TaLls1 in maintaining cell homeostasis to adapt in various stresses, and shed new light on the role of TaLls1 in cell death regulation. Furthermore, silencing of TaLls1 in wheat did not change the disease symptoms but enhanced tolerance to Pst via an significant increase in H2O2 generation, elevated cell death occurrence, and upregulation of pathogenesis-related genes.
Key words: cell death, disease resistance, pheophorbide a oxygenase, Puccinia striiformis f.sp. tritici, wheat.
Introduction
Plants mount elaborate defence mechanisms to protect themselves from various environmental stresses (Hutcheson, 1998). Programmed cell death is one key defence mechanism in plants, and is an important process involved in development and defence responses against biotic and abiotic stresses (Dangl et al., 1996). In plant–microbe interactions, programmed cell death occurs during the hypersensitive response (HR) to avirulent pathogens, as well as in response to an attack by a virulent pathogen (Greenberg, 1997). As a typical resistant response, HR is characterized as a rapid and localized cell death that occurs at or around the infection sites that are caused by avirulent pathogens, and its purpose is to protect plants from further pathogen colonization. HR is triggered by the recognition of a plant resistance (R) gene product and a pathogen avirulence (Avr) gene product in a gene-for-gene manner (Greenberg et al., 2000).
In addition to the HR-induced cell death, a class of lesion-mimic (Les/les) mutants is ubiquitous in plants, characterized by misregulated cell death phenotypes that mimic the HR. Unlike the HR, the Les/les mutant spontaneously forms lesions in the absence of any injuries, stresses, or pathogen infections (Neuffer and Calvert, 1975; Walbot et al., 1983). A large number of lesion-mimic mutants have been isolated in higher plants, and some have been characterized in detail. Lesion mimics are sometimes associated with enhanced disease resistance, including elevated expression of pathogenesis-related (PR) genes and other markers that are indicative of defence system activation (Wolter et al., 1993; Dietrich et al., 1994).
So far, many genes that are responsible for lesion-mimic phenotypes have been identified. Some of these genes encode enzymes that are involved in chlorophyll metabolism. Maize mutants deficient in les22, which encodes a key enzyme in the biosynthetic pathway of chlorophyll and heme, develop minute necrotic spots on leaves from the accumulation of photo-excitable uroporphyrin (Hu et al., 1998). In addition to the chlorophyll synthesis mutants, the Arabidopsis acd2 mutant defective in the red chlorophyll catabolite reductase (RCCR), which can break down red chlorophyll catabolites (RCCs) in vitro, exhibits a light-dependent lesion phenotype due to the excessive accumulation of phototoxic RCCs (Matile et al., 1999; Mach et al., 2001).
Pheophorbide a oxygenase (PaO) catalyses the oxygenation of pheophorbide a. It is an important chlorophyll breakdown enzyme. PaO was first cloned from maize under the name ZmLls1 with two conserved motifs, namely a Rieske-type centre and a mononuclear non-heme Fe-binding site, which make it resemble aromatic ring-hydroylating dioxygenases (Gray et al., 1997, 2002). The absence of ZmLls1 in maize resulted in a chloroplast-mediated cell death phenotype (Gray et al., 1997, 2002) that was influenced by light (Close et al., 1995), physical wounding (Yang et al., 2004), and pathogen attack (Obanni et al., 1994). Furthermore, the maize lls1 mutant exhibited elevated resistance to fungal pathogens at the leaf epidermis by reducing the lesion ratio and/or causing fungal sterility. Meanwhile, plants with lls1-type lesions accumulate high levels of PR1 and chitinase proteins (Simmons et al., 1998). Cell death is more likely to be triggered in the lls1 mutant through the accumulation of phototoxic pheophorbide a than by interfering with the cell death suppression mechanism (Gray et al., 1997).
The Arabidopsis accelerated cell death 1 (ACD1) gene, which is similar to PaO, is involved in the oxygenation of pheophorbide a and breaks down chlorophyll (Pružinská et al., 2003). In comparison with the lls1 mutant, an excessive amount of pheophorbide a is accumulated when chlorophyll breakdown occurs in transgenic Arabidopsis As-ACD1 plants, and cell death is induced under both darkness and illumination. Therefore, it appears that the accumulation of pheophorbide a not only enhances the oxidation of cellular components but also functions as a signal molecule or is able to inhibit a specific enzyme that induces cell death (Hirashima et al., 2009). In addition to PaO mutants in maize and Arabidopsis, knockdown of OsPAO in rice results in the accumulation of pheophorbide a and triggers cell death in rice seedlings. OsPAO is constitutively expressed in rice plants but induced by natural senescence and physical wounding (Tang et al., 2011).
PaO has been shown to be a suppressor of cell death in plants as a consequence of its catalytic activity in chlorophyll degradation, but the exact mechanism of cell death as regulated by PaO remains elusive. In this study, we isolated one wheat PaO homologue and characterized its function in cell death and disease resistance to Puccinia striiformis f.sp. tritici (Pst). We showed that the wheat lethal leaf-spot 1 gene, TaLls1, was wound inducible. In the TaLls1-knockdown lines, silencing of TaLls1 led to leaf cell death, suggesting a cell death suppressor role of TaLls1. Meanwhile, cell death was triggered in TaLls1-overexpressed wheat and tobacco leaves, which further proved a positive correlation of cell death and TaLls1 accumulation. Thereby, we presumed a rheostat role of TaLls1 in cell death regulation with a delicate threshold to maintain cell homeostasis in adaption to various stress. Deletion mutant analyses revealed that the conserved Rieske domain in the TaLls1 protein was essential for the cell death induction in overexpressing leaves. Furthermore, during the wheat–Pst interaction, TaLls1-knockdown plants exhibited reduced susceptibility to virulent stripe rust fungus. Our results demonstrated a negative regulation of TaLls1 in wheat resistance to Pst and provide new insight for a role of TaLls1 in cell death regulation, not only as a cell death suppressor.
Materials and methods
Plant materials, growth conditions, and chemical treatments
A Suwon 11 wheat genotype containing YrSu, Puccinia striiformis f.sp. tritici (Pst) pathotype CYR23 (avirulent) and the Pst pathotype CYR31 (virulent) were used in the wheat–Pst interaction study. The plant growth conditions and inoculation of Pst were operated as described by Kang and Li (1984). To study TaLls1 expression levels in wheat leaves that were infected by Pst CYR31 or CYR23, leaf tissues were sampled at 0, 8, 12, 24, 48, 72, and 120h post-inoculation (p.i.). Parallel mock-inoculated control plants were brushed with sterile water. Three biological replicates were used for each assay.
Nicotiana benthamiana plants used for transient overexpression of TaLls1 by particle bombardment were grown at an ambient temperature of 25 °C and light conditions of 16h light/8h dark.
Chemical treatments were performed as described by Wang et al. (2012b ).To cause wounding, the first leaves were mechanically scraped with a needle. For the cold treatment, leaves of the same developmental stage were kept at 4 °C. Leaves that were treated with various chemicals and stress elicitors along with the control plants were harvested at 0, 2, 6, 12, 24, and 48h post-treatment (p.t.). All samples were rapidly frozen in liquid nitrogen and stored at –80 °C. Three biological replications were performed independently for each time point.
Total RNA extraction and quantitative reverse transcription-PCR (qRT-PCR)
Total RNA from wheat leaves treated with exogenous hormones, challenged by abiotic stresses, infected with stripe rust, and derived from different wheat organs was extracted using Trizol (Invitrogen, Carlsbad, CA, USA). Two micrograms of total RNA was reverse transcribed into cDNA with an oligo(dT)18 primer using an RT-PCR system (Promega, Madison, WI, USA). The expression pattern of TaLls1 under the different conditions as described above were detected by qRT-PCR following the procedure described by Wang et al. (2009) using a 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The wheat elongation factor TaEF-1a gene (GenBank accession no. Q03033) was used as the internal reference for each qRT-PCR. All reactions were performed in triplicate, and reactions with non-template were used as negative controls. The comparative 2–ΔΔCT method was used to quantify relative gene expression (Livak and Schmittgen, 2001). The primers used for qRT-PCR are listed in Supplementary Table S1 at JXB online.
Isolation of TaLls1 cDNA sequence
Following the known sequence of the partial 3′ end of the cDNA fragment, a set of Race-5R primers that were geared towards the 5′ end was designed. 5′ RACE was performed using a SMART RACE cDNA Amplification Kit (Clontech, Mountain view, CA, USA). The full-length sequence of TaLls1 was assembled using the CAP3 Sequence Assembly Program.
Sequencing and sequence analysis
The conserved motifs and domain structure of TaLls1 protein were analysed using InterPro Scan. TMpred was used for transmembrane analysis and the signal peptide was predicted using TargetIP. Multiple sequence alignments were carried out using DNAMAN software, and polygenetic relationships were inferred using the neighbour-joining method.
Plasmid construction
A barley stripe mosaic virus (BSMV) γRNA-based vector was constructed as previously described by Holzberg et al. (2002). cDNA fragments derived from the coding sequence and the 3′ untranslated region (150bp, nt 1512–1661) and from the coding sequence (360bp, nt 729–1088) were used to construct the recombinant TaLls1-as and TaLls1-as1 plasmids, respectively. To guarantee the specificity of gene silencing, fragments that showed the highest polymorphism within the gene family and the lowest sequence similarities with other genes using a BLASTN search in the NCBI database were chosen for constructing their γRNA-based derivative plasmids.
The plasmids that were used in the transient expression of TaLls1 were constructed as described by Dou et al. (2008). pUCTaLls1 was obtained by insertion of the entire TaLls1 coding sequence into the XmaI- and KpnI-digested pUCBAX plasmid. pUCTaLls11–178 and pUCTaLls1212–397 constructs were made in a similar manner, containing the amino acid sequence of TaLls1 from positions 1 to 178 and 212 to 397, respectively.
Primers for all plasmid constructions are documented in Supplementary Table 1.
BSMV-mediated TaLls1 silencing in the Suwon 11 wheat cultivar
Infectious BSMV RNAs were prepared from each linearized plasmid by in vitro transcription using a high-yield capped RNA transcription kit (mMESSAGE mMACHINE; Ambion). A total of 2.5 μl of each transcript, including the BSMV RNAs α, β and genetically modified γ, were combined with 42.5 μl of FES buffer (Pogue et al., 1998) and inoculated into the second leaves of wheat plants by gently rubbing the surface with a gloved finger at the two-leaf stage (Scofield et al., 2005). BSMV:TaPDS and BSMV:00 were used as controls for the BSMV infection. Mock inoculations were carried out using 1× FES buffer. Each assay consisted of 18 seedlings and was conducted at least three times. BSMV-infected wheat plants were kept in a growth chamber at 25±2 °C. The fourth leaves were further inoculated with fresh urediniospores of Pst CYR23 or CYR31 at 9 d after virus inoculation, and the plants were then maintained as described above. The phenotypes of the fourth leaves were observed and photographed 14 d after pathogen inoculation.
Evans blue staining to detect cell death
To determine whether the silencing of TaLls1 could result in lesion mimicking, the phenotype of the BSMV-infected wheat plants without Pst inoculation was continually observed until 6 weeks after virus inoculation. Segments of the fourth wheat leaves infected with BSMV were detached and stained with 0.5% Evans blue (Sigma Chemicals Co., St. Louis, MO, USA) as described by Wang et al. (2007) to identify the dead cells. Plants inoculated with BSMV:00 were used as controls. Eighteen seedlings were examined for each assay. Ten fields of view for each assay were observed, and three biological replications were performed. The cell death areas in each field were calculated with DP-BSW software, and Tukey’s test was used for statistical analysis.
RNA analysis by qRT-PCR
The fourth leaves inoculated with BSMV:00, BSMV:TaLls1-as, or BSMV:TaLls1-as1 were collected at 0, 24, 48, and 120h p.i with CYR23 or CYR31 separately in addition to the mock-inoculated leaves. qRT-PCR was performed to determine the silencing efficiency of TaLls1 for each assay. The relative transcript levels of the PR protein genes (PR1, AAK60565; PR2, DQ090946; and PR5, FG618781), reactive oxygen species-related genes (catalase, TaCAT, X94352; class III superoxidase, TaPOD, TC303653), and secondary metabolite genes (TaPAL, TC294834) were also confirmed using qRT-PCR.
Histological observation of TaLls1-knockdown wheat plants
During the interaction between the stripe rust fungus and the TaLls1-knockdown plants, fungal development and host response were observed microscopically. Wheat leaves infected with BSMV were sampled at 0, 24, 48, and 120h p.i. with stripe rust fungus. The staining and fixing of specimens were performed as described by Wang et al. (2007). The hyphal length of both the virulent and avirulent stripe rust pathogen during infection were viewed under differential interference contrast optics and measured using DP-BSW software, as well as the spread of the fungal growth. The autofluorescence of attacked mesophyll cells was observed to determine the necrotic cell area using a fluorescence microscope (excitation filter 485nm, dichromic mirror 510nm, barrier filter 520nm) and measured with DP-BSW software. The percentage of infection sites displaying host cell necrosis was recorded in the compatible interaction. All microscopic examinations were performed with an Olympus BX-51 microscope (Olympus Corporation, Japan). Observations of 50 infection sites on each of five randomly selected leaf segments per treatment were carried out.
H2O2 production was studied in TaLls1-knockdown leaves at 24h p.i. with either the virulent or avirulent Pst using 3,3′-diaminobenzidine (DAB; Amresco, Solon, OH, USA) staining (Wang et al., 2007). Standard deviations were determined and Tukey’s test was used for statistical analysis.
Particle bombardment assay for overexpression of TaLls1 in tobacco and wheat
Particle bombardment assays were performed using a Bio-Rad He/1000 particle delivery system with a double-barrelled extension for overexpression of TaLls1 in tobacco leaves (Dou et al., 2008). The leaves of 5–7-week-old N. benthamiana plants were used. For the bombardment, 9mg of M-10 tungsten particles (Bio-Rad) was combined with 50 μg of empty vector and 50 μg of β-glucuronidase (GUS) plasmid (pUCGUS) or a mixture of 50 μg of plasmids encoding the TaLls1 protein (pUCTaLls1) and 50 μg pUCGUS. The mixed tungsten particles and plasmids were prepared for 30 shots. In each shot, a mixture of 1.67 μg of empty vector and 1.67 μg of pUCGUS or 1.67 μg of pUCTaLls1 and 1.67 μg of pUCGUS were delivered into the host cell side by side via the double-barrelled gene gun. The bombarded leaves were incubated for 3 d at 28 °C in darkness, and subsequently stained and destained as described by Dou et al. (2008). For each paired shot (GUS+TaLls1 DNA versus GUS+control DNA), the log ratio number for the fusion protein was calculated for comparison with that of the control using the Wilcoxon signed-ranks test, and at least 16 pairs of shots were performed
For the particle bombardment assay in wheat leaves, the first leaves of 7-d-old Suwon 11 wheat were used. During the bombardment, the leaves were tightly placed in dishes and shot with a Bio-Rad He/1000 single-barrelled particle delivery system according to a protocol described previously (Schweizer et al., 2000; Douchkov et al., 2005; Wang et al., 2012a ). All of the plasmids were prepared at approximately 1 μg μl–1. Seven micrograms of GUS plasmid (pUCGUS) and 7 μg of empty vector or 7 μg pUCTaLls1 were mixed with 26.7 μl of 90mg ml–1 of tungsten particles in a 1.5ml Eppendorf tube. The DNA–tungsten mixtures were prepared as described by Wang et al. (2012a ) and 4 μl of the mixture was used for each shot. The leaves were kept at 28 °C in darkness for 2 d and then stained as described above for 16h and destained in 100% ethanol. Each assay consisted of seven shots and was conducted at least twice. The significant differences between the treatments were analysed with a paired sample t-test using SPSS software.
The two truncated TaLls1 constructs pUCTaLls11–178 and pUCTaLls1212–397, which contained the conserved Rieske [2Fe-2S] motif and the mononuclear iron-binding site, were tested individually in tobacco and wheat, as well as pUCTaLls1.
Chlorophyll catabolite extraction and high-performance liquid chromatography (HPLC) analysis
The fourth leaves of the BSMV-infected plants were sampled 14 d after inoculation and pheophorbide a was extracted according to the method described by Pružinská et al. (2005). Following the procedure as described (Roca et al., 2004; Jiang et al., 2007), pheophorbide a was determined by reverse-phase HPLC (HPLC/Waters 600 controller) equipped with a Diamonsil C18 5 μm (150 mm×4.6mm) column and Waters 2487 dual λ absorbance detector. The RCCs of the TaLls1-overexpressing wheat leaves by particle bombardment were extracted according to the method described by Pružinská et al. (2007).
Data analysis
Analysis of variance and Tukey’s test for statistical analysis were performed using SAS software 9.13.
Results
TaLls1 encodes a typical PaO protein
A fragment of 669bp from a wheat–Pst incompatible interaction cDNA library was isolated. BLASTX analysis revealed a 669bp 3′-end cDNA fragment with high similarity to the Zea mays lethal leaf-spot 1 gene (ZmLls1, GenBank Accession No. AAC49676.1). Subsequently a 1633bp 5′-end cDNA fragment was amplified by 5′-RACE and a full-length 1926bp cDNA fragment in size was obtained using the CAP3 Sequence Assembly Program and was designated TaLls1.
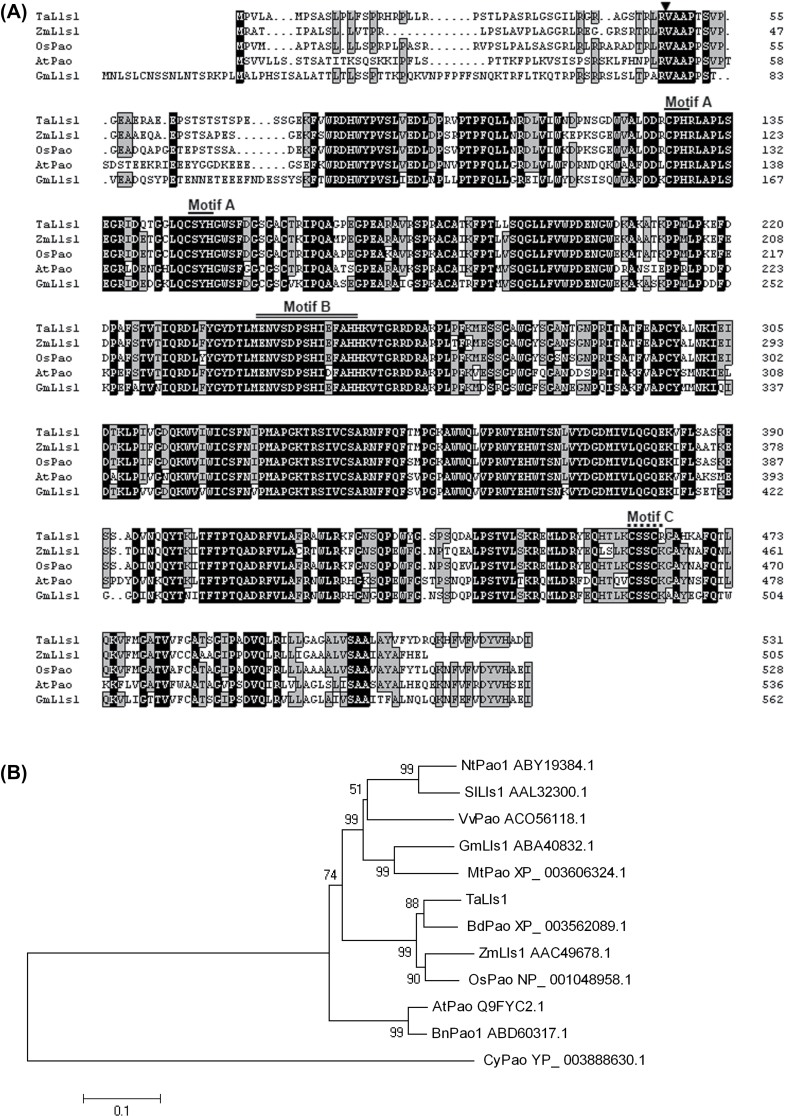
TargetP analysis indicated a chloroplast-targeting peptide at the N terminus, which was cleaved between aa 47 and 48 (R/V). In addition, a transmembrane helix from aa 490 to 511 was predicted by the TMpred program. The structural analysis of TaLls1 revealed a conserved Rieske [2Fe-2S] iron–sulphur motif, a mononuclear non-heme iron-binding motif, and a C-terminal CxxC motif. Alignment of TaLls1 with the Lls1 homologue from other species indicated that Lls1 proteins are conserved in mono- and dicotyledonous plants although residues at the N terminus were highly polymorphic (Fig. 1A).
Fig. 1.
Multi-sequence alignment and phylogenetic analysis of TaLls1 and other members of the PaO family. (A) Multiple alignment of amino acids. Identical and similar amino acid residues are shaded in black and light grey, respectively. The arrow indicates the cleavage site of the chloroplast peptide; Motif A, indicated by a single line, is the Rieske iron-binding motif; Motif B, indicated by a double line, is the mononuclear iron-binding site; Motif C, indicated by a dashed line, is the conserved CxxC sequence. (B) Phylogenetic analysis of TaLls1 and other PaO family members using MEGA 4.1 software. Branches are labelled with the protein names and GenBank accession numbers. Ta, Triticum aestivum L; Os, Oryza sativa; Zm, Zea mays; At, Arabidopsis thaliana; Nt, Nicotiana benthamiana; Sl, Solanum lycopersicum; Gm, Glycine max; Bn, Brassica napus; Mt, Medicago truncatula; Vv, Vitis vinifera; Cy, Cyanothece sp.
Phylogenetic analysis (Fig. 1B) revealed that TaLls1 homologous proteins from monocotyledonous plants were assembled in a large clade in which TaLls1 and BdPao were tightly clustered and OsPaO and ZmLls1 were assembled into the other tight cluster. Homologous proteins from dicotyledonous plants were clustered into another two large groups. The close relationship between TaLls1, OsPao, and ZmLls1 suggested a similar function for these genes.
TaLls1 expressed in different wheat tissues
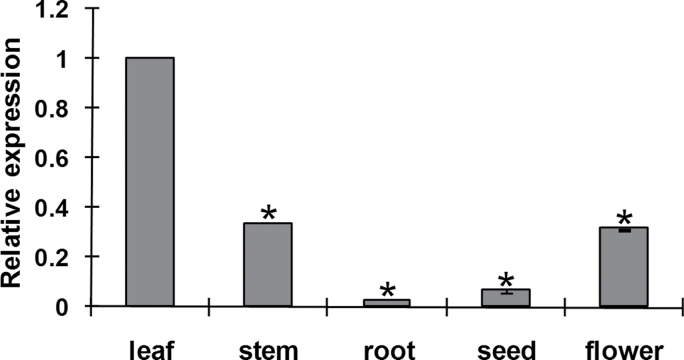
To determine the expression profiles of TaLls1, we examined the transcript of TaLls1 in different wheat tissues. qRT-PCR analyses revealed its abundance in green leaf, and showed that the level was twofold lower in the stem and flower than in the leaves (Fig. 2). TaLls1 expression was also detectable in wheat seeds and roots although at relatively low levels, about 15 and 49 times lower than that in the leaf.
Fig. 2.
Expression pattern of TaLls1 in different wheat tissues. Samples were collected from leaves, stems, roots, seeds, and flowers. Three independent biological replications were performed. Expression levels were normalized to TaEF-1a. Asterisks indicate a significant difference (P <0.05) from the leaf using Student’s t-test.
TaLls1 is upregulated following wounding, abscisic acid, and Pst treatments
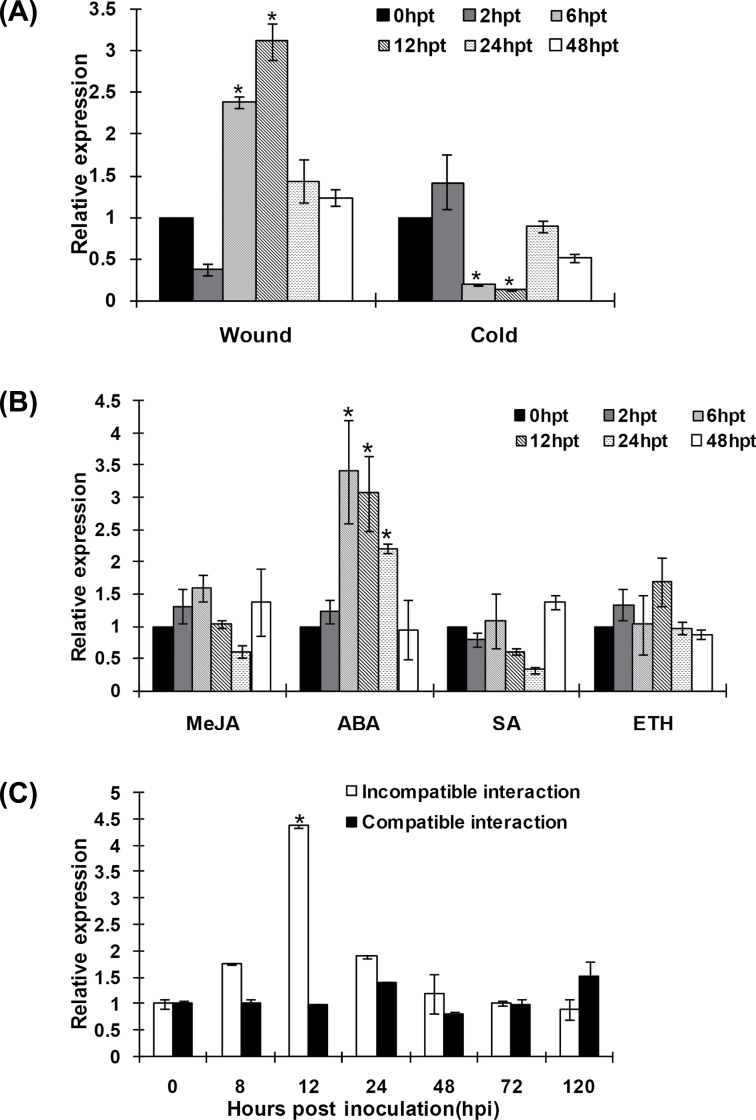
We further examined the expression level of TaLls1 in response to various abiotic and biotic stresses. The transcript level of TaLls1 increased twofold 6h after wounding treatment and reached a maximum of about threefold higher than that of the control at 12h (Fig. 3A). Under cold treatments, the expression of TaLls1 significantly decreased from 6 to 12h and then increased again to a normal level (Fig. 3A).
Fig. 3.
Expression profiles of TaLls1 in response to abiotic stresses (A), exogenous hormones (B), and pathogen attack (C). Expression levels were normalized to the wheat elongation factor TaEF-1a gene. Results are shown as means ±standard deviation of three biological replications. Asterisks indicate a significant difference (P <0.05) from 0h p.t. or 0h p.i. using Student’s t-test. ABA, abscisic acid; SA, salicylic acid; ETH, ethylene; MeJA, methyl jasmonate.
We also assayed TaLls1 expression in wheat leaves treated the exogenous hormones salicylic acid, ethylene, methyl jasmonate, and abscisic acid. As shown in Fig. 3B, TaLls1 transcription was transiently upregulated twofold at 6h p.t., with a gradual decrease from 12 to 24h p.t., and finally returned to a normal level at 48h p.t. following abscisic acid treatment. In contrast, salicylic acid, ethylene, and methyl jasmonate treatments did not cause significant changes in the expression level of TaLls1 (Fig. 3B).
During the wheat–Pst interactions, the transcription level of TaLls1 was sharply induced at 12h p.i. in wheat leaves challenged by the avirulent Pst race CYR23, reaching a level that was threefold higher than that of the control plants (Fig. 3C). In wheat leaves challenged by the virulent Pst race CYR31, the level of the TaLls1 transcription did not change significantly (Fig. 3C).
Knockdown of TaLls1 enhances wheat tolerance to virulent Pst
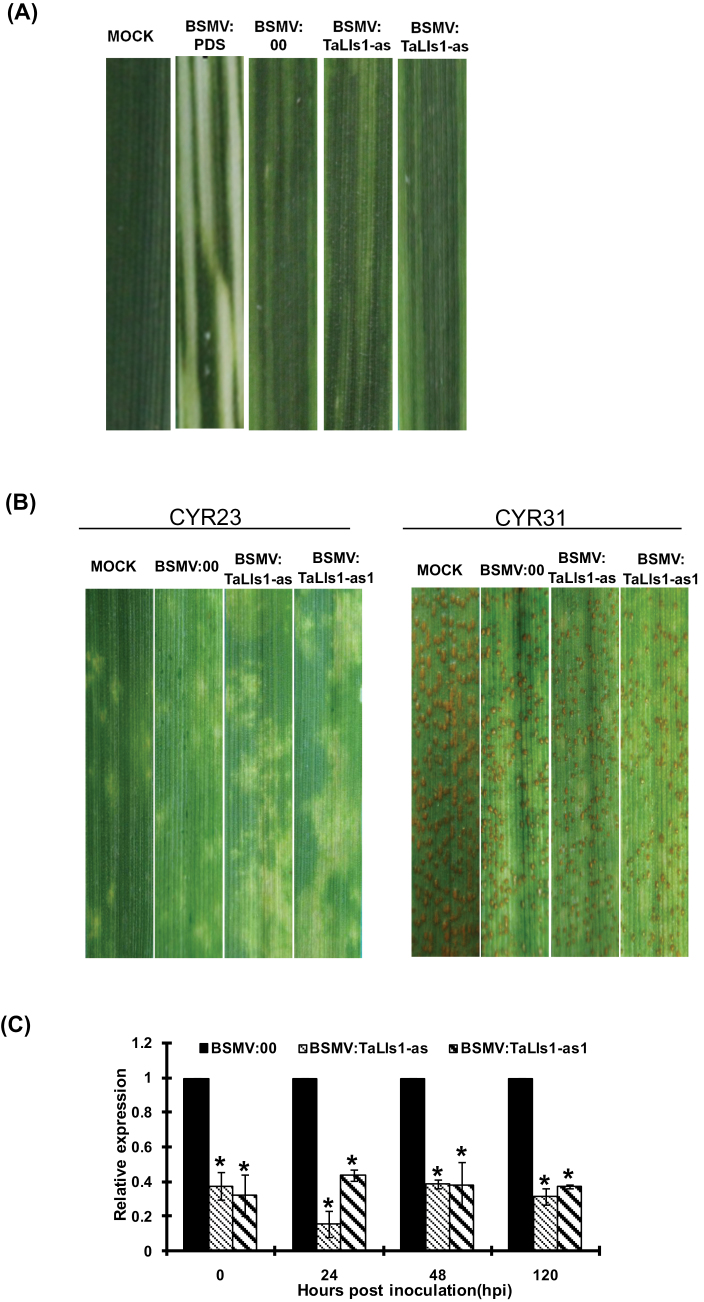
In order to get more details of the role of TaLls1 in wheat resistance to Pst, virus-induced gene silencing was used to silence the expression of TaLls1 during the wheat–Pst interaction. Mild chlorotic mosaic symptoms appeared on the fourth leaves at 9 d p.i. in all plants that were inoculated with BSMV (Fig. 4A), and the BSMV:TaPDS-inoculated plants exhibited strong photobleaching symptoms (Fig. 4A), suggesting that the BSMV virus-induced gene silencing system worked well. No obvious disease phenotypic changes were observed on wheat leaves from the TaLls1-knockdown plants inoculated with Pst race CYR23 or CYR31 when compared with the wild-type plants (Fig. 4B). A typical HR was observed on the CYR23-infected leaves, and all leaves challenged by CYR31 exhibited a fully susceptible phenotype. These results suggested that knockdown of TaLls1 was unable to alter the resistance or susceptibility phenotype of Suwon 11 to Pst.
Fig. 4.
Functional analyses of TaLls1 during the interaction between wheat and stripe rust using a BSMV-mediated virus-induced gene silencing system. (A) Mild chlorotic mosaic symptoms were observed on the fourth leaves of seedlings at 9 d p.i. with BSMV, and photobleaching was evident on the fourth leaves of plants infected with BSMV:TaPDS. CK, Wheat leaves with FES buffer. (B) Disease phenotypes of the fourth leaves pre-inoculated with BSMV:00 and then challenged with avirulent CYR23 or virulent CYR31. (C) Silencing efficiency assessment of TaLls1 in the fourth leaves of TaLls1-knockdown plants during a compatible interaction. Wheat leaves inoculated with BSMV:00 and sampled after inoculation with CYR23 were used as the controls. Data were normalized to the TaEF-1a expression level. Error bars represent variations among three independent replicates. Asterisks indicate a significant difference (P <0.05) from BSMV:00 using Student’s t-test. (This figure is available in colour at JXB online.)
To clarify whether TaLls1 was successfully silenced, qRT-PCR was carried out on RNA extracted from the fourth leaves of TaLls1-knockdown plants. As shown in Fig. 4C and Supplementary Fig. S1 (at JXB online), the abundance of TaLls1 transcripts was greatly reduced to different extents in TaLls1-knockdown plants when compared with the control.
Because no macroscopic phenotype changes in response to avirulent or virulent races of Pst were observed in TaLls1-knockdown wheat, we further studied the fungal development and host response during Pst infection through histological observations (Fig. 5). As shown in Table 1, the fungal hyphal length of CYR31 in TaLls1-knockdown plants was statistically (P >0.05) similar to the control as well as to that of Pst CYR23 (Supplementary Table S2 at JXB online). Additionally, the area infected by the pathogen at 120h p.i. also showed no significant difference between TaLls1-knockdown plants and the control (Table 1, Supplementary Table S2).
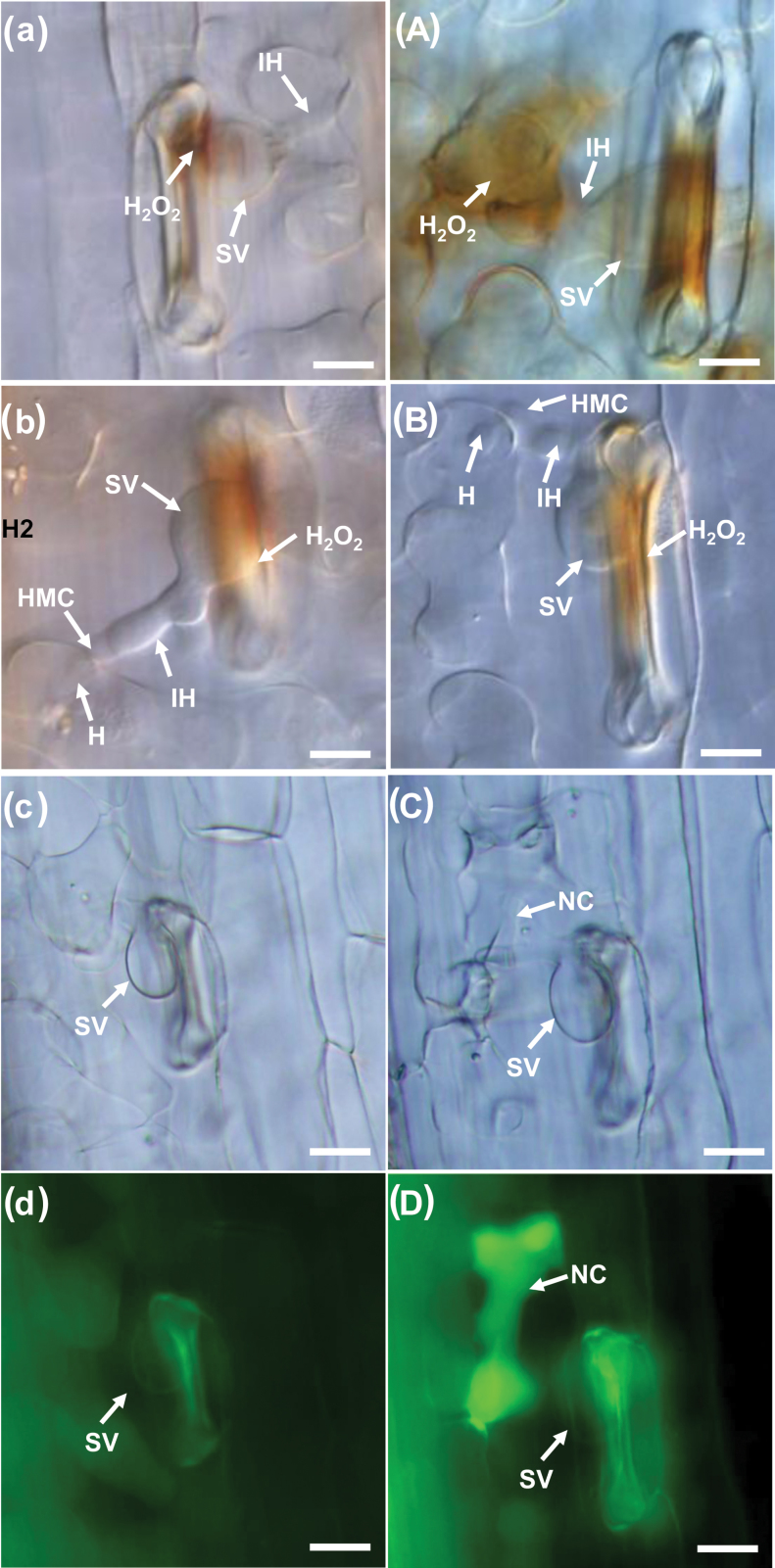
Fig. 5.
Histological observation of fungal growth and host response in wheat infected with BSMV: 00 and recombinant BSMV after inoculation with the virulent Pst pathotype CYR31. (a–d) Fungal growth and H2O2 accumulation in BSMV:00-infected plants 24h p.i. (a) or 48h p.i. (b), and the necrotic cells at 120h p.i. (c, d). (A–D) Fungal growth and H2O2 accumulation observed in BSMV:TaLls1-as-infected plants 24h p.i. (A) or 48h p.i. (B), and the necrotic cells at 120h p.i. (C, D). The photos of (c) and (d) were taken from the same infection site, as were (C) and (D). H2O2 accumulation was calculated by DAB staining. Bars, 20mm. SV, substomatal vesicle; HMC, haustorial mother cell; H, haustoria; IH, infection hypha; NC, necrosis cell. (This figure is available in colour at JXB online.)
Table 1.
Histological observations during the compatible interaction between the stripe rust fungus and TaLls1-knockdown wheat plants. Significance was measured according to a paired sample t-test method (different lower-case letters indicate a significant difference: b*, P <0.01; b, P <0.05).
| Treatmenta | Hyphal lengthb | Infected areac | H2O2 d | Ratio of cell death (%)e | ||
|---|---|---|---|---|---|---|
| 24h p.i. | 48h p.i. | 120h p.i. | 120h p.i. | 24h p.i. | 120h p.i. | |
| BSMV: 00 | 1.54a | 2.68a | 11.7a | 5.24a | 3.60a | 1.95±0.76 |
| BSMV:TaLls1-as | 1.72a | 2.62a | 11.9a | 5.76a | 7.59b* | 7.89±1.31 |
| BSMV:TaLls1-as1 | 1.50a | 2.62a | 13.4a | 7.07a | 5.32b | 7.02±0.62 |
a Wheat leaves pre-infected with BSMV: 00 or the recombinant BSMV: TaLls1-as and BSMV: TaLls1-as1 followed by inoculation with Pst CYR31 in the compatible interaction.
b The average distance from the junction of the substomatal vesicle and the hyphal tip calculated from at least 50 infection sites (×10 μm).
c The average infected area of expanding hyphae plus the host cells calculated from at least 50 infection sites (×1000 μm2).
d The amount of H2O2 production was identified by DAB staining and measured by calculating the area of H2O2 produced in at least 50 infection sites (×100 μm2).
e Ratio of cell death at the infection sites counted from at least 50 infection sites.
To analyse the host response, we measured the cell death areas per infection site at 120h p.i. in the incompatible interaction and determined the percentage of cell deaths resulting from successful infections in relation to the compatible interaction. In compatible controls, cell death was seldom observed around the infection sites, but in TaLls1-knockdown plants, the occurrence of necrotic cells that were caused by pathogen infection increased by approximately threefold (Table 1). In the incompatible interaction, the average cell death areas were 1.5–2-fold higher than those of the control (Supplementary Table S2). These results indicated that knockdown of TaLls1 resulted in an increase in the occurrence and area of cell death during Pst infection.
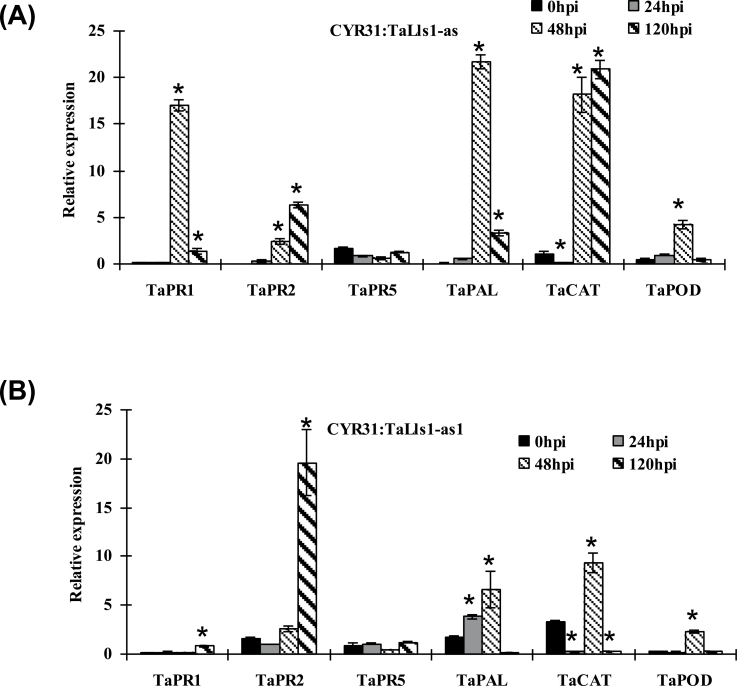
To confirm that the increase in cell death was involved in the resistance response, we examined the expression of PR proteins in TaLls1-knockdown plants by qRT-PCR. The expression of PR1 and PR2 was induced significantly in both TaLls1-knockdown plants when challenged by the virulent CYR31 race (Fig. 6) as well as the avirulent CYR23 (Supplementary Fig. S2 at JXB online). In contrast, the expression of PR5 showed no significant induction (Fig. 6, Supplementary Fig. S2). In additional, the expression level of phenylalanine ammonia-lyase (PAL), a defence-related gene, was dramatically upregulated as early as 24h p.i. in both the compatible (Fig. 6) and incompatible (Supplementary Fig. S2) interactions. These results indicated that knockdown of TaLls1 enhanced the tolerance of wheat to Pst to some degree but was insufficient to change the interaction type.
Fig. 6.
Transcriptional changes in defence-related genes in TaLls1-knockdown wheat seedlings using qRT-PCR. Leaves infected with BSMV:00 or the recombinant BSMV:00γ-infected seedlings were sampled at 0, 24, 48, and 120h p.i. Expression levels were normalized to the TaEF-1a gene. Error bars represent the variation among three independent replicates. Asterisks indicate a significant difference (P <0.05) from 0h p.i. using Student’s t-test.
To further clarify the resistance response in TaLls1-knockdown plants, H2O2 accumulation was detected by DAB staining. The results showed that H2O2 accumulation was almost doubled in TaLls1-knockdown seedlings compared with that in the control plants at 24h p.i. in the compatible interaction (Fig. 5a, A), and the amount of H2O2 generated in TaLls1-as knockdown plants (Table 1) was almost comparable to that of the incompatible interaction (Supplementary Table S2). The increased amounts of H2O2 were accompanied by transient expression of catalase (CAT) and superoxidase (POD) (Fig. 6), which might remove the excessive production of H2O2.
Knockdown of TaLls1 in wheat induces cell death without a pathogen attack
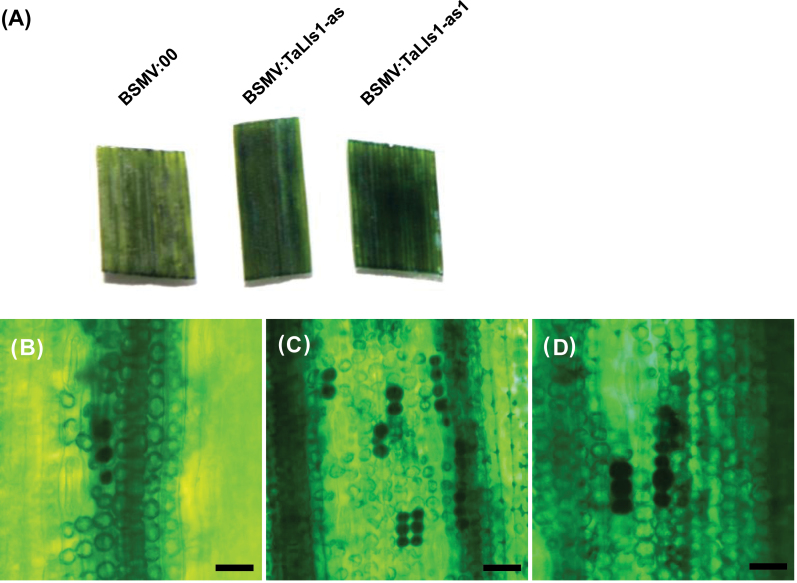
PaO has been reported to be a cell death suppressor in Arabidopsis and maize (Gray et al., 1997; Pružinská et al., 2003). To confirm its role in cell death in wheat, we carried out transcriptional suppression of TaLls1in wheat plants without Pst infection. As shown in Fig. 4A, the fourth leaves of TaLls1-knockdown seedlings exhibited mild chlorotic mosaic symptoms but no lesion formation. From the fourth leaves to the seventh leaves, mild chlorotic mosaic symptoms were constantly observed and no lesion-mimic formation was detected. Although no macroscopic lesion formations were observed, cell death was detected by Evans blue staining, whereby the dead cells were stained dark blue. The fourth leaves infected with recombinant BSMV showed a darker blue staining than those infected with BSMV:00 (Fig. 7A). Microscopic observation of the stained leaf segments revealed more dead cells stained dark blue in leaves infected with BSMV:TaLls1-as (Fig. 7C) and BSMV: TaLls1-as1(Fig. 7D) than those infected with BSMV:00 (Fig. 7B). Statistics analyses revealed that the average area of cell death in each field of view indicated by Evans blue staining in TaLls1-knockdown plants was one- to threefold greater than that in the control plants (Table 2).
Fig. 7.
Induction of cell death in TaLls1-knockdown wheat seedlings. (A) The fourth leaves of the wheat seedlings infected with BSMV:00 and the recombinant BSMV were stained with Evans blue and photographed. (B–D) Microscopic observation of dead cells stained with Evans blue in leaf segments infected with BSMV:00 (B), BSMV:TaLls1-as (C), and BSMV: TaLls1-as1 (D). Areas of cell death in each field were measured by DP-BSW software and calculated from at least ten fields for each segment. (This figure is available in colour at JXB online.)
Table 2.
Cell death induction in TaLls1-knockdown plants without pathogen attacks as measured by Evans blue staining.
| Treatmenta | BSMV:00 | BSMV:TaLls1-as | BSMV:TaLls1-as1 |
|---|---|---|---|
| Average area (×1000 μm2)b | 2.00±0.31 | 8.56±1.01 | 4.90±0.52 |
| P valuec | NA | <0.05 | <0.05 |
a The fourth wheat leaves inoculated with BSMV:00 and the recombinant BSMV:TaLls1-as, BSMV:TaLls1-as1 were sampled at 9 d p.i. and stained with Evans blue.
b Area of dead cells stained with Evans blue were counted from at least ten fields in each sample.
c Significance analysis was measured with the paired sample t-test method. NA, not applicable.
Overexpression of TaLls1 directly triggers cell death in tobacco and wheat
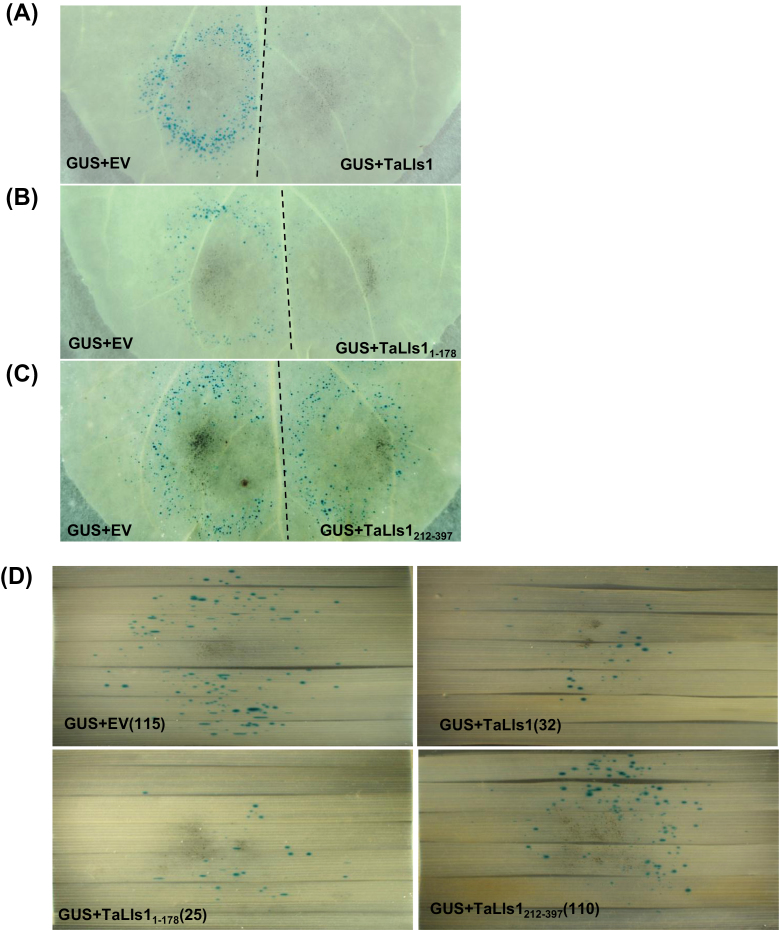
Besides the knockdown of TaLls1, we also investigated the involvement of TaLls1 in cell death regulation through transient expression in tobacco by particle bombardment. Because GUS is expressed only in living cells, fewer blue GUS spots indicate more cell death. On the half of tobacco leaves that were co-bombarded with GUS and empty vectors, numerous blue spots were observed (Fig. 8A), representing living tobacco cells. On the other half of tobacco leaves that were co-bombarded with GUS and TaLls1 vectors, much fewer GUS spots were observed (Fig. 8A), with a reduction of approximately 70% (Table 3), indicating that more cell death had occurred.
Fig. 8.
Overexpression of TaLls1 triggers cell death in N. benthamiana leaves and T. aestivum leaves through particle bombardment assay. (A–C) Transient expression of TaLls1 in N. benthamiana leaves using double-barrelled particle bombardment. Leaves were bombarded with the pairs of DNA mixtures indicated. The dotted line indicates the position of a divider that was used to prevent overlap of the two bombardment areas. (D) Overexpression of TaLls1 in T. aestivum leaves using a single-barrelled particle bombardment. DNA mixtures of different groups of bombarded leaves are indicated, and the number in the parentheses represents the number of GUS spots. Statistical analysis of results from 14 shots was conducted for each assay. EV, empty vector. (This figure is available in colour at JXB online.)
Table 3.
Induction of PCD by transient expression of TaLls1.
| Experiment | Barrel 1a | Barrel 2a | Direct ratiob | P valuec |
|---|---|---|---|---|
| A | TaLls1+GUS | EV+GUS | 0.42±0.08 | <0.01 |
| B | TaLls11–178+GUS | EV+GUS | 0.35±0.07 | <0.01 |
| C | TaLls1212–397+GUS | EV+GUS | 0.98±0.05 | >0.05 |
a Barrels 1 and 2 were physically identical and the masses of DNA in each barrel were identical. All of the replicates were conducted on the petiole-proximal half of the leaves and then the petiole-distal half. EV, empty vector.
b Ratios of blue spots that were counted in each barrel. Geometric averages and standard error were calculated from log ratios obtained from 16 pairs of shots.
c The P value for the direct comparison was calculated from the log ratios using a Wilcoxon signed-ranks test. A significant P value indicates that cell death was induced by TaLls1.
To learn more about the mechanism of cell death induced by TaLls1, we studied the activities of the two conserved motifs in TaLls1. The results (Table 3) showed that overexpression of PUCTaLls11–178, which contains the conserved Rieske [2Fe-2S] binding motif, induced cell death with a 68.9% reduction in GUS blue spots (Fig. 8B), whereas overexpression of PUCTaLls1212–397, including the mononuclear iron-binding motif, failed to induce cell death (Fig. 8C).
The transient expression of exogenous genes in tobacco has been widely used to study their role in cell death, but questions still remain, such as to what extent different plant species share similar cell death regulation mechanisms. Thus, we further studied the role of TaLls1 in cell death by the one-barrel co-bombardment system in wheat leaves. As shown in Fig. 8D, blue spots on the leaves that were co-bombarded with GUS and PUCTaLls1 or TaLls11–178 were significantly lower than in the control leaves (Supplementary Table S3 at JXB online), which is consistent with the results observed in tobacco leaves. On the other hand, overexpression of TaLls1212–397 in wheat leaves did not affect the number of blue spots as shown in tobacco leaves.
Pheophorbide a in TaLls1-knockdown plants and RCCs in TaLls1-overexpressing leaves are accumulated
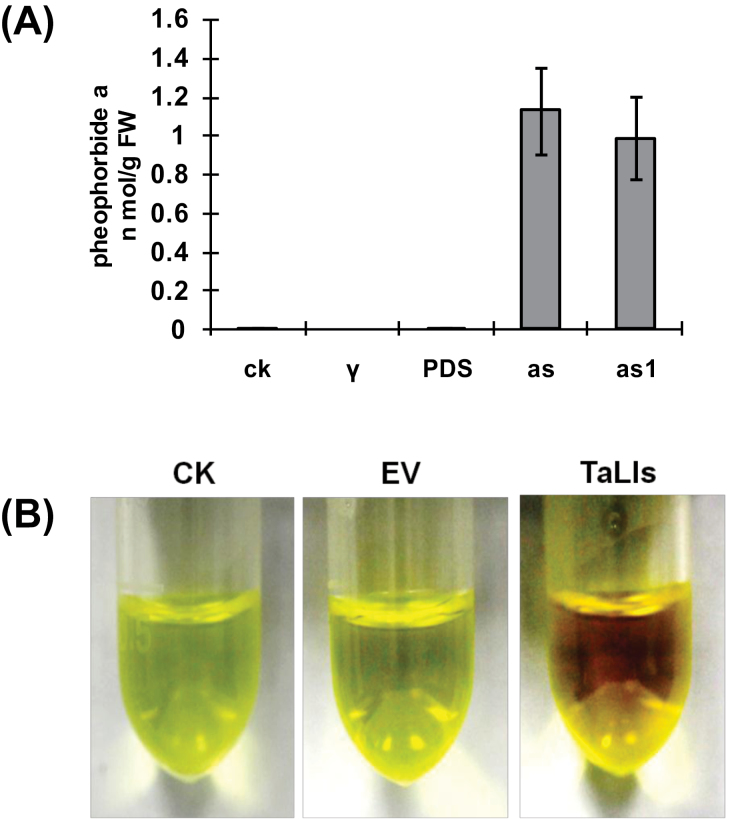
Recent reports have shown that defects in PaO in the Arabidopsis acd1 mutant and the maize lls1 mutant result in accumulation of pheophorbide a in leaves (Pružinská et al., 2003; Tanaka et al., 2003). HPLC analyses revealed that pheophorbide a accumulated to a concentration of 1.13 nmol g–1 and 0.99 nmol g–1 of fresh weight in TaLls1-as- and TaLls1-as1-knockdown plants, respectively, while it could not be detected in leaves infected with BSMV:00 or BSMV:TaPDS (Fig. 9A), suggesting that the accumulation of pheophorbide a was due to the downregulation of TaLls1.
Fig. 9.
Determination of pheophorbide a in TaLls1-knockdown plants and RCCs in TaLls1-overexpressing leaves. (A) Pheophorbide a determination in TaLls1-knockdown plants by HPLC. The fourth leaves of the plants infected with recombinant BSMV were sampled at 14 d p.i. to determine the pheophorbide a concentration. Significant accumulation of pheophorbide a was detected in plants infected with BSMV:TaLls1-as (as) and BSMV:TaLls1-as1 (as1), while none was detected in wheat plants inoculated with FES buffer (CK), BSMV:00 (γ) or BSMV:TaPDS (PDS). (B) RCC determination in wheat leaves overexpressing TaLls1. Methanol extracts of the TaLls1-overexpressing leaves exhibited deeper red compared with the leaves bombarded with empty vector (EV) and the healthy wild-type leaves (CK). (This figure is available in colour at JXB online.)
Defects in RCCR in the Arabidopsis acd2 mutant result in accumulation of RCCs accompanied by increased transcription of PaO (Pružinská et al., 2007). It is presumed that RCCs accumulate in TaLls1-overexpressing wheat leaves. We found that RCC extracts from TaLls1-overexpressing leaves were a deeper red compared with the control leaves and leaves bombarded with empty vector (Fig. 9B), as observed in RCCR1i rice plants (Tang et al., 2011). Thus, we assumed that the deep red colour was due to the accumulation of RCCs.
Discussion
In the present study, we have reported on the identification and functional analysis of a novel wheat Lls1 gene, which is homologous to PaO in maize and Arabidopsis. ZmLls1 was wound inducible in maize (Yang et al., 2004), and our qRT-PCR results also indicated the induction of TaLls1 upon wounding treatment. Giridhar and Thimann (1985) reported that wounding promotes chlorophyll loss in oat leaves, so we inferred that the elevated TaLls1 expression is responsible for the quick removal of chlorophyll in damaged cells. As for the decreased expression of TaLls1 under cold treatments, this may be associated with dampened PaO activity in the breakdown of chlorophyll. During seed development in canola, freezing delays the degradation of chlorophyll due to a reduced PaO activity (Chung et al., 2006). Our data suggest a regulated activity of TaLls1 in response to abiotic stresses, which may be related to its catalytic activity in chlorophyll breakdown.
During chlorophyll breakdown, chlorophyll is degraded to safe linear tetrapyrroles through a series of reactions catalysed by chlorophyllase, magnesium dechelatase, and PaO. PaO catalyses the oxygenic ring opening of pheophorbide a between C4 and C5 to RCCs (Pružinská et al., 2003, 2005; Tanaka et al., 2003). To clarify whether TaLls1 has this oxygenase activity, we assessed the content of pheophorbide a in TaLls1-knockdown plants. HPLC analyses revealed that pheophorbide a accumulated due to the silencing of TaLls1 in TaLls1-knockdown plants, implying that TaLls1 has PaO activity.
Accompanied by the accumulation of pheophorbide a, silencing of TaLls1 caused a significant increase in cell death, indicating a cell death suppressor role of TaLls1, as observed for ZmLls1 mutants (Gray et al., 1997) and Arabidopsis acd1 mutants (Hirashima et al., 2009). It has been reported that pheophorbide a, the substrate of PaO, accumulates and generates reactive oxygen species under light conditions, which lead to cell death (Pružinská et al., 2003, 2005; Tanaka et al., 2003). Thus, we infer that, in TaLls1-knockdown plants, the reduced activity of TaLls1 resulted in the accumulation of pheophorbide a, which would induce further cell death owing to the simulated H2O2 production. However, despite this phototoxic feature, pheophorbide a has been assumed to specifically inhibit the activity of channel proteins or other cellular components that are essential for membrane integrity and to cause the destruction of membrane systems (Hirashima et al., 2009). In addition, pheophorbide a has also been postulated to function as a signal molecule that regulates gene expression and induces programmes cell death (Hirashima et al., 2009). Therefore, in this study, we confirmed the cell death suppressor role of TaLls1, but the detailed mechanism of the cell death caused by pheophorbide a needs to be investigated further. Different from the lesion-mimic phenotype observed in ZmLls1 mutants, TaLls1 silencing was insufficient to cause visible necrosis in TaLls1-knockdown plants, which may due to the complicated regulatory mechanisms of plant cell death. In addition, the deficiency caused by TaLls1 silencing may be complemented by other genes with a similar function in wheat or genes from other pathways. Nevertheless, TaLls1 was demonstrated to partially contribute to cell death in wheat.
Although ZmLls1, AtAcd1, and OsPAO potentially are cell death suppressors according to the results of mutant and silencing assays (Gray et al., 1997; Pružinská et al., 2003; Tang et al., 2011), there is still no direct overexpression evidence for its role in cell death. Interestingly, in this study, the transient expression of TaLls1 also could induce cell death in both tobacco and wheat leaves instead of a cell death suppressor. In order to further determine the cause of this, we assessed the content of RCCs, which are the product of PaO, and found an accumulation of RCCs in wheat leaves overexpressing TaLls1. Previous studies have indicated that accumulation of RCCs is responsible for cell death in acd2 mutant and OsRCCRi rice plants (Pružinská et al., 2007; Tang et al., 2011). In addition, H2O2 is generated together with the accumulation of RCCs, which play an important role in acd2-induced cell death (Yao and Greenberg, 2006). Thus, we inferred that it was H2O2 generation by RCC accumulation that induced cell death in TaLls1-overexpressing wheat leaves. Here, our findings provide direct evidence of the correlation between cell death and PaO.
Taken together, the cell death induction activity of TaLls1 in either TaLls1-knockdown plants or transient overexpressing leaf tissues demonstrated the close relationship of cell death and H2O2 accumulation caused by the chlorophyll degradation catabolites resulting from the irregular TaLls1 activity. It appears that there is a threshold for TaLls1 in maintaining the balance of chlorophyll catabolism and cell homeostasis. In sum, TaLls1 seems to serve as a type of rheostat for cell death regulation in plants subjected to various stresses.
A deletion mutational analysis of the conserved structures revealed that Rieske [2Fe-2S] is responsible for cell death induction, while the mononuclear non-heme iron-binding domain was not. Previous reports have demonstrated that both the Rieske [2Fe-2S] and mononuclear non-heme iron-binding domains are essential for PaO catalytic activity (Ferraro et al., 2005), during which the electrons transfer from the Rieske domain to the active sites and subsequently activate the PaO activity (Gray et al., 2004). For the both truncated TaLls1 forms (TaLls1212–397 and TaLls1212–397), the oxygenase catalytic activity was unable to be activated. Thereby, we presume that it is not only the catalytic activity of TaLls1 that caused the induced cell death but other roles of TaLls1 produced by Rieske [2Fe-2S]. With the exception of their function as electron carriers in the photosynthetic electron transport chain and as electron donors to various cellular proteins, the iron–sulphur clusters were also found to serve a variety of biological functions, including regulatory and structural roles (Beinert et al., 1997). In the redox-sensing SoxR protein, the FeS centres allosterically link cellular oxidative stress to the expression of defence-related genes (Hidalgo et al., 1997). It has also been reported that the [2Fe-2S] cluster-binding site is involved in mitochondrial morphology and suppression of cell proliferation (Murata et al., 2011). Taken together, we speculate that the cell death induction activity may be related to other functions of TaLls1, such as a signal molecule in the cell death pathway or an electron transporter for other enzymes involved in cell death, rather than breaking down pheophorbide a alone.
After confirming the linkage between TaLls1 and cell death, we further studied the role of TaLls1 in the wheat–Pst interaction. TaLls1 was upregulated with the avirulent Pst pathogen infection at 12h p.i., when the pathogen had not yet formed haustoria (Wang et al., 2007). Our result is consistent with the induced expression of the Lls1 gene in the cowpea and cowpea rust interaction (Mould et al., 2003). These authors demonstrated that upregulation of PaO during the cowpea and cowpea rust interaction may reflect an anticipation of chlorophyll degradation related to HR (Mould et al., 2003). For the wheat–Pst interaction, Wang et al. (2007) reported that H2O2 rapidly increased at infection sites 12h after inoculation of avirulent Pst before HR, so we postulated that the early increase in TaLls1 in the incompatible interaction is associated with the generation of H2O2 at an early stage of HR initiation.
To get more information on the role of TaLls1 in the wheat–Pst interaction, TaLls1 was silenced through a BSMV-mediated gene silencing system. The TaLls1-knockdown plants exhibited no detectable change in disease symptoms, but did show a significant increase in cell death upon Pst infection. Generally, elevated cell death occurrence at the infection sites was proposed to inhibit the spread of pathogen infection, but growth of the virulent Pst CYR31 did not significantly change in TaLls1-knockdown plants in comparison with the control plants. It is reasonable to speculate that silencing of TaLls1 is able to increase cell death caused by Pst infection but insufficient to limit Pst growth. Aside from the significant upregulation of PR1, PR2, and PAL in response to Pst infection in the TaLls1-knockdown plants, the TaLls1-knockdown plants exhibited a sharply increased accumulation of H2O2 at the early stage of the virulent pathogen infection. Notably, the amounts of H2O2 generated in TaLls1-knockdown leaves in the compatible interaction reached approximately the same level as in the incompatible interactions, which is sufficient to trigger local cell death. During the interaction between wheat and the avirulent Pst, H2O2 generation in the early infection stage is associated with the occurrence of HR and the resistance response (Wang et al., 2007). Therefore, we assume that knockdown of TaLls1 is likely to result in HR elicitation during the compatible interaction at the early stage. Additionally, catalase and superoxidase are subsequently upregulated to eliminate the excessive H2O2, and the removal of H2O2 may lead to the failure of HR initiation. Hence, TaLls1-knockdown plants seemed to be susceptible to the virulent Pst. When combined with the induced cell death and elevated expression levels of defence-related genes, it is reasonable to assume that silencing of TaLls1 enhanced the tolerance of wheat to Pst but was not sufficient to alter the disease symptoms.
In conclusion, the present study revealed a self-regulation of TaLls1 in balancing cell death and a negative role in wheat disease tolerance to Pst, but it is not clear how they are related. Further studies of TaLls1-modulated HR resistance to Pst are needed with transgenic wheat plants.
Supplementary Material
Acknowledgements
We thank Dr Steven R. Scofield from Purdue University for providing BSMV vectors and Professor Heinrich Buchenauer (University of Hohenheim) and Jinrong Xu (Purdue University) for critical comments. This work was supported by grants from the National Basic Research Program of China (nos 2013CB127700 and 2012CB114001), the National Natural Science Foundation of China (no. 30930064 and 31000836), the 111 Project from the Ministry of Education of China (B07049).
Glossary
Abbreviations:
- BSMV
barley stripe mosaic virus
- DAB
3,3′-diaminobenzidine
- GUS
β-glucuronidase
- HR
hypersensitive response
- PaO
pheophorbide a oxygenase
- p.i.
post-inoculation
- p.t.
post-treatment
- PR
pathogenesis-related
- qRT-PCR
quantitative reverse transcription-PCR
- RCC
red chlorophyll catabolite.
References
- Beinert H, Holm RH, Münck E. 1997. Iron–sulfur clusters: nature’s modular, multipurpose structures. Science 277, 653–659. [DOI] [PubMed] [Google Scholar]
- Chung DW, Pruzinská A, Hörtensteiner S, Ort DR. 2006. The role of pheophorbide a oxygenase expression and activity in the canola green seed problem. Plant Physiology 142, 88–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close P, Gray J, Johal G. 1995. Observations on the effect of light on the progression of lethal leaf-spot lesions. Maize Genetics Cooperation News Letter 69, 48. [Google Scholar]
- Dangl JL, Dietrich RA, Richberg MH. 1996. Death don’t have no mercy: cell death programs in plant–microbe interactions. Plant Cell 8, 1793–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich RA, Delaney TP, Uknes SJ, Ward ER, Ryals JA, Dangl JL. 1994. Arabidopsis mutants simulating disease resistance response. Cell 77, 565–577. [DOI] [PubMed] [Google Scholar]
- Dou DL, Kale SD, Wang XL, et al. 2008. Conserved C-terminal motifs required for avirulence and suppression of cell death by Phytophthora sojae effector Avr1b . Plant Cell 20, 1118–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douchkov D, Nowara D, Zierold U, Schweizer P. 2005. A high-throughput gene-silencing system for the functional assessment of defense-related genes in barley epidermal cells. Molecular Plant–Microbe Interactions 18, 755–761. [DOI] [PubMed] [Google Scholar]
- Ferraro DJ, Gakhar L, Ramaswamy S. 2005. Rieske business: structure–function of Rieske non-heme oxygenases. Biochemical and Biophysical Research Communications 338, 175–190. [DOI] [PubMed] [Google Scholar]
- Giridhar G, Thimann KV. 1985. Interaction between senescence and wounding in oat leaves. Plant Physiology 78, 29–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J, Close PS, Briggs SP, Johal GS. 1997. A novel suppressor of cell death in plants encoded by the Lls1 gene of maize. Cell 89, 25–31. [DOI] [PubMed] [Google Scholar]
- Gray J, Janick-Buckner D, Buckner B, Close PS, Johal GS. 2002. Light-dependent death of maize lls1 cells is mediated by mature chloroplasts. Plant Physiology 130, 1894–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J, Wardzala E, Yang M, Reinbothe S, Haller S, Pauli F. 2004. A small family of LLS1-related non-heme oxygenases in plants with an origin amongst oxygenic photosynthesizers. Plant Molecular Biology 54, 39–54. [DOI] [PubMed] [Google Scholar]
- Greenberg JT, Silverman FP, Liang H. 2000. Uncoupling salicylic acid-dependent cell death and defense-related responses from disease resistance in the Arabidopsis mutant acd5 . Genetics 156, 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg JT. 1997. Programmed cell death in plant–pathogen interactions. Annual Review of Plant Biology 48, 525–545. [DOI] [PubMed] [Google Scholar]
- Hidalgo E, Ding H, Demple B. 1997. Redox signal transduction via iron–sulfur clusters in the SoxR transcription activator. Trends in Biochemical Sciences 22, 207–210. [DOI] [PubMed] [Google Scholar]
- Hirashima M, Tanaka R, Tanaka A. 2009. Light-independent cell death induced by accumulation of pheophorbide a in Arabidopsis thaliana . Plant Cell Physiology 50, 719–729. [DOI] [PubMed] [Google Scholar]
- Holzberg S, Brosio P, Gross C, Pogue GP. 2002. Barley stripe mosaic virus-induced gene silencing in a monocot plant. The Plant Journal 30, 315–327. [DOI] [PubMed] [Google Scholar]
- Hu G, Yalpani N, Briggs SP, Johal GS. 1998. A porphyrin pathway impairment is responsible for the phenotype of a dominant disease lesion mimic mutant of maize. Plant Cell 10, 1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutcheson SW. 1998. Current concepts of active defense in plants. Annual Review of Phytopathology 36, 59–90. [DOI] [PubMed] [Google Scholar]
- Jiang HW, Li MR, Liang NT, Yan HB, Wei YB, Xu XL, Liu J, Chen F, Wu GJ. 2007. Molecular cloning and function analysis of the stay green gene in rice. The Plant Journal 52, 197–209. [DOI] [PubMed] [Google Scholar]
- Kang ZS, Li ZQ. 1984. Discovery of a normal T type new pathogenic strain to Lovrin10. Acta Cllegii Septentrionali Occidentali Agriculturae 4, 18–28. [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Mach JM, Castillo AR, Hoogstraten R, Greenberg JT. 2001. The Arabidopsis-accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proceedings of the National Academy of Sciences, USA 98, 771–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matile P, Hörtensteiner S, Thomas H. 1999. Chlorophyll degradation. Annual Review of Plant Biology 50, 67–95. [DOI] [PubMed] [Google Scholar]
- Mould MJR, Xu T, Barbara M, Iscove NN, Heath MC. 2003. cDNAs generated from individual epidermal cells reveal that differential gene expression predicting subsequent resistance or susceptibility to rust fungal infection occurs prior to the fungus entering the cell lumen. Molecular Plant–Microbe Interactions 16, 835–845. [DOI] [PubMed] [Google Scholar]
- Murata Y, Furuyama I, Oda S, Mitani H. 2011. A novel Rieske-type protein derived from an apoptosis-inducing factor-like (AIFL) transcript with a retained intron 4 induces change in mitochondrial morphology and growth arrest. Biochemical and Biophysical Research Communications 407, 92–97. [DOI] [PubMed] [Google Scholar]
- Neuffer M, Calvert OH. 1975. Dominant disease lesion mimics in maize. Journal of Heredity 66, 265–270. [Google Scholar]
- Obanni M, Hipskind J, Tsai C, Nicholson R, Dunkle L. 1994. Phenylpropanoid accumulation and symptom expression in the lethal leaf spot mutant of maize. Physiological and Molecular Plant Pathology 44, 379–388. [Google Scholar]
- Pogue G, Lindbo J, Dawson W, Turpen T. 1998. Tobamovirus transient expression vectors: tools for plant biology and high-level expression of foreign proteins in plants. Plant Molecular Biology Manual 4, 1–27. [Google Scholar]
- Pružinská A, Anders I, Aubry S, Schenk N, Tapernoux-Lüthi E, Müller T, Kräutler B, Hörtensteiner S. 2007. In vivo participation of red chlorophyll catabolite reductase in chlorophyll breakdown. Plant Cell 19, 369–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pružinská A, Tanner G, Anders I, Roca M, Hörtensteiner S. 2003. Chlorophyll breakdown: pheophorbide a oxygenase is a Rieske-type iron–sulfur protein, encoded by the accelerated cell death 1 gene. Proceedings of the National Academy of Sciences, USA 100, 15259–15264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pružinská A, Tanner G, Aubry S, Anders I, Moser S, Müller T, Ongania KH, Kräutler B, Youn JY, Liljegren SJ. 2005. Chlorophyll breakdown in senescent Arabidopsis leaves. Characterization of chlorophyll catabolites and of chlorophyll catabolic enzymes involved in the degreening reaction. Plant Physiology 139, 52–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca M, Jamesa C, Pružinska A, Hörtensteiner S, Thomasa H, Ougham H. 2004. Analysis of the chlorophyll catabolism pathway in leaves of an introgression senescence mutant of Lolium temulentum . Phytochemistry 65, 1231–1238. [DOI] [PubMed] [Google Scholar]
- Schweizer P, Pokorny J, Schulze-Lefert P, Dudler R. 2000. Double-stranded RNA interferes with gene function at the single-cell level in cereals. The Plant Journal 24, 895–903. [DOI] [PubMed] [Google Scholar]
- Scofield SR, Huang L, Brandt AS, Gill BS. 2005. Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway. Plant Physiology 138, 2165–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons C, Hantke S, Grant S, Johal GS, Briggs SP. 1998. The maize lethal leaf spot 1 mutant has elevated resistance to fungal infection at the leaf epidermis. Molecular Plant–Microbe Interactions 11, 1110–1118. [Google Scholar]
- Tanaka R, Hirashima M, Satoh S, Tanaka A. 2003. The Arabidopsis-accelerated cell death gene ACD1 is involved in oxygenation of pheophorbide a: inhibition of the pheophorbide a oxygenase activity does not lead to the “stay-green” phenotype in Arabidopsis . Plant and Cell Physiology 44, 1266–1274. [DOI] [PubMed] [Google Scholar]
- Tang YY, Li MR, Chen YP, Wu PZ, Wu GJ, Jiang HW. 2011. Knockdown of OsPAO and OsRCCR1 cause different plant death phenotypes in rice. Journal of Plant Physiology 168, 1952–1959. [DOI] [PubMed] [Google Scholar]
- Walbot V, Hoisington DA, Neuffer M. 1983. Disease lesion mimic mutations. In: Kosuge T, Meredith CP, Hollaender A, eds. Genetic engineering of plants. New York: Plenum Publishing, 431–442. [Google Scholar]
- Wang CF, Huang LL, Buchenauer H, Han QM, Zhang HC, Kang ZS. 2007. Histochemical studies on the accumulation of reactive oxygen species (O2 – and H2O2) in the incompatible and compatible interaction of wheat–Puccinia striiformis f. sp. tritici . Physiological and Molecular Plant Pathology 71, 230–239. [Google Scholar]
- Wang XD, Feng H, Tang CL, Bai PF, Wei GR, Huang LL, Kang ZS. 2012a. TaMCA4, a novel wheat metacaspase gene functions in programmed cell death induced by the fungal pathogen Puccinia striiformis f. sp. tritici . Molecular Plant–Microbe Interactions 25, 755–764. [DOI] [PubMed] [Google Scholar]
- Wang XJ, Tang CL, Huang XL, Li FF, Chen XM, Zhang G, Sun YF, Han DJ, Kang ZS. 2012b. Wheat BAX inhibitor-1 contributes to wheat resistance to Puccinia striiformis . Journal of Experimental Botany 63, 4571–4584. [DOI] [PubMed] [Google Scholar]
- Wang XJ, Tang CL, Zhang G, et al. 2009. cDNA-AFLP analysis reveals differential gene expression in compatible interaction of wheat challenged with Puccinia striiformis f. sp. tritici . BMC Genomics 10, 289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolter M, Hollricher K, Salamini F, Schulze-Lefert P. 1993. The mlo resistance alleles to powdery mildew infection in barley trigger a developmentally controlled defence mimic phenotype. Molecular and General Genetics 239, 122–128. [DOI] [PubMed] [Google Scholar]
- Yang ML, Wardzala E, Johal GS, Gray J. 2004. The wound-inducible Lls1 gene from maize is an orthologue of the Arabidopsis Acd1 gene, and the LLS1 protein is present in non-photosynthetic tissues. Plant Molecular Biology 54, 175–191. [DOI] [PubMed] [Google Scholar]
- Yao N, Greenberg JT. 2006. Arabidopsis accelerated cell death 2 modulates programmed cell death. Plant Cell 18, 397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.