Abstract
Males of the guppy (Poecilia reticulata) vary tremendously in their ornamental patterns, which are thought to have evolved in response to a complex interplay between natural and sexual selection. Although the selection pressures acting on the color patterns of the guppy have been extensively studied, little is known about the genes that control their ontogeny. Over 50 years ago, two autosomal color loci, blue and golden, were described, both of which play a decisive role in the formation of the guppy color pattern. Orange pigmentation is absent in the skin of guppies with a lesion in blue, suggesting a defect in xanthophore development. In golden mutants, the development of the melanophore pattern during embryogenesis and after birth is affected. Here, we show that blue and golden correspond to guppy orthologs of colony-stimulating factor 1 receptor a (csf1ra; previously called fms) and kita. Most excitingly, we found that both genes are required for the development of the black ornaments of guppy males, which in the case of csf1ra might be mediated by xanthophore–melanophore interactions. Furthermore, we provide evidence that two temporally and genetically distinct melanophore populations contribute to the adult camouflage pattern expressed in both sexes: one early appearing and kita-dependent and the other late-developing and kita-independent. The identification of csf1ra and kita mutants provides the first molecular insights into pigment pattern formation in this important model species for ecological and evolutionary genetics.
Keywords: guppy, color pattern, pigment pattern development, Kita, Csf1ra
THE guppy (Poecilia reticulata) is thought to be among the most color-polymorphic vertebrates (Endler 1983). Male guppies have an outstanding degree of variation in their ornamental patterns, which are shaped by a complex interplay between natural and sexual selection in wild populations. Along with introduction experiments, studies on guppy life-history traits, mate choice behavior, and predator–guppy as well as guppy–environment interactions have demonstrated that guppy populations can adapt rapidly to new environments (for an overview, see Magurran 2005). The guppy is therefore a prime model organism for the study of “evolution in action.”
Despite our wealth of knowledge about the ecological importance of coloration, the genes and developmental pathways underlying guppy pigment pattern formation are unknown. Both forward and reverse genetic studies are hampered by the fact that guppies are livebearers with internal fertilization, an average gestation period of 3–4 weeks, and a relatively small brood size (Houde 1997). The genetic basis of sex determination is highly variable within the Poeciliid family, to which the guppy belongs. The guppy itself has incipient X and Y chromosomes that include a nonrecombining part (Traut and Winking 2001). Only males develop highly polymorphic ornaments during puberty, which are under hormonal control (Houde 1997). The genetic analysis of male guppy ornaments first attracted attention >80 years ago, when Winge described a total of 18 putative ornamental loci, of which 17 showed sex-linked inheritance and 9 were strictly Y-linked (Winge 1922, 1927). Many more pigment pattern loci, which can be Y-linked, X-linked, XY-linked, or autosomal, have since been described (Lindholm and Breden 2002). Ornamental traits linked to the sex chromosomes are typically expressed only in males, but females can develop some male color patterns when treated with testosterone (Clemens et al. 1966; Lindholm and Breden 2002). An analysis of quantitative trait loci (QTL) has confirmed that most male color traits are controlled by multiple genes, including genes on autosomes (Tripathi et al. 2009b). In contrast to the sex-specific genes, several autosomal color factors behave as ordinary Mendelian recessive genes and are expressed in both sexes (Goodrich et al. 1944; Dzwillo 1959; Lindholm and Breden 2002).
The pigment pattern of the guppy consists of three to four different types of neural crest-derived chromatophores: black melanophores, yellow/orange to reddish xanthophores, blue iridescent iridophores, and, possibly, white leukophores (Takeuchi 1976; Tripathi et al. 2008). Guppy embryos are staged according to the differentiation of their eyes. In the middle-eyed stage, the retina is fully pigmented and the first melanophores differentiate on the head above the midbrain (Martyn et al. 2006). More melanophores appear during the late-eyed stage and form dark stripes along the lateral midline, on the back, and on the belly (Martyn et al. 2006). In the very late-eyed stage shortly before birth, a rhombic reticulate pattern consisting of melanophores emerges on the trunk (Martyn et al. 2006). It has also been referred to as a ground, diamond, or camouflage pattern (Goodrich et al. 1944; Martyn et al. 2006; Tripathi et al. 2008). All pigment cell types are present in wild-type embryos at birth and contribute to the newborn pattern (Figure 1A); e.g., the yolk is partially covered by iridophores and melanophores, while xanthophores are widely dispersed (Martyn et al. 2006).
Figure 1.
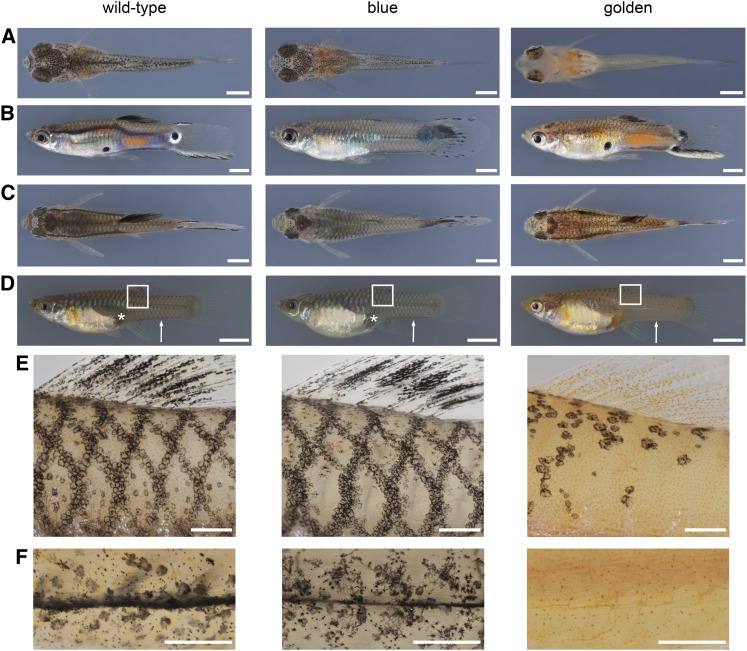
Blue and golden phenotypes. (A) Dorsal aspect of newborns. (B) Lateral aspect of adult males. (C) Dorsal aspect of adult males. (D) Lateral aspect of adult females. (E) Details of areas boxed in D showing the reticulate pattern. (F) Ventral view of the caudal peduncle of females (indicated by white arrows in D). golden mutants of both sexes lack a ventral black stripe and have only a few melanophores on the anterior head, including the choroid of the eyes. Golden females lack the female pigment spot above the anal fin (white asterisks in D). Individuals shown are from the BDZW1 (wild type, golden) and BDZW2 (blue) background. Bars: (A) 1 mm; (B and C) 2 mm; (D) 5 mm; (E and F) 0.5 mm.
The reticulate pattern of very late-eyed embryos and newborn guppies appears to persist into adulthood (Tripathi et al. 2008). This pattern is expressed in both sexes, but becomes overlain in males by male-specific ornaments (Figure 1, B–E). Two different melanophore types occur in adult wild-type guppies: large, roundish corolla and more heterogeneously shaped dendritic melanophores (Goodrich et al. 1944). The reticulate pattern is composed of corolla melanophores in deep skin layers, which are additionally arranged irregularly over the whole body, whereas dendritic melanophores are distributed superficially and are associated with the scales (Figure 1E) (Goodrich et al. 1944; Winge and Ditlevsen 1947).
Two autosomal color loci that are expressed in male and female guppies are blue and golden [also known as fredlini (Haskins and Druzba 1938; Winge and Ditlevsen 1947) and not to be confused with the zebrafish (Danio rerio) golden locus (Lamason et al. 2005)]. The mutations in both genes occurred spontaneously and act recessively (Goodrich et al. 1944; Dzwillo 1959). Blue mutants lack orange pigmentation in their skin, indicating a xanthophore defect (Figure 1) (Dzwillo 1959).
In golden mutants, the development of the melanophore pattern during embryogenesis and after birth is affected (Haskins and Druzba 1938; Goodrich et al. 1944; Winge and Ditlevsen 1947). Golden guppies of both sexes lack melanophores in the skin at birth and have only a few peritoneal melanophores above the swim bladder (Figure 1A), but eventually develop an incomplete reticulate pattern, which gives them a “coarsely mottled appearance” (Goodrich et al. 1944; Winge and Ditlevsen 1947). Corolla melanophores are restricted to the reticulate pattern in golden mutants, which also have only a very few dendritic melanophores (Figure 1E) (Haskins and Druzba 1938; Goodrich et al. 1944; Winge and Ditlevsen 1947). golden mutant females have only half of the normal number of melanophores in total (Goodrich et al. 1944). golden mutant males develop male-specific ornaments (Figure 1, B and C) (Haskins and Druzba 1938).
Among teleost fish, pigment pattern formation has been most extensively studied in zebrafish. Zebrafish undergo a complex pigment pattern metamorphosis during the transition from the embryonic/early larval to the juvenile/early adult stage (Johnson et al. 1995; Parichy and Turner 2003a,b; Kelsh et al. 2009; Parichy et al. 2009; Budi et al. 2011). The two type III receptor tyrosine kinases encoded by kita and its ancient paralog colony-stimulating factor 1 receptor a (csf1ra; previously called fms) (Braasch et al. 2006) play key roles during the establishment of the adult zebrafish pigment pattern: early metamorphic melanophores require kita for their development, while late metamorphic melanophores depend on csf1ra and endothelin receptor b1a (ednrb1a) (Parichy et al. 1999, 2000a,b; Parichy and Turner 2003b). Csf1ra is also crucial for xanthophore development (Parichy et al. 2000a; Parichy and Turner 2003b). In other teleost species, the functions of kita and csf1ra are less well understood; comparative studies including interspecies hybrids suggest some functional diversification of both receptor tyrosine kinases even within the Danio genus (Quigley et al. 2005; Parichy 2006; Mills et al. 2007).
Here, we present evidence that golden and blue correspond to guppy orthologs of kita and csf1ra. Both an early kita-dependent and a later-appearing kita-independent melanophore population contribute to the adult reticulate pattern in this species. In contrast to zebrafish, csf1ra is not required for the development of the late-appearing kita-independent melanophores. Csf1ra, however, is essential for xanthophore development and the formation of the male-specific melanophore pattern, which also requires kita.
Materials and Methods
Fish husbandry
All fish were maintained at 25° in a 12-hr light and dark cycle and fed 6 days a week with Artemia. No more than eight fish were kept per 1.5-liter tank. We used virgins for crosses, as guppy females are capable of storing sperm.
Strains
Guppies of the following inbred strains were used in this study; available phenotypes other than wild-type are shown in parentheses: Maculatus (MAC) (golden) (Tripathi et al. 2008); BDZW1 (golden, golden blue) and BDZW2 (blue only) (Dzwillo 1959); Armatus (golden) (Winge 1927); Guanapo and Quare6 (Reznick and Endler 1982); Quare6 family II 215-3 (Tripathi et al. 2008); and Cumaná (Alexander and Breden 2004). Maculatus, BDZW1, BDZW2, and Armatus are domesticated strains that have been bred in captivity for >50 years [in our laboratory since 2003 (Maculatus, BDZW1) and 2011 (Armatus, BDZW2)]; the others are derived from wild individuals caught in Trinidad (Guanapo and Quare rivers) and Venezuela (Cumaná region). Quare and Cumaná guppies were obtained in 2003 and Lower Guanapo (Twin Bridge) fish in 2009. All guppy strains are kept separately in small groups, usually consisting of two to three females and four to five males per tank and are allowed to reproduce freely, except for the Guanapo fish that are inbred by brother–sister mating.
blue was found in two strains that likely both were derived from the original population of blue mutants described by Dzwillo in 1959. We renamed them BDZW1 and BDZW2 for clarity. BDZW1 was obtained under the name “Blau” and comprised individuals heterozygous for golden and blue; BDZW2 (“Dzwillo 1959 Blau”) fish were all homozygous for blue.
Phylogenetic analyses
Only unambiguously annotated sequences were used for the analyses (ORF begins with a start codon; number of exons are as predicted for other species; and introns begin and end with splice sites). Sequences were aligned with Clustal Omega (v1.1.0) (Goujon et al. 2010; Sievers et al. 2011). Maximum-parsimony and maximum-likelihood phylogenetic trees were constructed by the PhylipParsimony algorithm (Felsenstein 1989) via the SplitsTree4 (v4.12.8) interface (Huson and Bryant 2006) and by PhyML (Guindon et al. 2010), respectively. The topologies of the maximum-likelihood trees were not fully resolved (therefore not shown), but, as the maximum-parsimony trees, they suggest that the sequenced guppy ORFs are most similar to kita, kit ligand a (kitla), and csf1ra of other teleost species.
Complementary DNA analyses
We used polymerase chain reaction (PCR) to amplify genes of interest from first-strand complementary DNA (cDNA). Total RNA was extracted from 10 to 15 pooled early to very-late eyed embryos with TRIzol (Invitrogen) according to the manufacturer’s instructions. Total RNA was then directly used for first-strand cDNA synthesis using PrimeScript Reverse Transcriptase (TaKaRa) and the oligo(dT) primer 5′-ATTCTAGAGGCCGAGGCGGCCGACATGT(18)VN-3′. We added 1 U/µl SUPERaseIn RNase Inhibitor (Ambion) to each reaction. First-strand cDNA was used as template for PCR, which was carried out with Advantage 2 Polymerase Mix (Clontech) according to the manufacturer’s instructions. PCR program was 10 cycles touchdown [95° for 15 sec, Tm (melting temperature) of primers (lower one) + 5° −0.5°/cycle for 30 sec, 68° for 2–5 min) followed by 27 cycles without touchdown (95° for 15 sec, Tm for 30 sec, 68° for 2–5 min]. Elongation time was adapted to the length of the expected product (∼1 min/kb). Primer sequences and methods used for full-ORF amplification of candidate genes are listed in Supporting Information, Table S1. PCR products were analyzed by gel electrophoresis, purified with MinElute Gel Extraction Kit (QIAGEN), and subsequently cloned into pGEM-T Easy vector (Promega) following the manufacturer’s instructions. Plasmid DNA was purified with Wizard Plus SV Minipreps DNA Purification System (Promega) according to the instruction manual and sequenced.
To investigate whether the kita transcripts V1–V6 cosegregate with the golden phenotype, we prepared first-strand cDNA from adult individuals as described before. Total RNA for cDNA synthesis was extracted separately from liver tissue of the parental male and the parental female and from pooled liver tissue of seven wild-type N2 fish and 11 golden N2 fish. Liver tissue was used as total RNA isolated from liver usually is of very high quality (personal observation). We used primers in exon 5 (forward: 5′-GATGCTGGGAGTTACAAATGCGTAG-3′) and exon 9 (reverse: 5′-AAACAGTATGTAGGCTTGCTCTCC-3′) for PCR amplification with Advantage 2 Polymerase Mix (Clontech) and cloned the products into pGEM-T Easy vector (Promega). We sequenced the purified plasmid DNA of 24 colonies per parent and N2 pool to identify wild-type and golden mutant kita transcripts.
Real-time quantitative PCR
Total RNA for real-time quantitative PCR was prepared as described above from skin of adult wild-type, golden, and blue females that were 6–9 months old. Following DNaseI treatment, first-strand cDNA was prepared from 830 ng of total RNA primed with oligo(dT)18 using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) according to the manufacturer’s instructions. First-strand cDNA was diluted 10-fold for real-time quantitative PCR that was conducted with Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen) on a CFX384 Touch Real-Time PCR Detection System (Bio-Rad) according to the instruction manuals provided by the manufacturers. PCR program was 95° for 3 min, 40 times (95° for 10 sec, 60° for 10 sec, 72° for 5 sec), and 95° for 10 sec. Expression of csf1ra, csf1rb, kita, and kitb was determined by using three biological replicates with three technical repetitions each. Expression of the target genes was normalized to glyceraldehyde-3-phosphate dehydrogenase expression. Standard deviation and normalized expressions (ΔΔCq) were calculated with CFX Manager software. Primer sequences and efficiencies (Pfaffl 2001; Vandesompele et al. 2002) are given in Table S2.
Genomic DNA analyses
Genomic DNA was prepared with DNeasy Blood and Tissue Kit (Qiagen) from trunk tissue of adult guppies. We used 100 ng of DNA per PCR reaction. If not mentioned otherwise, Advantage 2 Polymerase Mix (Clontech) was used to carry out the PCRs. Kitainsert was first amplified using a forward primer in exon 6 (5′-TGTCTCTGAACGTTAGCATGGAG-3′) and a reverse primer in exon 7 (5′-ACACGGAGAAGTTCTGCTTTACC-3′) of kita (elongation time: 5 min). To test whether kitainsert and the golden phenotype are associated, we designed PCR assays for kitainsert and kitawt using Phusion High-Fidelity DNA Polymerase (New England Biolabs) according to the manufacturer’s instructions. Details can be found in Table S3. PCR products were analyzed by gel electrophoresis. PCR products of csf1rawt and csf1raindel were purified with 0.2 U/µl FastAP Thermosensitive Alkaline Phosphatase (Fermentas) and 2 U/µl exonuclease I (Fermentas) (37° for 15 min, 85° for 15 min) and subsequently sequenced. Primer sequences are given in Table S3.
Sequence analysis
Purified plasmid DNA and PCR products were sequenced with BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) on a DNA Analyzer ABI Prism 3730XL (Applied Biosystems). Sequences were analyzed using the Staden package (Pregap4 v1.5 and Gap4 v4.10; http://staden.sourceforge.net/) and inspected manually. Exon–intron structures were predicted according to the gene structure of kitla, kita, and csf1ra of other teleost species (for species, see Table S1; exon–intron structure was inferred from http://www.ensembl.org).
Imaging
Photos of whole embryos or details of adult fish were taken with a Leica MZFLIII dissecting microscope connected to a Zeiss AxioCam HRc color camera and processed with AxioVision Software Release 4.7.2. Photos of fish after birth and adult fish were taken with a Canon EOS 10D digital camera using a Canon Macro Photo Lens MP-E 65 mm or Canon Macro Lens EF 100 mm. Adult males were at least 4 months old to ensure that their pigment pattern was fully developed. All photos were taken under incident light conditions. Fish were anesthetized in 0.1% (w/v) tricaine (ethyl 3-aminobenzoate methanesulfonate salt) solution (pH 7) before imaging. The background of most images was equalized with Adobe Photoshop software version 12.1; all original images are available upon request.
Analysis of melanophores
To analyze melanophore pattern development, photos of the same fish were taken every 3 days (day 1–40 after birth). Each fish was kept separately in a 1.5-liter tank. Fish were briefly anesthetized in 0.04% tricaine solution before imaging. Their siblings were kept as control; none of the imaged fish showed retarded development. Newly arising melanophores were detected by comparing consecutive images to each other.
To analyze the number of melanophores, fish were immersed in standard E2 solution (Nusslein-Volhard and Dahm 2002) containing 2.4 mg/ml epinephrine for ∼5 min to contract melanosomes. Afterward, fish were anesthetized, and the right side under the dorsal fin, as well as the complete fish, were imaged. Melanophores were counted manually utilizing Adobe Photoshop software version 12.1. The number of melanophores was plotted against the area in square millimeters using Microsoft Excel for Mac 2011 version 14.2.3.
Results
Identification of golden as kita:
We used a candidate gene approach to identify golden at the molecular level. Based on a comparison with zebrafish pigment pattern mutants, we investigated two potential candidates for golden, kita, and kitla (also called stem cell/steel factor). Kitla is the ligand for the Kita receptor, and together the two are required for melanophore migration and survival in zebrafish (Hultman et al. 2007). While only one copy of kit and kitl exists in mouse and humans, two copies of each gene are present within the Teleostei. These copies can be considered as “ohnologs” since they are derived from ancestral kit and kitl genes that were duplicated during the teleost-specific whole-genome duplication event (Mellgren and Johnson 2005; Braasch et al. 2006; Hultman et al. 2007). Mutations in zebrafish kita, as in golden, greatly reduce melanophore number (Parichy et al. 1999). Kita is located on guppy autosomal linkage group 4 (Tripathi et al. 2008). Kitla has not been mapped so far.
To amplify the guppy orthologs of kita and kitla, we used rapid amplification of cDNA ends (Table S1). Phylogenetic analyses demonstrated that the obtained full-ORF sequences are orthologous to kita (GenBank accession no. KC143124) and kitla (GenBank accession no. KC143125) of other teleost species (Figure 2A and Figure S1). We also identified a potential guppy kitb ortholog by performing BLAST searches of zebrafish kitb against a preliminary genome and transcriptome assembly of the guppy (E. Sharma, A. Künstner, B. A. Fraser, M. Hoffmann, V. A. Kottler, G. Zipprich, D. Weigel, and C. Dreyer, unpublished data) (Figure 2A) (Altschul et al. 1990). We could not determine whether a guppy kitlb ortholog exists, as BLAST yielded no significant results.
Figure 2.
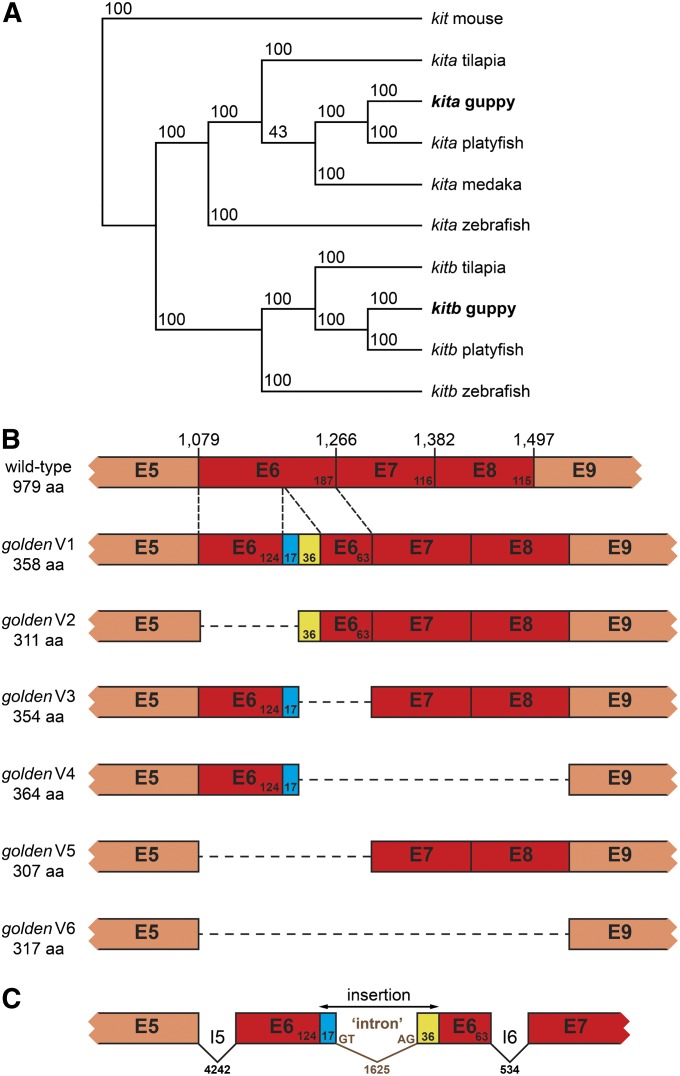
golden is an ortholog of kita. (A) Maximum-parsimony phylogenetic tree of kita and kitb ORF sequences. Mouse kit was used as an outgroup. Bootstrap support values from 100 replicates are shown. Accession numbers of sequences are the following: guppy kita, KC143124; medaka (Oryzias latipes) kita, ENSORLT00000000707; mouse (Mus musculus) kit, NM_021099; platyfish (Xiphophorus maculatus) kita, ENSXMAT00000013579; platyfish kitb, ENSXMAT00000017731; tilapia (Oreochromis niloticus) kita, ENSONIT00000003735; tilapia kitb, XM_003452144; zebrafish kita, NM_131053; and zebrafish kitb, NM_001143918. Guppy kitb sequence is available upon request. (B) Exons 5–9 of kita cDNAs sequenced from wild-type and golden mutant [variants (V) 1–6] backgrounds; exons affected by splicing defects are shown in dark red. Length in amino acids (aa) of the respective predicted protein is shown on the left. Numbers above wild type refer to the wild-type kita cDNA sequence deposited at GenBank (KC143124) and demarcate the last nucleotide of each exon. (C) Part of kita genomic locus in golden mutants (kitainsert) (GenBank accession no. KC143126). Numbers refer to base pairs. Exons (E), introns (I), and inserted sequence are not drawn to scale.
Golden mutant guppies did not have any obvious polymorphisms in the kitla ORF. In contrast, among cDNAs from golden fish of the MAC and BDZW1 backgrounds, we found a total of six different kita splice variants, none of which was wild type. Most of these variants contained inserts with a length of 17, 36, or 53 bp and/or lacked parts of or the complete exon 6 (Figure 2B and Table S4). Exons 7 and 8 were additionally absent in two variants (Figure 2B and Table S4). In all cases, these alterations cause frameshifts and truncate the ORF. These kita variants are likely nonfunctional since the encoded proteins would lack the transmembrane, juxtamembrane, and split kinase domains, all of which are required for normal protein function (Mol et al. 2003).
Analysis of genomic DNA indicated that kita exon 6 of golden mutants has an insertion of 1678 bp. This insertion consists of two potential short exons of 17- and 36-bp length that surround a 1625-bp intron delineated by 5′ GT-AG 3′ canonical splice sites (Figure 2C). The splice sites within the novel intron seem to be recognized by the spliceosome and lead to mis-splicing of the kita pre-messenger RNAs (mRNAs) in the golden mutants. Depending on which splice sites are used, this leads to short insertions or removal of parts of exon 6 or of exons 6–8 from the mature transcript. Comparison with a preliminary genome assembly of the guppy (A. Künstner, E. Sharma, B. A. Fraser, M. Hoffmann, V. A. Kottler, D. Weigel, and C. Dreyer, unpublished data) suggests that >50 copies similar to the 1678-bp insertion, which all include a long terminal repeat of ∼300 bp, exist (data not shown).
To confirm that kitainsert corresponds to golden, we tested for genetic linkage. Forty golden mutant fish from the MAC, BDZW1, and Armatus (ARM) backgrounds were homozygous for kitainsert and complementation test crosses between the strains demonstrated allelism (Figure S2B and S1C). In 28 wild-type MAC, BDZW1, and ARM individuals, only kitawt could be detected. This indicates that kitainsert was likely introduced into several guppy strains by breeders because of the golden coloration. Next, we investigated whether kitainsert is also linked to the golden phenotype in a segregating backcross N2 population: we crossed a golden blue double-mutant female of the BDZW1 strain to a wild-type male from the Guanapo river in Trinidad; F1 males were then backcrossed to produce XBDZW1XBDZW1/XBDZW1YGU N2 individuals. Forty-six golden and golden blue N2 fish were homozygous for kitainsert, while 12 wild-type and blue fish were heterozygous kitainsert/kitawt. In addition, we could amplify only kitainsert variants from pooled cDNA of golden mutant N2 offspring. Taken together, all of these results make it very likely that guppy golden is allelic to kita.
Identification of blue as csf1ra:
We hypothesized csf1ra to be a candidate for blue, since blue mutants lack xanthophores as do csf1ra mutants of zebrafish (Dzwillo 1959; Parichy et al. 2000a). Csf1ra and csf1rb ohnologs have been identified in several teleost species (Braasch et al. 2006). csf1ra has previously been mapped to guppy autosomal linkage group 10 (Tripathi et al. 2008).
Rapid amplification of cDNA ends was used to amplify the guppy csf1ra ortholog (Table S1). Phylogenetic analyses showed that the sequenced cDNA is closely related to csf1ra of other teleost species (Figure 3A). We also identified a presumptive guppy csf1rb ortholog within a preliminary genome and transcriptome assembly of the guppy (E. Sharma, A. Künstner, B. A. Fraser, M. Hoffmann, V. A. Kottler, G. Zipprich, D. Weigel, and C. Dreyer, unpublished data) (Figure 3A).
Figure 3.
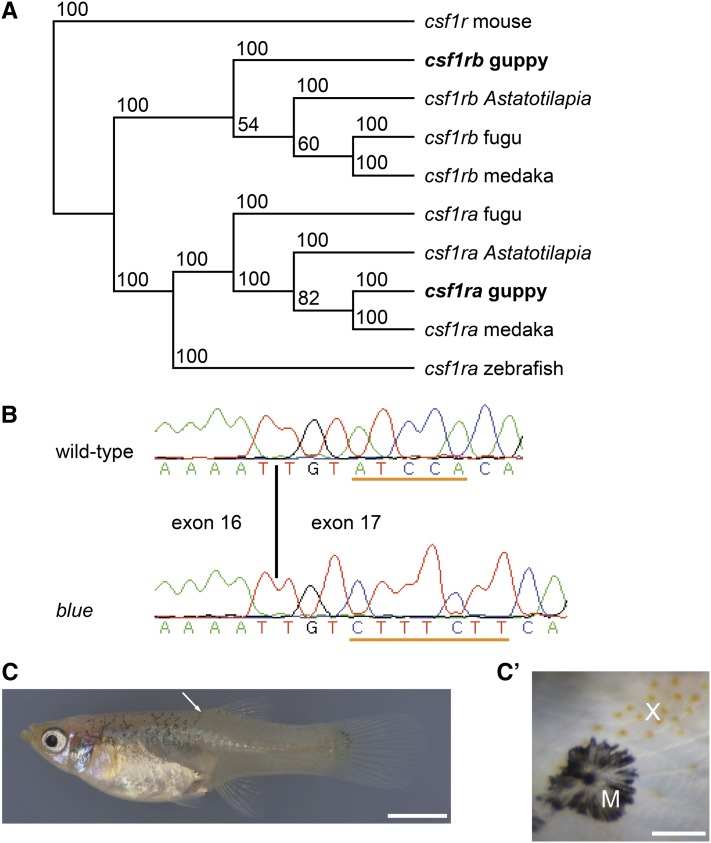
blue is an ortholog of csf1ra. (A) Maximum-parsimony phylogenetic tree of csf1ra and csf1rb ORF sequences. Mouse csf1r was used as an outgroup. Bootstrap support values from 100 replicates are shown. Accession numbers of sequences are the following: Astatotilapia burtoni csf1ra, DQ386648; A. burtoni csf1rb, DQ386647; fugu (Takifugu rubripes) csf1ra, U63926; fugu csf1rb, AF411927; guppy csf1ra, KC143122; medaka csf1ra, ENSORLT00000006111; medaka csf1rb, XM_004076731; mouse csf1r, NM_001037859; and zebrafish csf1ra, AF240639. Guppy csf1rb sequence is available upon request. (B) Partial sequence electropherograms of cDNAs from wild-type and blue mutant fish, which carry a deletion of 5 nt in exon 17 (underlined in wild-type sequence) that simultaneously contains a 7-nt insertion (underlined in blue sequence). The length of the predicted protein produced by blue mutants is 794 aa, with the last 12 new. The wild-type protein is 978 aa long. The first nucleotide of exon 17 corresponds to nucleotide 2392 in the 3084-bp wild-type csf1ra sequence (GenBank accession numbers of cDNAs: wild-type, KC143122; blue, KC143123). (C) golden blue mutant female; white arrow points to detail shown in C′. (C′) Small patch of xanthophores (X) and melanophore (M) on the back close to the dorsal fin of the female. Variation in the number of xanthophores was high in blue and golden blue mutant fish. Xanthophores were abundant and evenly distributed in wild-type and golden mutant females (Figure 1, E and F). Bars: (C) 5 mm; (C′) 50 µm.
Blue mutant guppies carry a complex change in exon 17 of csf1ra, with a deletion of 5 bp and an insertion of 7 bp (Figure 3B). This indel causes a frameshift and truncates the ORF. The predicted protein lacks part of the second half of the split kinase domain, which is required for the activity of type III receptor tyrosine kinases (Yarden and Ullrich 1988; Mol et al. 2003). A similar mutation in zebrafish, fmsj4blue, inactivates csf1ra and leads to loss of xanthophores (Parichy et al. 2000a).
We used the same cross as above to test for linkage of csf1ra and blue. Forty-six blue and golden blue N2 fish were homozygous for csf1raindel, while 12 wild-type and golden N2 individuals tested were heterozygous csf1raindel/csf1rawt. Consistent with this, 24 golden blue mutant guppies of the BDZW1 strain were homozygous for csf1raindel, whereas only csf1rawt could be identified in 12 wild-type and golden fish of the same population. Another blue strain obtained from a hobby breeder, BDZW2, was also homozygous for the csf1raindel allele, and complementation test crosses to BDZW1 confirmed allelism (Figure S2A). blue is therefore likely to be the guppy ortholog of csf1ra, with the same csf1raindel mutation present in both strains tested.
In zebrafish, csf1ra promotes the migration of the xanthophore precursors from the neural crest (Parichy et al. 2000a), and both embryonic and metamorphic xanthophore populations require csf1ra activity (Parichy and Turner 2003b). To investigate whether csf1ra mutant guppies entirely lack xanthophores, we thoroughly inspected 7-month-old golden blue and blue fish that shared the same grandparents. In contrast to a previous study (Dzwillo 1959), we found a small number of xanthophores in 19 of 20 golden blue mutant individuals and in 5 of 17 blue mutants (Figure 3, C and C′). Most of the xanthophores were arranged on scale margins close to the dorsal fin. We could not detect any xanthophores on the lateral or ventral side of adult golden blue or blue fish or in blue embryos and newborns (we here refer to guppies as newborns 1–3 days after birth). A small number of xanthophores can also be found in zebrafish larvae, but not in adults, which are homozygous for salztl41a or pfeffertm36b, two alleles that fail to complement fmsj4blue (Maderspacher and Nusslein-Volhard 2003).
Expression of kita, csf1ra, and their ohnologs in female skin
We could detect kita, kitb, csf1ra, and csf1rb expression in the skin of adult wild-type, golden, and blue females by Reverse Transcriptase-PCR (data not shown). To assess whether Kitb and Csf1rb might be upregulated to compensate for the loss of Kita and Csf1ra function in the golden and blue mutants, respectively, we investigated the expression levels of these genes and their a-paralogons by real-time quantitative PCR. We found that csf1ra expression is downregulated in the skin of blue mutant females compared with the wild type, while there was no significant difference in the expression levels of csf1rb (Figure S3). In zebrafish, csf1ra is expressed in the xanthophore, macrophage, and osteoclast lineages (Parichy et al. 2000a). Our data suggest that the guppy ortholog of csf1ra is expressed in guppy xanthophores, as the almost complete absence of these cells in blue skin coincides with a low csf1ra expression level. Additionally, the blue transcript of csf1ra might undergo nonsense-mediated mRNA decay triggered by the premature termination stop codon. In contrast, the expression levels of kita and kitb were not significantly different in golden skin compared with wild type, respectively (Figure S3), which suggests that the golden kita transcripts are not subjected to nonsense-mediated decay. In conclusion, no significantly elevated expression of the kit and csf1r b-paralogons could be detected by real-time quantitative PCR in the skin of golden and blue mutants (Figure S3). Yet we cannot exclude that less efficient salvage pathways mediated by Kitb and Csf1rb compensate for the loss of Kita and Csf1ra function in the mutants based on these findings.
Contribution of distinct melanophore populations to the adult reticulate pattern
Kita-dependent and -independent metamorphic melanophores contribute to the adult pigment pattern in zebrafish (Parichy et al. 1999; Parichy and Turner 2003b). Additionally, Kita activity is required for the dispersal of melanoblasts from the neural crest in zebrafish embryos (Parichy et al. 1999). To investigate whether temporally and genetically distinct melanophore populations exist in the guppy, we examined golden mutant guppies at different developmental stages.
First, we explanted wild-type and golden embryos 12 days after last parturition, which corresponds to the middle-eyed stage, at which the first melanophores differentiate in the skin above the midbrain in wild-type embryos (Martyn et al. 2006). golden mutant embryos lacked melanophores on the head and the trunk at this stage (Figure 4A). Second, we investigated the development of the pigment pattern between the 1st and the 40th day after birth by taking photographs of the same individuals every 3 days.
Figure 4.
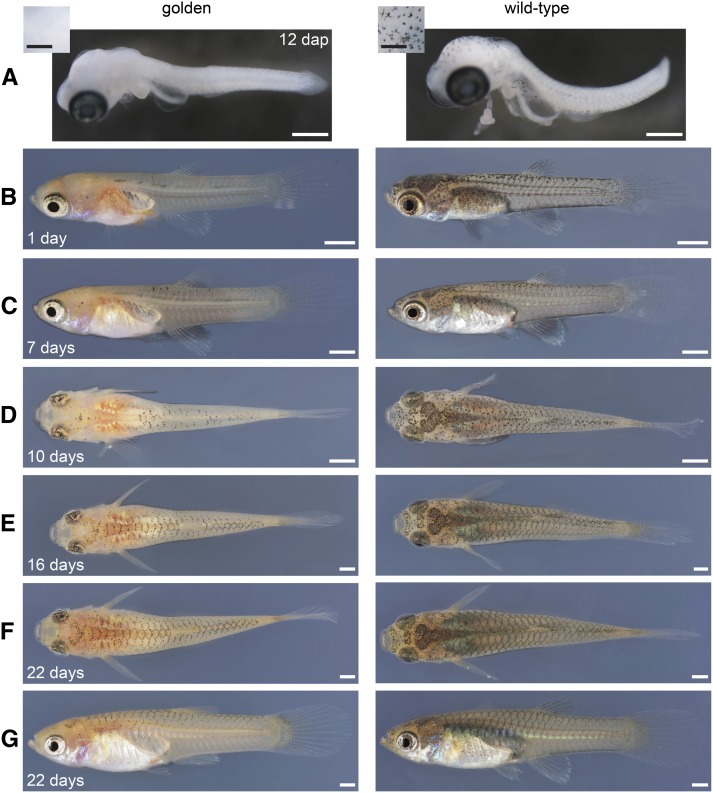
Melanophore pattern development in BDZW1 wild-type and golden mutant fish. (A) Embryos explanted 12 days after last parturition (dap). Yolk sacs were removed and embryos were fixed overnight at 5° in 4% paraformaldehyde and 1% dimethyl sulfoxide. Insets: Dorsal aspect of the midbrain region with individual melanophores apparent in wild type. (B, C, and G) Lateral, (D–F) dorsal aspects of the same females over a 3-week time course (days are after birth). All images taken, including the ones of males, can be found in File S1. Bars: (A) 500 µm, (insets) 250 µm; (B–G) 1 mm.
Immediately after birth, golden mutants had few patches of peritoneal melanophores, as described previously (Goodrich et al. 1944; Winge and Ditlevsen 1947). Additionally, we found some melanophores in their skin and close to the neural tube (Figure 4B). Four and 7 days after birth, few scattered melanophores were present close to the dorsal midline and on the head of golden mutants (Figure 4C and File S1). After 10 days, melanophores had become more abundant on the dorsal part of golden mutant fish and formed an incipient reticulate pattern, which became quite prominent after 16 days (Figure 4, D and E). The formation of the reticulate pattern seemed to be completed ∼22 days after birth in the mutants (Figure 4, F and G), although we detected a few newly arising melanophores even in 40-day-old golden individuals (File S1). This suggests that two melanophore populations that are temporally and genetically distinct contribute to the adult reticulate pattern: one that develops early and depends on Kita and one that appears late and is independent of Kita. In wild-type fish, melanophores were abundant and distributed over the whole body from the day of birth onward (Figure 4, B–G).
Adult golden mutant females have about half the number of skin melanophores of wild-type females (Goodrich et al. 1944) and lack the pigment spot by the anal fin (Figure 1, D and E). Additionally, the amount of superficial dendritic melanophores associated with the scales is strongly reduced (Haskins and Druzba 1938; Goodrich et al. 1944; Winge and Ditlevsen 1947), and black pigment is scarce on the choroid of the eyes as well as on the anterior head in golden fish, indicating that these pigmentation traits depend on Kita (Figure 1, B–E). The arrangement of the melanophores along the scales seems to be independent of Kita, as the size of the reticulate pattern (diameter of rhombi) was similar in golden and wild-type fish (data not shown). Onset of puberty in golden and wild-type males was observed between day 19 and 28 (File S1); the development of the reticulate pattern in golden males was similar to that of golden females (File S1).
csf1ra-independent melanophore populations
In zebrafish, early kita-dependent metamorphic melanophores are independent of Csf1ra, whereas late-differentiating kita-independent melanophores require Csf1ra and Ednrb1a activity for their differentiation (Parichy et al. 2000a,b; Parichy and Turner 2003b). kita csf1ra double-mutant zebrafish lack almost all melanophores because of the strong additive effect of mutations in these two genes (Parichy et al. 2000a). Since we identified a kita-dependent and -independent melanophore population in the guppy, we asked whether any of them requires csf1ra.
Inspection of embryos and newborns revealed no major differences between the blue mutant and wild-type melanophore patterns (Figure S4A), although we cannot exclude that a small subset of the early appearing melanophores depends on Csf1ra signaling since we could not count these early cells (see Figure S4 legend). To investigate whether the late-differentiating melanophores depend on csf1ra, we compared an area below the dorsal fin in golden and golden blue adult females (Figure S4, E and E′). We found that both single- and double-mutant fish have similar numbers of melanophores per area (Figure S4F). Therefore, unlike in zebrafish, mutations in kita and csf1ra have no additive effect on the reticulate pattern of the guppy, which is further supported by the observation that the reticulate pattern of blue mutant guppies develops as in the wild type until at least day 40 after birth (Figure S4, A–D; File S1). Consequently, the late-appearing melanophores of the guppy reticulate pattern are independent of both Kita and Csf1ra signaling.
Requirement of Kita and Csf1ra signaling for male-specific ornaments
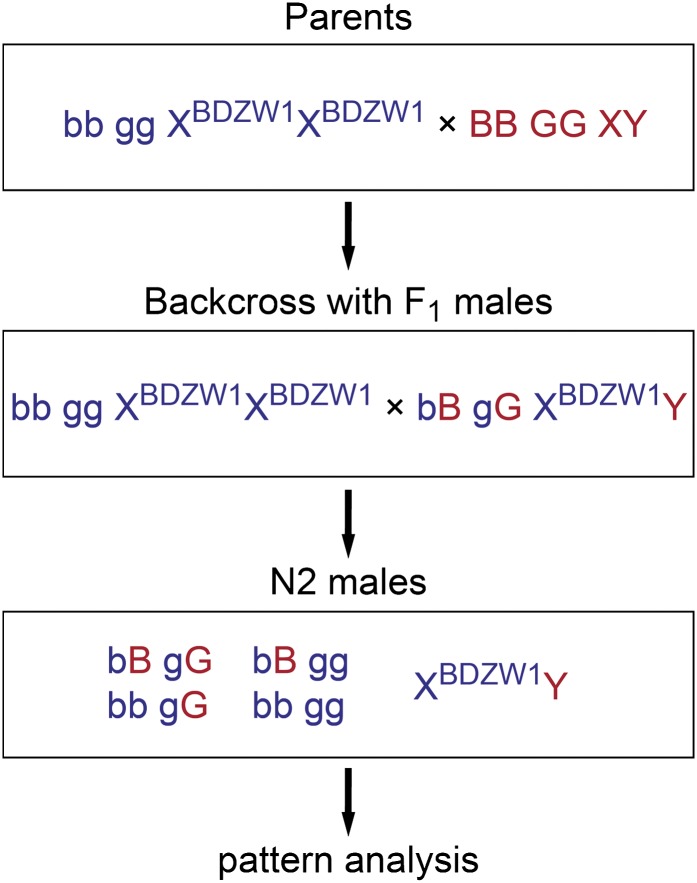
Guppy male-specific pigmentation patterns vary tremendously within and among populations, yet the within-population variance decreases considerably with inbreeding (personal observation). To investigate the roles of Kita and Csf1ra in male color pattern formation, we compared the ornamental patterns of related wild-type and mutant males. We crossed golden blue mutant BDZW1 females with wild-type males from the inbred wild-derived Cumaná (CUM), Guanapo (GU), Quare6 family II 215-3 (QUII), and Quare6 (QU) strains. The phenotypically wild-type F1 males were backcrossed to golden blue mutant BDZW1 females to produce a N2 generation (Figure 5). As a result of the backcross, the grandfather’s Y chromosome was combined with an X chromosome of the BDZW1 strain in all N2 males. Since recombination frequency of sex chromosomes is comparatively low in male meiosis (Tripathi et al. 2009a), this experimental design minimized the amount of pattern variation caused by X chromosomes derived from different strains, thereby allowing us to study the influence of the mutant autosomal genes on the pattern directed by a given Y chromosome. The number of male offspring derived from each cross is given in Table 1, and all images of the backcrosses can be found in File S2.
Figure 5.
Crossing scheme that was used in the case of the CUM, GU, QUII, and QU strains. The color patterns of the wild-type, golden, blue, and golden blue N2 males were analyzed. blue (b) and golden (g) are located on different autosomes and can therefore recombine freely. “B” and “G” indicate wild-type alleles. BDZW1 and MAC males were crossed with females of the same strain (for details see Figure 8).
Table 1. Male offspring obtained from each cross.
| Male parent | F1 | N2 | |||
|---|---|---|---|---|---|
| Wild type | Golden | Blue | Golden blue | ||
| Cumaná (CUM) | 55 | 39 | 33a | 52 | 53 |
| Guanapo (GU) | 30 | 24 | 25 | 18 | 21 |
| Quare6 family II 215-3 (QUII) | 34 | 16 | 16 | 11 | 10 |
| Quare6 (QU) | 43 | 17 | 18 | 25 | 14 |
| F2 | |||||
| Wild type | Golden | ||||
| BDZW1 | 24 | 56 | 18 | ||
| Maculatus (MAC) | 12 | 30 | 13 | ||
Actual number was 34, but one male had a BDZW1 instead of a CUM-like color pattern (see text).
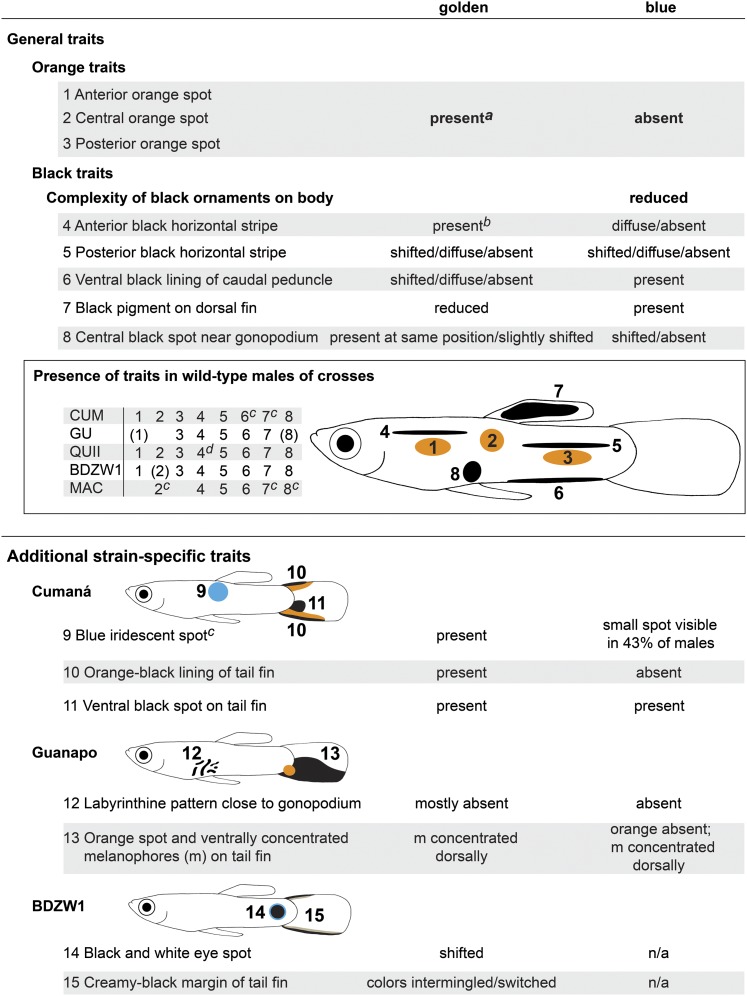
For our analysis, we focused on the most prevalent characteristic traits of each cross, as seen on the grandfather and its wild-type male offspring (Figure 6, Figure 7, Figure 8 and Figure 9; Figure S5). Furthermore, we tried to homologize the male ornaments of the different strains based on their color, shape, and approximate positions. A summary of all traits and generalization of our results is shown in Figure 9, which should facilitate tracking of single traits within the complex male patterns. We are, however, aware of the fact that superficially similar-looking traits need neither be directed by the same developmental pathways nor be derived from the same putative cell precursor pool.
Figure 6.
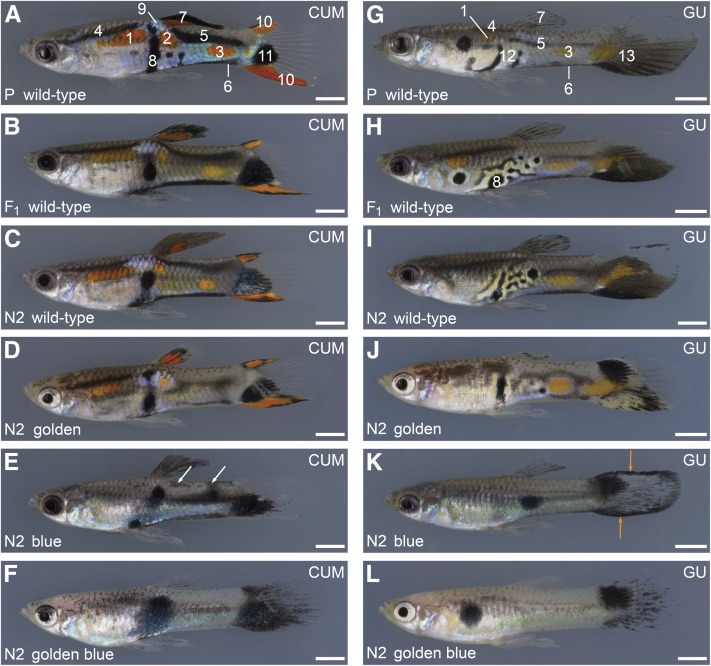
Ornaments in golden and blue mutant males from the Cumaná and Guanapo backgrounds. (A–F) Cross between a golden blue mutant BDZW1 female and a wild-type Cumaná male (A). Representative F1 (B) and N2 males with different phenotypes (C–F) are shown. Cumaná traits are highlighted in A. White arrows in E show incoherent patches of melanophores described in the text. (G–L) Cross between a golden blue mutant BDZW1 female and a wild-type Guanapo male from a F5 Guanapo laboratory inbreeding population (G). Representative F1 (H) and N2 males with different phenotypes (I–L) are shown. Guanapo traits are highlighted in G and H. I–L lack the anterior orange spot. Orange arrows in K show the black margins of the tail fin described in the text. Traits are labeled with numbers that correspond with numbers used in Figure 9: 1, 2, and 3: anterior, central, and posterior orange spot; 4 and 5: anterior and posterior black horizontal stripes; 6: ventral black lining of caudal peduncle; 7: black pigment on dorsal fin (in CUM in combination with orange); 8: central black spot near gonopodium; 9: blue iridescent spot; 10: orange-black lining of tail fin; 11: ventral black spot on tail fin; 12: black crescents forming a labyrinthine pattern close to the gonopodium; and 13: orange spot and ventrally concentrated melanophores on tail fin. N2 males shown for each backcross are siblings or cousins. All photos taken from the backcrosses can be found in File S1. Bottom left in each panel: generation (P, grandfather; F1; N2) and phenotype. Top right in each panel: Y chromosome origin. Bars: 2 mm.
Figure 7.
Ornaments in golden and blue mutant males from the Quare6 family II 215-3 background. Cross between a golden blue mutant BDZW1 female and a wild-type Quare6 family II 215-3 male (A). Representative F1 (B) and N2 males with different phenotypes (C–F) are shown. Quare traits are highlighted in A. Yellow arrow points to black spot associated with anterior black horizontal stripe. Blue arrow points to black spot close to the tail fin that was not included in the analysis as its location was highly variable in wild-type fish. The green arrowhead in E shows a black spot close to the operculum described in the text. Traits are labeled with numbers corresponding with numbers used in Figure 6 and Figure 9. N2 males shown are cousins. All photos from this backcross and from the Quare6 backcross that was very similar regarding the male patterns can be found in File S2. Bottom left: generation (P, grandfather; F1; N2) and phenotype. Bars: 2 mm.
Figure 8.
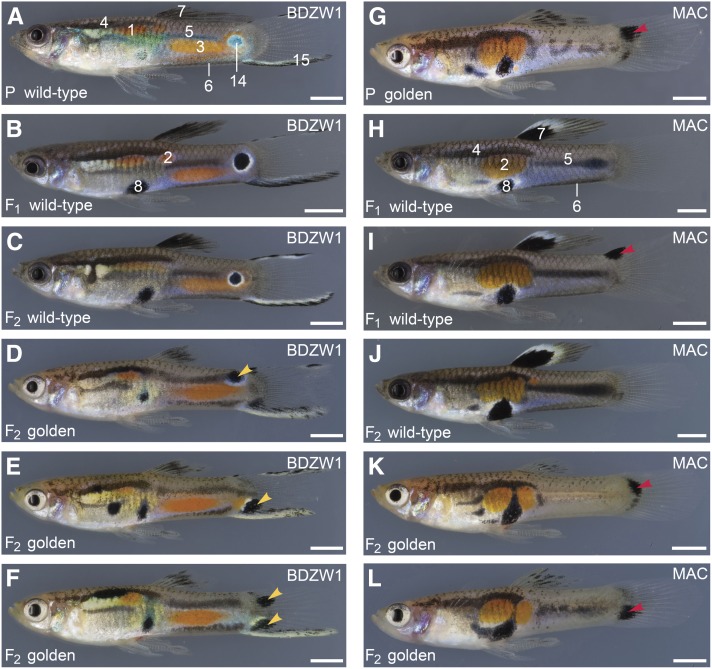
Ornaments in golden mutant males from the BDZW1 and Maculatus backgrounds. (A–F) Cross between a golden mutant BDZW1 female and a wild-type BDZW1 male (A). F1 siblings were crossed to produce a F2 generation. Representative F1 (B) and F2 males (C–F) are shown. BDZW1 traits are highlighted in A and B. All wild-type F2 and 92% of the wild-type F1 males had a central black-and-white eye spot on the caudal peduncle. Instead of this eye spot, one-half of the golden mutant F2 males had a spot located more dorsally on the trunk, while the other half had a spot at a more ventral position, mostly on the tail fin (yellow arrowheads in D and E). Three males had two spots (yellow arrowheads in F). (G–L) Cross between a wild-type Maculatus female and a golden mutant Maculatus male (G). F1 siblings were crossed to produce a F2 generation. Representative F1 (H and I) and F2 males with different phenotypes (J–L) are shown. Maculatus traits are highlighted in H. Red arrowheads in (G, I, K, and L) indicate untypical black spots on the tail fin. Only one of the F1 males showed such a black spot (I). It was not seen in any of the wild-type F2 males, but it was present in 85% of the golden mutant F2 males (K and L). Traits are labeled with numbers corresponding with numbers in Figure 6 and Figure 9. Additional traits to those shown in Figure 6: 14, black-and-white eye spot; 15, creamy-black margin of tail fin. F2 males shown for each cross are siblings or cousins. Bottom left: generation (P, grandfather; F1; F2) and phenotype. Top right: Y chromosome origin. Bars: 2 mm.
Figure 9.
Summary and generalization of the results regarding the male-specific pattern. Both Csf1ra and Kita are required for the formation of male ornaments of the guppy. The black pigment on the dorsal fin can be a distinct patch or diffuse. Numbers in parentheses refer to traits that were not present in all wild-type males of the specific strain. Traits that were too variable in wild-type males were not included into the analysis. More details are described in the text and shown in Figure 6, Figure 7, and Figure 8 and in Figure S5. aException: a concise posterior orange spot was absent in 94% of the golden N2 males with a YCUM background. bException: often discontinuous in golden N2 males with a YGU background. cY-linked in respective strain. dAssociated with a black spot in 83% of the wild-type males with a YQUII background.
Regardless of the origin of the Y chromosome, blue mutant N2 males lacked all orange color traits, indicating that Csf1ra activity is also required for the dispersal or differentiation of male-specific xanthophores (Figure 6, Figure 7, and Figure 9). In addition, the location and shape of the black male-specific ornaments were modified in the mutants.
Previous studies of the male-specific color pattern of the Cumaná guppy have shown that the blue iridescent spot on the trunk close to the dorsal fin, the combination of black and orange on the dorsal fin, and the ventral black lining of the caudal peduncle constitute strictly Y-linked traits (Figure 6A and Figure 9) (Tripathi et al. 2008, 2009a,b). The male-specific ventral black lining is more pronounced than the ventral black stripe of both sexes described in Figure 1F. Typically, two thicker black horizontal stripes, an anterior and a posterior one, are present on the trunk of Cumaná males (Figure 6A and Figure 9). The orange-black lining of the tail fin, which is often more pronounced on the ventral margin, is another male-specific Cumaná trait (Figure 6A and Figure 9). The prominent ventral black spot on the tail fin of many Cumaná males (Figure 6A and Figure 9; Figure S5) is very likely directed by one or more dominant factors located in the pseudo-autosomal region of the Cumaná sex chromosomes (Tripathi et al. 2008).
Most Cumaná male-specific traits were fully developed in the grandfather, in all F1, and in all wild-type N2 males (Figure 6, A–C; for the ventral black spot on the tail fin, see Figure S5). Of the golden mutant N2 males, all but one showed all of these traits as well (Figure 6D and Figure 9; the pattern of the one exceptional individual might be the outcome of a rare recombination event). Comparisons between wild-type and golden males revealed that (i) golden mutants had less black on the dorsal fin; (ii) the ventral black lining of the caudal peduncle was shifted upward and was often discontinuous in golden fish; (iii) the posterior black stripe of golden mutants was shifted downward and was often discontinuous (Figure 6D and Figure 9). In addition, a concise posterior orange spot on the caudal peduncle was absent in 94% of the golden mutants (Figure 6D). The anterior black stripe, the anterior orange spot, the blue iridescent spot, the orange-black lining, and the ventral black spot on the tail fin were not obviously changed by the mutation in kita (Figure 6D and Figure 9). The blue mutant N2 males not only lacked all orange traits, but also lost the black components of the orange-black lining of the tail fin, whereas the ventral black spot persisted (Figure 6E and Figure 9). Other defects of the blue mutant fish were the following: (i) the anterior black stripe was absent or very diffuse; (ii) the posterior black stripe was shifted downward and seemed more diffuse, and small incoherent patches of melanophores were often found on the dorsal trunk; (iii) in only 43% of the blue males a comparatively small blue iridescent spot was visible close to the dorsal fin (Figure 6E and Figure 9). The phenotype of the golden blue mutant N2 males resembled the blue mutant fish, but they showed fewer melanophores on the dorsal fin and trunk (Figure 6F).
The pattern of the Guanapo grandfather was characterized by several black crescents forming a labyrinthine pattern in proximity to the gonopodium (Figure 6G and Figure 9). The melanophores on the proximal and distal parts of the tail fin were concentrated on the ventral half of the fin, and an orange spot was located close by (Figure 6G and Figure 9). The anterior and posterior black horizontal stripes, the ventral black lining of the caudal peduncle, and the anterior and posterior orange spots on the trunk appeared weaker compared with the Cumaná male (Figure 6G). The posterior black stripe approximately demarcated the lateral midline in the Guanapo grandfather.
All of the F1 and wild-type N2 males showed the Guanapo traits described above, but only 41% of the wild-type males had an anterior orange spot (Figure 6, H and I). Furthermore, the F1 and wild-type N2 males tended to have more complex black labyrinthine ornaments close to the gonopodium than the Guanapo grandfather, suggesting that cofactors derived from the autosomes or X chromosome of the BDZW1 strain modulate this trait (Figure 6, H and I). The black pigment pattern of the golden mutant N2 males was substantially changed in the following ways: (i) black pigment in the dorsal fin was reduced; (ii) the labyrinthine black ornaments close to the gonopodium were mostly lost; (iii) black pigment in the proximal part of the tail fin was concentrated dorsally; (iv) anterior and posterior black stripes were often discontinuous; and (v) the ventral black lining was shifted upward (Figure 6J and Figure 9). The orange ornaments persisted, and an anterior orange spot was found in 32% of the golden N2 males (Figure 6J and Figure 9). The blue mutant N2 males had an even more severe phenotype: (i) only one to two round black spots were present on the trunk that were in most individuals not located near the gonopodium; (ii) as in golden N2, the black in the proximal part of the tail fin was located dorsally, and several males had black tail-fin margins; and (iii) the anterior black stripe was absent or diffuse (Figure 6K and Figure 9). The golden blue mutant N2 males had fewer melanophores on the fins and trunk than the blue mutants (Figure 6L).
The pigmentation traits of the Quare6 family II 215-3 grandfather are shown in Figure 7A. Compared with the Cumaná grandfather, the black pigment on the dorsal fin and the ventral black lining of the caudal peduncle of the Quare6 family II 215-3 grandfather appeared weak. It had a black spot associated with its anterior black horizontal stripe, which we found in 83% of its wild-type and in 56% of its golden N2 male offspring (Figure 7, A–D). We found that golden mutant N2 males of this backcross had (i) less black on the dorsal fin, (ii) a more diffuse posterior black horizontal stripe, (iii) an upwardly shifted or no ventral black lining of the caudal peduncle, and (iv) a slightly upwardly shifted central black spot (Figure 7D and Figure 9). In blue mutant N2 males, black ornaments on the trunk were mostly reduced to a few spots, and in some individuals a spot appeared at an unusual position just behind the operculum (Figure 7E).
Fewer black ornaments on the dorsal fin were also observed in golden mutant males of the highly inbred BDZW1 and Maculatus strains (Figure 8). The black spot in the dorsal fin of the Maculatus strain is considered to be a strictly Y-linked trait (Winge 1922), which illustrates that the expression of such traits nevertheless depends on autosomal cofactors. Wild-type males of the BDZW1 strain have a creamy-black margin of the tail fin and a prominent black-and-white eye spot on the caudal peduncle (Figure 8, A–C, and Figure 9). The colors of the creamy-black tail-fin margin were intermingled or switched in golden mutant BDZW1 males, and, instead of the eye spot, they had black spots at a more dorsal position on the trunk or ventral position on the tail fin (Figure 8, D–F, and Figure 9; occasionally two spots were present). Hence, the position of the eye spot is more variable in golden mutant BDZW1 males compared with the wild type. The posterior black horizontal stripe of golden BDZW1 males was often diffuse (Figure 8, D–F, and Figure 9), while it was curved, absent, or diffuse in golden Maculatus males (Figure 8, G, K, and L, respectively, and Figure 9). The ventral black lining of the caudal peduncle was shifted upward (BDZW1, Figure 8, D–F, and Figure 9) or absent (MAC, Figure 8, G, K, and L, and Figure 9). golden mutant Maculatus males showed dorsal or ventral black spots on the tail fin as well, which were only rarely seen in wild-type males (Figure 8, G–L).
Discussion
How the extreme variation in colorful ornaments satisfies the conflicting demands of predator evasion and mate attraction in male guppies has fascinated scientists for almost a century (Winge 1922, 1927; Lindholm and Breden 2002; Magurran 2005). Importantly, despite the extreme interindividual differences, many color traits are highly heritable, and sons greatly resemble their fathers (Winge 1922, 1927; Dzwillo 1959; Lindholm and Breden 2002). A better understanding of these issues requires better knowledge of the ontogeny of guppy pigmentation. This has unfortunately been challenging, due to the intrinsic difficulties of working with a livebearer for which many standard techniques that can be utilized in model organisms are not available. Here, we have exploited the power of forward genetics to advance the understanding of pigmentation in guppies. We have discovered that mutations in the two type III receptor tyrosine kinase genes kita and csf1ra underlie the guppy golden and blue phenotype, respectively, and have studied the effects of the mutations on both the reticulate pattern shared by females and males and the male-specific ornaments. We found that the mutations in kita and csf1ra have strong effects on the expression of male-specific color patterns. In general, the mutation in kita made none or only minor changes in orange ornaments and affected the male melanophore pattern more subtly and in a more reproducible manner than the mutation in csf1ra. The salient feature of the csf1ra mutant males was the absence of all orange traits, with concomitant severe changes in black ornaments.
In many teleost species, including zebrafish, medaka (Oryzias latipes), stickleback (Gasterosteus aculeatus), and Japanese flounder (Paralichthys olivaceus), larvae and adults differ in their pigmentation patterns (Johnson et al. 1995; Parichy and Turner 2003a,b; Lynn Lamoreux et al. 2005; Kelsh et al. 2009; Yamada et al. 2010; Budi et al. 2011; Greenwood et al. 2012). Our study shows that the camouflage reticulate pattern of newborn guppies is not yet completely developed and that it is fully elaborated only during the first month after birth. Absence of most melanophores in embryonic and newborn golden mutants suggests that Kita is essential for the differentiation of an early melanophore population. A second melanophore population arises after birth and remains restricted to the dorsal half of the body in golden mutant fish, indicating that Kita signaling is not required for the differentiation of this melanophore subpopulation, yet that it might be essential for its proper migration. Alternatively, the migratory behavior of this subpopulation might be normal in golden mutants, with the later-differentiating melanophores enhancing the camouflage in wild-type fish on the dorsal side only. This could be investigated in the future by selective labeling and tracing of the kita-independent population or by finding another mutation that eliminates this population. Independently of these details, we conclude that the guppy has an early-appearing kita-dependent and a later-developing partially or fully kita-independent melanophore population and that both populations are required to form the non-sex-specific reticulate pattern. Whole-mount in situ hybridization experiments turned out to be extremely difficult in guppy embryos due to their size and very low permeability (U. Martyn, and C. Dreyer, unpublished data), but could potentially help to determine the developmental time point at which the first melanophores differentiate in guppy embryos, especially as the melanization of these melanophores might be delayed in a livebearing fish like the guppy. The tracking of putative pigment cell precursors for adult ornaments, however, would require analysis of serial sections of specimens from early embryogenesis to puberty.
Our study suggests that Kita functions have at least partially been conserved across teleosts. The last common ancestor of zebrafish and guppies lived probably >300 million years ago (Kasahara et al. 2007); nevertheless, the adult pigment pattern of both species depends on an early kita-dependent and a late kita-independent melanophore population, and loss-of-function mutations in kita decrease the amount of melanophores, including scale melanophores, in both species (Haskins and Druzba 1938; Goodrich et al. 1944; Winge and Ditlevsen 1947; Parichy et al. 1999; Parichy et al. 2000a). Yet the teleost-specific whole-genome duplication likely also facilitated subfunctionalization and phenotypic diversification, as revealed in different Danio species (Braasch et al. 2006; Mills et al. 2007).
An early xanthophore population contributes as regularly spaced cells to the reticulate pattern of guppy juveniles and adults (Figure 1, E and F). Our study shows that these non-sex-specific xanthophores depend on Csf1ra signaling. The presence of isolated dorsal clusters of xanthophores in blue mutant fish suggests that Csf1ra activity might not be absolutely required for differentiation, but for proliferation and dispersal of guppy xanthoblasts. We do not know yet when during ontogeny csf1ra acts and whether different xanthophore populations exist, but we showed that Csf1ra is not required for the formation of the non-sex-specific black reticulate pattern of the guppy. In contrast, csf1ra mutant zebrafish cannot form a complete non-sex-specific stripe pattern (Parichy et al. 2000a; Parichy and Turner 2003b).
Guppies are conspicuously sexually dimorphic in their pigmentation, and mate-choice experiments have demonstrated that females prefer males with pronounced orange and blue iridescent ornamentation (Endler 1983; Kodric-Brown 1985). Males are also able to intensify their black ornaments while courting (Endler 1983). Crosses between golden blue mutant females with male guppies of geographically and genetically diverse origins (Willing et al. 2010) gave us the opportunity to study the influence of Kita and Csf1ra loss-of-function on the diverse male-specific patterns of the guppy. The males originated from West Trinidad (Guanapo), East Trinidad (Quare), and Venezuela (Cumaná). The latter two strains had previously been used for genetic mapping and QTL analysis (Tripathi et al. 2009b). Our study demonstrates that the male-specific xanthophores of the guppy, whose development is induced during puberty like the one of male-specific melanophores and iridophores, also depend on csf1ra and that loss of Csf1ra and Kita function substantially changes the formation of male ornaments.
Comparison between wild-type and golden males showed that black stripes and spots appeared ectopically in golden mutants, although in a manner that varied between populations and individuals; e.g., the ventral melanophores on the Guanapo tail fin were shifted to a more dorsal position, and golden mutant Maculatus males had novel black spots on their tail fins. In the BDZW1 strain, the arrangement of the creamy-black tail-fin margin, which might involve iridophores (personal observation), appeared reversed. In contrast, loss of Kita function did not change the dorso-ventral arrangement of the orange-black lining of the tail fin of golden N2 from the Cumaná cross. Interestingly, the marginal black components of the Cumaná tail-fin ornaments were lost together with the orange in blue mutants, whereas the major black spot on the tail fin persisted. In zebrafish, kita is expressed in melanoblasts, while csf1ra acts nonautonomously via short- and long-range xanthophore–melanophore interactions to promote melanophore migration and death during adult stripe formation (Parichy et al. 1999; Parichy and Turner 2003b; Nakamasu et al. 2009; Inaba et al. 2012). As transplantation experiments are not yet possible in the guppy, we could not determine whether Kita and Csf1ra act cell-autonomously or non-cell-autonomously during male pattern formation. However, as downregulation of csf1ra expression coincides with the absence of almost all xanthophores in the skin of blue fish, it is likely that csf1ra acts cell-autonomously within guppy xanthophores. Kita is an early melanoblast marker not only in zebrafish, but also in mice, and therefore most likely is expressed in guppy melanophores as well (Kelsh et al. 2009). Our observations suggest that some pattern elements in guppy males depend on coordinate expression of different cell types and that the formation of some of these pattern elements requires interactions between, or joint contribution from, different cell types. For example, xanthophore–melanophore interactions might underlie the formation of the orange-black lining of the tail fin of Cumaná males, but not the development of the black spot on the tail fin. Kita might directly affect the migration and/or survival of a subset of male melanophores. Terminal deoxynucleotidyl transferase-dUTP Nick End Labeling (TUNEL) assays could reveal whether cell apoptosis plays a role during male pattern formation in the guppy.
Comparisons between wild-type and blue offspring of the backcrosses suggest that compact black spots can form in absence of Csf1ra, most probably without interactions between xanthophores and melanophores, yet the final positions of these spots appear unpredictable. The labyrinthine ornaments close to the gonopodium in Guanapo males provide an example of interacting genetic cofactors in that backcross and potentially also of interactions between different cell types. Compared with wild type, these ornaments were greatly reduced in complexity in golden mutant N2 and lost in blue and golden blue mutant N2. Since only faint yellow pigment is seen in this area in wild-type fish, the contribution of xanthophores and their interactions is here hard to assess. The loss of the labyrinthine ornaments in golden mutant N2 might be explained by a reduced migratory potential of the kita-independent melanophores or their precursors in golden N2. We can, however, not exclude that a kita-dependent subpopulation of late melanophores contributes to this complex trait in wild-type fish.
golden blue double-mutant guppies from four different backcrosses always had less total black than blue mutants, but much more than zebrafish kita csf1ra double mutants (Parichy 2006). This might reflect different requirements for Kita and Csf1ra signaling: while xanthophores enhance the survival of adult stripe melanophores in zebrafish (Parichy and Turner 2003b), the survival of male hormone-induced melanophores in the guppy might not require xanthophores; yet the ability of these melanophores to form some of the complex traits might depend on interactions with these cells.
Taken together, we conclude that at least three temporally and genetically distinct melanophore populations occur in the guppy: first, a kita-dependent population differentiating during embryogenesis; second, a partially or fully kita-independent population mostly differentiating after birth; and third, a male-specific melanophore population whose differentiation, migration, and proliferation might be induced by testosterone during puberty. It remains to be resolved where the precursors of these male-specific pigment cells reside and whether they might be derived from the same pool as the late non-sex-specific kita-independent melanophores. Only a few recent publications have addressed the routes and fates of pigment cell precursor pools destined for delayed differentiation in other vertebrates (Watanabe et al. 2008; Adameyko et al. 2009; Yamada et al. 2010; Budi et al. 2011). While the embryonic and early larval pigment pattern of zebrafish is formed by melanoblasts that are derived directly from the neural crest, later-appearing metamorphic melanophores of zebrafish develop from post-embryonic extrahypodermal pigment cell precursors, which migrate to the hypodermis during pigment pattern metamorphosis (Budi et al. 2011). These precursors are associated with nerves and depend on Erbb3b and Tubulin α 8-like 3a signaling (Budi et al. 2011). Improvement of in vitro culture methods of guppy embryos (Martyn et al. 2006) may facilitate the ability to treat, and to subsequently raise, explanted guppy embryos with an ErbB inhibitor, which might reveal whether ErbB signaling is required to establish melanophore stem cells in the guppy as well.
In stickleback, regulatory mutations in kitla are associated with the light coloration of gills and ventral skin in several freshwater populations (Miller et al. 2007). This indicates that differential distribution of Kitla can lead to distinct pigmentation patterns in natural populations. In contrast to the receptor Kita, the functions of Csf1ra seem to be less conserved even among species in the genus Danio, as indicated by limited complementation of csf1ra loss of function in interspecies hybrids (Quigley et al. 2005). Furthermore, a study with haplochromine cichlids has suggested that positive selection has acted on csf1ra, which is expressed in the yellow egg spots of these fish (Salzburger et al. 2007). QTL mapping with higher marker density may reveal whether or not Kita and Csf1ra, and/or their ligands, also affect natural variation of guppy ornaments. For this purpose, we are generating a denser genetic map of the guppy based on Restriction-site Associated DNA (RAD) markers. Combined with the ongoing whole-genome sequencing, these experiments will further enhance our efforts to unravel the network of genetic factors that cooperatively maintain the highly complex male ornaments of the guppy.
Supplementary Material
Acknowledgments
We thank Harald Auer for the donation of the ARM and BDZW2 guppy strains; Axel Meyer for the BDZW1 guppy strain; David Reznick for guppies from the Quare and Guanapo rivers; Christopher Dooley for general advice on zebrafish pigment pattern mutants; Tobias Langenecker for assistance with real-time quantitative PCR; Gertrud Scheer, Alexandra Schnell, and Philipp Vollmer for guppy images; Richard Neher for help with data analysis; Joffrey Fitz for technical support; and Bonnie Fraser, Felicity Jones, and Axel Künstner for helpful suggestions on the manuscript. This work was supported by a Gottfried Wilhelm Leibniz Award of the Deutsche Forschungsgemeinschaft and by funds from the Max Planck Society to D.W.
Footnotes
Communicating editor: D. Parichy
Literature Cited
- Adameyko I., Lallemend F., Aquino J. B., Pereira J. A., Topilko P., et al. , 2009. Schwann cell precursors from nerve innervation are a cellular origin of melanocytes in skin. Cell 139: 366–379 [DOI] [PubMed] [Google Scholar]
- Alexander H. J., Breden F., 2004. Sexual isolation and extreme morphological divergence in the Cumana guppy: a possible case of incipient speciation. J. Evol. Biol. 17: 1238–1254 [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Braasch I., Salzburger W., Meyer A., 2006. Asymmetric evolution in two fish-specifically duplicated receptor tyrosine kinase paralogons involved in teleost coloration. Mol. Biol. Evol. 23: 1192–1202 [DOI] [PubMed] [Google Scholar]
- Budi E. H., Patterson L. B., Parichy D. M., 2011. Post-embryonic nerve-associated precursors to adult pigment cells: genetic requirements and dynamics of morphogenesis and differentiation. PLoS Genet. 7: e1002044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens H. P., McDermitt C., Inslee T., 1966. The effects of feeding methyl testosterone to guppies for sixty days after birth. Copeia 1966: 280–284 [Google Scholar]
- Dzwillo M., 1959. Genetic investigations of domesticated strains of Lebistes reticulatus Peters Mitt. Hamburg. Zool. Mus. Inst. 57: 143–186 (in German) [Google Scholar]
- Endler J. A., 1983. Natural and sexual selection on color patterns in poeciliid fishes. Environ. Biol. Fishes 9: 173–190 [Google Scholar]
- Felsenstein J., 1989. PHYLIP-phylogeny inference package (version 3.2). Cladistics 5: 164–166 [Google Scholar]
- Goodrich H. B., Josephson N. D., Trinkaus J. P., Slate J. M., 1944. The cellular expression and genetics of two new genes in Lebistes reticulatus. Genetics 29: 584–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon M., McWilliam H., Li W., Valentin F., Squizzato S., et al. , 2010. A new bioinformatics analysis tools framework at EMBL–EBI. Nucleic Acids Res. 38: W695–W699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood A. K., Cech J. N., Peichel C. L., 2012. Molecular and developmental contributions to divergent pigment patterns in marine and freshwater sticklebacks. Evol. Dev. 14: 351–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., et al. , 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59: 307–321 [DOI] [PubMed] [Google Scholar]
- Haskins C. P., Druzba J. P., 1938. Note on anomalous inheritance of sex-linked color factors in the Guppy. Am. Nat. 72: 571–574 [Google Scholar]
- Houde A. E., 1997. Sex, Color, and Mate Choice in Guppies. Princeton University Press, Princeton, NJ [Google Scholar]
- Hultman K. A., Bahary N., Zon L. I., Johnson S. L., 2007. Gene duplication of the zebrafish kit ligand and partitioning of melanocyte development functions to kit ligand a. PLoS Genet. 3: e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D. H., Bryant D., 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23: 254–267 [DOI] [PubMed] [Google Scholar]
- Inaba M., Yamanaka H., Kondo S., 2012. Pigment pattern formation by contact-dependent depolarization. Science 335: 677. [DOI] [PubMed] [Google Scholar]
- Johnson S. L., Africa D., Walker C., Weston J. A., 1995. Genetic control of adult pigment stripe development in zebrafish. Dev. Biol. 167: 27–33 [DOI] [PubMed] [Google Scholar]
- Kasahara M., Naruse K., Sasaki S., Nakatani Y., Qu W., et al. , 2007. The medaka draft genome and insights into vertebrate genome evolution. Nature 447: 714–719 [DOI] [PubMed] [Google Scholar]
- Kelsh R. N., Harris M. L., Colanesi S., Erickson C. A., 2009. Stripes and belly-spots: a review of pigment cell morphogenesis in vertebrates. Semin. Cell Dev. Biol. 20: 90–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodric-Brown A., 1985. Female preference and sexual selection for male coloration in the guppy (Poecilia reticulata). Behav. Ecol. Sociobiol. 17: 199–205 [Google Scholar]
- Lamason R. L., Mohideen M. A., Mest J. R., Wong A. C., Norton H. L., et al. , 2005. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science 310: 1782–1786 [DOI] [PubMed] [Google Scholar]
- Lindholm A., Breden F., 2002. Sex chromosomes and sexual selection in Poeciliid fishes. Am. Nat. 160: S214–S224 [DOI] [PubMed] [Google Scholar]
- Lynn Lamoreux M., Kelsh R. N., Wakamatsu Y., Ozato K., 2005. Pigment pattern formation in the medaka embryo. Pigment Cell Res. 18: 64–73 [DOI] [PubMed] [Google Scholar]
- Maderspacher F., Nusslein-Volhard C., 2003. Formation of the adult pigment pattern in zebrafish requires leopard and obelix dependent cell interactions. Development 130: 3447–3457 [DOI] [PubMed] [Google Scholar]
- Magurran A. E., 2005. Evolutionary Ecology: The Trinidadian Guppy. Oxford University Press, Oxford [Google Scholar]
- Martyn U., Weigel D., Dreyer C., 2006. In vitro culture of embryos of the guppy, Poecilia reticulata. Dev. Dyn. 235: 617–622 [DOI] [PubMed] [Google Scholar]
- Mellgren E. M., Johnson S. L., 2005. kitb, a second zebrafish ortholog of mouse Kit. Dev. Genes Evol. 215: 470–477 [DOI] [PubMed] [Google Scholar]
- Miller C. T., Beleza S., Pollen A. A., Schluter D., Kittles R. A., et al. , 2007. cis-regulatory changes in Kit ligand expression and parallel evolution of pigmentation in sticklebacks and humans. Cell 131: 1179–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills M. G., Nuckels R. J., Parichy D. M., 2007. Deconstructing evolution of adult phenotypes: genetic analyses of kit reveal homology and evolutionary novelty during adult pigment pattern development of Danio fishes. Development 134: 1081–1090 [DOI] [PubMed] [Google Scholar]
- Mol C. D., Lim K. B., Sridhar V., Zou H., Chien E. Y., et al. , 2003. Structure of a c-kit product complex reveals the basis for kinase transactivation. J. Biol. Chem. 278: 31461–31464 [DOI] [PubMed] [Google Scholar]
- Nakamasu A., Takahashi G., Kanbe A., Kondo S., 2009. Interactions between zebrafish pigment cells responsible for the generation of Turing patterns. Proc. Natl. Acad. Sci. USA 106: 8429–8434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusslein-Volhard C., Dahm R., 2002. Zebrafish: A Practical Approach. Oxford University Press, Oxford [Google Scholar]
- Parichy D. M., 2006. Evolution of danio pigment pattern development. Heredity (Edinb) 97: 200–210 [DOI] [PubMed] [Google Scholar]
- Parichy D. M., Turner J. M., 2003a Zebrafish puma mutant decouples pigment pattern and somatic metamorphosis. Dev. Biol. 256: 242–257 [DOI] [PubMed] [Google Scholar]
- Parichy D. M., Turner J. M., 2003b Temporal and cellular requirements for Fms signaling during zebrafish adult pigment pattern development. Development 130: 817–833 [DOI] [PubMed] [Google Scholar]
- Parichy D. M., Rawls J. F., Pratt S. J., Whitfield T. T., Johnson S. L., 1999. Zebrafish sparse corresponds to an orthologue of c-kit and is required for the morphogenesis of a subpopulation of melanocytes, but is not essential for hematopoiesis or primordial germ cell development. Development 126: 3425–3436 [DOI] [PubMed] [Google Scholar]
- Parichy D. M., Ransom D. G., Paw B., Zon L. I., Johnson S. L., 2000a An orthologue of the kit-related gene fms is required for development of neural crest-derived xanthophores and a subpopulation of adult melanocytes in the zebrafish, Danio rerio. Development 127: 3031–3044 [DOI] [PubMed] [Google Scholar]
- Parichy D. M., Mellgren E. M., Rawls J. F., Lopes S. S., Kelsh R. N., et al. , 2000b Mutational analysis of endothelin receptor b1 (rose) during neural crest and pigment pattern development in the zebrafish Danio rerio. Dev. Biol. 227: 294–306 [DOI] [PubMed] [Google Scholar]
- Parichy D. M., Elizondo M. R., Mills M. G., Gordon T. N., Engeszer R. E., 2009. Normal table of postembryonic zebrafish development: staging by externally visible anatomy of the living fish. Dev. Dyn. 238: 2975–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W., 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley I. K., Manuel J. L., Roberts R. A., Nuckels R. J., Herrington E. R., et al. , 2005. Evolutionary diversification of pigment pattern in Danio fishes: differential fms dependence and stripe loss in D. albolineatus. Development 132: 89–104 [DOI] [PubMed] [Google Scholar]
- Reznick D., Endler J. A., 1982. The impact of predation on life history evolution in Trinidadian guppies (Poecilia reticulata). Evolution 36: 160–177 [DOI] [PubMed] [Google Scholar]
- Salzburger W., Braasch I., Meyer A., 2007. Adaptive sequence evolution in a color gene involved in the formation of the characteristic egg-dummies of male haplochromine cichlid fishes. BMC Biol. 5: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., et al. , 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7.: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi I. K., 1976. Electron microscopy of two types of reflecting chromatophores (iridophores and leucophores) in the guppy, Lebistes reticulatus Peters. Cell Tissue Res. 173: 17–27 [DOI] [PubMed] [Google Scholar]
- Traut W., Winking H., 2001. Meiotic chromosomes and stages of sex chromosome evolution in fish: zebrafish, platyfish and guppy. Chromosome Res. 9: 659–672 [DOI] [PubMed] [Google Scholar]
- Tripathi N., Hoffmann M., Dreyer C., 2008. Natural variation of male ornamental traits of the guppy, Poecilia reticulata. Zebrafish 5: 265–278 [DOI] [PubMed] [Google Scholar]
- Tripathi N., Hoffmann M., Weigel D., Dreyer C., 2009a Linkage analysis reveals the independent origin of Poeciliid sex chromosomes and a case of atypical sex inheritance in the guppy (Poecilia reticulata). Genetics 182: 365–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi N., Hoffmann M., Willing E. M., Lanz C., Weigel D., et al. , 2009b Genetic linkage map of the guppy, Poecilia reticulata, and quantitative trait loci analysis of male size and colour variation. Proc. Biol. Sci. 276: 2195–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., et al. , 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Washio Y., Fujinami Y., Aritaki M., Uji S., et al. , 2008. Adult-type pigment cells, which color the ocular sides of flounders at metamorphosis, localize as precursor cells at the proximal parts of the dorsal and anal fins in early larvae. Dev. Growth Differ. 50: 731–741 [DOI] [PubMed] [Google Scholar]
- Willing E. M., Bentzen P., van Oosterhout C., Hoffmann M., Cable J., et al. , 2010. Genome-wide single nucleotide polymorphisms reveal population history and adaptive divergence in wild guppies. Mol. Ecol. 19: 968–984 [DOI] [PubMed] [Google Scholar]
- Winge Ö., 1922. One-sided masculine and sex-linked inheritance in Lebistes reticulatus. J. Genet. 12: 145–162 [Google Scholar]
- Winge Ö., 1927. The location of eighteen genes in Lebistes reticulatus. J. Genet. 18: 1–43 [Google Scholar]
- Winge Ö., Ditlevsen E., 1947. Colour inheritance and sex determination in Lebistes. Heredity 1: 65–83 [Google Scholar]
- Yamada T., Okauchi M., Araki K., 2010. Origin of adult-type pigment cells forming the asymmetric pigment pattern, in Japanese flounder (Paralichthys olivaceus). Dev. Dyn. 239: 3147–3162 [DOI] [PubMed] [Google Scholar]
- Yarden Y., Ullrich A., 1988. Growth factor receptor tyrosine kinases. Annu. Rev. Biochem. 57: 443–478 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.