Abstract
Our understanding of the development of intrinsic reproductive isolation is still largely based on theoretical models and thorough empirical studies on a small number of species. Theory suggests that reproductive isolation develops through accumulation of epistatic genic incompatibilities, also known as Bateson–Dobzhansky–Muller (BDM) incompatibilities. We can detect these from marker transmission ratio distortion (TRD) in hybrid progenies of crosses between species or populations, where TRD is expected to result from selection against heterospecific allele combinations in hybrids. TRD may also manifest itself because of intragenomic conflicts or competition between gametes or zygotes. We studied early stage speciation in Arabidopsis lyrata by investigating patterns of TRD across the genome in F2 progenies of three reciprocal crosses between four natural populations. We found that the degree of TRD increases with genetic distance between crossed populations, but also that reciprocal progenies may differ substantially in their degree of TRD. Chromosomes AL6 and especially AL1 appear to be involved in many single- and two-locus distortions, but the location and source of TRD vary between crosses and between reciprocal progenies. We also found that the majority of single- and two-locus TRD appears to have a gametic, as opposed to zygotic, origin. Thus, while theory on BDM incompatibilities is typically illustrated with derived nuclear alleles proving incompatible in hybrid zygotes, our results suggest a prominent role for distortions emerging before zygote formation.
Keywords: Bateson–Dobzhansky–Muller incompatibilities, cytonuclear interactions, gametic selection, hybrid incompatibility, speciation
COMPLETE reproductive isolation between natural populations is usually achieved through several reproductive barriers, acting at different developmental stages. In plants prezygotic reproductive isolation acts either prepollination (e.g., habitat, temporal, or pollinator isolation) or postpollination (e.g., unilateral incompatibility or conspecific pollen precedence). Postzygotic isolating mechanisms either reduce viability, fertility, or fitness of hybrids or prevent development of a hybrid altogether. The effect of postzygotic reproductive barriers may be environment dependent (e.g., Hatfield and Schluter 1999) or independent of the environment. The latter, intrinsic postzygotic barriers, cause reduced viability or fertility of hybrids and have been studied extensively, e.g., in Drosophila (for a review see Coyne and Orr 2004; Presgraves 2010).
Despite the importance of development of reproductive isolation as an evolutionary process, and despite considerable study, precise molecular genetic mechanisms of postzygotic isolation have rarely been described (Presgraves 2010; Rieseberg and Blackman 2010). It is generally assumed, however, that intrinsic postzygotic isolation is due to Bateson–Dobzhansky–Muller incompatibilities (BDMI; Orr 1995; Orr and Turelli 2001). These incompatibilities emerge when in (usually) isolated populations new alleles arise, which upon secondary contact do not function together. In a few cases single genes involved in BDMIs have been identified (reviewed by Presgraves 2010) and from Drosophila even a pair of loci is known (Brideau et al. 2006), but generally the mechanism by which BDMI cause postzygotic isolation is unknown.
Genic incompatibilities that cause reduced hybrid viability or fertility may lead to non-Mendelian segregation of alleles and genotypes (transmission ratio distortion, TRD) in hybrid progenies of experimental crosses. Thus, BDMI manifest themselves by distorting transmission ratios of alleles, a phenomenon commonly observed in experimental crosses between species (Jenczewski et al. 1997; Rieseberg et al. 2000). Loci involved in BDMI have been mapped by analyzing TRD of alleles in the offspring of experimental crosses between (sub)species or populations (Vogl and Xu 2000; Harushima et al. 2001). In plants, locations of transmission ratio-distorted loci (TRDL) have been used as indicators of genetic incompatibilities in Mimulus (Fishman et al. 2001), rice (Harushima et al. 2001), Eucalyptus (Myburg et al. 2004), tomato (Moyle and Graham 2006), Ceratodon moss (McDaniel et al. 2008), Ceratopteris fern (Nakazato et al. 2007), and Arabidopsis thaliana (Salomé et al. 2011).
TRD is not always a sign of BDMI, but may also arise for various other reasons during development of gametes or after the gametes are formed but before fertilization. The former could be a selfish segregation distorter or meiotic drive system where a certain allele gets transmission advantage over the other, e.g., in meiosis of germ cells (reviewed by Lyttle 1991; Werren 2011). Selfish genetic changes have been found, surprisingly often, to be involved in formation of hybrid dysfunction (Presgraves 2010). Selfish genetic elements and the genomic conflicts they cause have been recognized as important factors in promoting evolutionary change (Werren 2011). Presgraves (2010) concluded that intrinsic genomic instability underlies evolution of speciation genes (genes causing hybrid sterility or inviability) more often than classical adaptive incompatibilities developing in both diverging populations. Currently most of the examples derive from a few model species and more investigations are needed (Johnson 2010).
TRD could also be due to other prefertilization events such as competition between gametes, e.g., variable pollen tube growth rate (reviewed by Howard 1999). In angiosperms, competition would be more likely to occur between male gametes, because of their greater number and longer haploid phase compared to female gametes. TRD could also be due solely to factors acting after fertilization, such as differential viability of hybrid genotypes, competition between zygotes on maternal resources (Korbecka et al. 2002), or as a combination of both gametic and zygotic factors.
Transmission ratios may be distorted due to epistatic interactions not only between nuclear genes, but also between nuclear genes and cytoplasmic factors, such as mitochondrial or chloroplast genes. To be able to investigate the role of cytoplasmic factors in generating TRD, one must perform reciprocal crosses, so that the two reciprocal hybrid progenies share the same nuclear genome but differ in cytoplasm. Reciprocal crosses are feasible with hermaphroditic species and have therefore been most commonly conducted in plants. The role of cytonuclear interactions in performance of reciprocal hybrids has been demonstrated in many species (reviewed by Levin 2003). Apart from cytoplasmic male sterility, little is known about the role of cytonuclear interactions in causing postzygotic reproductive isolation (Rieseberg and Blackman 2010).
Designs of experimental crosses differ, but studies of TRD are usually started with two parents representing different populations, subspecies, or species. However, these parents have often been derived from selfed lines. While this simplifies mapping algorithms and increases statistical power to detect TRDL, it provides little information about allelic variation within natural populations (except if several independent crosses were produced). Only a few studies have examined variation in incompatibility alleles within populations in nature (e.g., Christie and Macnair 1987; Reed and Markow 2004; Sweigart et al. 2007; Koide et al. 2008a; Martin and Willis 2010; Gérard and Presgraves 2012), but the importance of polymorphic hybrid incompatibility is becoming more acknowledged (Cutter 2012).
In addition to the interest in understanding how, and at what developmental stage, intrinsic postzygotic reproductive isolation acts, interesting questions that remain to be answered concern the rate of development of postzygotic reproductive isolation. Earlier studies on Drosophila, Lepidoptera, frogs, and some plant genera (reviewed in Coyne and Orr 2004) imply that intrinsic postzygotic isolation develops gradually over time. If transmission ratio distortion reflects reproductive isolation, then the degree of TRD should also increase with the genetic distance between populations or species in experimental crosses. It has been predicted that the number of genic incompatibilities increases between taxa faster than linearly with time (Orr 1995; Orr and Turelli 2001). This seemingly simple prediction has been surprisingly difficult to test in practice: even if genetic distances between populations or species are known, it is difficult to compare the degree of TRD between crosses that differ in statistical power to detect and locate TRD loci because of different numbers, locations, and types of genetic markers and different numbers of hybrid individuals. The same applies to mapping of quantitative trait loci for reproductive isolation. Hence, the first empirical efforts supporting this prediction were published only recently (Matute et al. 2010; Moyle and Nakazato 2010), and it remains poorly known whether and how the degree of TRD increases with genetic distance between populations.
Here we investigate TRD in crosses between isolated populations of A. lyrata and map transmission ratio distorting loci. Earlier studies in this species indicated that TRD is common in crosses of genetically distant populations (Kuittinen et al. 2004) but rare in crosses within populations (Leppälä et al. 2008). We compare three experimental crosses, at different genetic distances as assessed with microsatellite (Muller et al. 2008) and sequence data (Wright et al. 2003; Ross-Ibarra et al. 2008; Pyhäjärvi et al. 2012). The most distant populations crossed (FST = 0.62; Pyhäjärvi et al. 2012) represent the subspecies lyrata and petraea. The intermediate (FST = 0.35) and closely (FST = 0.20) related crosses are between populations of ssp. petraea. This allows us to investigate whether TRD increases with genetic distance between the populations crossed.
We used novel algorithms that allow us to distinguish gametic and zygotic TRD, as well as to detect epistatic two-locus TRD from crosses of outbred individuals. The outbred F2 design of our crosses (Figure 1), with two differently heterozygous F1, allows us to evaluate the roles of within-population allelic variation, male and female function, and cytoplasm for TRD loci. Because one of the populations is involved in all of the three crosses, we can also examine whether the same or different genomic regions are associated with TRD when crossed with different populations.
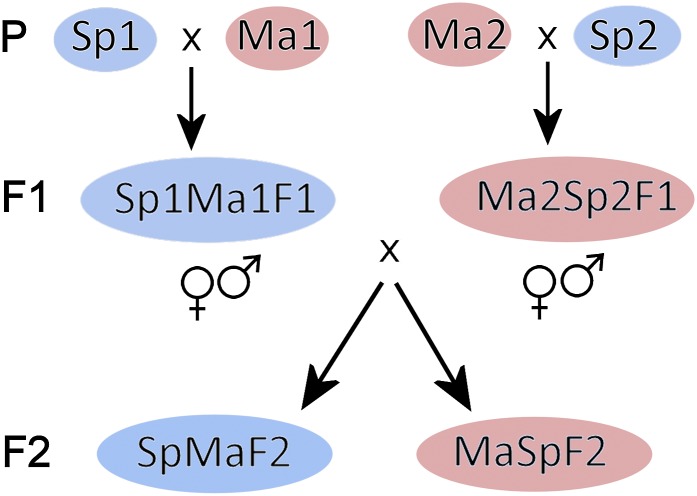
Figure 1.
The crossing design for all three reciprocal crosses of A. lyrata examplefied by the Sp × Ma cross. The four parents (P), two F1 individuals, and F2 progenies are shown. The blue background indicates Spiterstulen and red Mayodan cytoplasm. Note that the F1 individuals were crossed reciprocally (Sp1Ma1 × Ma2Sp2 and Ma2Sp2 × Sp1Ma1) so that four different alleles (Sp1, Sp2, Ma1, Ma2) could segregate in both reciprocal F2 progenies. The image has been modified from Leppälä and Savolainen (2011).
Materials and Methods
Crosses
To study transmission ratio distortion we used genetic marker data from three crosses between different A. lyrata populations. For a cross between Spiterstulen, Norway (Sp), and Mayodan, North Carolina (Ma), crossing details and plant growth conditions are described in Leppälä and Savolainen (2011). In brief, two unrelated plants from both populations were crossed to produce F1 hybrids (Figure 1). The crosses were done reciprocally so that F1’s carry different cytoplasm. From both crosses one F1 was used to produce the F2 generation. Again the crosses were conducted reciprocally to gain F2 progenies with different cytoplasmic backgrounds (referred to as SpMaF2 when cytoplasm from Sp and MaSpF2 when cytoplasm from Ma).
The same crossing design was used when crossing Spiterstulen with Stubbsand, Sweden (Stu). To make crosses comparable, the same plants from Spiterstulen were used as the parents for the F1. They were crossed in the same manner, so that the Sp individual that acted as a pollen recipient in the Sp × Ma cross also acted as a pollen recipient in the Sp × Stu cross. The F2 individuals from the Sp × Stu cross were grown in the same greenhouse at the same time as the Sp × Ma F2.
The third cross was conducted between Spiterstulen and Plech, Germany (Pl). For this cross, different Sp individuals were used as parents for the F1 than those used in the crosses described above. Here the crossing design also differed in that the two Sp parents were used as pollen recipients and the two Pl individuals as pollen donors in both initial crosses to achieve F1. The F1 were crossed as described above to get two F2 populations, both carrying Sp cytoplasm. To distinguish between the F2 reciprocal progenies, they are referred to as SpPl2F2 and SpPl3F2. The growth conditions of the F2 are described in Quilot-Turion et al. (2013).
Genotyping
DNA was isolated and genotyping performed for Sp × Ma and Sp × Stu F2 as described in Leppälä and Savolainen (2011) and for Sp × Pl F2 as in Kemi et al. (2013). For Sp × Ma 391 F2 individuals were genotyped (204 for Sp cytoplasm and 187 for Ma cytoplasm) with 76 SNP, microsatellite, and Cleaved Amplified Polymorphic Sequences (CAPS) markers (for details see Leppälä and Savolainen 2011). Sp × Stu was genotyped with the same methods as Sp × Ma. For Sp × Stu, 389 F2 plants were genotyped (195 for Sp cytoplasm and 194 for Stu cytoplasm) with 65 SNP, microsatellite, and CAPS markers. The markers used to genotype Sp × Stu F2 and the origins of the primer sequences are in Supporting Information, Table S1. In the Sp × Pl cross, 529 F2 individuals were genotyped (326 SpPl2F2 and 203 SpPl3F2). For the Sp × Pl cross, 40 microsatellite and CAPS markers were genotyped. For details see Kemi et al. (2013).
Analysis of transmission ratio distortion
Each of the n F2 offspring of F1 heterozygotes ab and cd must have one of the four possible genotypes ac, ad, bc, and bd. Because the genotype of each offspring is independent of that of the other offspring, the log-likelihood of the observed numbers of genotypes x is given by the multinomial distribution
| (1) |
where Σx = n and p denotes the expected frequency of a genotype.
Under Mendelian segregation we expect the four genotypes in equal frequencies (i.e., all p = 0.25) and substituting these values in Equation 1 we obtain the likelihood of the observed numbers of genotypes in the absence of transmission ratio distortion, L0. Deviations of observed genotype numbers x from their expectations p may be due to chance, due to differential transmission of alleles in gametes or due to differential survival of genotypes in the zygotic phase. Let us first consider differential gametic transmission of alleles. If the male parent transfers allele a with probability qm, then the probability that allele b is transferred is 1 − qm. Similarly, we denote the probability that the female transmits allele c qf, so that allele d is transmitted with probability 1− qf. Given qm and qf, the expected frequencies of the genotypes are: pac= qmqf, pad= qm(1 − qf), pbc= (1 − qm)qf, and pbd= (1 − qm)(1 − qf). These expected frequencies can be substituted in Equation 1 to obtain the likelihood, and we can use standard algorithms to find the values of qm and qf that maximize the likelihood of the observed genotype frequencies under gametic phase transmission ratio distortion, Lgametic. Finally, differential zygotic survival or fertilization success may cause unequal genotype frequencies. In that case, the only constraint on p is Σp = 1, and the likelihood of the observed genotype frequencies Lzygotic is maximized when expected frequencies are equal to observed frequencies, i.e., pac= xac/n (and similarly for the other genotypes).
Distinguishing gametic from zygotic TRD
We can evaluate which of the above models best explains the observed numbers of genotypes x using likelihood ratio tests. Let L0 and La denote the maximized log-likelihoods of the observed genotype frequencies under the null and alternative hypothesis, respectively. The sampling distribution of the logarithmic-likelihood ratio −2(L0− La) follows a χ2-distribution with degrees of freedom equal to the difference in the number of unknown parameters under the hypotheses being compared. Under Mendelian segregation all p = 0.25 so that there are no free parameters. With gametic phase TRD there are two unknown parameters (qm and qf) that determine p, and if TRD arises at the zygotic stage there is only one constraint (Σp = 1) so that there are three unknown parameters. Thus, to test for gametic TRD, we evaluate the χ2 distribution with 2 −0 = 2 degrees of freedom at −2(L0/Lgametic). To test for zygotic TRD, we evaluate the χ2 distribution with 3 − 0 = 3 degrees of freedom at −2(L0/Lzygotic). In case of gametic phase TRD also Lzygotic (and not only Lgametic) will be substantially higher than L0. Therefore we evaluate the χ2 distribution with 3 − 2 = 1 degrees of freedom at −2(Lgametic/Lzygotic). If zygotic TRD explains the data better than gametic TRD, while gametic TRD explains the data better than Mendelian segregation, this indicates zygotic TRD in addition to gametic phase TRD. Thus, comparing likelihoods of the observed genotype frequencies under different hypotheses as illustrated in Figure S1, we can identify loci with gametic, zygotic, and with both gametic and zygotic TRD. We considered genomic regions as transmission ratio distorted when P < 0.001.
Epistatic TRD
Transmission ratio distortion can be caused by single-locus factors, but also by interactions between two (or more) loci. To detect two-locus epistatic TRD we calculated two-locus genotype frequencies for all pairs of markers (excluding markers at the same chromosome), calculated expected frequencies by cross tabulation, and evaluated the probability that observed and expected genotype frequencies differ by chance using χ2 tests. Because we calculated expected frequencies using cross tabulation, observed single-locus genotype frequencies are taken into account when detecting epistatic TRD.
Just like single-locus TRD, also epistatic TRD may emerge both in the gametic and in the zygotic phase. To detect gametic phase epistatic TRD we calculated expected two-locus allelic combinations by cross-tabulation from observed single-locus allelic frequencies. For example, if the pollen donor (male parent) transmitted allele a at locus 1 with observed frequency qm1, and allele a at locus 2 with observed frequency qm2, then the probabilities of observing two-locus allelic combinations a1a2, a1b2, a2b1, and a2b2 are qm1 qm2, qm1 (1 − qm2), (1 − qm1) qm2, and (1 − qm1)(1 − qm2), respectively. We evaluated deviation of observed two-locus allelic combinations from their expectations using χ2 tests. Parents were analyzed separately since at the gametic stage interaction between parents is not yet plausible.
Missing genotypes and informativeness
Above we assumed that genotypes are known with certainty for each individual, but at several loci with partially informative markers or pseudomarkers (see below) that is not the case. We first inferred missing genotypes using Wijsman’s (1987) genotyping rules. Subsequently, flanking markers were used to infer genotype probabilities at partly informative genotyped loci and at pseudomarker loci. For single-locus analysis of TRD, individuals were assigned genotypes so as to maximize the sum of the log-likelihood from Equation 1, plus the log-likelihood of the assigned genotypes. For analyses of epistatic TRD this was computationally not feasible, so individuals were simply assigned the genotype that was most likely based on the flanking markers. Details on the algorithms we used for TRD mapping are in File S1.
Comparing TRD between crosses
Because numbers and positions of markers differ between crosses, we cannot compare the degree of TRD between crosses and reciprocal progenies directly by calculating the number or percentage of distorted markers. As a simple alternative, we used the percentage of the genome showing single locus TRD. We inserted pseudomarkers so as to have intermarker distances <1 cM and then approximated the percentage of the genome showing TRD as the percentage of distorted (pseudo)markers.
As we use many markers, using the conventional P-value of 0.05 as a threshold may result in a high false-positive rate. However, using a standard correction for multiple testing based on the number of markers would lead to unacceptable false-negative rates, not only because markers are statistically dependent through linkage, but also because the markers do not provide repeated tests of the same hypothesis. Therefore we made a somewhat arbitrarily trade-off between avoiding type I and type II errors and used a threshold of P = 0.001, but we confirmed that results are qualitatively similar when using different thresholds in the range 0.05–0.0001.
Results
All three reciprocal F2 progenies had transmission ratio distorted markers. The reciprocal progenies of each cross were examined separately for TRD because of possible cytoplasmic effects, or differences in heterogeneous F1 parental plants when acting as pollen donors or recipients. In the progeny of the cross between the most distant populations (FST = 0.62) SpMaF2, 33 of 76 markers (43% of χ2 tests at α = 0.05, not corrected for multiple testing) deviated from expected Mendelian genotype frequencies, and in the reciprocal MaSpF2 progeny 26 of 76 markers had non-Mendelian inheritance (34% at α = 0.05). With a more conservative threshold of α = 0.001 the percentages of distorted markers remained high (SpMaF2 20% and MaSpF2 23%). At intermediate genetic distance between populations (FST = 0.35), in the SpPl2F2 progeny 13 of the 40 markers (33% at α = 0.05) showed non-Mendelian segregation and in the reciprocal SpPl3F2 9 of the 40 markers had distorted segregation ratios (23% at α = 0.05). Finally, at the smallest genetic distance between populations (FST = 0.20), the progeny had a lower percentage of transmission ratio distorted markers on average. However, only 8 of 65 deviated from Mendelian segregation in the SpStuF2 (12% at α = 0.05), while the reciprocal StuSpF2 progeny had 20 transmission ratio distorted markers (31% at α = 0.05).
To more precisely identify the genomic regions showing TRD and to investigate at which level (gametic, zygotic, or both) the distortions likely developed, we applied TRD mapping methods, to compare observed genotype frequencies to expectations under the null model (Mendelian segregation), and under models allowing for gametic and zygotic phase distortion. In all crosses, most of the TRD regions were best explained by the gametic model (Table 1), but locations and types of TRD differ between progenies (Figure 2, Figure S2, Figure S3, Figure S4, Figure S5, Figure S6, and Figure S7).
Table 1. The number of single-locus TRD regions and the percentage of markers in TRD (P < 0.001, or P < 0.01 in parentheses) in three reciprocal crosses of A. lyrata.
| Measurement | Phase | Sp × Ma | Sp × Pl | Sp × Stu | ||||
|---|---|---|---|---|---|---|---|---|
| Fst between populationsa | 0.62 | 0.35 | 0.20 | |||||
| Total lenght of the genetic map (cM) | 514 | 511 | 500 | |||||
| Number of markers | 76 | 40 | 65 | |||||
| Reciprocal progenies | SpMaF2 | MaSpF2 | SpPl2F2 | SpPl3F2 | SpStuF2 | StuSpF2 | ||
| F2 progeny size | 204 | 187 | 326 | 203 | 195 | 194 | ||
| Single-locus TRD regions at P < 0.001 (0.01) | Gametic | 2 (6) | 3 (6) | 3 (5) | 1 (4) | 1 (1) | 2 (6) | |
| Zygotic | — | — | — | — | — | — | ||
| Both | 1(1) | 1(1) | — | — | 0 (1) | 0 (1) | ||
| % of genome distorted at P < 0.001 (0.01) | Gametic | 23 (38) | 23 (34) | 14 (18) | 10 (19) | 2 (2) | 9 (23) | |
| Zygotic | 1 (3) | 1 (1) | 0.7 (4) | 0 (0.3) | 0.6 (1) | 0.6 (2) | ||
FST values from Pyhäjärvi et al. (2012).
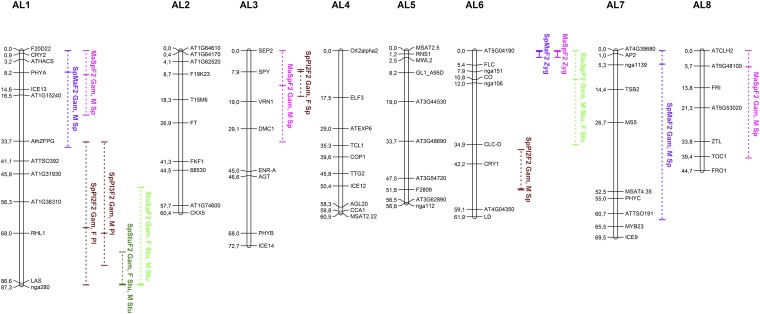
Figure 2.
Locations of TRD regions in A. lyrata crosses on a Sp × Ma map modified from Leppälä and Savolainen (2011). Vertical lines indicate regions with significant (P < 0.001) gametic (dashed line) or zygotic (solid line) TRD. Horizontal ticks mark the likelihood peaks of TRD within such regions. The text indicates which reciprocal progeny was involved and whether gametic (Gam) or zygotic (Zyg) TRD was inferred. With gametic TRD the parent (F, female; M, male) with most distorted allelic ratios and the preferentially transferred allele (e.g., Sp or Ma) is marked.
Single-locus gametic TRD
We examined more closely the transmission of the two F1 parents’ alleles in the regions showing TRD. To visualize how the F1 parents transmitted their alleles, we plotted observed allele frequencies for each parent over the whole chromosome (Figure 3). As can be seen from Figure 3, gametic TRD can potentially arise from four different sources (male and female function in F1 parent 1 and in F1 parent 2) in a single cross, because both F1 parents were used as pollen donors and recipients.
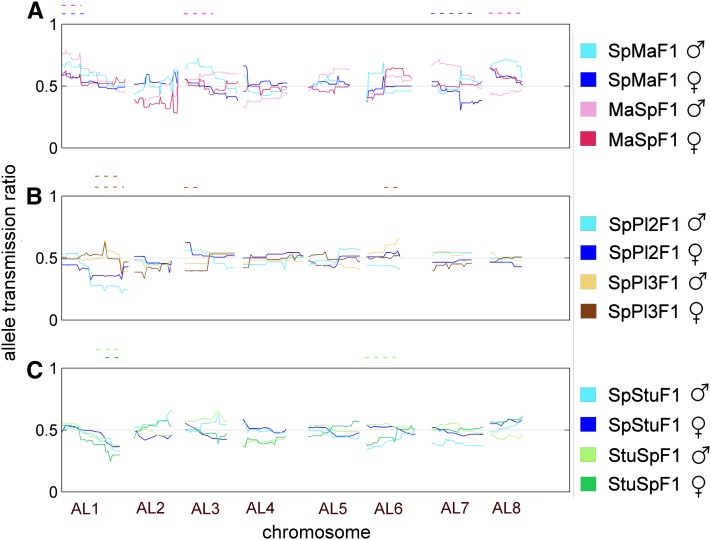
Figure 3.
Allele transmission ratio of Spiterstulen alleles from the F1 to the F2 for each parent, separately in male (light color) and female function (dark color), over all eight chromosomes. (A) Sp × Ma, (B) Sp × Pl, (C) Sp × Stu. F1 parents with Sp cytoplasm are in blue (SpMaF1, Sp1Pl1F1, and SpStuF1). MaSpF1 is in red (A) and StuSpF1 in green (C). The Sp2Pl2F1 parent had cytoplasm from Sp but is represented in brown (B). The Mendelian expectation of equal segregation of both alleles, 0.5, is marked with a gray dashed line. The significant gametic TRD regions have been marked above the chromosomes as in Figure 2.
In the SpMaF2, three regions on three chromosomes experienced strong TRD (Figure S2), of which two (on chromosomes AL1 and AL7) were best explained as gametic. On AL1 excessive transmission of the Sp allele by the pollen donor appeared to be the source of most of the distortion (Figure 3A), although transmission of the pollen recipient’s alleles was biased in the same direction. On chromosome AL7 at least two loci were involved because the F1 pollen donor transmitted more of its Sp allele on the upper arm of the chromosome, while the pollen recipient F1 transmitted more of its Ma allele at the lower chromosome arm (Figure 3A). Two of the three TRD regions observed in the SpMaF2, namely those on AL1 and AL6, were distorted also in the reciprocal MaSpF2 cross. Just like in the SpMaF2, TRD on chromosome AL1 in MaSpF2 was largely due to excess transmission of the Sp allele by the F1 pollen donor, but in the MaSpF2 a similar excess transmission of the Sp allele was observed in the pollen recipient, although less severe than in the pollen donor (Figure 3A). The other TRD region that the reciprocal progenies had in common (on AL6) showed single-locus zygotic TRD. The two other gametic TRD regions in MaSpF2, which were not shared by SpMaF2, were on chromosomes AL3 and AL8, and in both these regions the pollen donor transmitted predominantly its Sp allele.
In the Sp × Pl cross (FST = 0.35) more TRD regions were observed in the SpPl2F2 progeny than in its reciprocal. In SpPl2F2 three regions (AL1, AL3, and AL6) were found to be involved in formation of TRD at gametic level. The region on AL1 was also found in the reciprocal SpPl3F2 where it was the only region that showed significant TRD. Distortion in this region was most pronounced around the marker RHL1 and appears to be due in both reciprocal progenies only to a single F1 parent (Sp1Pl1F1) transmitting its Pl allele in excess. (Sp1Pl1F1 acted as a pollen donor for SpPl3F2 progeny and as pollen recipient for SpPl2F2 progeny.) The two additional TRD regions found in SpPl2F2, on AL3 and AL6, were also apparently of gametic origin, with the Sp allele of Sp1Pl1F1 (AL3) and Sp alleles of both F1 parents (AL6) overrepresented (Figure 3B).
In the Sp × Stu cross (FST = 0.20) one gametic TRD region was observed on AL1 in the SpStuF2 progeny and two regions in its reciprocal progeny, on AL1 and AL6. The region at the lower arm of chromosome AL1 was common for both reciprocal progenies, with an excess of Stu alleles transmitted by both parents in both reciprocal crosses (Figure 3C). The other TRD region in StuSpF2 was on AL6 where both parents transmitted an excess of Stu alleles.
Two-locus gametic TRD
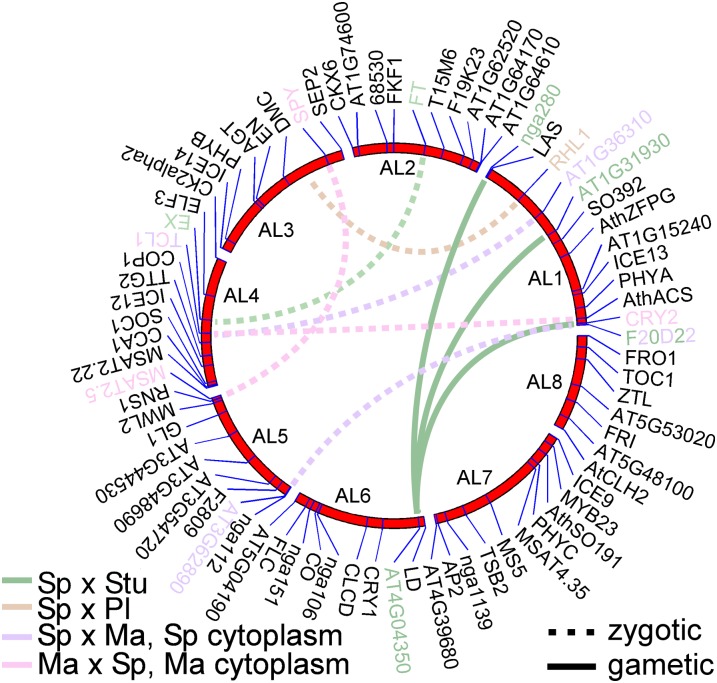
We also examined two-locus genotypes for TRD at gametic and zygotic level. We describe here the most significant epistatic interactions where P < 0.0001. In the SpMaF2, one epistatic gametic interaction was observed in the pollen donor and one in the pollen recipient. The interaction in the pollen recipient (SpMaF1) was between AL1 and AL5 (P = 3.1 × 10−5; Figure S8). The region of AL1 involved in this epistatic interaction was the same as that seen at the single locus level. However, while at the single locus level in AL1 excess transmission of Sp alleles was observed, here the combinations of Sp at AL1 and Sp at AL5 and Ma at AL1 and Ma at AL5 were less frequent than expected. For the pollen donor (MaSpF1) the epistatic interaction was between AL1 and AL4 (P = 1.2 × 10−9; Figure S8). The region in AL1 was not the same as that for single-locus TRD in this progeny and also not for the two-locus interaction in the pollen recipient (SpMaF1), but the pattern was the same; combinations with alleles from the same population were less frequent than expected and gametes combining alleles from different populations were more common.
For the reciprocal progeny (MaSpF2) also two significant gametic two-locus interactions were observed. The interactions were different from those in the reciprocal progeny, and both were observed in the pollen donor (SpMaF1) (Figure S8). The interactions were between AL1 and AL4 and between AL3 and AL5 (Figure 4). Both interactions showed a similar pattern; gametes combining alleles from the different populations (at these interacting loci) were less frequent than expected, and gametes combining alleles from the same population were more common. This was exactly opposite to the pattern observed in the two-locus gametic interactions in the reciprocal F2 progeny.
Figure 4.
Loci experiencing epistatic two-locus interactions are marked with color and connected with a dashed (gametic interaction) or solid line (zygotic interaction).
In the Sp × Pl cross only one, gametic, two-locus interaction (P = 7.7 × 10−7; Figure S9) was found, namely between AL1 and AL3 in the SpPl2F2. The interacting regions were also observed to be in TRD at the gametic single-locus level. The interaction was seen in the pollen recipient (Sp1Pl1F1) and was also observed but weaker (P = 0.001; Figure S9) in the reciprocal cross where Sp1Pl1F1 was acting as a pollen donor. In both reciprocal crosses AL1–AL3 combinations of parental alleles (SpSp or PlPl) were in excess and the combinations of alleles from different populations on the same gamete were less frequent than expected.
In the Sp × Stu cross one gametic two-locus interaction was found in the pollen donor for SpStuF2. The interaction was between AL2 and AL4 (P = 3.3 × 10−5; Figure S10), neither of which showed strong single-locus TRD. Just as in the Sp × Pl cross, within-population combinations between the loci were in excess (SpSp and StuStu) and between-population allelic combinations were less frequent than expected.
Zygotic TRD
Zygotic TRD was rarely detected in these crosses. The strongest single-locus zygotic TRD was detected in both Sp × Ma reciprocal progenies, in the top of chromosome AL6 (Figure S2 and Figure S3). Both heterozygotes were less frequent than expected while parental population homozygotes were in excess (Figure S2 and Figure S3). There might have been some gametic contribution to this case of TRD as the SpMaF1 parent, when acting as a pollen donor, predominantly transmitted its Sp allele, whereas otherwise allele frequencies were as expected (Figure 3).
The other zygotic TRD was a two-locus interaction observed in both reciprocal progenies of the Sp × Stu cross. Upon closer examination of two-locus genotype ratios, it appeared that likely two or three loci from AL1 were interacting with one locus at AL6. Of the interacting regions on chromosomes AL1 and AL6, only that on the lower arm of AL1 also showed single-locus TRD. The two-locus distortion maps to the end of the lower arm of AL1 in both reciprocal crosses, but in StuSpF2 also the upper arm seems to be involved in the interaction (Figure 4). Some TRD was already observed in the gametes of SpStuF1 (both as pollen donor and pollen recipient; Figure S10). In the F2 generation one two-locus genotype was entirely absent from the progeny: no F2 individual was Sp homozygote at AL1 (marker nga280/LAS) and Stu homozygote at AL6 (AT4G04350) (for two-locus genotype counts see Table S2). Similarly no F2 individuals in SpStuF2 were Stu homozygotes at AL1 (AT1G31930) and Sp homozygotes at AL6 (AT4G04350) or in StuSpF2 Stu homozygotes at AL1 (F20D22) and Sp homozygotes at AL6 (AT4G04350).
Comparisons between the crosses
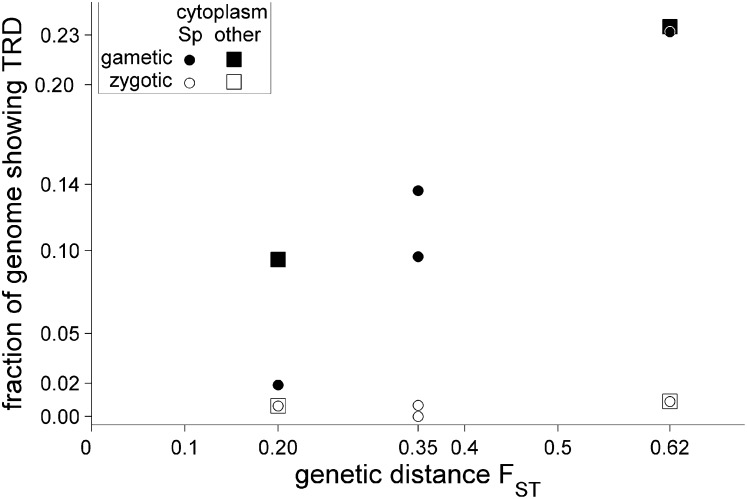
TRD was clearly more prevalent in the distant Sp × Ma cross than in the crosses between the less diverged populations, at gametic as well as zygotic, one-locus and two-locus levels. We compared the percentage of the genome showing single-locus distortion (either gametic or zygotic, at P < 0.001) between different crosses (Figure 5). (Using a different significance threshold for P did not alter the results qualitatively; see Table 1.) On average, the degree of distortion increased with genetic distance between populations. However, between the Sp × Pl and Sp × Stu crosses the difference in degree of TRD was not clear. This was mainly due to a large difference in the degree of TRD between the Sp × Stu reciprocal progenies. While the Sp × Pl reciprocal progenies (both with the Sp cytoplasm) showed an approximately equal degree of TRD, the StSpF2 was distinctly more distorted than its reciprocal F2 progeny, where transmission showed almost no deviation from the Mendelian expectation.
Figure 5.
The fraction of the hybrid genome showing single-locus TRD (P < 0.001) as a function of genetic distance between crossed populations (FST, estimated from nucleotide sequences by Pyhäjärvi et al. (2012). The crosses from left to right: Sp × Stu, Sp × Pl, Sp × Ma.
It has been suggested that the number of genic incompatibilities increases between taxa faster than linearly with time (Orr 1995; Orr and Turelli 2001). To test that “snowballing” prediction, we used linear and exponential functions to describe the increase of the degree of TRD with genetic distance (Figure 5). As is evident from Figure S11, the increase is best described by linear functions, irrespective of whether we measure the degree of TRD as the proportion of the genome showing TRD or as the number of TRD regions. Formal model comparison using Akaike’s information criterion confirms this visual impression.
As may be apparent from what was written above, the regions showing TRD and the inferred origin of distortion in these regions were mostly different between different crosses. Nevertheless, some patterns emerge: both single-locus and epistatic TRD was often found arising from chromosomes AL1 and AL6. All crosses showed TRD on AL1, and in the Sp × Pl and Sp × Stu crosses this TRD maps onto the same chromosomal region, at the end of the lower chromosome arm (Figure 2). In both crosses, at AL1, the Sp allele had been transmitted less frequently than expected, although in the Sp × Pl cross this happened in only one of the F1 parents (Sp1Pl1) whereas in the Sp × Stu cross both parents were transmitting fewer Sp alleles—both when acting as pollen donor and recipient. The other chromosome that commonly showed TRD was AL6, but the strongest TRD did not map onto the same regions of the chromosome, suggesting that the loci causing TRD on AL6 differ between crosses.
Discussion
Gametic vs. zygotic TRD
In all three crosses between A. lyrata populations, most observations of non-Mendelian segregation are best explained by processes acting at the gametic stage. This could be partly due to hybrids experiencing genic incompatibilities or genomic conflicts already in the F1 generation. Alternatively, it is possible that processes acting at the zygotic stage result in a pattern of distortion that mimics gametic TRD. For single-locus zygotic TRD to look like gametic TRD, selection must act against at least one of the genotypes combining parental population alleles (e.g., Sp1Sp2) and against one of the genotypes combining alleles from different populations (e.g., Sp1Ma2). (In this example, selection against these genotypes would mimic selection against the Sp1 allele.) If zygotic TRD were common in these crosses, it would need to be of the kind that mimics gametic TRD, as we detected only two clear examples of zygotic-stage TRD. It seems unlikely that most of the zygotic TRD would mimic gametic TRD. Therefore, we believe that most of the gametic TRD that we reported could really be due to events before zygote formation.
Of the two cases of zygotic TRD, one region in the Sp × Ma cross was best explained by the zygotic single-locus model. The other zygotic TRD region was one interacting pair of chromosomes in the Sp × Stu cross where certain genotypic combinations were completely absent from the F2 progeny. The latter case seems like a classical example of a recessive BDM incompatibility, with selection against individuals that are homozygous at both loci but for alleles from different populations. We also detected two (or three) regions in AL1 interacting with the same locus in AL6, but because our analysis was limited to two-locus interactions we could not detect whether the interaction was between more than two loci. In A. thaliana a similar two-locus interaction was found to cause hybrid embryo lethality because functional copies of a duplicate gene were not at the same locus in different accessions (Bikard et al. 2009). Most TRD loci in A. lyrata, however, appeared to be the best explained by the gametic model.
It should be emphasized that the F2 seeds, once formed, had high viability. This high viability of F2 hybrids has been observed earlier in the cross between subspecies (Sp × Ma), where F2 viability was good but male fertility was reduced (Leppälä and Savolainen 2011). Additional F2 plants from this cross were grown in their native habitats (Norway and North Carolina). In North Carolina, hybrid fitness was intermediate to the parental populations but in Norway the F2 hybrids surprisingly showed heterosis (Leinonen et al. 2011). Thus, the observed TRD in F2 would not be expected to be due to low seed germination success or high mortality in later life stages.
The origin of gametic TRD was traced back to the F1 parents, to see which parent (if not both) caused the distortion and which of the alleles were under- or overrepresented. Most commonly both single-locus and two-locus TRD was caused by a single F1 parent or both F1 parents when acting as a pollen donor (half of the gametic single locus TRD regions and four out of six gametic two-locus regions). Thus, reduced male fertility or pollen competition would be a likely source of TRD. Further support for a possible role of reduced male fertility in causing TRD comes from analysis of fertility of the Sp × Ma F2 progeny (Leppälä and Savolainen 2011). While F2 hybrids did not appear to suffer reduced female fertility, male fertility was strongly affected, and three out of five male fertility QTL were located in TRD regions. In this cross, male fertility was observed to be reduced also in the F1 generation, but unfortunately the male fertility of the two parental F1 plants was not studied. Thus both TRD and fertility reductions in A. lyrata appear to be due mostly to problems in male function. This may be connected to haploid (postmeiotic) gene expression which is abundant during male gametophyte development (reviewed in Borg et al. 2009). Whether transcription of diploid sporocytes (by masking effects of recessive deleterious incompatibilities) is more important for female than male gametophyte development has not been thoroughly studied yet (Muralla et al. 2011).
It is important to note that TRD in male gametes may be due to BDM incompatibilities that reduce fertility, but also to other processes such as pollen competition or action of segregation distorters or some other meiotic drivers. We cannot distinguish with certainty what process was responsible for TRD in male gametes in our study. The role of pollen competition as a source of gametic TRD could be assessed in future studies with pollen competition or pollen tube growth rate assays.
We also detected three sex-independent single-locus transmission ratio distorters where the F1 parents in male as well as female function transmitted alleles from the same population in excess. These cases were observed on the lower arm of AL1 in the Sp × Stu and Sp × Pl cross (but only in one of the F1 parents) and on the upper arm of AL1 in the Sp × Ma cross (although here male function was a source of higher TRD; Figure 3). These could have been caused by sex-independent segregation distorter loci that are fixed but suppressed in their population of origin. This type of segregation distorter advances its own transmission in the naïve hybrid background without suppressors. Sex-independent TRD systems have been identified in tomato (Rick 1969) and rice (Sano et al. 1979; Sano 1992), where one such system is mainly caused by a single locus (Koide et al. 2008b) with some unlinked modifiers (Koide et al. 2012). The classic meiotic drive systems known from mouse and Drosophila are restricted to males (reviewed in Lyttle 1991) but some female segregation distorters are now known, e.g., the knob chromosomes in maize (Buckler et al. 1999), B chromosomes (reviewed in Palestis et al. 2004), and female meiotic drive in Mimulus (Fishman and Saunders 2008). Sex-independent TRD systems seem to be rarer than those affecting just male or female function. The connection between hybrid incompatibility and meiotic drive (and other genomic conflicts) has become recently better established (reviewed by Johnson 2010; McDermott and Noor 2010). To determine whether this type of segregation distorters is involved in causing TRD in F2 hybrid progenies of A. lyrata would require further study.
The role of cytoplasmic factors
Genetic incompatibilities affecting hybrid fitness are often thought to develop between nuclear genes, but they may as well develop between nuclear and cytoplasmic factors. Reciprocal crosses allow us to investigate the role of cytoplasmic factors, but such crosses require hermaphrodite organisms and have therefore been restricted largely to studies in plants. In our crosses the F1 parents had different nuclear genotypes, so we could not distinguish between cytoplasmic effects and nuclear polymorphisms within populations. However, the present results suggest that either cytoplasm or allelic polymorphisms (or a combination of these) play a major role, as the extent and especially location of TRD differ substantially between reciprocal crosses. Kuittinen et al. (2004) studied a cross between A. lyrata populations and reached the same conclusion: only one TRD region of the seven observed was shared between their reciprocal progenies.
Here, the difference between reciprocal progenies was most pronounced in the Sp × Stu cross, where 2% of the genome (P < 0.01, gametic model vs. null model) showed TRD in the SpStuF2, against 23% in the StuSpF2 reciprocal. In the more distant St × Ma cross the difference between reciprocals in the degree of TRD was less pronounced, but in both crosses the nature and location of TRDL differed substantially between reciprocal progenies. A similar result was obtained from analysis of male fertility of the Sp × Ma F2: both the extent of fertility reduction and the underlying QTL differed between reciprocal F2 progenies (Leppälä and Savolainen 2011). In addition to the role of cytoplasmic factors, alleles within a population also had different effects on male fertility of Sp × Ma cross (Leppälä and Savolainen 2011).
In one of our reciprocal crosses (Sp × Pl, at intermediate genetic distance), the role of cytoplasm vs. within population allelic variation could be evaluated because the cytoplasm was from the same population. In this cross the degree of TRD differed relatively little between the reciprocal F2 (18% vs. 19% of genome in TRD, P < 0.01, gametic vs. null model), but within-population variation was also found: two of the TRD regions were found in only one of the progenies (SpPl2F2) and the shared TRD region was sex independent, i.e., the source of TRD could be traced back to a single F1 parent. However, based on a single cross we cannot conclude whether one mechanism is more likely to be involved in TRD than the other.
TRD increases with genetic distance
As an indicator of genic incompatibility, the degree of TRD is expected to increase with genetic distance between populations. It is, however, difficult to compare the degree of TRD between crosses from different populations or species. As the results of the present study illustrate, TRD may be a single-locus or multiple-locus phenomenon, and TRD at any specific locus may emerge from pollen donor, pollen recipient, or both parents or in the hybrid zygotes. Furthermore, TRD at any locus may differ in severity, from a slight distortion of transmission ratios to complete absence of some genotypes. For these reasons it is difficult to compare the degree of TRD even between reciprocal crosses from the same parents. Comparisons involving distant populations or species may be further complicated by different numbers, types, and positions of marker loci, making it hard to tell whether a difference in degree of TRD is a real property of the populations studied or the result of difference in statistical power to detect TRD. Consequently, it is difficult to determine from comparison of published studies whether the degree of TRD increases with genetic distance.
Because of the difficulties associated with expressing the degree of TRD as a scalar value, we decided to use a simple measure for the sake of tractability: the percentage of the genome showing distortion. This measure does not solve the problems described above, but it is tractable, it can be calculated for almost any study, and it has one attractive property: a larger number of TRDL increases the percentage of the genome showing TRD, and further, more severe TRD will be apparent over a larger part of a chromosome because of linkage disequilibrium. Thus, measuring the degree of TRD as a percentage of the genome showing TRD hides many interesting aspects of the underlying data, but it does provide a figure that naturally integrates the number of TRDL and the severity of their effects in a simple measure.
In line with expectation, TRD has been found to increase from intraspecific to interspecific experimental crosses (Jenczewski et al. 1997; Rieseberg et al. 2000). By contrast, in natural populations of Mimulus, the level of TRD did not differ between intra- and interspecific crosses (Hall and Willis 2005). However, because of lack of genetic data, Hall and Willis (2005) were unable to conclude whether the Mimulus guttatus populations they studied were less diverged than the species pair used in an earlier study (Fishman et al. 2001). Good estimates of genetic distance are needed for testing directly whether crosses between more distant populations show higher TRD. A recent study by Salomé et al. (2011) did not find any relation between sequence divergence of parental accessions and F2 segregation distortion in A. thaliana. For the A. lyrata populations studied here, good microsatellite and sequence-based estimates of genetic divergence were available (Muller et al. 2008; Ross-Ibarra et al. 2008; Pyhäjärvi et al. 2012). We found that the proportion of the genome showing TRD increased with genetic distance between populations and was clearly the highest in the most distant pair of populations crossed. This is in line with earlier studies in A. lyrata, which showed that the level of within-population TRD is very low (Leppälä et al. 2008) while several TRDL were mapped from a cross between genetically differentiated populations (Kuittinen et al. 2004) and from a backcross population between A. lyrata and its close relative A. halleri (Willems et al. 2007). Similarly, intraspecific rice crosses suggest increasing number of TRDL with increasing genetic distance between parental accessions (Matsubara et al. 2011). Our results do not support the snowballing hypothesis that the degree of TRD increases exponentially with time (Orr 1995; Orr and Turelli 2001): both the number of TRD regions and the percentage of the genome showing TRD increase approximately linearly with genetic distance.
An interesting feature of the overview of TRDL in the three crosses studied here was the uneven distribution of TRD over the genome, and in particular the degree of distortion of chromosomes AL1 and AL6: all three experimental crosses had TRDL on chromosomes 1 and 6. The same chromosomes showed TRD in an A. lyrata cross studied by Kuittinen et al. (2004) and in an interspecific cross between A. lyrata and A. halleri (Willems et al. 2007). In addition, in A. thaliana F2 progenies, Salomé et al. (2011) found most TRD on chromosomes 1 and 5, in areas that locate to chromosomes 1 and 6 in A. lyrata. We do not currently have an explanation for this observation, but it does suggest that the degree of TRD varies across the genome and that in the genus Arabidopsis especially some regions of chromosome 1 seem to be involved. This suggests that we may learn more about the processes causing TRD, and its relation with genetic distance, by comparing nucleotide variation, gene content, and other variation between genomic regions with no TRD and regions with high numbers of TRD loci.
Supplementary Material
Acknowledgments
The authors thank M. Otsukka and A. Airikainen for their help in genotyping the Sp x Stu F2 progeny. This study was supported by the Swedish Research Council and Umeå University (FB); work by J.L. and O.S. was supported by the Academy of Finland’s Research Council for Biosciences and Environment (O.S.), by Biocenter Oulu (O.S.), and by the Finnish Population Genetics Graduate School (J.L.).
Footnotes
Communicating editor: S. I. Wright
Literature Cited
- Bikard D., Patel D., Le Mette C., Giorgi V., Camilleri C., et al. , 2009. Divergent evolution of duplicate genes leads to genetic incompatibilities within A. thaliana. Science 323: 623–626 [DOI] [PubMed] [Google Scholar]
- Borg M., Brownfield L., Twell D., 2009. Male gametophyte development: a molecular perspective. J. Exp. Bot. 60: 1465–1478 [DOI] [PubMed] [Google Scholar]
- Brideau N. J., Flores H. A., Wang J., Maheshwari S., Wang X., et al. , 2006. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science 314: 1292–1295 [DOI] [PubMed] [Google Scholar]
- Buckler E. S., Phelps-Durr T. L., Buckler C. S. K., Dawe R. K., Doebley J. F., et al. , 1999. Meiotic drive of chromosomal knobs reshaped the maize genome. Genetics 153: 415–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie P., Macnair M. R., 1987. The distribution of postmating reproductive isolating genes in populations of the yellow monkey flower, Mimulus guttatus. Evolution 41: 571–578 [DOI] [PubMed] [Google Scholar]
- Coyne J. A., Orr H. A., 2004. Speciation. Sinauer Associates, Sunderland, MA [Google Scholar]
- Cutter A. D., 2012. The polymorphic prelude to Bateson-Dobzhansky-Muller incompatibilities. Trends Ecol. Evol. 27: 209–218 [DOI] [PubMed] [Google Scholar]
- Fishman L., Saunders A., 2008. Centromere-associated female meiotic drive entails male fitness costs in monkeyflowers. Science 322: 1559–1562 [DOI] [PubMed] [Google Scholar]
- Fishman L., Kelly A. J., Morgan E., Willis J. H., 2001. A genetic map in the Mimulus guttatus species complex reveals transmission ratio distortion due to heterospecific interactions. Genetics 159: 1701–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gérard P. R., Presgraves D. C., 2012. Abundant genetic variability in Drosophila simulans for hybrid female lethality in interspecific crosses to Drosophila melanogaster. Genet. Res. 94: 1–7 [DOI] [PubMed] [Google Scholar]
- Hall M. C., Willis J. H., 2005. Transmission ratio distortion in intraspecific hybrids of Mimulus guttatus: implications for genomic divergence. Genetics 170: 375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harushima Y., Nakagahra M., Yano M., Sasaki T., Kurata N., 2001. A genome-wide survey of reproductive barriers in an intraspecific hybrid. Genetics 159: 883–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield T., Schluter D., 1999. Ecological speciation in sticklebacks: environment-dependent hybrid fitness. Evolution 53: 866–873 [DOI] [PubMed] [Google Scholar]
- Howard D. J., 1999. Conspecific sperm and pollen precedence and speciation. Annu. Rev. Ecol. Syst. 30: 109–132 [Google Scholar]
- Jenczewski E., Gherardi M., Bonnin I., Prosperi J. M., Olivieri I., et al. , 1997. Insight on segregation distortions in two intraspecific crosses between annual species of Medicago (Leguminosae). Theor. Appl. Genet. 94: 682–691 [Google Scholar]
- Johnson N. A., 2010. Hybrid incompatibility genes: remnants of a genomic battlefield? Trends Genet. 26: 317–325 [DOI] [PubMed] [Google Scholar]
- Kemi U., Niittyvuopio A., Toivainen T., Pasanen A., Quilot-Turion B., et al. , 2013. Role of vernalization and of duplicated FLOWERING LOCUS C in the perennial Arabidopsis lyrata. New Phytol. 197: 323–335 [DOI] [PubMed] [Google Scholar]
- Koide Y., Onishi K., Nishimoto D., Baruah A. R., Kanazawa A., et al. , 2008a Sex-independent transmission ratio distortion system responsible for reproductive barriers between Asian and African rice species. New Phytol. 179: 888–900 [DOI] [PubMed] [Google Scholar]
- Koide Y., Ikenaga M., Sawamura N., Nishimoto D., Matsubara K., et al. , 2008b The evolution of sex-independent transmission ratio distortion involving multiple allelic interactions at a single locus in rice. Genetics 180: 409–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide Y., Shinya Y., Ikenaga M., Sawamura N., Matsubara K., et al. , 2012. Complex genetic nature of sex-independent transmission ratio distortion in Asian rice species: the involvement of unlinked modifiers and sex-specific mechanisms. Heredity 108: 242–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbecka G., Klinkhamer P. G. L., Vrieling K., 2002. Selective embryo abortion hypothesis revisited: a molecular approach. Plant Biol. 4: 298–310 [Google Scholar]
- Kuittinen H., de Haan A. A., Vogl C., Oikarinen S., Leppälä J., et al. , 2004. Comparing the linkage maps of the close relatives Arabidopsis lyrata and A. thaliana. Genetics 168: 1575–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen P. H., Remington D. L., Savolainen O., 2011. Local adaptation, phenotypic differentiation and hybrid fitness in diverged natural populations of Arabidopsis lyrata. Evolution 65: 90–107 [DOI] [PubMed] [Google Scholar]
- Leppälä J., Savolainen O., 2011. Nuclear-cytoplasmic interactions reduce male fertility in hybrids of Arabidopsis lyrata subspecies. Evolution 65: 2959–2972 [DOI] [PubMed] [Google Scholar]
- Leppälä J., Bechsgaard J. S., Schierup M. H., Savolainen O., 2008. Transmission ratio distortion in Arabidopsis lyrata: effects of population divergence and the S-locus. Heredity 100: 71–78 [DOI] [PubMed] [Google Scholar]
- Levin D. A., 2003. The cytoplasmic factor in plant speciation. Syst. Bot. 28: 5–11 [Google Scholar]
- Lyttle T. W., 1991. Segregation distorters. Annu. Rev. Genet. 25: 511–557 [DOI] [PubMed] [Google Scholar]
- Martin N. H., Willis J. H., 2010. Geographical variation in postzygotic isolation and its genetic basis within and between two Mimulus species. Phil. Trans. R. Soc. B. 365: 2469–2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara K., Ebana K., Mizubayashi T., Itoh S., Ando T., et al. , 2011. Relationship between transmission ratio distortion and genetic divergence in intraspecific rice crosses. Mol. Genet. Genomics 286: 307–319 [DOI] [PubMed] [Google Scholar]
- Matute D. R., Butler I. A., Turissini D. A., Coyne J. A., 2010. A test of the snowball theory for the rate of evolution of hybrid incompatibilities. Science 329: 1518–1521 [DOI] [PubMed] [Google Scholar]
- McDaniel S. F., Willis J. H., Shaw A. J., 2008. The genetic basis of developmental abnormalities in interpopulation hybrids of the moss Ceratodon purpureus. Genetics 179: 1425–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott S. R., Noor M. A. F., 2010. The role of meiotic drive in hybrid male sterility. Phil. Trans. R. Soc. B. 365: 1265–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle L. C., Graham E. B., 2006. Genome-wide associations between hybrid sterility QTL and marker transmission ratio distortion. Mol. Biol. Evol. 23: 973–980 [DOI] [PubMed] [Google Scholar]
- Moyle L. C., Nakazato T., 2010. Hybrid incompatibility “snowballs” between Solanum species. Science 329: 1521–1523 [DOI] [PubMed] [Google Scholar]
- Muller M. H., Leppälä J., Savolainen O., 2008. Genome-wide effects of postglacial colonization in Arabidopsis lyrata. Heredity 100: 47–58 [DOI] [PubMed] [Google Scholar]
- Muralla R., Lloyd J., Meinke D., 2011. Molecular foundations of reproductive lethality in Arabidopsis thaliana. PLoS ONE 6: e28398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myburg A. A., Vogl C., Griffin A. R., Sederoff R. R., Whetten R. W., 2004. Genetics of postzygotic isolation in eucalyptus: whole-genome analysis of barriers to introgression in a wide interspecific cross of Eucalyptus grandis and E. globulus. Genetics 166: 1405–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazato T., Jung M.-K., Housworth E. A., Rieseberg L. H., Gastony G. J., 2007. A genomewide study of reproductive barriers between allopatric populations of a homosporous fern, Ceratopteris richardii. Genetics 177: 1141–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A., 1995. The population-genetics of speciation: the evolution of hybrid incompatibilities. Genetics 139: 1805–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr A. H., Turelli M., 2001. The evolution of postzygotic isolation: accumulating Dobzhansky–Muller incompatibilities. Evolution 55: 1085–1094 [DOI] [PubMed] [Google Scholar]
- Palestis B. G., Trivers R., Burt A., Jones R. N., 2004. The distribution of B chromosomes across species. Cytogenet. Genome Res. 106: 151–158 [DOI] [PubMed] [Google Scholar]
- Presgraves D. C., 2010. The molecular evolutionary basis of species formation. Nat. Rev. Genet. 11: 175–180 [DOI] [PubMed] [Google Scholar]
- Pyhäjärvi T., Aalto E., Savolainen O., 2012. Time scales of divergence and speciation among natural populations and subspecies of Arabidopsis lyrata (Brassicaceae). Am. J. Bot. 99: 1–9 [DOI] [PubMed] [Google Scholar]
- Quilot-Turion B., Leppälä J., Leinonen P. H., Waldmann P., Savolainen O., et al. , 2013. Genetic changes in flowering and morphology in response to adaptation to a high-latitude environment in Arabidopsis lyrata. Ann. Bot. 111: 957–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L. K., Markow T. A., 2004. Early events in speciation: polymorphism for hybrid male sterility in Drosophila. Proc. Natl. Acad. Sci. USA 101: 9009–9012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rick C. M., 1969. Controlled introgression of chromosomes of Solanum pennellii into Lycopersicon esculentum: segregation and recombination. Genetics 62: 753–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg L. H., Blackman B. K., 2010. Speciation genes in plants. Ann. Bot. 106: 439–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg L. H., Baird S. J. E., Gardner K. A., 2000. Hybridization, introgression, and linkage evolution. Plant Mol. Biol. 42: 205–224 [PubMed] [Google Scholar]
- Ross-Ibarra J., Wright S. I., Foxe J. P., Kawabe A., DeRose-Wilson L., et al. , 2008. Patterns of polymorphism and demographic history in natural populations of Arabidopsis lyrata. PLoS ONE 3: e2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé P. A., Bomblies K., Fitz J., Laitinen R. A. E., Warthmann N., et al. , 2011. The recombination landscape in Arabidopsis thaliana F2 populations. Heredity 108: 447–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano Y., 1992. Genetic comparisons of chromosome 6 between wild and cultivated rice. Jpn. J. Breed 42: 561–572 [Google Scholar]
- Sano Y., Chu Y.E., Oka H. I., 1979. Genetic studies of speciation in cultivated rice, 1. Genic analysis for the F1 sterility between O. sativa L. and O. glaberrima Steud. Jpn. J. Genet 54: 121–132 [Google Scholar]
- Sweigart A. L., Mason A. R., Willis J. H., 2007. Natural variation for a hybrid incompatibility between two species of Mimulus. Evolution 61: 141–151 [DOI] [PubMed] [Google Scholar]
- Vogl C., Xu S. Z., 2000. Multipoint mapping of viability and segregation distorting loci using molecular markers. Genetics 155: 1439–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren J. H., 2011. Selfish genetic elements, genetic conflict, and evolutionary innovation. Proc. Natl. Acad. Sci. USA 108: 10863–10870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijsman E. M., 1987. A deductive method of haplotype analysis in pedigrees. Am. J. Hum. Genet. 41: 356–373 [PMC free article] [PubMed] [Google Scholar]
- Willems G., Drager D. B., Courbot M., Gode C., Verbruggen N., et al. , 2007. The genetic basis of zinc tolerance in the metallophyte Arabidopsis halleri ssp halleri (brassicaceae): an analysis of quantitative trait loci. Genetics 176: 659–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., Lauga B., Charlesworth D., 2003. Subdivision and haplotype structure in natural populations of Arabidopsis lyrata. Mol. Ecol. 12: 1247–1263 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.